Abstract
Bacterial lipoproteins represent a subset of membrane-associated proteins that are covalently modified with lipids at the N-terminal cysteine. The final step of lipoprotein modification, N-acylation of apolipoproteins, is mediated by apolipoprotein N-acyltransferase (Lnt). Examinations with reconstituted proteoliposomes and a conditional mutant previously indicated that N-acylation of lipoproteins is required for their efficient release from the inner membrane catalyzed by LolA and LolCDE, the lipoprotein-specific chaperone and ABC transporter, respectively. Because Lnt is essential for Escherichia coli, a mutant lacking Lnt activity has not been isolated. However, we report here that lnt-null strains can be constructed when LolCDE is overproduced in strains lacking either the major outer membrane lipoprotein Lpp or transpeptidases that cross-link Lpp with peptidoglycan. Lipoproteins purified from the lnt-null strain exhibited increased mobility on SDS-PAGE compared to those from wild-type cells and could be sequenced by Edman degradation, indicating that lipoproteins in this mutant exist as apolipoproteins that lack N-acylation. Overexpression of Lpp in the lnt-null strain resulted in the accumulation of apoLpp in the inner membrane and caused growth arrest. In contrast to the release of mature Lpp in the presence of LolA and LolCDE, that of apoLpp from the inner membrane was significantly retarded. Furthermore, the amount of lipoproteins copurified with LolCDE was significantly reduced in the lnt-null strain. These results indicate that the affinity of LolCDE for apolipoprotein is very low, and therefore, overexpression of LolCDE is required for its release and sorting to the outer membrane.
INTRODUCTION
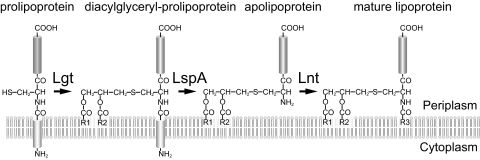
Bacterial lipoproteins are synthesized in the cytoplasm as precursors with N-terminal signal peptides. Modification of lipoproteins occurs after lipoprotein precursors have been translocated from the cytosol to the periplasmic side of the inner membrane through the action of Sec translocase. The modification reaction comprises three steps in Gram-negative bacteria: (i) formation of a thioether linkage between a cysteine in the N-terminal region and diacylglycerol by phosphatidylglycerol-prolipoprotein diacylglyceryl transferase (Lgt); (ii) cleavage of a signal peptide by signal peptidase II (LspA), which turns the S-lipidated cysteine into the N-terminal residue of the mature protein; and (iii) N-acylation of this cysteine by apolipoprotein N-acyltransferase (Lnt) (Fig. 1) (30). Thus, lipoproteins have an N-terminal glycerylcysteine containing two ester-linked acyl chains and one amide-linked acyl chain. Gram-positive bacteria, except for actinomycetes, lack a homologous gene for Lnt. It is therefore considered that lipoproteins exist as diacylated forms without amide-linked acyl chains in most Gram-positive bacteria, though exceptions have been recently reported for several Gram-positive bacteria (1, 18, 47, 49). In Gram-negative bacteria, mature lipoproteins are further targeted to the outer membrane by the Lol system, which comprises the inner membrane ABC transporter LolCDE, the periplasmic lipoprotein carrier protein LolA, and the outer membrane receptor LolB (30). LolCDE binds a lipoprotein in the inner membrane and transfers it to LolA to form a soluble lipoprotein-LolA complex that traverses the periplasm to the outer membrane, where LolB accepts the lipoprotein and incorporates it into the lipid layer of the outer membrane. The residue next to the lipid-modified Cys functions as the sorting signal for lipoproteins in enterobacteria (20, 39, 45, 53). When Asp is present at this position (position 2), the lipoprotein remains in the inner membrane. Asp at position 2 is considered to make the N-terminal structure of lipoproteins distinct from those with other residues, thereby functioning as a “LolCDE avoidance signal” (10, 22).
Fig. 1.
Biosynthetic pathway for lipoproteins. After being translocated across the inner membrane by Sec translocase, prolipoproteins are processed into mature lipoproteins through the sequential actions of three inner membrane enzymes, phosphatidylglycerol-prolipoprotein diacylglyceryl transferase (Lgt), signal peptidase II (LspA), and apolipoprotein N-acyltransferase (Lnt).
The importance of triacylation for the outer membrane targeting of lipoproteins was first demonstrated by an in vitro study. It was shown that apoPal, an immediate precedent of mature Pal, was not released from proteoliposomes even in the presence of LolCDE and LolA (7). Because it is essential, only conditional-lethal mutants of lnt have been isolated. A temperature-sensitive mutant of Salmonella enterica that accumulated apolipoproteins at nonpermissive temperature was shown to have a mutation in the lnt gene (9). More recently, Robichon et al. (35) constructed a conditional mutant of E. coli in which the chromosomal lnt gene was placed under the arabinose-inducible promoter and showed that apolipoproteins synthesized in the absence of arabinose remained in the inner membrane. The functions of Lnt mutants have been studied by using a conditional lnt strain to identify essential residues, including the catalytic triad E267-K335-C387 (48). It has also been recently established that Lnt forms a thioester acyl-enzyme intermediate between C387 and an acyl group derived from phospholipids (3). On the other hand, it remained unclear how the lack of Lnt activity affects the functions of Lol proteins. In this study, we report that lnt-null mutants of E. coli that grow despite the absence of N-acylated lipoproteins can be constructed under specific conditions.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The Escherichia coli K-12 strains and plasmids used in this study are listed in Table 1. Plasmid pKD46 (5) was obtained from the E. coli Genetic Stock Center (Yale University, New Haven, CT). Plasmid pCP20 (4) and the Keio collection (2) were from the NBRP (National Institute of Genetics, Mishima, Japan).
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| BW25113 | lacIqrrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 5 |
| DLP79-22 | HfrC pps man | 14 |
| DLP79-36 | DLP79-22 lpp | 45 |
| SN495 | BW25113 ΔybiS ΔerfK ΔycfS | This study |
| LNT0654 | BW25113 Δlnt::FRT-kan-FRT | This study |
| LNT6541 | DLP79-36 Δlnt::FRT-kan-FRT, constructed in the presence of pKM001 | This study |
| LNT6542 | DLP79-36 Δlnt::FRT-kan-FRT, constructed in the presence of pNASCDEH2 | This study |
| LNT6543 | SN495 Δlnt::FRT-kan-FRT, constructed in the presence of pNASCDEH2 | This study |
| Plasmids | ||
| pKD46 | pSC101 derivative; Rep(Ts) bla PBAD-gam-bet-exo | 5 |
| pCP20 | pSC101 derivative; Rep(Ts) bla cat λcI857 λPRFLP+ | 4 |
| pTLH631 | pTTQ18 derivative; Ptac-lnt-his6bla | 7 |
| pBLH631 | pMAN885EH derivative; PBAD-lnt-his6cat | This study |
| pKM001 | pUC19 derivative; Plac-lolCDEbla | 52 |
| pNASCDEH2 | pNAS885 derivative; PBAD (ΔaraC araICXC)-lolCDE-his6cat | This study |
| pAM201 | pMAN885EH derivative; PBAD-lolA-his6cat | 26 |
| pJY811 | pMAN885EH derivative; PBAD-lppcat | 50 |
| pJY851 | pMAN885EH derivative; PBAD-lppSRcat | 50 |
| pJY856 | pMAN885EH derivative; PBAD-lppDRcat | 50 |
| pLMALE2-His | pUCP20 derivative; Plac-lipomalE(S) bla | 29 |
| pLMALE2(S2D)-His | pUCP20 derivative; Plac-lipomalE(D) bla | 29 |
Construction of plasmids.
pBLH631, encoding Lnt-His, was constructed by subcloning the EcoRI-HindIII fragment derived from pTLH631 (7) into the same sites of pMAN885EH (50). pNASCDEH2, which constitutively overexpresses LolCDE-His, was constructed as follows. A DNA fragment encoding LolC, LolD, and LolE-His was amplified by PCR with pKM001 (52) as a template and a pair of primers, LolC-3 and LolE-4 (Table 2). The amplified DNA fragment was digested with EcoRI and HindIII and then cloned into the same sites of pNAS885, a derivative of pMAN885EH in which the araC gene was deleted by removing a 0.5-kb EcoRV-EcoRV fragment inside araC, and araI(Con) araX(Con) mutations (12) were introduced into the araBAD promoter by site-directed mutagenesis with a pair of complementary primers, pBAD-f and pBAD-r (Table 2).
Table 2.
Primers used in this work
| Primer | Sequence |
|---|---|
| LolC-3 | 5′-GATGAATTCGGAGGTTTAAATTTATGTACCAACCTGTCGCTCTATTTA-3′ |
| LolE-4 | 5′-CAATTCAAGCTTAATGATGATGATGATGATGCTCCAGCTGGCCGCTAAGGACTCGCGCAG-3′ |
| pBAD-f | 5′-AGCGGATCCTACCTGGCGCTTTTTATCGCAACTCTCTACTATTTCTCCATACCCGT-3′ |
| pBAD-r | 5′-ACGGGTATGGAGAAATAGTAGAGAGTTGCGATAAAAAGCGCCAGGTAGGATCCGCT-3′ |
| lnt-1 | 5′-CCCAGCCGAAGCTGGATGAATAAAACCGAAACTGGATAGATAACTACATGATTCCGGGGATCCGTCGACC-3′ |
| lnt-2 | 5′-GGGATGTATTCCGGCACGATAAGAAGGGATTATTTACGTCGCTGACGCAGTGTAGGCTGGAGCTGCTTCG-3′ |
| lnt-3 | 5′-TCCCGGATGACTCACCCCAG-3′ |
Construction of lnt-null strains.
The chromosomal lnt gene was replaced with a kanamycin resistance cassette flanked by FLP recognition target (FRT) sites as described by Datsenko and Wanner (5). PCR was performed by using the chromosomal DNA of JW1105 (ΔnagK::FRT-kan-FRT) (2) as a template and a pair of primers, lnt-1 and lnt-2 (Table 1), that contained 50 nucleotides homologous to each side of the lnt gene and 20 nucleotides for priming upstream and downstream of the FRT sites. BW25113 carrying pKD46 and pBLH631 was grown in LB medium supplemented with 0.02% arabinose and electroporated with the PCR fragment, and then the cells were spread onto LB agar containing kanamycin and incubated overnight at 37°C. A kanamycin-resistant colony was selected, and chromosomal deletion of the lnt gene was verified by PCR with primers lnt-2 and lnt-3 (Table 2). The resultant lnt-null strain was named LNT0654. The Δlnt::FRT-kan-FRT allele was transduced by P1-mediated transduction at 30°C (25) into DLP79-36 harboring either the LolCDE-overexpressing plasmid pKM001 or pNASCDEH2, yielding LNT6541 and LNT6542, respectively. A ΔybiS ΔerfK ΔycfS triple mutant, SN495, was constructed by successively transducing the kanamycin resistance cassette from one corresponding clone in the Keio collection (2) into BW25113, followed by transformation with pCP20 to excise the kanamycin resistance cassette (4) and transduction of another kanamycin resistance cassette. The Δlnt::FRT-kan-FRT allele was transduced into SN495 harboring pNASCDEH2, yielding LNT6543.
Partial purification and Edman degradation of lipoproteins.
Lpp was purified as previously described (11). DLP79-36 or LNT6541 cells harboring pKM001 and pJY811 (50) were cultivated in LB medium at 30°C. When the culture optical density (OD) reached 0.2, expression of Lpp was induced for 3 h by the addition of 0.2% l-arabinose. Cells were passed through a French press twice at 10,000 lb/in2. After removal of unbroken cells by centrifugation at 10,000 × g for 10 min, membranes were recovered by centrifugation at 100,000 × g for 1 h and then solubilized with 10 mM Tris-HCl, pH 7.5, containing 5 mM MgCl2 and 1% Triton X-100 at 4°C for 10 min, followed by centrifugation at 100,000 × g for 30 min. The precipitate was solubilized with 10 mM Tris-HCl, pH 7.5, containing 1% SDS and then boiled for 5 min, followed by centrifugation at 100,000 × g for 30 min. Trichloroacetic acid (TCA) was added to the supernatant to a final concentration of 10%. Lpp was obtained in the TCA-soluble fraction after centrifugation at 5,000 × g for 10 min and then precipitated with 9 volumes of acetone overnight at −20°C. Pal was purified from DLP79-36 or LNT6541 cells harboring pKM001 as reported previously (27). Partially purified Pal and Lpp were subjected to SDS-PAGE, followed by transfer to a polyvinylidene difluoride membrane and Coomassie brilliant blue (CBB) staining. Bands were excised from the membrane, and N-terminal amino acid sequences were determined by Edman degradation using a pulsed-liquid sequencer (477A/120A; Applied Biosystems).
Separation of inner and outer membranes.
The membrane localization of LppSR, a derivative of Lpp devoid of the C-terminal Lys, or its derivative LppDR, with a Ser-to-Asp substitution at position 2, was determined as follows. DLP79-36 or LNT6541 cells harboring pKM001 were transformed with pJY851 or pJY856, encoding LppSR or LppDR, respectively, and then grown on LB medium at 30°C. When the culture OD reached 0.6, expression of LppSR or LppDR was induced for 30 min by the addition of 0.1% l-arabinose. To determine the membrane localization of lipoMalE, DLP79-36 or LNT6542 cells harboring pNASCDEH2 were transformed with pLMALE2-His or pLMALE2(S2D)-His (29) encoding the model lipoprotein lipoMalE(S) or lipoMalE(D), respectively, and then grown on LB medium at 37°C. Cells were harvested and then converted into spheroplasts as described previously (23). The spheroplasts were disrupted by a single passage through a French pressure cell at 10,000 lb/in2. A total membrane fraction was recovered by centrifugation at 100,000 × g for 1 h at 4°C, suspended in 50 mM Tris-HCl (pH 7.5) containing 1 mM EDTA, and then layered on a 30 to 55% (wt/wt) sucrose gradient. After centrifugation at 60,000 × g for 12 h at 4°C, fractions were collected from the gradient with a Piston Gradient Fractionator (BioComp Instruments, Fredericton, Canada) and then analyzed by SDS-PAGE and immunoblotting.
Assay for lipoprotein release from spheroplasts.
DLP79-36 or LNT6541 cells harboring pKM001 and pJY851 were grown in M63 medium (50) at 30°C. When the culture OD reached 0.8, expression of LppSR was induced for 5 min by the addition of 0.2% l-arabinose. Cells were harvested and converted into spheroplasts as previously described (23). Aliquots (300 μl) of suspensions containing 5 × 108 spheroplasts were incubated with or without LolA (10 μg) at 30°C for 1 min. M63 medium (750 μl) containing 0.25 M sucrose and 370 kBq of Tran35S-label (MP Biomedicals, Solon, OH) was then added to label LppSR for 2 min, followed by the addition of nonradioactive Met and Cys (each at 12 mM) to chase the labeling. Aliquots were withdrawn after 2, 5, or 10 min and chilled on ice to terminate the release of LppSR. Spheroplasts and medium were separated by centrifugation at 16,000 × g for 2 min. Each fraction was immunoprecipitated with anti-Lpp antibodies, and then the radiolabeled LppSR was analyzed by SDS-PAGE and phosphorimaging with a Storm 820 (GE Healthcare).
Isolation of liganded LolCDE.
DLP79-36 or LNT6542 cells harboring pNASCDEH2 were grown on LB medium at 30°C until the culture OD reached 1.5. A total membrane fraction (10 mg protein) was prepared as described above and then solubilized with 10 ml of 20 mM Tris-HCl (pH 7.5) containing 1% DDM (n-dodecyl-β-o-maltopyranoside), 5 mM MgCl2, and 10% glycerol for 30 min on ice. A supernatant was obtained by centrifugation at 100,000 × g for 30 min and then applied on a 0.25-ml Talon column preequilibrated with buffer A (20 mM Tris-HCl, pH 7.5, containing 300 mM NaCl, 0.01% DDM, and 10% glycerol). After washing with 10 ml of buffer A, the protein was eluted using a stepwise imidazole gradient comprising 10-ml each of buffer A containing 1, 2, 5, 10, 20, and 50 mM imidazole. Where specified, 2 mM ATP was included in all purification steps.
RESULTS
Overproduction of LolCDE allowed the construction of an lnt-null strain.
Although Lnt is essential for the growth of both S. enterica (9) and E. coli (35), the lack of the major outer membrane lipoprotein Lpp renders these strains less dependent on Lnt activity. This is most likely caused by the fact that an extraordinarily large number of Lpp molecules are present in a single cell and the fact that mislocalized Lpp in the inner membrane forms a lethal covalent linkage with peptidoglycans, resulting in cell lysis (50). It was shown by both in vivo and in vitro examinations that lipoproteins lacking N-acylation were scarcely released from the inner membrane (7, 35), which is dependent on both LolCDE (52) and LolA (23). Based on these observations, we attempted to disrupt the lnt gene in an E. coli mutant that lacks Lpp and overexpresses LolCDE or LolA, or both.
We first replaced the chromosomal lnt gene with a kanamycin resistance gene (kan) in the presence of Lnt expressed from a plasmid, pBLH631. The Δlnt::kan allele was then transferred by P1 transduction from the Δlnt strain to either an lpp+ or lpp mutant strain, which harbored either pKM001 encoding LolCDE or pUC19, the empty vector. DLP79-22 (lpp+) cells generated a small number of kanamycin-resistant colonies, but they were nonviable irrespective of the plasmid species. In contrast, more than 1,000 kanamycin-resistant colonies were derived from DLP79-36 (lpp mutant) cells harboring either plasmid. However, colonies derived from DLP79-36/pUC19 were nonviable on new plates. In marked contrast, colonies derived from DLP79-36 cells harboring pKM001 were viable even after purification. When DLP79-36 cells were grown in the absence of ampicillin at 37°C, they were readily cured of pKM001. On the other hand, the plasmid could not be deleted from the Δlnt::kan transductant LNT6541. These results are partly consistent with the observation that the absence of Lpp lowers the level of Lnt necessary for growth, though it remains essential (35). Furthermore, the results mentioned above strongly indicate that Lnt activity is dispensable in the absence of Lpp when LolCD is overexpressed. It should be noted that pKM001 does not carry any suppressor mutations because pKM001 isolated from LNT6541 did not affect the frequency of the Δlnt::kan transduction when transformed into DLP79-36.
Growth phenotype of the lnt-null strain.
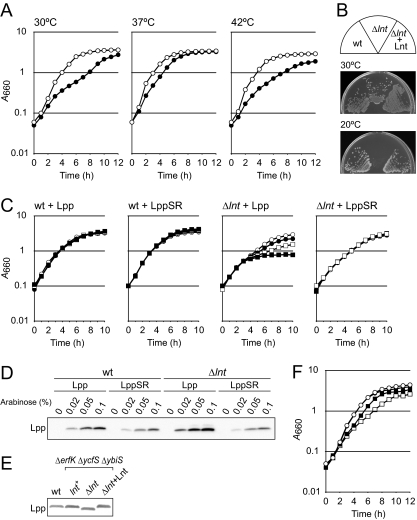
Growth of LNT6541 (Δlnt) cells harboring pKM001 was slower than that of the parental strain harboring the same plasmid at both 30°C and 37°C and severely defective at 42°C (data not shown). Since the growth of the DLP79-36 strain was also retarded when the strain harbored pKM001, it seemed likely that the growth defect of LNT6541 at high temperature is caused by the toxic effect of LolCDE overexpression from the pUC-derived high-copy-number plasmid. To address these issues, another Δlnt::kan transductant, LNT6542, was constructed from DLP79-36 cells harboring pNASCDEH2, a low-copy-number plasmid that carries a p15A replicon and constitutively overexpresses LolCDE-His. Transformation of DLP79-36 with pNASCDEH2 caused no obvious growth defect at 42°C (Fig. 2 A). The growth of LNT6542 harboring pNASCDEH2 was significantly slower than that of DLP79-36/pNASCDEH2. However, LNT6542/pNASCDEH2 continued to grow at 30°C, 37°C, and 42°C, indicating that the Δlnt mutation can be suppressed by the overproduction of LolCDE even at high temperature. On the other hand, the lnt-null strain did not form colonies at 20°C on LB agar even after incubation for 3 days (Fig. 2B). This cold-sensitive growth was corrected by plasmid pTLH631 encoding Lnt, suggesting that the Lnt activity is more critical at lower temperature. Lnt has been inferred to be important for the copper tolerance of E. coli (9, 36). Indeed, LNT6541 harboring pKM001 failed to form colonies on LB agar containing 2 mM CuSO4, while DLP79-36 harboring pKM001 and LNT6541 harboring both pKM001 and pBLH631, encoding Lnt, formed colonies (data not shown). We also found that lnt-null strains are more sensitive than the parental strain to several structurally and functionally unrelated antibiotics, such as vancomycin and erythromycin, or the dye ethidium bromide (data not shown), suggesting that the integrity of the outer membrane is perturbed by the Δlnt mutation, which makes cells sensitive to copper.
Fig. 2.
Growth of the lnt-null strain. (A) DLP79-36 (open circles) and its Δlnt::kan derivative, LNT6542 (closed circles), both harboring pNASCDEH2, were grown in LB medium at the indicated temperatures. (B) DLP79-36 (wild type [wt]), LNT6542 harboring pNASCDEH2 (Δlnt), and LNT6542 harboring both pNASCDEH2 and pTLH631 encoding Lnt-His (Δlnt + Lnt) were grown on LB agar at the indicated temperatures. (C) DLP79-36 (wt) and LNT6541 (Δlnt), both harboring pKM001, were transformed with a plasmid encoding Lpp or LppSR. Growth without arabinose (open circles) or with arabinose concentrations of 0.02% (closed circles), 0.05% (open squares), and 0.1% (closed squares) was examined. (D) Aliquots in panel C were withdrawn at 4 h, and expression of Lpp was examined by SDS-PAGE (13) and immunoblotting. (E) Cell extracts of BW25113 (wt), SN495 (BW25113 ΔybiS ΔerfK ΔycfS) (lnt+), LNT6543 (SN495 Δlnt) (Δlnt), and LNT6543 harboring pTLH631 (Δlnt + Lnt), all of which contained pNASCDEH2, were subjected to 16% acrylamide Tricine-SDS-PAGE (38), followed by immunoblotting with anti-Lpp antibodies. (F) DLP79-36 and LNT6541, both harboring pKM001, were transformed with pAM201 encoding LolA-His. Growth in the absence or presence of 0.2% arabinose was examined. Closed and open circles, DLP79-36 with and without arabinose, respectively; closed and open squares, LNT6541 with and without arabinose, respectively.
The C-terminal Lys of Lpp covalently cross-links with a peptidoglycan. This cross-linking contributes to the integrity of the outer membrane when Lpp is present in it. However, the cross-linking becomes lethal to E. coli when Lpp is mislocalized to the inner membrane (50). It has been reported that deletion of the C-terminal Lys makes E. coli cells resistant to both mislocalization and globomycin, an inhibitor of lipoprotein-specific signal peptidase (50), and less dependent on Lnt activity (35). We examined whether the C-terminal Lys of Lpp is important for growth inhibition by the lnt-null mutation. Lpp or its mutant LppSR, lacking the C-terminal Lys, was expressed in DLP79-36 and LNT6541 from plasmids carrying the respective genes under the control of the araBAD promoter (Fig. 2C). Growth of LNT6541 was increasingly inhibited when Lpp, but not LppSR, was expressed on the addition of arabinose. The expression level of LppSR, as judged on Western blotting with anti-Lpp antiserum, was lower than that of Lpp (Fig. 2D), presumably because the absence of the C-terminal Lys decreased the reactivity with antibodies.
Three different l,d-transpeptidases, encoded by ybiS, erfK, and ycfS, are capable of catalyzing cross-linking between Lpp and peptidoglycans, while YbiS is the main transpeptidase whose inactivation almost completely suppresses the covalent anchoring of Lpp to the peptidoglycans (21). The Δlnt::kan allele could be transduced by P1 into a triple mutant of ybiS, erfK, and ycfS, in which LolCDE-His was overexpressed. Immunoblotting with anti-Lpp antibodies revealed that Lpp in the resultant lnt-null strain was as abundant as in wild-type cells and migrated faster on Tricine-SDS-PAGE (Fig. 2E), presumably because it lacked N-acylation. These results support the above prediction that the formation of a covalent linkage between mislocalized Lpp and peptidoglycans is the main cause of toxicity upon Lnt inactivation.
The release of lipoproteins from the inner membrane requires both LolCDE and LolA. It was shown previously that LolA did not cause the release of apolipoproteins from proteoliposomes reconstituted with LolCDE (7). We next examined whether overexpression of LolA also allows the construction of an lnt-null strain. The Δlnt::kan allele could be transferred to DLP79-36 cells harboring pAM201 encoding LolA-His (26). However, the purified transductants formed no colony after overnight incubation even when LolA-His was induced (data not shown). On the other hand, introduction of pAM201 into the LNT6541 strain slightly improved its growth (Fig. 2F). These results indicate that overexpression of LolA does not support but improves the growth of the lnt-null strain.
Lipoproteins in the lnt-null strain lack N-acylation.
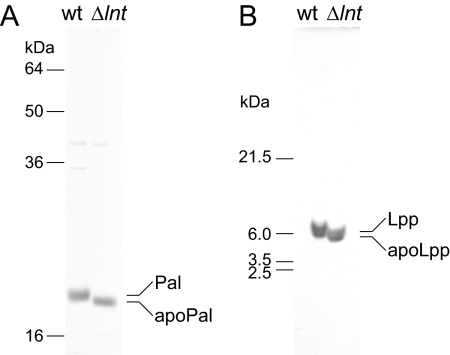
Pal and transiently expressed Lpp were partially purified from the lnt-null strain as described in Materials and Methods. Both Pal and Lpp derived from the lnt-null strain migrated slightly faster than those from wild-type cells (Fig. 3). These bands were excised and subjected to N-terminal sequence analysis by Edman degradation. Both Pal and Lpp prepared from wild-type cells gave no signal because their N termini are blocked. In contrast, Pal and Lpp prepared from the lnt-null strain possessed modified residues at the N terminus that were followed by the sequence Ser-Ser-Asn-Lys and Ser-Ser-Asn-Ala, respectively, which coincided with the N-terminal second to fifth residues of the respective lipoproteins. These results indicate that lipoproteins in the lnt-null strain are present as apolipoproteins.
Fig. 3.
SDS-PAGE analysis of lipoproteins purified from the lnt-null strain. (A) Pal was purified from DLP79-36 (wt) and LNT6541 (Δlnt), both of which harbored pKM001, and then subjected to 17.5% acrylamide SDS-PAGE (19), followed by staining with CBB. (B) Lpp was expressed from a plasmid, pJY811, in the strains described for panel A, purified, and analyzed by SDS-PAGE (13) and CBB staining. The indicated bands were excised, and the N-terminal amino acid sequences were determined by Edman degradation.
Sorting of apolipoproteins to the outer membrane.
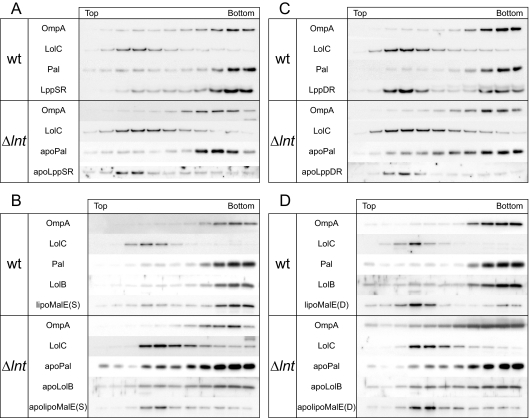
N-acylation is essential for the release of lipoproteins from proteoliposomes reconstituted with LolCDE (7). Moreover, lipoproteins expressed under Lnt-depleted conditions remained in the inner membrane (35). Based on these observations, we examined the membrane localization of apolipoproteins in the lnt-null strain overexpressing LolCDE. DLP79-36 and LNT6541, both harboring pKM001, were transformed with pJY851 encoding LppSR. Membranes were prepared from these cells after 30-min expression of LppSR induced by the addition of arabinose. An outer membrane protein, OmpA, and an inner membrane protein, LolC, were localized in the correct membrane fractions of not only the wild-type, but also the lnt-null strain (Fig. 4). Pal and LppSR were localized in the outer membranes of wild-type cells (Fig. 4A). ApoPal was also found in the outer membrane of the lnt-null strain, indicating that apoPal can be correctly sorted to the outer membrane when the Δlnt mutation is suppressed by the overexpression of LolCDE. However, most apoLppSR molecules remained in the inner membrane of the lnt-null strain (Fig. 4A), presumably because the amount of apoLppSR exceeds the capacity of LolCDE. Alternatively, because Lpp loses its release competence soon after maturation on the outer leaflet of the inner membrane, presumably due to trimerization, while Pal retains its release competence for at least 15 min after maturation (51), decreased release activity would have preferentially caused accumulation of release-incompetent Lpp trimers in the inner membrane of the lnt-null strain. In any case, these results reveal why the Δlnt::kan allele could not be transferred to wild-type cells expressing Lpp. Accumulation of apolipoproteins in the inner membrane was also observed for lipoMalE(S), a model lipoprotein that has a lipoprotein-specific signal peptide fused to the mature region of maltose-binding protein (29). LipoMalE(S), which has Ser at position 2, was sorted to the outer membrane of wild-type cells as previously observed (29), whereas it remained in the inner membrane when expressed in the lnt-null strain (Fig. 4B). While apoPal was correctly sorted to the outer membrane, sorting of another authentic lipoprotein, LolB, was less efficient in the lnt-null strain (Fig. 4B). Thus, the efficiency of outer membrane targeting of each apolipoprotein varies depending on the nature of its protein moiety. It is assumed that a considerable amount of apolipoproteins could be located in the inner membrane of the lnt-null strain, despite their outer membrane-targeting signals.
Fig. 4.
Membrane localization of lipoproteins in the lnt-null strain. The wild type (wt) and the lnt-null strain (Δlnt) were transformed with a plasmid encoding LppSR (A), lipoMalE(S) (B), or their derivatives with Asp at position 2 (C and D). Expression of LppSR and LppDR was induced by the addition of arabinose for 30 min. Membranes were separated with a sucrose density gradient into inner and outer membranes. Proteins were detected by immunoblotting using antibodies raised against the respective proteins. LipoMalE was expressed without induction and was detected with anti-maltose-binding protein.
Asp at position 2 of lipoproteins functions as an inner membrane retention signal because it prevents the recognition of lipoproteins by LolCDE (10, 22). The membrane localization of LppDR, which has a Ser-to-Asp substitution at position 2 while the C-terminal Lys has been deleted to avoid cross-linking with peptidoglycans, was determined. Most LppDR molecules with Asp in place of Ser at position 2 accumulated in the inner membranes of wild-type cells (Fig. 4C), although an appreciable amount of LppDR was also found in the outer membranes of wild-type cells, presumably because of the overproduction of LolCDE, as observed previously (28). LipoMalE(D), a derivative of lipoMalE(S) with Asp at position 2, was mainly accumulated in the inner membranes of wild-type cells (Fig. 4D). In the lnt-null strains, both apoLppDR and apolipoMalE(S) were exclusively detected in the inner membrane fraction (Fig. 4C and D). Because the sorting of these apolipoproteins was inefficient, it is unclear whether Asp at position 2 functions as a LolCDE avoidance signal without N-acylation.
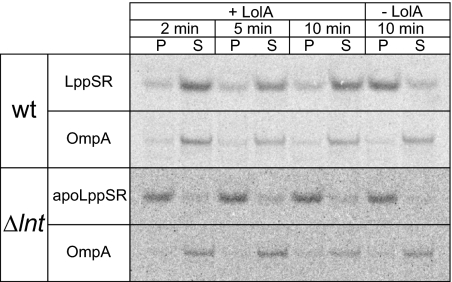
LolA-dependent release of LppSR and apoLppSR was examined in spheroplasts prepared from DLP79-36 and LNT6541 cells, both of which were overproducing LolCDE. LppSR was efficiently released from spheroplasts prepared from wild-type cells in a LolA-dependent manner (Fig. 5). In contrast, only a marginal amount of apoLppSR was released from spheroplasts of LNT6541 cells in the presence of LolA, indicating that the efficiency of apoLppSR that was released from the inner membrane was much lower than that of mature LppSR.
Fig. 5.
Release of lipoproteins from spheroplasts of the lnt-null strain. DLP79-36 (wt) and LNT6541 (Δlnt), both harboring pKM001 and pJY851, were converted into spheroplasts. Release of 35S-labeled LppSR from the spheroplasts was examined in the presence and absence of LolA. The spheroplasts were subjected to labeling for 2 min, followed by chasing for the indicated periods. LppSR and OmpA in the spheroplast (P) and supernatant (S) fractions were immunoprecipitated and analyzed by SDS-PAGE, followed by phosphorimaging.
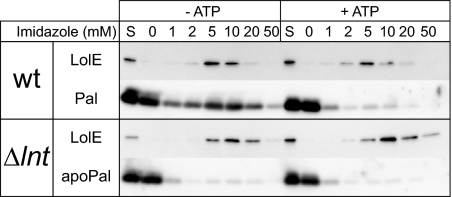
When LolCDE was purified in the absence of ATP after solubilization of membranes with a detergent, outer-membrane-specific, but not inner-membrane-specific, lipoproteins were copurified (15). This LolCDE-lipoprotein complex represents the intermediate of the LolCDE-mediated release reaction (15, 44). We examined complex formation between LolCDE and apoPal. Membrane fractions were prepared from the wild-type and lnt-null strains, both overexpressing LolCDE with a hexahistidine tag at the C terminus of LolE, and solubilized with DDM, followed by immobilized metal affinity chromatography in the presence and absence of ATP. LolE-His was always eluted from the metal affinity resin with a buffer containing 5 to 20 mM imidazole, indicating that LolCDE is eluted in these fractions (Fig. 6). Immunoblotting with anti-Pal antibodies showed that Pal was copurified with LolCDE-His that was purified from wild-type membranes in the absence of ATP. In contrast, apoPal was hardly copurified with LolCDE-His solubilized from membranes of the lnt-null strain (Fig. 6). Because it was recently shown that Pal derivatives with Asp at position 2 were not copurified with LolCDE (37), it is unlikely that mature Pal exhibits intrinsic affinity for the metal affinity resin. Rather, these results indicate that the affinity of LolCDE for apoPal is low in regard to the formation of a stable complex.
Fig. 6.
N-acylation is required for formation of the lipoprotein-LolCDE complex. LolCDE was solubilized with DDM from membranes of DLP79-36 and LNT6542, both harboring pNASCDEH2. LolCDE-His was adsorbed to a metal affinity resin and eluted with the indicated concentrations of imidazole. Each fraction was analyzed by SDS-PAGE and immunoblotting. S, 1/10 the amount of solubilized membrane proteins.
DISCUSSION
It is known that a lack of Lpp decreases the requirement for the activities of both lipoprotein-processing enzymes (8, 9) and the Lol system (41, 43, 52). Lpp is the most abundant protein in E. coli, and its mislocalization to the inner membrane causes the formation of lethal cross-linking with peptidoglycans (50). As demonstrated here, the lnt gene could be deleted when LolCDE was overexpressed in cells lacking Lpp or l,d-transpeptidases that cross-link Lpp with peptidoglycans. Unless LolCDE was overexpressed, the lnt gene was still required even in the absence of Lpp. This might indicate that N-acylation is important to ensure the efficient sorting of lipoproteins, especially ones such as LolB, BamD, and LptE, which are involved in outer membrane biogenesis. Alternatively, some other lipoproteins, like Pal and LolB, might be very deleterious when they remain in the inner membrane (54). In any case, we concluded that by means of overexpessed LolCDE, non-N-acylated lipoproteins can be released from the inner membrane, transported across the periplasm by LolA, and then incorporated into the outer membrane by LolB.
Suppression of the lnt-null mutation by LolCDE overexpression resembles the suppression of the htrB-null mutation by the overexpression of MsbA (16). The htrB (later renamed lpxL) gene encodes an enzyme that transfers an acyl chain to the tetra-acylated precursor of di[3-deoxy-d-manno-octulosonyl]-lipid A (Kdo2-lipid A), a core component of essential lipopolysaccharides (LPS) (40). The htrB-null strain accumulates the tetra-acylated LPS precursor in the inner membrane under nonpermissive conditions (32, 55). Overexpression of MsbA translocates the tetra-acylated LPS precursor to the outer membrane and allows the growth of the htrB-null strain. The tetra-acylated form of Kdo2-lipid A is a poor substrate for MsbA and is sorted to the outer membrane when a large amount of MsbA is present. Although Kdo is required to stimulate HtrB activity, it was recently reported that overexpression of MsbA also compensates for a Kdo deficiency (24, 34).
In contrast to Pal, affinity purification of LolCDE in the absence of ATP revealed that apoPal was not copurified with LolCDE (Fig. 6), indicating that N-acylation is important for the intimate interaction between lipoproteins and LolCDE. It is not known at present whether N-acylation of lipoproteins also affects the interaction with LolA and LolB. However, since apolipoproteins are localized to the outer membrane of the lnt-null strain, both Lol proteins should be able to recognize apolipoproteins. Indeed, overproduction of LolA improved the growth of the lnt-null strain, but LolCDE overexpression remained essential (Fig. 2E). Taken together, these results indicate that the lipoprotein release step catalyzed by LolCDE is the most critical step of the lipoprotein-sorting pathway and therefore requires ATP energy.
The crystal structures of LolA and LolB revealed that these proteins comprise similar hydrophobic cavities that are most likely the binding sites for the acyl chains of lipoproteins (42). However, the size of the cavities is not large enough to accommodate all three acyl chains. The LolA homolog of Pseudomonas aeruginosa was recently reported to possess hydrophobic surface patches that might accommodate two acyl chains (33). On the other hand, LprG is a lipoprotein of Mycobacterium tuberculosis and has a LolA/LolB-like hydrophobic cavity, whose size (∼1,500 Å3) is large enough to bind three acyl chains (6). LprG was speculated to carry triacylated lipoproteins to Toll-like receptor 2 (TLR2), a member of the TLR family that recognizes various microbial molecules and plays a crucial role in innate immunity (17). It was recently found that the LolA cavity undergoes opening and closing upon the binding and release of lipoproteins, respectively (31). The cavity size of the open form of LolA was speculated to be ∼1,700 Å3 (Y. Oguchi and H. Tokuda, unpublished data). Therefore, both LolA and LolB might be able to accommodate all three acyl chains in their cavities, although the crystal structure of LolA or LolB bound to a lipoprotein has not been solved yet.
The S. enterica mutant with a point mutation of lnt was temperature sensitive (9). Amino acid substitutions in Lnt of E. coli that result in temperature-sensitive complementation of a conditional lnt mutant have been also reported (48). In contrast, the lnt-null strain exhibited a cold-sensitive phenotype, indicating that the absence of N-acylation itself has a more deleterious effect on the growth of E. coli at lower temperatures. This might indicate that the activity of LolCDE is low in the cold, where the membrane fluidity decreases. On the other hand, the cold sensitivity might be due to the impaired function of the outer membrane. The E. coli chromosome encodes at least 80 species of outer membrane lipoproteins (46). In the lnt-null strain, they are expected to exist as diacylated lipoproteins in the outer membrane. The way in which diacylated lipoproteins affect the integrity of the outer membrane is of great interest.
ACKNOWLEDGMENTS
We thank an anonymous reviewer for suggesting the construction of an lnt-null strain in the absence of l,d-transpeptidases, Shigeo Tomioka for determination of the N-terminal amino acid sequence, and Yoshinori Akiyama for the laboratory resources.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Asanuma M., et al. 2011. Structural evidence of α-aminoacylated lipoproteins of Staphylococcus aureus. FEBS J. 278:716–728 [DOI] [PubMed] [Google Scholar]
- 2. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buddelmeijer N., Young R. 2010. The essential Escherichia coli apolipoprotein N-acyltransferase (Lnt) exists as an extracytoplasmic thioester acyl-enzyme intermediate. Biochemistry 49:341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cherepanov P. P., Wackernagel W. 1995. Gene disruption in Escherichia coli: Tcr and Kmr cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 5. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drage M. G., et al. 2010. Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2. Nat. Struct. Mol. Biol. 17:1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fukuda A., et al. 2002. Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J. Biol. Chem. 277:43512–43518 [DOI] [PubMed] [Google Scholar]
- 8. Gan K., Gupta S. D., Sankaran K., Schmid M. B., Wu H. C. 1993. Isolation and characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in prolipoprotein modification. J. Biol. Chem. 268:16544–16550 [PubMed] [Google Scholar]
- 9. Gupta S. D., Gan K., Schmid M. B., Wu H. C. 1993. Characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in apolipoprotein N-acyltransferase. J. Biol. Chem. 268:16551–16556 [PubMed] [Google Scholar]
- 10. Hara T., Matsuyama S., Tokuda H. 2003. Mechanism underlying the inner membrane retention of Escherichia coli lipoproteins caused by Lol avoidance signals. J. Biol. Chem. 278:40408–40414 [DOI] [PubMed] [Google Scholar]
- 11. Hirashima A., Wu H. C., Venkateswaran P. S., Inouye M. 1973. Two forms of a structural lipoprotein in the envelope of Escherichia coli. Further characterization of the free form. J. Biol. Chem. 248:5654–5659 [PubMed] [Google Scholar]
- 12. Horwitz A. H., Morandi C., Wilcox G. 1980. DNA sequence of araBAD promoter mutants of Escherichia coli. J. Bacteriol. 142:659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hussain M., Ichihara S., Mizushima S. 1980. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J. Biol. Chem. 255:3707–3712 [PubMed] [Google Scholar]
- 14. Ito H., et al. 2007. A new screening method to identify inhibitors of the Lol (localization of lipoproteins) system, a novel antibacterial target. Microbiol. Immunol. 51:263–270 [DOI] [PubMed] [Google Scholar]
- 15. Ito Y., Kanamaru K., Taniguchi N., Miyamoto S., Tokuda H. 2006. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins. Mol. Microbiol. 62:1064–1075 [DOI] [PubMed] [Google Scholar]
- 16. Karow M., Georgopoulos C. 1993. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 7:69–79 [DOI] [PubMed] [Google Scholar]
- 17. Kawai T., Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384 [DOI] [PubMed] [Google Scholar]
- 18. Kurokawa K., et al. 2009. The triacylated ATP binding cluster transporter substrate-binding lipoprotein of Staphylococcus aureus functions as a native ligand for Toll-like receptor 2. J. Biol. Chem. 284:8406–8411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 20. Lewenza S., Vidal-Ingigliardi D., Pugsley A. P. 2006. Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae. J. Bacteriol. 188:3516–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Magnet S., Dubost L., Marie A., Arthur M., Gutmann L. 2008. Identification of the l,d-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J. Bacteriol. 190:4782–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Masuda K., Matsuyama S., Tokuda H. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. U. S. A. 99:7390–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsuyama S., Tajima T., Tokuda H. 1995. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 14:3365–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meredith T. C., Aggarwal P., Mamat U., Lindner B., Woodard R. W. 2006. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Biol. 1:33–42 [DOI] [PubMed] [Google Scholar]
- 25. Miller J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Miyamoto A., Matsuyama S., Tokuda H. 2001. Mutant of LolA, a lipoprotein-specific molecular chaperone of Escherichia coli, defective in the transfer of lipoproteins to LolB. Biochem. Biophys. Res. Commun. 287:1125–1128 [DOI] [PubMed] [Google Scholar]
- 27. Mizuno T. 1979. A novel peptidoglycan-associated lipoprotein found in the cell envelope of Pseudomonas aeruginosa and Escherichia coli. J. Biochem. 86:991–1000 [DOI] [PubMed] [Google Scholar]
- 28. Narita S., Kanamaru K., Matsuyama S., Tokuda H. 2003. A mutation in the membrane subunit of an ABC transporter LolCDE complex causing outer membrane localization of lipoproteins against their inner membrane-specific signals. Mol. Microbiol. 49:167–177 [DOI] [PubMed] [Google Scholar]
- 29. Narita S., Tokuda H. 2007. Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J. Biol. Chem. 282:13372–13378 [DOI] [PubMed] [Google Scholar]
- 30. Narita S., Tokuda H. October 2010, posting date Chapter 4.3.7, Biogenesis and membrane targeting of lipoproteins. In Böck A., et al.(ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: http://www.ecosal.org/ [DOI] [PubMed] [Google Scholar]
- 31. Oguchi Y., et al. 2008. Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release. J. Biol. Chem. 283:25414–25420 [DOI] [PubMed] [Google Scholar]
- 32. Polissi A., Georgopoulos C. 1996. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 20:1221–1233 [DOI] [PubMed] [Google Scholar]
- 33. Remans K., et al. 2010. Hydrophobic surface patches on LolA of Pseudomonas aeruginosa are essential for lipoprotein binding. J. Mol. Biol. 401:921–930 [DOI] [PubMed] [Google Scholar]
- 34. Reynolds C. M., Raetz C. R. 2009. Replacement of lipopolysaccharide with free lipid A molecules in Escherichia coli mutants lacking all core sugars. Biochemistry 48:9627–9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robichon C., Vidal-Ingigliardi D., Pugsley A. P. 2005. Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J. Biol. Chem. 280:974–983 [DOI] [PubMed] [Google Scholar]
- 36. Rogers S. D., Bhave M. R., Mercer J. F., Camakaris J., Lee B. T. 1991. Cloning and characterization of cutE, a gene involved in copper transport in Escherichia coli. J. Bacteriol. 173:6742–6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sakamoto C., Satou R., Tokuda H., Narita S. 2010. Novel mutations of the LolCDE complex causing outer membrane localization of lipoproteins despite their inner membrane-retention signals. Biochem. Biophys. Res. Commun. 401:586–591 [DOI] [PubMed] [Google Scholar]
- 38. Schägger H., von Jagow G. 1987. Tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis for the separation of proteins in the range from 1-100 kDalton. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 39. Seydel A., Gounon P., Pugsley A. P. 1999. Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34:810–821 [DOI] [PubMed] [Google Scholar]
- 40. Sperandeo P., Dehò G., Polissi A. 2009. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta 1791:594–602 [DOI] [PubMed] [Google Scholar]
- 41. Tajima T., Yokota N., Matsuyama S., Tokuda H. 1998. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett. 439:51–54 [DOI] [PubMed] [Google Scholar]
- 42. Takeda K., et al. 2003. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 22:3199–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tanaka K., Matsuyama S., Tokuda H. 2001. Deletion of lolB, encoding an outer membrane lipoprotein, is lethal for Escherichia coli and causes accumulation of lipoprotein localization intermediates in the periplasm. J. Bacteriol. 183:6538–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taniguchi N., Tokuda H. 2008. Molecular events involved in a single cycle of ligand transfer from an ATP binding cassette transporter, LolCDE, to a molecular chaperone, LolA. J. Biol. Chem. 283:8538–8544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Terada M., Kuroda T., Matsuyama S., Tokuda H. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 276:47690–47694 [DOI] [PubMed] [Google Scholar]
- 46. Tokuda H., Matsuyama S., Tanaka-Masuda K. 2007. Structure, function and transport of lipoproteins in Escherichia coli, p. 67–79 In Ehrmann M.(ed.),The periplasm. ASM Press, Washington, DC [Google Scholar]
- 47. Tschumi A., et al. 2009. Identification of apolipoprotein N-acyltransferase (Lnt) in mycobacteria. J. Biol. Chem. 284:27146–27156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vidal-Ingigliardi D., Lewenza S., Buddelmeijer N. 2007. Identification of essential residues in apolipoprotein N-acyl transferase, a member of the CN hydrolase family. J. Bacteriol. 189:4456–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Widdick D. A., et al. 2011. Dissecting the complete lipoprotein biogenesis pathway in Streptomyces scabies. Mol. Microbiol. 80:1395–1412 [DOI] [PubMed] [Google Scholar]
- 50. Yakushi T., Tajima T., Matsuyama S., Tokuda H. 1997. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J. Bacteriol. 179:2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yakushi T., Yokota N., Matsuyama S., Tokuda H. 1998. LolA-dependent release of a lipid-modified protein from the inner membrane of Escherichia coli requires nucleoside triphosphate. J. Biol. Chem. 273:32576–32581 [DOI] [PubMed] [Google Scholar]
- 52. Yakushi T., Masuda K., Narita S., Matsuyama S., Tokuda H. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212–218 [DOI] [PubMed] [Google Scholar]
- 53. Yamaguchi K., Yu F., Inouye M. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53:423–432 [DOI] [PubMed] [Google Scholar]
- 54. Yokota N., Kuroda T., Matsuyama S., Tokuda H. 1999. Characterization of the LolA-LolB system as the general lipoprotein localization mechanism of Escherichia coli. J. Biol. Chem. 274:30995–30999 [DOI] [PubMed] [Google Scholar]
- 55. Zhou Z., White K. A., Polissi A., Georgopoulos C., Raetz C. R. 1998. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273:12466–12475 [DOI] [PubMed] [Google Scholar]