Abstract
The alternative sigma factor σB of Staphylococcus aureus is involved in the coordination of the general stress response, expression of virulence determinants, and modulation of antibiotic resistance levels. It controls a large regulon, either directly by recognizing conserved σB promoter sequences or indirectly via σB-dependent elements. The σB-controlled yabJ-spoVG operon encodes two such putative downstream elements. We report here transcriptome analysis in S. aureus Newman, showing that inactivation of the yabJ-spoVG operon had primarily a repressing effect on a small subregulon encoding mainly virulence factors, including a nuclease (nuc), a protease (splE) and a lipase (lip). As a consequence, extracellular nuclease, protease, and lipase activities were reduced in a ΔyabJ-spoVG mutant. trans-complementation by SpoVG was sufficient to restore their reduced phenotypic expression and lowered transcription due to the yabJ-spoVG deletion. It did not restore, however, the changes triggered by σB inactivation, indicating that both regulons only partially overlap, despite the σB dependency of the yabJ-spoVG expression. Thus, σB is likely to control additional, SpoVG-independent factors affecting the expression of numerous hydrolytic enzymes. SpoVG, on the other hand, seems to fine-tune the σB-dependent regulation of a subset of virulence factors by antagonizing the σB effect.
INTRODUCTION
Staphylococcus aureus secretes a wide array of extracellular virulence factors that enable it to cause superficial skin infections or severe invasive infections, such as bacteremia, endocarditis, and osteomyelitis (4, 9, 18). The growth- and environment-dependent expression of virulence factors is a prerequisite for the survival of S. aureus in the host and the pathogenesis of staphylococcal infections. During initial colonization steps, the expression of cell surface adhesins promotes the binding to human extracellular matrix components (16). Extracellular enzymes, such as nucleases, lipases, and proteases, are, on the other hand, produced mainly in later infection stages. They destroy the local host tissue to access nutrients required for growth and spreading within the host (18) and facilitate immune evasion by interfering with the host's innate immune system (5, 41).
The coordinated expression of virulence determinants in S. aureus is controlled by a complex network of regulatory elements, including two-component regulatory systems, DNA-binding proteins, and alternative sigma factors (8, 13, 32, 53). σB is the best characterized alternative sigma factor of S. aureus. It is involved in general stress responses (11, 22, 28, 32, 38) and part of the regulatory network controlling the expression of virulence determinants (25, 28, 31, 54) and modulates antibiotic resistance levels (6, 37, 46, 48, 53). σB activity is growth phase regulated, peaking toward early stationary phase (47), and controls a large regulon of genes involved in many different cellular processes by recognizing and binding to a conserved nucleotide consensus sequence (GTTTAA-12-15-GGGTAT) in the upstream region of the respective genes/operons (7, 27, 38). However, many of the genes that are regulated by σB are not preceded by such a σB promoter. It is therefore thought that σB-dependent regulation of those genes occurs by downstream regulatory elements (7, 38).
The bicistronic yabJ-spoVG operon, whose transcription depends heavily on σB activity, was hypothesized to play a role in the indirect σB-dependent regulation of genes lacking an obvious σB promoter (7, 36, 46). The molecular mechanisms of YabJ, which belongs to the YjgF protein family of unknown biochemical function (49), and of the stage V sporulation protein G SpoVG (30), originally identified in Bacillus subtilis as being involved in the formation of the spore cortex, are yet unknown for the nonsporulating S. aureus. However, the finding that inactivation of the staphylococcal yabJ-spoVG operon, like inactivation of σB, reduces the transcription of the capA gene, which is not preceded by a σB promoter (36), and reduces the resistance level of methicillin-resistant S. aureus (MRSA) and glycopeptide intermediate-resistant S. aureus (GISA) (46) support the hypothesis that the yabJ-spoVG operon may encode a σB downstream regulatory element.
Early work on σB activity in S. aureus showed that extracellular nuclease and lipase activities are negatively regulated by the alternative sigma factor σB via a yet unknown indirect mechanism (31). In this study, we show that deletion of the σB-dependent yabJ-spoVG operon reduces nuclease, lipase, and protease activities in S. aureus Newman. We present transcriptomic data allowing the comparison of the yabJ-spoVG regulon with the σB regulon, and we identify a set of genes that is influenced by σB as well as by yabJ-spoVG. We demonstrate that SpoVG, and not YabJ, is the major regulator of the yabJ-spoVG operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown on Luria Bertani (LB) agar (Becton Dickinson, Allschwil, Switzerland) or in LB broth with shaking (180 rpm) at 37°C. Where required, media were supplemented with either 20 μg chloramphenicol or 10 μg tetracycline per ml.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Reference or source |
|---|---|---|
| S. aureus strains | ||
| Newman | Clinical isolate, ATCC 25904, saeS constitutively active | 1, 20 |
| SM148 | Newman ΔyabJ-spoVG::erm(B); Emr | 46 |
| MS64 | Newman sigB1(Am); Tcr | 47 |
| IK184 | Newman ΔrsbUVW-sigB::erm(B); Emr | 32 |
| Coln | Tcs derivative of S. aureus Col; Mcr | 29 |
| SM165 | Coln ΔyabJ-spoVG::erm(B); Emr | 46 |
| BS128 | Coln ΔrsbUVW-sigB::erm(B); Emr | 46 |
| Plasmids | ||
| pMGS100 | Overexpression plasmid containing the bacA promoter of E. faecalis; Cmr | 21 |
| pBus1 | E. coli-S. aureus shuttle plasmid with multicloning site from pBlue-script II SK (Stratagene) and the rrnT14 terminator sequence from pLL2443; Tcr | 43 |
| pSTM08 | pBus1 with a 2.1-kb PCR fragment covering yabJ, spoVG, and the preceding σB-dependent promoter PyabJ; Tcr | 46 |
| p08-yabJ-His | pSTM08 with a 6-histidine tag added to yabJ; Tcr | This study |
| p08-spoVG-His | pSTM08 with a 6-histidine tag added to spoVG; Tcr | This study |
| pyabJ | pBus1 containing a bacA promoter-yabJ ORF fusion construct; Tcr | This study |
| pyabJ-His | pyabJ with a 6-histidine tag added to yabJ; Tcr | This study |
| pspoVG | pBus1 containing a bacA promoter-spoVG ORF fusion construct; Tcr | This study |
| pspoVG-His | pspoVG with a 6-histidine tag added to spoVG; Tcr | This study |
| pspoVG-Bsu | pBus1 containing the bacA promoter fused to the spoVG ORF of B. subtilis; Tcr | This study |
| pyabJspoVG | pBus1 containing a bacA promoter-yabJ-spoVG operon fusion construct; Tcr | This study |
Abbreviations are as follows: Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Mcr, methicillin resistant; Tcr, tetracycline resistant; Tcs, tetracycline sensitive.
Molecular biological methods.
General molecular biology techniques were performed according to standard protocols described by Sambrook et al. (44) and Ausubel et al. (2). Sequencing was done using the BigDye Terminator cycle sequencing ready reaction kit and an ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, CA). Sequences were analyzed with the Lasergene software package (DNASTAR, Inc., Madison, WI).
Plasmid construction.
For the construction of the complementing plasmids pyabJ, pspoVG, and pyabJspoVG, DNA fragments covering yabJ, spoVG, or the whole yabJ-spoVG operon were amplified from S. aureus Newman genomic DNA using primer pairs oSTM51/oBS09, oBS11/oBS10, and oSTM51/oBS10, respectively. These fragments were digested with NruI and EclXI and inserted into plasmid pMGS100 downstream of the bacA promoter. The promoter-gene fusions were amplified from the resulting plasmids with primer pair oDB2/oBS30 or oBS28/oBS30, digested with PstI and XbaI, and cloned into Escherichia coli-S. aureus shuttle vector pBus1 to give pyabJ, pspoVG, and pyabJspoVG.
pspoVG-Bsu was constructed by amplifying spoVG with primers Bsu-spoVG-f and Bsu-spoVG-r from Bacillus subtilis ATCC 6633 genomic DNA, digesting the resulting fragment with Eco521 and SacI and cloning it into Eco521/SacI-digested pyabJ.
A 6-histidine tag was added to the 3′ end of yabJ and spoVG in pSTM08 by amplifying it with primer pairs YabJ-his/oBS33 and SpoVG-his/oBS34, respectively, and ligating it to give plasmids p08-yabJ-His and p08-spoVG-His. Similarly, a 6-histidine tag was added to pyabJ and pspoVG by amplifying the respective plasmid with primer pair YabJ-his/oBS33 for pyabJ and SpoVG-his/oBS34 for pspoVG and ligating it to give plasmids pyabJ-His and pspoVG-His.
All plasmid constructs were confirmed by sequence analyses.
Northern blot analysis.
Overnight cultures were diluted 1:100 into LB broth, grown for 2 h, and then used to inoculate 100 ml prewarmed LB to an optical density at 600 nm (OD600) of 0.05. Cell samples were taken at the time points indicated and centrifuged at 9,000 × g and 4°C for 2 min, the culture supernatants discarded, and the cell sediments snap-frozen in liquid nitrogen. Total RNA was isolated according to the method of Cheung et al. (14). RNA samples (8 μg) were separated in a 1.5% agarose gel containing 20 mM guanidine thiocyanate in 1× Tris-borate-EDTA buffer (24). RNA transfer and detection were performed as previously described (35). Digoxigenin (DIG)-labeled probes were amplified using the PCR DIG probe synthesis kit (Roche, Basel, Switzerland). The primer pairs used for amplification of the hybridization probes are listed in Table 2. All Northern blots were repeated at least twice, using independently isolated RNA samples.
Table 2.
Oligonucleotide primers used in this study
| Primer name | Sequence (5′-3′)a | Source or reference |
|---|---|---|
| DIG probes | ||
| NWMN_0095-DIG f | CAGTTTATGGCGCAAGAGT | This study |
| NWMN_0095-DIG r | GAACCCAATACAGGCAATCC | This study |
| NWMN_0218-DIG f | CCTGCCCAGTTTTTAGCATC | This study |
| NWMN_0218-DIG r | AACAACACGCCAAACAACAA | This study |
| NWMN_0219-DIG f | TCCAGAGGAAATCAGAGCAAA | This study |
| NWMN_0219-DIG r | CTTGTTCTTGAACGGCATCA | This study |
| NWMN_0394-DIG f | ACATCAGAAGGCCAAGCAGT | This study |
| NWMN_0394-DIG r | TCTTGCTTTTCTCCATCTTTCA | This study |
| NWMN_0655-DIG f | TTTCATCAAATGCATGAATGACT | This study |
| NWMN_0655-DIG r | CAATGCTCAAAGACAAGTTAATCG | This study |
| NWMN_0760-DIG f | GAAAGGGCAATACGCAAAGA | This study |
| NWMN_0760-DIG r | TAGCCAAGCCTTGACGAACT | This study |
| NWMN_1560-DIG f | GATTAACCCCTCCTGGTGCT | This study |
| NWMN_1560-DIG r | AGCTTAACGCAAACATCACG | This study |
| NWMN_1702-DIG f | TTGCCTGGTTCTACGAAAGC | This study |
| NWMN_1702-DIG r | GGCACAACAGTGGTTGAGG | This study |
| NWMN_1718-DIG f | AGTTACCACGCGCCAATAA | This study |
| NWMN_1718-DIG r | GCAGCCGGAAACATTAATTC | This study |
| NWMN_2569-DIG f | AAATTTCGCCACCATGTTTC | This study |
| NWMN_2569-DIG r | CCAAACAAAGGGACAAAGGA | This study |
| oSTM16 (glmU f) | GAAGACACGCGATAATTTTGG | This study |
| oSTM17 (glmU r) | CAGTTGCTTGAGCATTAGCATC | This study |
| SAasp23+ | ATGACTGTAGATAACAATAAAGC | 22 |
| SAasp23− | TTGTAAACCTTGTCTTTCTTGG | 22 |
| Plasmid constructions | ||
| oDB02 | CGCTCTAGATTATAATTCCTTAATTTTACTTAC | This study |
| oBS09 | TTTTTCGCGATAATTCCTTAATTTTACTTACTAATTC | This study |
| oBS10 | TTTTTCGCGAAGCTTCTTCTGAATCTTCTGATGTAG | This study |
| oBS11 | TTTTTCGGCCGGCATGAAAGTGACAGATGTAAGAC | This study |
| oBS28 | CGTTCTAGATTAAGCTTCTTCTGAATCTTCTG | This study |
| oBS30 | GGGCTGCAGTGATTGAAACTCAAGATAGATATG | This study |
| oBS33 | TTTTCGATTAATATGTTTAATC | This study |
| oBS34 | TCAATTTTATATTTAGCGATG | This study |
| oSTM51 | TTTTTCGGCCGGCATGAAAATGATTAACACAACAAG | This study |
| SpoVG-his | GCGGTAGGTACCTTAATGATGATGATGATGATGAGCTTCTTCTGAATCTTCTG | This study |
| YabJ-his | GTAGGTACCTTAATGATGATGATGATGATGTAATTCCTTAATTTTACTTAC | This study |
| Bsu-spoVG-f | GCACGGCCGGCATGGAAGTTACTGACGTAAG | This study |
| Bsu-spoVG-r | GCAGAGCTCTTAAGAAGCTCCAGCTTCTTC | This study |
Restriction sites are underlined.
Microarray design and manufacturing.
The microarray was manufactured by in situ synthesis of 60-base-long oligonucleotide probes (Agilent, Palo Alto, CA), selected as previously described (12). It covers approximately >98% of all open reading frames (ORFs) annotated for strains N315 and Mu50 (33), MW2 (3), COL (23), NCTC8325 (accession no. NC_007795), USA300 (17), and MRSA252 and MSSA476 (26), including their respective plasmids.
Expression microarrays.
Total RNA was purified from bacteria grown in LB broth for 5 h as described for the Northern blot analysis. DNA-free total RNA was obtained after DNase treatment on RNeasy columns (Qiagen). For each strain, RNA of three independently grown cultures was analyzed. The absence of remaining DNA traces was confirmed by quantitative PCR (SDS 7700; Applied Biosystems, Framingham, MA) with assays specific for 16S rRNA genes (40, 45). Batches of 5 μg of total S. aureus RNA were labeled by Cy-3-dCTP using the SuperScript II (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Labeled products were purified onto QiaQuick columns (Qiagen).
Purified genomic DNA from the different sequenced strains used for the design of the microarray was extracted (DNeasy, Qiagen), labeled with Cy5-dCTP using the Klenow fragment of DNA polymerase I (BioPrime; Invitrogen) (12), and used for the normalization process (50). Cy5-labeled DNA (500 ng) and Cy3-labeled cDNA mixture were diluted in 50 μl Agilent hybridization buffer and then hybridized at a temperature of 60°C for 17 h in a dedicated hybridization oven (Robbins Scientific, Sunnyvale, CA). Slides were washed with Agilent proprietary buffers, dried under nitrogen flow, and scanned (Agilent) using 100% photon multiplier tube power for both wavelengths.
Microarray analysis.
Fluorescence intensities were extracted using the Feature extraction software (Agilent, version 8). Local background-subtracted signals were corrected for unequal dye incorporation or unequal loading of the labeled product. The algorithm consisted of a rank consistency filter and a curve fit using the default Lowess (locally weighted linear regression) method. Data consisting of three independent biological experiments were analyzed using GeneSpring 8.0 (Silicon Genetics, Redwood City, CA). A filter was applied to select oligonucleotides mapping ORFs in the Newman genome, yielding approximately 92% coverage. Statistical significance of differentially expressed genes was identified by analysis of variance (ANOVA) (15, 45), using GeneSpring, including the Benjamini and Hochberg false discovery rate correction of 5% (P value cutoff of 0.05) and an arbitrary cutoff of 2-fold for expression ratios. Microarray data have been posted on the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GPL7137 for the platform and GSE30115 for the complete experimental data set.
Enzyme activity assays. (i) Protease.
The proteolytic activity of S. aureus was determined on skim milk agar plates (75 g liter−1 skim milk [Becton Dickinson]) as clear zones surrounding the colonies. The strains were grown on skim milk plates for 24 h at 37°C and incubated for 72 h at room temperature.
(ii) Nuclease.
The nuclease activity was measured as described by Berends et al. (5). Supernatants were harvested from early stationary-phase cultures (OD600=4) after centrifugation for 5 min at 2,000 × g.
(iii) Lipase.
Lipase activity was tested by monitoring hydrolysis of 4-nitrophenyl octanoate by supernatants of cultures grown to an OD600 of 7 (31). A volume of 50 μl of the supernatant was incubated with 1.2 ml of 4-nitrophenyl octanoate (Sigma) solution (1 mg ml−1 in 20 mM Tris-HCl [pH 9.0], 10 mM CaCl2, and 0.1% [vol/vol] Triton X-100) for 15 min at 37°C. The absorption at 405 nm was measured, and the lipase activity was expressed as a percentage of that of the wild-type strain. Values of four independent assay were statistically evaluated using a paired, two-tailed Student t test.
Western blot analysis.
Cytoplasmic protein extracts were obtained by lysing S. aureus, grown in LB broth at 37°C for 5 h, in phosphate-buffered saline (pH 7.4) containing 2 mM phenylmethylsulfonyl fluoride and 40 μg each of lysostaphin, lysozyme, DNase, and RNase per ml. Protein fractions (10 to 15 μg/lane) were separated using SDS-PAGE, blotted onto a nitrocellulose membrane, and subjected to Western blot analysis using rabbit anti-6×His tag antibodies (Abcam, Cambridge, MA).
Population analysis profiles.
Oxacillin resistance profiles were determined by plating aliquots of overnight cultures on LB agar plates containing increasing concentrations of oxacillin and determining the number of CFU after 48 h of incubation at 37°C.
RESULTS AND DISCUSSION
Transcriptional profiling of genes depending on the yabJ-spoVG operon.
The σB-controlled yabJ-spoVG operon has pleiotropic effects in S. aureus, stimulating cap operon transcription and capsule formation, and supporting resistance to cell wall antibiotics (36, 46). To explore the extent of the yabJ-spoVG regulon, we used full-genome DNA microarrays and compared the transcriptional patterns of S. aureus Newman and its isogenic ΔyabJ-spoVG mutant, SM148, in the early stationary growth phase, when σB activity and spoVG transcription are known to be maximal (7, 36, 47). Forty-three genes belonging to 27 predicted transcriptional units were found to be differently regulated. Nineteen operons were downregulated, and 8 were upregulated 2-fold or more in SM148 (Tables 3 and 4), suggesting that the yabJ-spoVG operon had primarily activating functions. The majority of the differentially expressed genes belong to the group of virulence factors, including those that encode members of the capsular polysaccharide synthesis (cap) operon, members of the staphylococcal ESX-1 secretion system (esxA and esaA), leukotoxins (lukED), exotoxins (set1/6/7nm), a nuclease (nuc), a serine protease (splE), a lipase (lip) and an adhesin (sdrE).
Table 3.
Genes with ≥2 times higher expression in wild-type S. aureus Newman than in the ΔyabJ-spoVG mutant SM148
| Newman TIGR locus namea | Gene symbolb | Descriptionb | Newman/SM148 fold changec | Newman/MS64 fold changec | Reference(s) with reported σB dependence | σB promoterd | Transcription unit 5′-3′e | Patternf |
|---|---|---|---|---|---|---|---|---|
| NWMN_0095* | capA | Capsular polysaccharide biosynthesis protein Cap5A | 9.3 | 2.8 | 7, 25, 36, 38 | No | nwmn_0095-0110 | i |
| NWMN_0096 | capB | Capsular polysaccharide biosynthesis protein Cap5B | 11.0 | 2.8 | 7, 38 | No | nwmn_0095-0110 | |
| NWMN_0097 | capC | Capsular polysaccharide biosynthesis protein Cap5C | 9.5 | 2.5 | 7, 38 | No | nwmn_0095-0110 | |
| NWMN_0098 | capD | Capsular polysaccharide biosynthesis protein Cap5D | 12.1 | 2.8 | 7, 38 | No | nwmn_0095-0110 | |
| NWMN_0099 | capE | Capsular polysaccharide biosynthesis protein Cap5E | 7.2 | 2.4 | 7, 38 | No | nwmn_0095-0110 | |
| NWMN_0100 | capF | Capsular polysaccharide biosynthesis protein Cap5F | 7.5 | 7, 38 | No | nwmn_0095-0110 | ||
| NWMN_0101 | capG | Capsular polysaccharide biosynthesis protein Cap5G | 6.0 | 7, 38 | No | nwmn_0095-0110 | ||
| NWMN_0102 | capH | Capsular polysaccharide biosynthesis protein Cap5H | 4.4 | 2.1 | 7, 38 | No | nwmn_0095-0110 | |
| NWMN_0103 | capI | Capsular polysaccharide biosynthesis protein Cap5I | 4.9 | 2.4 | 7, 38 | No | nwmn_0095-0110 | |
| NWMN_0104 | capJ | Capsular polysaccharide biosynthesis protein Cap5J | 3.1 | 2.1 | 7, 38 | No | nwmn_0095-0110 | |
| NWMN_0105 | capK | Capsular polysaccharide biosynthesis protein Cap5K | 2.7 | 2.1 | 7, 38 | No | nwmn_0095-0110 | |
| NWMN_0106 | capL | Capsular polysaccharide biosynthesis protein Cap5L | 2.1 | 7, 38 | No | nwmn_0095-0110 | ||
| NWMN_0107 | capM | Capsular polysaccharide biosynthesis protein Cap5M | 2.2 | 2.1 | 7, 38 | No | nwmn_0095-0110 | |
| NWMN_0108 | capN | Capsular polysaccharide biosynthesis protein Cap5N | 2.0 | 7, 38 | No | nwmn_0095-0110 | ||
| NWMN_0152 | Maltose ABC transporter substrate-binding protein, putative | 2.8 | 25 | No | nwmn_0151-0156 | iii | ||
| NWMN_0218* | Staphyloxanthin biosynthesis protein, putative | 2.9 | 0.5 | 7, 25 | No | nwmn_0218 | ii | |
| NWMN_0219* | esxA | Virulence factor EsxA | 2.7 | Yes | nwmn_0219 | iii (ii) | ||
| NWMN_0220 | esaA | Membrane protein EsaA | 2.2 | 7, 25 | No | nwmn_0220-0227 | iii | |
| NWMN_0451 | Conserved hypothetical protein | 2.1 | No | nwmn_0451-0452 | iii | |||
| NWMN_0460 | yabJ | YabJ protein | 254.0 | 3.6 | 7, 36, 46 | Yes | nwmn_0460-0461 | − |
| NWMN_0461 | spoVG | Regulatory protein SpoVG | 77.6 | 4.1 | 7, 36, 46 | No | nwmn_0460-0461 | − |
| NWMN_0525 | sdrE | Ser-Asp rich fibrinogen/bone sialoprotein-binding protein SdrE | 2.2 | 0.4 | No | nwmn_0525 | ii | |
| NWMN_0559 | Conserved hypothetical protein | 2.3 | No | nwmn_0559-0560 | iii | |||
| NWMN_0753 | Lipoprotein, putative | 2.1 | No | nwmn_0753 | iii | |||
| NWMN_0760* | nuc | Thermonuclease precursor | 3.0 | 7, 25, 38 | No | nwmn_0760 | iii (ii) | |
| NWMN_1279 | trpE | Anthranilate synthase component I | 2.1 | No | nwmn_1279-1285 | iii | ||
| NWMN_1560* | abrB | Ammonia monooxygenase | 2.4 | 3.3 | No | nwmn_1560 | i | |
| NWMN_1702* | splE | Serine protease SplE | 2.1 | 0.1 | 7, 54 | No | nwmn_1702 | ii |
| NWMN_1708 | Conserved hypothetical protein | 2.0 | No | nwmn_1708 | iii | |||
| NWMN_1718* | lukD | Leucotoxin LukD | 2.9 | 0.1 | No | nwmn_1719-1718 | ii | |
| NWMN_1719 | lukE | Leucotoxin LukE | 2.7 | 0.1 | No | nwmn_1719-1718 | ||
| NWMN_2061 | fmtB | Methicillin resistance determinant FmtB protein | 2.5 | No | nwmn_2061 | iii | ||
| NWMN_2365 | Conserved hypothetical protein | 2.2 | 38 | No | nwmn_2367-2365 | iii | ||
| NWMN_2553 | hsa | LPXTG cell wall surface anchor family protein | 2.1 | 7 | Yes | nwmn_2553 | iii | |
| NWMN_2569* | lip | Lipase | 2.3 | 0.3 | 7, 54 | No | nwmn_2569 | ii |
Locus number for S. aureus Newman (http://www.tigr.org). *, genes selected for Northern blot analysis.
Gene symbol and description of the gene based on the TIGR annotation (http://www.tigr.org).
Fold change of gene transcription comparing S. aureus Newman with either SM148 or MS64.
ORFs preceded by a promoter sequence that resembles the σB consensus sequence of S. aureus described by Homerova et al. (27). Sequences matching the GNNTNN-12-15-NGGTAN consensus, with no more than three mismatches of the degenerated bases from the GTTTAA-12-15-GGGTAT sequence and lying within a range of 500 bp upstream of the ORF were accepted.
Transcription units were deduced from Northern blot analyses done in this study or published by Pané-Farré et al. (38) or Meier et al. (36). Where no experimental data were available, the operons are indicated as predicted by Wang et al. (52).
Assignment of transcription units into regulatory patterns: i, upregulated by YabJ/SpoVG and σB; ii, upregulated by YabJ/SpoVG, downregulated by σB; iii, upregulated by YabJ/SpoVG; −, deleted yabJ-spoVG operon, upregulated by σB; in parentheses, assignment to class ii according to Northern blot analyses.
Table 4.
Genes with ≥2 times lower expression in wild-type S. aureus Newman than in the ΔyabJ-spoVG mutant SM148
| Newman TIGR locus namea | Gene symbolb | Descriptionb | Newman/SM148 fold changec | Newman/MS64 fold changec | Reference(s) with reported σB dependence | σB promoterd | transcription unit 5′-3′e | Patternf |
|---|---|---|---|---|---|---|---|---|
| NWMN_0388 | set1nm | Superantigen-like protein, exotoxin NM1 | 0.4 | No | nwmn_0388 | v | ||
| NWMN_0393 | set6nm | Superantigen-like protein, exotoxin NM6 | 0.5 | 2.6 | No | nwmn_0393 | iv | |
| NWMN_0394* | set7nm | Superantigen-like protein, exotoxin NM7 | 0.3 | 2.2 | No | nwmn_0394 | iv | |
| NWMN_0462* | glmU | UDP-N-acetylglucosamine pyrophosphorylase | 0.2 | No | nwmn_0462-0464 | v | ||
| NWMN_0463 | prsA | Ribose-phosphate pyrophosphokinase | 0.3 | No | nwmn_0462-0464 | |||
| NWMN_0464 | rplY | 50S ribosomal protein L25, Ctc form | 0.4 | No | nwmn_0462-0464 | |||
| NWMN_0655* | norR | HTH-type transcriptional regulator MgrA | 0.4 | No | nwmn_0655 | v (iv) | ||
| NWMN_2074 | Conserved hypothetical protein | 0.5 | No | nwmn_2074 | v | |||
| NWMN_2086* | asp23 | Alkaline shock protein 23 | 0.3 | 33.9 | 7, 22, 38 | Yes | nwmn_2088-2086 | iv |
| NWMN_2187 | yut | Urea transporter | 0.4 | No | nwmn_2187 | v |
Locus number for S. aureus Newman (http://www.tigr.org). *, genes selected for Northern blot analysis.
Gene symbol and description of the gene based on the TIGR annotation (http://www.tigr.org).
Fold change of gene transcription comparing S. aureus Newman with either SM148 or MS64.
ORFs preceded by a promoter sequence that resembles the σB consensus sequence of S. aureus described by Homerova et al. (27). Sequences matching the GNNTNN-12-15-NGGTAN consensus, with no more than three mismatches of the degenerated bases from the GTTTAA-12-15-GGGTAT sequence and lying within a range of 500 bp upstream of the ORF were accepted.
Transcription units were deduced from Northern blot analyses done in this study or published by Pané-Farré et al. (38) or Meier et al. (36). Where no experimental data were available, the operons are indicated as predicted by Wang et al. (52).
Assignment of transcription units into regulatory pattern: iv, downregulated by YabJ/SpoVG, upregulated by σB; v, downregulated by YabJ/SpoVG; in parentheses, assignment to class iv according to Northern blot analyses.
Overlap of the yabJ-spoVG and the σB regulons.
To identify the overlap of the σB and yabJ-spoVG regulons, we performed in parallel differential transcriptome microarrays of S. aureus Newman and its isogenic ΔsigB mutant, MS64. This analysis identified 191 genes to be differentially influenced by σB 2-fold or more (see Table S1 in the supplemental material). The intersection of the yabJ-spoVG and the σB regulons covered 10 transcriptional units summarized in Tables 3 and 4. The microarrays revealed five different regulatory patterns (i to v) of the YabJ/SpoVG subregulon: pattern i contained genes upregulated by both YabJ/SpoVG and σB; pattern ii contained genes upregulated by YabJ/SpoVG but downregulated by σB; pattern iii contained genes upregulated by YabJ/SpoVG; pattern iv contained genes that were downregulated by YabJ/SpoVG but upregulated by σB; and pattern v contained genes that were downregulated by YabJ/SpoVG. Interestingly, 8 operons affected by σB activity were regulated by YabJ/SpoVG in an opposite way, indicating that YabJ/SpoVG might antagonize the σB activity to fine-tune the σB-driven expression of a subset of σB-dependent genes/operons.
Computer-based searches for putative σB-binding sites in the region upstream of the translational start site of the ORFs of the yabJ-spoVG regulon were done using the σB promoter consensus sequence determined by Homerova et al. (27) (Tables 3 and 4). Most genes were not preceded by a σB consensus sequence, suggesting an indirect σB-dependent regulation for the majority of the genes of the yabJ-spoVG regulon. The only exceptions were the genes asp23, which is known to be σB dependent (7, 22, 38), and, esxA and hsa. The latter are preceded by putative σB promoter sequences, but their functionality remains to be confirmed.
Confirmation of microarray results.
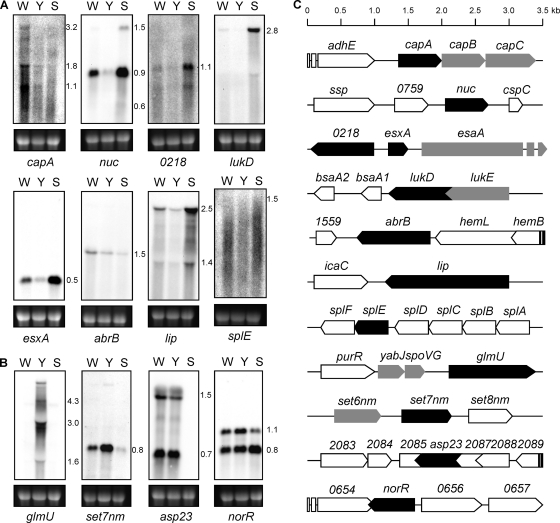
To confirm the microarray results, the transcriptional profiles of a selection of ORFs found to be regulated by the yabJ-spoVG operon were verified by Northern blot analyses. The transcription levels of eight down- and four upregulated ORFs were compared for the wild-type strain Newman, the ΔyabJ-spoVG mutant, SM148, and the ΔsigB mutant, MS64, selecting those genes with the highest differential expression upon yabJ-spoVG inactivation and one representative per predicted operon. The Northern blots of the selected ORFs as well as their genomic context are shown in Fig. 1. All downregulated ORFs tested, including capA, nuc, nwmn_0218, which encodes a putative staphyloxanthin biosynthesis protein, lukD, esxA, abrB, which encodes an ammonia monooxygenase, lip, and splE, produced weaker transcripts in the ΔyabJ-spoVG mutant, SM148, compared to the wild type. In the ΔsigB mutant, MS64, which consequently does not express the σB-dependent yabJ-spoVG operon, two different responses were observed for these eight genes: (i) capA and abrB transcripts were reduced, while (ii) the remaining six ORFs were overexpressed, confirming the microarray data for splE, lukD, and lip. The Northern blot analyses also suggested that σB downregulated esxA, nwmn_0218, and nuc, despite the fact that the σB microarray data did not reach the threshold of 2 for these last genes, but downregulation of nwmn_0218 and nuc by σB was reported previously (7). Taking into account these results, the predominant regulatory pattern for genes influenced by YabJ/SpoVG and σB was pattern ii, in which YabJ/SpoVG acts as activator and counteracts the repressing effect of σB.
Fig. 1.
Northern blot analysis of yabJ-spoVG-dependent genes. Northern blots of total RNA from S. aureus Newman (W), ΔyabJ-spoVG mutant SM148 (Y), and ΔsigB mutant MS64 (S) for selected genes that were found to be down- (A) or upregulated (B) in the microarray in SM148. Transcript masses are indicated in kb. The ethidium bromide-stained 16S rRNA pattern is shown as an indication for the RNA loading. (C) Locus maps of ORFs selected for Northern blots (black). ORFs appearing in the microarray are highlighted in gray. adhE, encoding iron-containing alcohol dehydrogenase; ssp, encoding extracellular matrix and plasma binding protein; cspC, encoding cold-shock protein CSD family protein; bsaA1/2, encoding lantibiotic precursors; hemL, encoding glutamate-1-semialdehyde-2,1-aminomutase; hemB, encoding delta-aminolevulinic acid dehydratase; icaC, encoding intercellular adhesion protein IcaC; splABCDF, encoding serine proteases; purR, encoding pur operon repressor; set8 nm, encoding exotoxin NM8.
Only two of the four genes predicted by the microarray data to be upregulated in the ΔyabJ-spoVG mutant, SM148, could be confirmed by Northern blot analyses, namely, glmU, encoding a UDP-N-acetylglucosamine pyrophosphorylase, and set7nm, encoding an exotoxin. Transcription of the genes encoding the alkaline shock protein 23 (asp23) and the transcriptional regulator NorR (norR) apparently did not respond to yabJ-spoVG inactivation. The reasons for this discrepancy need further exploration.
Complementation analyses.
Since the yabJ-spoVG operon is under σB control and therefore not transcribed in ΔsigB mutants (36, 46), we fused yabJ and spoVG to the bacA promoter of Enterococcus faecalis, which is expressed constitutively in S. aureus (21). The resulting plasmids pyabJ, pspoVG, and pyabJspoVG allowed a σB activity-independent complementation of both the ΔyabJ-spoVG and ΔsigB mutants.
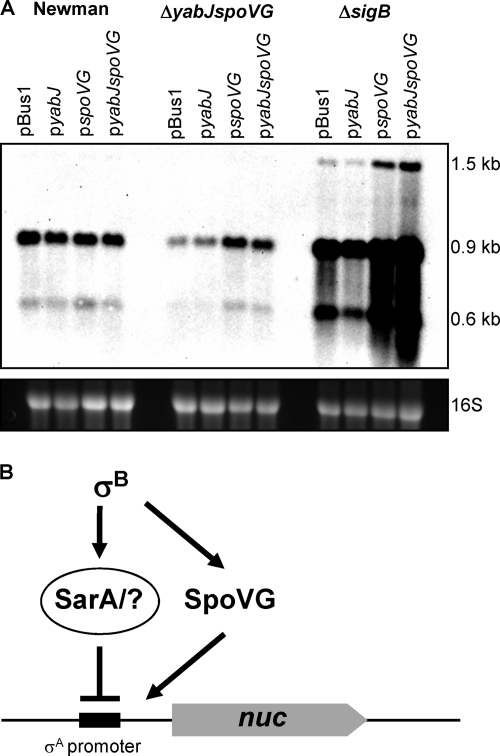
The typical and most common regulatory pattern, ii, namely, upregulation by YabJ/SpoVG and downregulation by σB as exemplified by nuc (Fig. 1) was used to analyze if YabJ and/or SpoVG was required for restoring nuc expression in the ΔyabJ-spoVG mutant SM148 and to assess their effect on nuc regulation in the absence of σB. For this purpose we complemented SM148 and the σB mutant IK184 with either gene separately or both genes together, expressed from the σB-independent constitutive promoter. In analogy to the complementation of capA transcription in a ΔyabJ-spoVG background, where SpoVG was shown to be the effector of the yabJ-spoVG operon (36, 46), the plasmids pspoVG and pyabJspoVG did largely restore nuc expression in the ΔyabJ-spoVG mutant SM148 to the level seen with the wild type, while pyabJ did not complement this phenotype (Fig. 2). In line with previous findings (32), the σB mutant showed a higher transcription of nuc than the wild type. Since the nuc gene and its vicinity do not contain any σB consensus sequences, the high nuc transcriptional response must be under indirect σB-dependent negative control. The σB-dependent repression of nuc transcription in the wild type functions presumably over σB-activated SarA, reported to downregulate nuc (10, 19). Since transcription of yabJ-spoVG is virtually abolished in σB mutants (7, 36, 46), SpoVG could not contribute to nuc regulation in the σB mutant. When SpoVG, however, was expressed constitutively from plasmid pspoVG or pyabJspoVG, it was able to further activate nuc transcription in the σB mutant. These data suggest that SpoVG has an activating effect on nuc transcription that is antagonized and/or overruled by the repressing effect of σB (Fig. 2B).
Fig. 2.
trans-complementation of nuc expression. (A) Northern blot analysis of nuc in S. aureus Newman, its isogenic ΔyabJ-spoVG mutant SM148, and ΔrsbUVW-sigB mutant IK184 with pBus1 (empty plasmid), pyabJ (PbacA-yabJ), pspoVG (PbacA-spoVG), and pyabJspoVG (PbacA-yabJ-spoVG) after 5 h of growth in LB broth. Transcript masses are indicated. The ethidium bromide-stained 16S rRNA pattern is shown as an indication of the RNA loading. (B) Postulated regulatory pattern of nuc in S. aureus Newman. σB downregulates through activation of SarA and/or other repressors the transcription of nuc. At the same time σB activates SpoVG expression, which in turn counteracts SarA and activates nuc transcription.
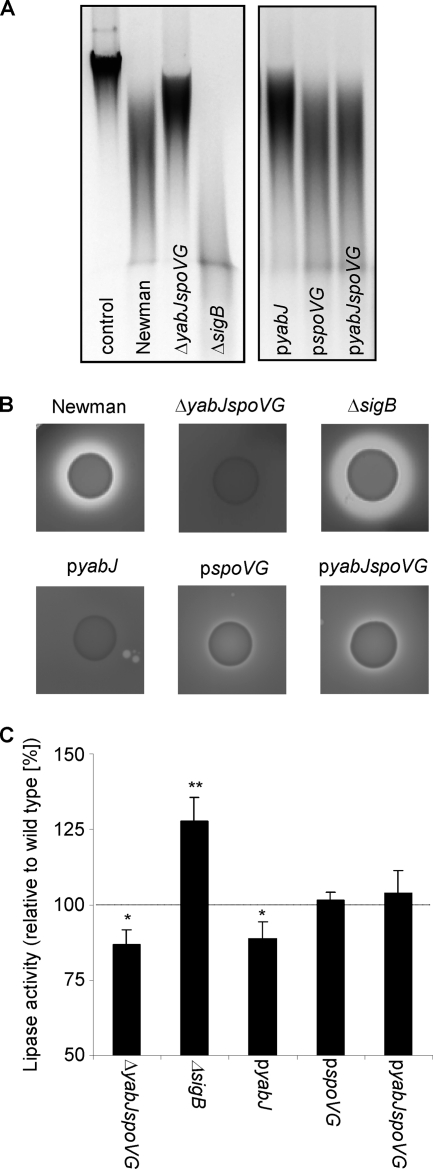
SpoVG inactivation reduces excreted virulence factors.
The transcriptional response of nuc was mirrored by the nuclease activity measured in bacterial supernatants (Fig. 3A). The ΔyabJ-spoVG mutant expressed lower nuclease activity than the wild type, while the ΔsigB mutant exhibited higher nuclease activity. Again, nuclease activity in the ΔyabJ-spoVG mutant was increased by complementation with SpoVG with either plasmid pspoVG or pyabJspoVG, but not by pyabJ. The extracellular lipase and protease activities followed the same pattern, being reduced in the ΔyabJ-spoVG mutant and enhanced in the ΔsigB mutant (Fig. 3B and C). As described above, complementation of the ΔyabJ-spoVG mutant with SpoVG increased their activity. Interestingly, the nuclease, lipase, and protease activities in the supernatant of a ΔsigB mutant were not further enhanced by any of the three complementing plasmids, as would have been expected from the transcriptional data (data not shown), possibly due to a bottleneck between transcription and secretion.
Fig. 3.
Influence of σB and the σB-dependent yabJ-spoVG operon on excreted virulence factors. The nuclease, protease, and lipase activities of S. aureus Newman, the ΔyabJ-spoVG mutant SM148, the ΔsigB mutant MS64, and SM148 complemented with pyabJ, pspoVG, and pyabJspoVG were compared. (A) Representative agarose gels of calf thymus DNA after incubation with bacterial culture supernatants to detect nuclease activity. Whereas the DNA is not degraded with sterile LB broth (control), samples incubated with culture supernatants show different stages of DNA degradation. (B) Proteolytic activity of the strains after growth on skim milk agar plates. (C) Lipase activity monitored by hydrolysis of 4-nitrophenyl octanoate by culture supernatants. Data shown are the means ± SD of four independent experiments and are presented as percentage of lipase activity compared with that of the wild-type Newman (100%). Statistical significances between wild-type and mutant supernatants were assessed with a paired, two-tailed Student t test (*, P < 0.05; **, P < 0.01).
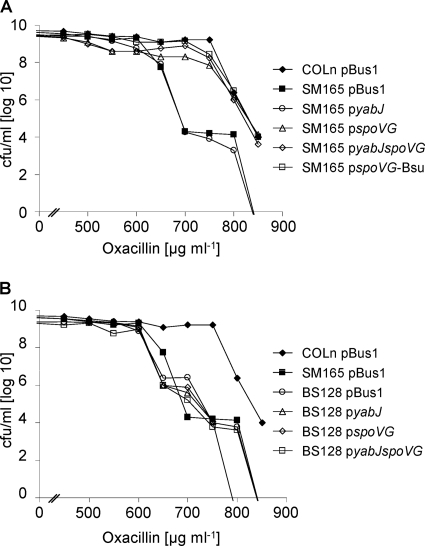
Role of σB and the yabJ-spoVG operon in oxacillin resistance.
We previously showed that deletion of the yabJ-spoVG operon reduces oxacillin and glycopeptide resistance of MRSA and GISA strains to the same extent as σB inactivation, which led to the assumption that σB might affect antibiotic resistance of MRSA and GISA via the expression of YabJ and/or SpoVG (46). However, the plasmids used in our previous study to complement the ΔyabJ-spoVG mutants harbored yabJ-spoVG under the control of their native σB-dependent promoter and thus did not allow the complementation of a ΔsigB mutant to support this hypothesis. Here, we complemented the MRSA ΔyabJ-spoVG mutant SM165 with plasmids pyabJ, pspoVG, and pyabJspoVG (Fig. 4 A), expressing yabJ and spoVG from the heterologous σB-independent bacA promoter, which also allowed us now to complement the ΔsigB mutant MRSA strain BS128 (Fig. 4B). In line with our previous findings, SpoVG was sufficient to complement the SM165 resistance phenotype, while YabJ had no apparent effect on the resistance level. However, neither YabJ nor SpoVG, nor both YabJ and SpoVG, were able to restore the oxacillin resistance of the ΔsigB mutant, indicating that SpoVG is not the only mediator of the σB effect on antibiotic resistance in MRSA.
Fig. 4.
Role of YabJ and SpoVG in oxacillin resistance in MRSA strain COLn. Population analysis profiles for oxacillin are shown for COLn and its isogenic ΔyabJ-spoVG (SM165) (A) and ΔrsbUVW-sigB (BS128) (B) mutants with pBus1 (empty plasmid), pyabJ (PbacA-yabJ), pspoVG (PbacA-spoVG), pyabJspoVG (PbacA-yabJ-spoVG), and pspoVG-Bsu (PbacA-spoVG from B. subtilis).
Complementation of the S. aureus yabJ-spoVG deletion mutant with B. subtilis spoVG.
SpoVG in S. aureus was originally discovered and named after its sequence homologue in B. subtilis, showing an amino acid identity of 61% and a similarity of 76%. To test whether B. subtilis spoVG can functionally complement the spoVG deletion in S. aureus, we constructed a fusion plasmid which harbors B. subtilis spoVG under the control of the constitutive bacA promoter (pspoVG-Bsu). trans-complementation of the MRSA ΔyabJ-spoVG mutant SM165 with this plasmid restored the oxacillin resistance to the wild-type level (Fig. 4A), signaling that not only the sequence but also functional characteristics were conserved.
Only SpoVG appears translated under in vitro conditions.
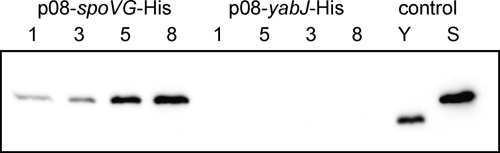
To analyze the expression profiles of YabJ and SpoVG, we fused a C-terminal 6-histidine tag to yabJ and spoVG, respectively, in pSTM08, expressing the yabJ-spoVG operon from its native promoter. The resulting constructs p08-yabJ-His and p08-spoVG-His were expressed in S. aureus SM148 during growth over 8 h in LB broth. While the SpoVG-His6 expression followed the observed transcriptional profile and increased within time, no signal for YabJ-His6 could be detected (Fig. 5), indicating that YabJ was not expressed or expressed only in very small amounts under these in vitro growth conditions.
Fig. 5.
Expression of YabJ and SpoVG. Western blot analysis of SpoVG-His6 and YabJ-His6 in cytoplasmic protein fractions of strain SM148 with p08-spoVG-His and p08-yabJ-His, respectively, grown for 1, 3, 5, and 8 h (indicated at top of lanes) in LB broth at 37°C and as a control of strain SM148 with pyabJ-His (Y) or pspoVG-His (S) after 5 h of growth. SpoVG-His6 has a molecular mass of 12.3 kDa (43). YabJ-His6 was smaller, although its theoretical molecular mass (14.8 kDa) was higher than that of SpoVG-His6, most likely due to a degradation of YabJ-His6 at the N terminus.
The possibility that YabJ-His6 might be unstable or not accessible by anti-his6 antibodies was ruled out by expressing YabJ-His6 and as a control also SpoVG-His6 from the constitutive bacA promoter (constructs pyabJ-His and pspoVG-His). These plasmids allowed us to detect YabJ-His6 and SpoVG-His6 in the cell extracts obtained from the respective derivatives (Fig. 5). A direct fusion of the luciferase reporter gene luc+ to PyabJ exhibited luciferase activity (46), excluding further that elements required for translation initiation were missing in the region upstream of the yabJ ORF. However, mRNA structure predictions of the sequences around the yabJ and spoVG ribosome binding site (RBS) indicate that the yabJ RBS might be sequestered by a hairpin structure, whereas the spoVG RBS is freely accessible (data not shown), suggesting that the mRNA secondary structure preceding the yabJ ORF might impede the initiation of YabJ translation (51). The conditions under which YabJ is produced, if at all, as well as the function of YabJ, remain to be identified.
Conclusion.
We showed that the deletion of the σB-dependent yabJ-spoVG operon in S. aureus Newman strongly reduced the expression and activities of important virulence determinants, such as extracellular nuclease, protease, and lipase. Complementation experiments confirmed SpoVG as the main effector of the yabJ-spoVG operon, as shown earlier for capsule production and antibiotic resistance (46). Although the crystal structure of SpoVG from S. aureus has recently been solved, its mode of action remains to be elucidated (30). Transcriptomic analysis of the role of the yabJ-spoVG operon indicated that SpoVG counteracts a subset of σB-regulated genes. In B. subtilis, for which SpoVG was originally described, it is characterized as a regulator of sporulation and asymmetric septation (34, 42). Despite the functional discrepancy between SpoVG from the sporulating B. subtilis and the nonsporulating S. aureus, there seems to be a conserved underlying molecular function, as spoVG inactivation in B. subtilis also influenced gene expression directed by an alternative sigma factor, however, not by the respective σB homolog (39). This hypothesis is further supported by our finding that SpoVG from B. subtilis, like SpoVG from S. aureus, could complement the reduced methicillin resistance level of a ΔyabJ-spoVG MRSA mutant. Future studies will have to reveal the mode of action of SpoVG as well as the function of YabJ and the environmental conditions required for its expression.
Supplementary Material
ACKNOWLEDGMENTS
This study was carried out with financial support from the Forschungskredit of the University of Zurich to B.S., from the Swiss National Science Foundation grant 31-117707 to B.B.-B., and from the HOMFOR grant T201000365 to M.B.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Adhikari R. P., Novick R. P. 2008. Regulatory organization of the staphylococcal sae locus. Microbiology 154:949–959 [DOI] [PubMed] [Google Scholar]
- 2. Ausubel F. M., et al. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 3. Baba T., et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 4. Becher D., et al. 2009. A proteomic view of an important human pathogen—towards the quantification of the entire Staphylococcus aureus proteome. PLoS One 4:e8176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berends E. T., et al. 2010. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2:576–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bischoff M., Berger-Bächi B. 2001. Teicoplanin stress-selected mutations increasing σB activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1714–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bischoff M., et al. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bronner S., Monteil H., Prevost G. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183–200 [DOI] [PubMed] [Google Scholar]
- 9. Burlak C., et al. 2007. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol. 9:1172–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cassat J., et al. 2006. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 152:3075–3090 [DOI] [PubMed] [Google Scholar]
- 11. Chan P. F., Foster S. J. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232–6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Charbonnier Y., et al. 2005. A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics 6:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung A. L., Bayer A. S., Zhang G., Gresham H., Xiong Y. Q. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1–9 [DOI] [PubMed] [Google Scholar]
- 14. Cheung A. L., Eberhardt K. J., Fischetti V. A. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511–514 [DOI] [PubMed] [Google Scholar]
- 15. Churchill G. A. 2004. Using ANOVA to analyze microarray data. Biotechniques 37:173–175, 177 [DOI] [PubMed] [Google Scholar]
- 16. Clarke S. R., Foster S. J. 2006. Surface adhesins of Staphylococcus aureus. Adv. Microb. Physiol. 51:187–224 [DOI] [PubMed] [Google Scholar]
- 17. Diep B. A., et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 18. Dinges M. M., Orwin P. M., Schlievert P. M. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dunman P. M., et al. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duthie E. S., Lorenz L. L. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95–107 [DOI] [PubMed] [Google Scholar]
- 21. Fujimoto S., Ike Y. 2001. pAM401-based shuttle vectors that enable overexpression of promoterless genes and one-step purification of tag fusion proteins directly from Enterococcus faecalis. Appl. Environ. Microbiol. 67:1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Giachino P., Engelmann S., Bischoff M. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gill S. R., et al. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goda S. K., Minton N. P. 1995. A simple procedure for gel electrophoresis and northern blotting of RNA. Nucleic Acids Res. 23:3357–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hempel K., et al. 2010. Quantitative cell surface proteome profiling for σB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach. J. Proteome Res. 9:1579–1590 [DOI] [PubMed] [Google Scholar]
- 26. Holden M. T., et al. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Homerova D., Bischoff M., Dumolin A., Kormanec J. 2004. Optimization of a two-plasmid system for the identification of promoters recognized by RNA polymerase containing Staphylococcus aureus alternative sigma factor σB. FEMS Microbiol. Lett. 232:173–179 [DOI] [PubMed] [Google Scholar]
- 28. Horsburgh M. J., et al. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katayama Y., Zhang H. Z., Chambers H. F. 2004. PBP 2a mutations producing very-high-level resistance to beta-lactams. Antimicrob. Agents Chemother. 48:453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim H. H., Lee B. J., Kwon A. R. 2010. Expression, crystallization, and preliminary X-ray crystallographic analysis of putative SpoVG from Staphylococcus aureus. Arch. Pharm. Res. 33:1285–1288 [DOI] [PubMed] [Google Scholar]
- 31. Kullik I., Giachino P., Fuchs T. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kullik I. I., Giachino P. 1997. The alternative sigma factor σB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167:151–159 [DOI] [PubMed] [Google Scholar]
- 33. Kuroda M., et al. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 [DOI] [PubMed] [Google Scholar]
- 34. Matsuno K., Sonenshein A. L. 1999. Role of SpoVG in asymmetric septation in Bacillus subtilis. J. Bacteriol. 181:3392–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCallum N., et al. 2006. In vivo survival of teicoplanin-resistant Staphylococcus aureus and fitness cost of teicoplanin resistance. Antimicrob. Agents Chemother. 50:2352–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meier S., et al. 2007. σB and the σB-dependent arlRS and yabJ-spoVG loci affect capsule formation in Staphylococcus aureus. Infect. Immun. 75:4562–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morikawa K., et al. 2001. Overexpression of sigma factor σB urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem. Biophys. Res. Commun. 288:385–389 [DOI] [PubMed] [Google Scholar]
- 38. Pané-Farré J., Jonas B., Forstner K., Engelmann S., Hecker M. 2006. The σB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296:237–258 [DOI] [PubMed] [Google Scholar]
- 39. Perez A. R., Abanes-De Mello A., Pogliano K. 2006. Suppression of engulfment defects in Bacillus subtilis by elevated expression of the motility regulon. J. Bacteriol. 188:1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Renzoni A., et al. 2006. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3048–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rollof J., Braconier J. H., Soderstrom C., Nilsson-Ehle P. 1988. Interference of Staphylococcus aureus lipase with human granulocyte function. Eur. J. Clin. Microbiol. Infect. Dis. 7:505–510 [DOI] [PubMed] [Google Scholar]
- 42. Rosenbluh A., Banner C. D., Losick R., Fitz-James P. C. 1981. Identification of a new developmental locus in Bacillus subtilis by construction of a deletion mutation in a cloned gene under sporulation control. J. Bacteriol. 148:341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rossi J., Bischoff M., Wada A., Berger-Bächi B. 2003. MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob. Agents Chemother. 47:2558–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Scherl A., et al. 2006. Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics 7:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schulthess B., et al. 2009. Functional characterization of the σB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob. Agents Chemother. 53:1832–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Senn M. M., et al. 2005. Molecular analysis and organization of the σB operon in Staphylococcus aureus. J. Bacteriol. 187:8006–8019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh V. K., Schmidt J. L., Jayaswal R. K., Wilkinson B. J. 2003. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int. J. Antimicrob. Agents 21:256–261 [DOI] [PubMed] [Google Scholar]
- 49. Sinha S., et al. 1999. Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family. Proc. Natl. Acad. Sci. U. S. A. 96:13074–13079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Talaat A. M., et al. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Unoson C., Wagner E. G. 2007. Dealing with stable structures at ribosome binding sites: bacterial translation and ribosome standby. RNA Biol. 4:113–117 [DOI] [PubMed] [Google Scholar]
- 52. Wang L., Trawick J. D., Yamamoto R., Zamudio C. 2004. Genome-wide operon prediction in Staphylococcus aureus. Nucleic Acids Res. 32:3689–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu S., de Lencastre H., Tomasz A. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ziebandt A. K., et al. 2004. The influence of agr and σB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics 4:3034–3047 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.