Abstract
The intermolecular interactions of the mycobacteriophage Ms6 secretion chaperone with endolysin were characterized. The 384-amino-acid lysin (lysin384)-binding domain was found to encompass the N-terminal region of Gp1, which is also essential for a lysis phenotype in Escherichia coli. In addition, a GXXXG-like motif involved in Gp1 homo-oligomerization was identified within the C-terminal region.
TEXT
Mycobacteriophages, phages that specifically infect mycobacteria, have evolved remarkable and sophisticated lysis mechanisms to overcome the disadvantage that the mycobacterial complex cell envelope represents for a successful infective cycle (1, 11). Mycobacteriophage Ms6 is a temperate double-stranded DNA bacteriophage (15) with an unusual lysis cassette: in addition to the endolysin-holin lysis system encoded by genes lysA (gp2) and gp4 and gp5 (2, 7), the Ms6 lytic region comprises two accessory lysis proteins encoded by genes gp1 and gp3 (lysB). The lysB gene encodes a lipolytic enzyme that was shown to hydrolyze the ester bond between the mycolic acids and the arabinogalactan in the mycolyl-arabinogalactan-peptidoglycan complex, compromising the stability of the mycobacterial outer membrane (8, 9, 13).
The Ms6 lysis mechanism is also unique in that the endolysin (LysA) does not possess an N-terminal signal sequence which would allow the transport via the host Sec system, as it happens with endolysins exported in a holin-independent manner. Our previous results have shown that translocation of Ms6 LysA is assisted by the gene product of gp1, a chaperone-like protein. Gp1 specifically binds the N-terminal 60 amino acids of the Ms6 endolysin and allows the enzyme access to its substrate, the peptidoglycan, by somehow facilitating its secretion across the cytoplasmic membrane independently of Ms6 holin-like protein activity (1, 2).
Even though lysA was shown to encode two proteins (3), the 384-amino-acid lysin (lysin384) and lysin241, with lysin241 resulting from the use of an internal, in-frame translation initiation site within the lysA gene, only lysin384 interacts with Gp1 (1). However, both forms of the endolysin are essential for the normal timing, progression, and completion of host cell lysis. The secretion state seems to rely on specific binding of Gp1 to the endolysin, as in the absence of the chaperone, the enzyme stability is severely affected during Mycobacterium smegmatis infection (3).
The central and C-terminal regions of Gp1 are involved in homo-oligomerization.
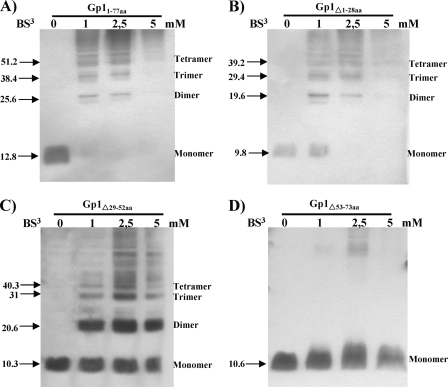
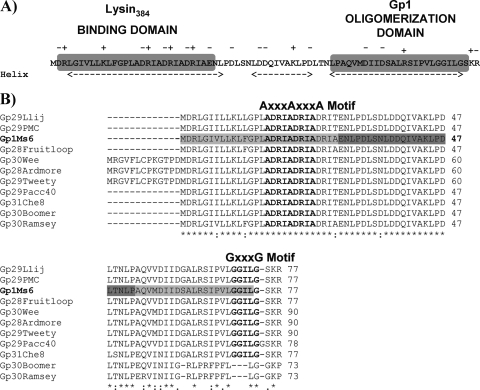
In a previous study, we observed that Gp1 oligomerizes up to pentamers, an important feature for its role in mycobacterial lysis, as Gp1 homo-oligomers interact with Ms6 endolysin during phage infection of M. smegmatis (1). To determine the region involved in homo-oligomerization, deletions of the N-terminal, central, or C-terminal Gp1 region were generated by a QuikChange site-directed mutagenesis protocol for large deletions (Stratagene) (16, 20). Deletions were performed on a pET29 derivative plasmid that harbors the gp1 gene (pMJC1) (1), yielding plasmids pMJC1Δ1-84bpgp1, pMJC1Δ85-157bpgp1, and pMJC1Δ158-221bpgp1 (Table 1). Loss of the first 28 amino acids (Gp1Δ1-28) was not sufficient to abrogate oligomerization, and a cross-linking pattern identical to the wild-type Gp1 was observed (Fig. 1A and B). However, removal of the central 24 amino acids (Gp1Δ29-52) resulted in an altered cross-linking pattern with monomeric and dimeric form accumulation (Fig. 1C). In addition, deletion of the 21 amino acids (Gp1Δ53-73) at the C terminus eliminated the interaction (Fig. 1 D), which suggests that both the central and C-terminal domains are important for Gp1 oligomerization. Both experimental and statistical searches for specific motifs that mediate transmembrane helix-helix interactions showed that motifs of two small amino acids can assist but are not sufficient to mediate transmembrane helix interactions: two glycine residues separated by three intervening residues (GXXXG) are often found to mediate specific interactions of α-helices (6, 18, 19). The C-terminal regions of Gp1 and related proteins of F1 subcluster mycobacteriophages except for phages Boomer and Ramsey (Fig. 2A) have a potential α-helix with a GXXXG-like motif (G70GILG74) that might serve as the basis for homotypic helix interactions and stabilization of intramolecular interactions within Gp1 (Fig. 2B).
Table 1.
Strains, oligonucleotides, and plasmids used in this study
| Strain, plasmid, or oligonucleotide | Description or sequence (5′-3′)a | Reference or source |
|---|---|---|
| Bacteria (Escherichia coli) | ||
| JM109 | recA1 endA1 gyr96 thi hsdR17 supE44 relA1Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Stratagene |
| BL21 | F−ompThsdSB (rB− mB−) gal dmc | Novagen |
| BL21(DE3) | F−ompThsdSB (rB− mB−) gal dmc (DE3) | Novagen |
| XL1-Blue | endA1 gyrA96(Nalr) thi-1recA1relA1lacglnV44 F′[::Tn10 proAB+lacIqΔ(lacZ)M15] hsdR17 (rK− mK+) | Stratagene |
| Plasmids | ||
| pQE30 | Expression vector; T5 promoter; Ampr, lacIq | Qiagen |
| pET29b(+) | Expression vector; T7 promoter; Kanr | Novagen |
| pMJC1 | gp1 cloned in pET29b | 1 |
| pMJC1Δ1-84bpgp1 | gp1 with 84-bp deletion at the 5′ end cloned in pET29b | This study |
| pMJC1Δ85-157bpgp1 | gp1 with 72-bp central deletion cloned in pET29b | This study |
| pMJC1Δ158-221bpgp1 | gp1 with 63-bp deletion at the 3′ end cloned in pET29b | This study |
| pMJC3 | gp1 and lysA cloned in pET29b | 1 |
| pMJC3Δ1-84bpgp1 | gp1 with 84-bp deletion at the 5′ end and lysA cloned in pET29b | This study |
| pMJC3Δ85-157bpgp1 | gp1 with 72-bp central deletion and lysA cloned in pET29b | This study |
| pMJC3Δ158-221bpgp1 | gp1 with 63-bp deletion at the 3′ end and lysA cloned in pET29b | This study |
| pMP320 | gp1 and lysA cloned in pQE30 | 1 |
| pMP320Δ1-84bpgp1 | gp1 with 84-bp deletion at the 5′ end and lysA cloned in pQE30 | This study |
| pMP320Δ85-157bpgp1 | gp1 with 72-bp central deletion and lysA cloned in pQE30 | This study |
| pMP320Δ158-221bpgp1 | gp1 with 63-bp deletion at the 3′ end and lysA cloned in pQE30 | This study |
| Oligonucleotides | ||
| PrΔ1-84bpgp1fwd | GGTACCCTGGTGCCACGCGGTTCCATGGCGATATCGGATCCCGAGAACCTGCCCGACCTGTCCAAC | This study |
| PrΔ1-84bpgp1rv | GTTGGACAGGTCGGGCAGGTTCTCGGGATCCGATATCGCCATGGAACCGCGTGGCACCAGGGTACC | This study |
| PrΔ85-157bpgp1fwd | CGACCGAATCGCCGACAGGATCGCCGCGCAAGTCATGGACATCATCGACAGCG | This study |
| PrΔ85-157bpgp1rv | CGCTGTCGATGATGTCCATGACTTGCGCGGCGATCCTGTCGGCGATTCGGTCG | This study |
| PrΔ158-221bpgp1fwd | GCGAAACTCCCCGACCTGACCAACCTTCCAGGGAGCAAACGGTGACC | This study |
| PrΔ158-221bpgp1rv | GGTCACCGTTTGCTCCCTGGAAGGTTGGTCAGGTCGGGGAGTTTCGC | This study |
The GenBank accession number for Ms6 lysis genes is AF319619.
Fig. 1.
Ms6 Gp1 homo-oligomerization. Protein extracts from of E. coli BL21(DE3) expressing Gp1 (Gp11-77aa)or Gp1 deletion proteins were cross-linked with different bis(sulfosuccinimidyl)suberate (BS3) concentrations as previously described (1). Proteins were detected by Western blotting with an anti-His6 antibody. Predicted molecular masses (in kDa) of the Gp1 homo-oligomers are indicated to the left of the panels. Oligomerization bands are indicated by arrows.
Fig. 2.
(A) Gp1 secondary structure protein prediction according to the Chou and Fasman algorithm (4, 5; http://www.biogem.org/tool/chou-fasman/). Charged residues in the Gp1 amino acid sequence are indicated by + or −. N-terminal lysin384-binding and C-terminal homo-oligomerization domains on Gp1 are indicated in gray boxes. (B) CLUSTALW alignment of Ms6 Gp1 (AAG48317) with similar sequences of members included in subcluster F1, namely, Llij Gp29 (ABD58248), PMC Gp29 (ABE67530), Fruitloop Gp28 (YP002241713), Wee Gp30 (YP004123852), Ardmore Gp28 (ACY39910), Tweety Gp29 (YP001469262), Pacc40 Gp29 (YP002241613), Che8 Gp31 (NP817369), Boomer Gp30 (YP002014246), and Ramsey Gp30 (YP002241817) (the primary accession numbers in the UniProtKB/TrEmb1 database are given in parentheses). Identical (*), highly similar (:), and similar (·) amino acids are indicated. Numbers refer to the amino acids positions. The AXXXAXXXA and GXXXG motifs are indicated in bold above the protein amino acid sequence. N-terminal/C-terminal and central Gp1 deletions are indicated by pale and dark gray shading, respectively.
LysA binding requires the N-terminal region of Gp1.
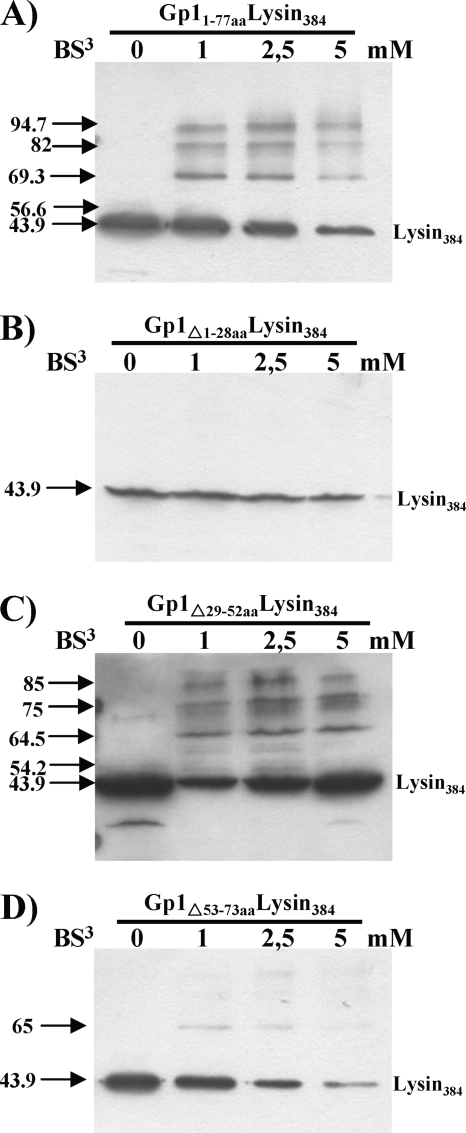
The region of binding of Ms6 lysin384 to its chaperone has been already determined (Fig. 3A) and encompasses the N-terminal region of the enzyme, which is sufficient for the interaction (1). To further characterize this interaction, we deleted the same regions in gp1, as described above but using plasmid pMJC3, which harbors the gp1 and lysA genes. The shortened proteins were expressed from the generated plasmids pMJC3Δ1-84bpgp1, pMJC3Δ85-157bpgp1, and pMJC3Δ158-221bpgp1 (Table 1) and cross-linked with the wild-type lysin384 as previously described (1). We observed that neither the loss of the central 24 amino acids (Gp1Δ29-52) (Fig. 3C) nor the loss of the C-terminal 21 amino acids (Gp1Δ53-73) of Gp1 (Fig. 3D) was sufficient to abolish the interaction, even though deletion of the Gp1 C-terminal domain decreased lysin384 binding (Fig. 3D), presumably because this region is involved in Gp1 homo-oligomerization. Conversely, removal of the N-terminal 28 amino acids of Gp1 (Gp1Δ1-28) resulted in loss of Gp1 and lysin384 hetero-oligomerization, which indicates that lysin384 no longer binds (Fig. 3B). In addition to the GXXXG motif, an AXXXA motif has been also identified in α-helical interactions in soluble proteins (12, 17). Of note is the fact that the N-terminal regions of Gp1 and related proteins also possess an AXXXAXXXA sequence (A16DRIA20DRIA24) within the α-helix (Fig. 2A and B), which led us to think that it may stabilize intermolecular interactions between the endolysin and Gp1 subunits.
Fig. 3.
Interaction of different regions of S-tagged Gp1 with lysin384. BS3 cross-linking of E. coli lysates expressing lysin384 and wild-type Gp1 (Gp11-77aa) (A), Gp1Δ1-28 (B), Gp1Δ29-52 (C), or Gp1Δ53-73 (D). Cross-linking experiments were performed as previously described (1). The numbers to the left are molecular masses (in kDa).
E. coli lysis requires the N-terminal region of Gp1.
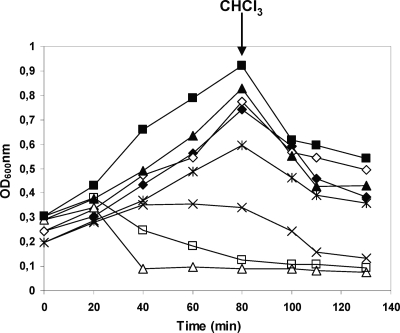
Besides determination of the Gp1 regions that interact with LysA, investigation of its contribution to the lysis phenotype is also important. Wild-type Gp1 and mutant proteins were coexpressed with wild-type lysin384 in Escherichia coli, under the control of a T5 promoter by using derivative plasmids of pMP320 (a pQE30 [Qiagen] derivative that harbors gp1 and lysA genes) (2). Deletions of the N-terminal, central, and C-terminal Gp1 regions were made as described above, generating plasmids pMP320Δ1-84bpgp1, pMP320Δ85-157bpgp1, and pMP320Δ158-221bpgp1 (Table 1). E. coli JM109 carrying plasmid pQE30 or the derivative plasmids was grown in LB broth at 37°C until the optical density at 600 nm (OD600) was 0.2 to 0.3. At time zero, transcription of cloned genes was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Expression of wild-type Gp1 and lysin384 resulted in cell lysis, beginning 20 min after induction (Fig. 4). However, this phenotype was not observed when the N-terminal region of Gp1 was absent (Fig. 4). This result strengthens the hypothesis that Gp1 binding to lysin384 is required for a lysis phenotype. Interestingly, removal of the central 24 amino acids of Gp1 results in an accentuated lysis phenotype in E. coli cells, presumably due to an increase of the Gp1 hydrophobicity (Fig. 4). In addition, deletion of the C-terminal Gp1 region results in growth inhibition; however, a lysis phenotype could not be observed (Fig. 4), suggesting that Gp1 homo-oligomerization is required for LysA access to the peptidoglycan.
Fig. 4.
Effect of lysin384 expression with Gp1 and Gp1 deletion derivatives on E. coli growth. E. coli JM109 carrying plasmid pQE30 with no insert or cloned genes was grown in LB broth at 37°C to an OD600 of 0.2. At time zero, transcription of cloned lysis genes was induced with 1 mM IPTG. Culture turbidity was monitored at 600 nm. At the time indicated by the arrow, 2% CHCl3 was added to cultures. ✦, Gp1Δ1-28–lysin384, uninduced; ◊, Gp1Δ1-28–lysin384, induced; ▴, Gp1Δ29-52–lysin384, uninduced; ▵, Gp1Δ29-52–lysin384, induced; *, Gp1Δ53-73–lysin384, uninduced; ×, Gp1Δ53-73-lysin384, induced; ▪, wild-type Gp1-lysin384, uninduced; □, wild-type Gp1-lysin384, induced.
The data presented herein indicate that Gp1 interaction with lysin384 requires the N-terminal region of the chaperone, and the relevant region has been narrowed to amino acids 1 to 28: deletion of this region completely abolishes hetero-oligomerization between the two proteins and also hinders E. coli lysis. However, additional studies in mycobacteria are needed due to the lack of information regarding the physiological levels and localization of Ms6 lysis proteins in M. smegmatis and the limitations that exist when employing overexpression systems. To date, at least 80 mycobacteriophage genome sequences are available in GenBank (14); however, excepting the study on mycobacteriophage Ms6 endolysin translocation (1, 2), little is known about how the mycobacteriophage lytic enzymes, the endolysin LysA, and the lipolytic enzyme LysB are localized to their substrates. Concerning the sequence similarity of the lysis regions of mycobacteriophages grouped in subcluster F1 and Ms6, we conceived that the molecular mechanisms underlying F1 mycobacteriophage lysis of mycobacteria are identical. We propose that N-terminal interactions linking lysin384 and the secretion chaperone Gp1 allow the enzyme to be exported to the periplasm during phage assembly in order to guarantee rapid cell lysis when lysis is triggered.
Acknowledgments
We thank the Fundação para a Ciência e Tecnologia (FCT) (PTDC/SAU-FCF/73017/2006) for financial support. M. J. Catalão (SFRH/BD/24452/2005) and F. Gil (SFRH/BD/29167/2006) are recipients of Ph.D. fellowships from FCT.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Catalão M. J., Gil F., Moniz-Pereira J., Pimentel M. 2010. The mycobacteriophage Ms6 encodes a chaperone-like protein involved in the endolysin delivery to the peptidoglycan. Mol. Microbiol. 77:672–686 [DOI] [PubMed] [Google Scholar]
- 2. Catalão M. J., Gil F., Moniz-Pereira J., Pimentel M. 2011. Functional analysis of the holin-like proteins of mycobacteriophage Ms6. J. Bacteriol. 193:2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Catalão M. J., Gil F., Moniz-Pereira J., Pimentel M. 2011. A second endolysin gene is fully embedded in-frame with the lysA gene of mycobacteriophage Ms6. PLoS One 6:e20515 doi:10.1371/journal.pone.0020515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chou P. Y., Gerald D. F. 1974. Prediction of protein conformation. Biochemistry 13:222–245 [DOI] [PubMed] [Google Scholar]
- 5. Chou P. Y., Gerald D. F. 1974. Conformational parameters for amino acids in helical, β-sheet, and random coil regions calculated from proteins. Biochemistry 13:211–222 [DOI] [PubMed] [Google Scholar]
- 6. Curran A. R., Engelman D. M. 2003. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 13:412–417 [DOI] [PubMed] [Google Scholar]
- 7. Garcia M., Pimentel M., Moniz-Pereira J. 2002. Expression of mycobacteriophage Ms6 lysis genes is driven by two sigma (70)-like promoters and is dependent on a transcription termination signal present in the leader RNA. J. Bacteriol. 184:3034–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gil F., et al. 2008. The lytic cassette of mycobacteriophage Ms6 encodes an enzyme with lipolytic activity. Microbiology 154:1364–1371 [DOI] [PubMed] [Google Scholar]
- 9. Gil F., et al. 2010. The mycobacteriophage Ms6 LysB specifically targets the outer membrane of Mycobacterium smegmatis. Microbiology 156:1497–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reference deleted.
- 11. Hatfull G. F., et al. 2010. Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J. Mol. Biol. 397:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kleiger G., Grothe R., Mallick P., Eisenberg D. 2002. GxxxG and AxxxA: common alpha-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry 41:5990–5997 [DOI] [PubMed] [Google Scholar]
- 13. Payne K., Sun Q., Sacchettini J., Hatfull G. F. 2009. Mycobacteriophage lysin B is a novel mycolylarabinogalactan esterase. Mol. Microbiol. 73:367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pope W. H., et al. 2011. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS One 6:e16329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Portugal I., Anes E., Moniz-Pereira J. 1989. Temperate mycobacteriophage from M. smegmatis. Acta Leprol. 7:243–244 [PubMed] [Google Scholar]
- 16. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17. Schneider D., Engelman D. M. 2004. Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J. Mol. Biol. 343:799–804 [DOI] [PubMed] [Google Scholar]
- 18. Senes A., Gerstein M., Engelman D. M. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighbouring positions. J. Mol. Biol. 296:921–936 [DOI] [PubMed] [Google Scholar]
- 19. Senes A., Engel D. E., DeGrado W. F. 2004. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14:465–479 [DOI] [PubMed] [Google Scholar]
- 20. Wang W., Malcolm B. A. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. Biotechniques 26:680–682 [DOI] [PubMed] [Google Scholar]