Abstract
Heat-resistant agglutinin 1 (Hra1) is an accessory colonization factor of enteroaggregative Escherichia coli (EAEC) strain 042. Tia, a close homolog of Hra1, is an invasin and adhesin that has been described in enterotoxigenic E. coli. We devised a PCR-restriction fragment length polymorphism screen for the associated genes and found that they occur among 55 (36.7%) of the enteroaggregative E. coli isolates screened, as well as lower proportions of enterotoxigenic, enteropathogenic, enterohemorrhagic, and commensal E. coli isolates. Overall, 25%, 8%, and 3% of 150 EAEC strains harbored hra1 alone, tia alone, or both genes, respectively. One EAEC isolate, 60A, produced an amplicon with a unique restriction profile, distinct from those of hra1 and tia. We cloned and sequenced the full-length agglutinin gene from strain 60A and have designated it hra2. The hra2 gene was not detected in any of 257 diarrheagenic E. coli isolates in our collection but is present in the genome of Salmonella enterica serovar Heidelberg strain SL476. The cloned hra2 gene from strain 60A, which encodes a predicted amino acid sequence that is 64% identical to that of Hra1 and 68% identical to that of Tia, was sufficient to confer adherence on E. coli K-12. We constructed an hra2 deletion mutant of EAEC strain 60A. The mutant was deficient in adherence but not autoaggregation or invasion, pointing to a functional distinction from the autoagglutinin Hra1 and the Tia invasin. Hra1, Tia, and the novel accessory adhesin Hra2 are members of a family of integral outer membrane proteins that confer different colonization-associated phenotypes.
INTRODUCTION
Enteroaggregative Escherichia coli (EAEC) strains are increasingly implicated in human diarrhea, especially among children living in developing countries (18, 33). EAEC strains are exceptional colonizers and are defined by a characteristic stacked-brick adherence pattern (43). This aggregative pattern of adherence is a convergent phenotype produced in different lineages by a variety of adhesins, only some of which have been described (35). To date, most studies of EAEC adherence have focused on structural adhesins known as aggregative adherence fimbriae. However, recent research has shown that EAEC strains also harbor an expanding repertoire of nonstructural outer membrane proteins that contribute to colonization (1, 4, 14, 28).
One such outer membrane protein is heat-resistant agglutinin 1 (Hra1), an accessory adhesin that we recently characterized in EAEC strain 042 (1). The hra1 gene (along with its 90% identical allelic variant hek, reported from uropathogenic E. coli and neonatal meningitic E. coli [11, 39]) is predicted to encode a 29-kDa precursor, which is processed to a 25-kDa outer membrane protein. The 792-bp hra1 gene is sufficient to confer agglutination of human erythrocytes, bacterial autoaggregation, enhanced biofilm formation, and aggregative adherence to cultured HEp-2 cells (1, 25). Hra1 shares 67% identity with the previously characterized outer membrane invasin and adhesin Tia (12, 13). The tia gene has been reported as widely disseminated; however, it is now known that many of the strains initially thought to carry tia, including genome-sequenced EAEC strain 042, actually have the hra1 gene (1, 13).
The distribution of hra1 (hek) among extraintestinal E. coli strains was studied by Dobrindt et al. (9) and by Cooke et al. (5). Dobrindt et al. reported that 43.5% of 62 uropathogenic E. coli isolates and 32% of 28 human and animal E. coli isolates from meningitis and sepsis had the 5′ end of uropathogenic E. coli strain 536 pathogenicity island II, which contains the hra1 (hek) gene. Using a specific PCR protocol, they did not detect this locus in any of 18 diarrheagenic pathogens (9). Cooke et al. also employed specific primers, but their focus was bloodstream isolates, of which 34% were positive for hra1 (hek). Srinivasan et al. (39) reported hra1 from 52% of E. coli isolates from urinary tract infections, 65% of pyelonephritis isolates, and 22% of rectal isolates. However, they used a hybridization screen which would most probably have cross-reacted with tia and they did not investigate diarrheagenic E. coli. Having identified hra1 in an enteroaggregative E. coli strain (1), and with the foreknowledge that this gene and/or tia was present in at least some diarrheagenic E. coli isolates from an earlier report by Fleckenstein et al. (13), we studied the distribution of agglutinin genes hra1 and tia among diarrheagenic E. coli isolates. Using a purpose-devised PCR-restriction fragment length polymorphism (PCR-RFLP) test, we found that hra1 and tia are common among enteric bacteria that colonize the lumen, particularly those that are enteroaggregative. We additionally identified a novel member of the family, hra2, which we hypothesized also codes for an afimbrial integral outer membrane colonization factor.
MATERIALS AND METHODS
Strains.
Wild-type EAEC strain 60A was originally isolated from a child with diarrhea in Mexico. This strain, its genetic derivatives constructed in this study, and wild-type strains used as controls are listed in Table 1. We additionally screened 288 fecal E. coli and 23 Providencia isolates from our collection (32, 34, 35) for agglutinin genes as described below. Unless otherwise indicated, strains were cultured in Luria broth (LB) or LB agar at 37°C and maintained at −80°C in LB-glycerol (1:1). Where applicable, media were supplemented with antibiotics at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; neomycin, 50 μg/ml; tetracycline, 25 μg/ml.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant featuresa | Source or reference(s) |
|---|---|---|
| E. coli strains | ||
| H10407 | ETEC isolate from Bangladesh | 10 |
| 042 | EAEC isolate from Peru; Tcr Cmr | 4, 31 |
| 60A | EAEC isolate from Mexico; Apr Tmr | 7 |
| JM1 | 60AΔhra2; Apr Tmr Kmr | This study |
| HS | Normal flora isolate validated as nonpathogenic by human challenge | 24 |
| TOP10 | E. coli K-12 F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697galUgalKrpsL (Strr) endA1nupG | Invitrogen |
| Plasmids | ||
| pJM1A | 1,511-bp clone of hra2 in pGEM-T Easy; Apr | This study |
| pJM2A | 1,511-bp clone of hra2 in pACYC184; Tcr | This study |
| pJM2B | hra2::aphA-3 (1,919 bp) deletion construct in pACYC184; Kmr Tcr | This study |
| pGEM-T Easy | TA cloning vector; Apr | Promega |
| pACYC184 | Low-copy-no. cloning vector; Cmr Tcr | New England Biolabs |
| pUC18K | aphA-3 cassette in pUC18; Kmr | 26 |
| pKM200 | λ Red recombinase system; Cmr; temp-sensitive origin of replication | 29 |
| pBAD102 | Arabinose-inducible expression vector | Invitrogen |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin/neomycin resistance; Strr, streptomycin resistance, Tcr, tetracycline resistance; Tmr, trimethoprim resistance.
Routine molecular biology procedures.
Standard molecular biology procedures were employed (38). DNA amplification was performed using Platinum PCR Supermix (Invitrogen) and 1 μM oligonucleotide primer for each reaction. PCR templates were boiled bacterial colonies or plasmid or genomic DNA. Oligonucleotide primer sequences are listed in Table S1 in the supplemental material. All amplifications began with a 2-min hot start at 94°C followed by 30 cycles of denaturing at 94°C for 30 s, annealing for 30 s at 5°C below the primer annealing temperature, and extending at 72°C for 1 min for every kilobase of DNA. Unless otherwise stated, ligations were performed using Quick T4 ligase enzyme (New England BioLabs), and plasmids were transformed into chemically competent E. coli K-12 TOP10 cells. Transformation of plasmids into EAEC strains was accomplished by electroporation using a Micropulser (Bio-Rad) according to the manufacturer's instructions.
PCR-RFLP for agglutinin genes.
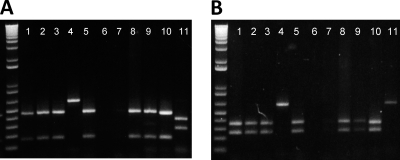
Boiled bacterial colonies or genomic DNA of test strains was subjected to PCR using primers agglF and agglR (see Table S1 in the supplemental material) in PCR Supermix (Invitrogen) or using recombinant Taq polymerase and PCR buffer from New England BioLabs according to the manufacturer's recommendations. All amplifications began with a 2-min hot start at 94°C followed by 35 cycles of denaturing at 94°C for 30 s, annealing at 58°C for 30 s, and extending at 72°C for 20 s. In the presence of an agglutinin gene, the expected 0.6-kb product that was obtained was digested separately with the restriction enzymes EcoRI and HincII, both of which produce different restriction profiles for hra1 and tia. Digested DNA was electrophoretically resolved on 1.5% Tris-acetate-EDTA (TAE)-agarose gels.
Cloning and characterization of hra2.
The central part of hra2 was amplified by PCR using the agglF and agglR primers, then was cloned into the pGEM-T Easy vector and sequenced. The 3′ end of the hra2 gene, downstream of primer agglR, was amplified and cloned by chromosomal walking from the central region of the gene using the TOPO Walker kit (Invitrogen) according to the manufacturer's instructions. Briefly, genomic DNA was digested with PstI, which produces a 3′ TGCA overhang. The digested DNA was dephosphorylated, and primer extension was performed using the agglF primer and Taq polymerase. The extended DNA was ligated to the TOPO Walker kit 58-bp DNA linker using topoisomerase I. The 3′ end was then amplified by PCR with primers agglFGSP2 (which is the reverse complement of aggR) and LinkAmp Primer 1 (a TOPO Walker kit primer that anneals to the linker). 5′ walking of the hra2 sequence upstream of primer agglF by the same method with complementary primers was not successful. As the central and 3′ regions of hra2 proved identical to open reading frame (ORF) SeHA_C4728 in the in-process genome of Salmonella enterica serovar Heidelberg strain SL476 (GenBank accession no. CP001120), the upstream sequence of this open reading frame was used as a template for primer design to capture the 5′ end of hra2 in E. coli 60A. Oligonucleotides JMhra2upF3 and JMhra2upF4 were designed to prime 321 bp and 181 bp, respectively, upstream of the hra2 gene. PCR amplification with primer JMregR, located 426 bp downstream of hra2, produced expected amplicons of 1,371 and 1,511 bp, respectively, in 60A but not commensal strain E. coli HS. The 1,511-bp PCR amplification product was TA cloned into pGEM-T Easy to produce pJM1A, which was sequenced before its insert was subcloned into the EcoRI site of pACYC184 to produce pJM2A.
In-frame expression vectors of agglutinin genes were constructed using the pBAD102 vector in accordance with the manufacturer's instructions. hra1, hra2, and tia were cloned from EAEC strain 042, EAEC strain 60A, and enterotoxigenic E. coli (ETEC) strain H10407, respectively, using primers pBADhra1F and -R (hra1), tiapBADF and -R (tia), and JLGHra2001F and -R (hra2) (see Table S1 in the supplemental material). Expression and outer membrane presentation from these clones in E. coli TOP10 upon induction with arabinose and repression by glucose were verified by SDS-PAGE analysis of outer membrane fractions of harvested bacteria purified by the method of Chart et al. (3).
Construction of an hra2 mutant of EAEC strain 60A.
DNA comprising nucleotides 221 to 662 (the central portion of the gene, containing 84% of the predicted surface-exposed region of hra2) was excised from pJM2A with the restriction enzymes StuI and PsiI. This region was replaced with a promoterless aphA-3 cassette excised from pUC18K (26). Because the aphA-3 gene is preceded by translational stop codons in all three reading frames and is followed by a consensus ribosomal binding site, the cassette produces nonpolar insertions. The resulting deletion construct, pJM2B, was used as template DNA for PCRs with primer pair JMhra2upF3-JMregR. The purified PCR product was electroporated into EAEC strain 60A carrying lambda Red vector pKM200 as recommended by Murphy and Campellone (30) with slight modifications to optimize for strain 60A (21). Successful allelic exchange and loss of plasmid pKM200 were verified by plasmid and resistance profile analysis as well as PCR with four separate primer pairs. The mutant was complemented in trans with hra2 using pJM2A.
Autoaggregation assay.
We quantified bacterial settling rates in overnight cultures of LB and in high-glucose Dulbecco's minimum essential medium (DMEM; Invitrogen) over time as a measure of autoaggregation (17). All assays were performed in duplicate. Cultures of each strain were adjusted to the same optical density at 600 nm (OD600). For each assay, 5 ml of each adjusted culture was placed into two separate tubes. One tube remained static and the other was vortexed before each optical density measurement. The tubes were incubated without shaking at 37°C. At various time points, 0.5 ml was removed from within 2 cm of the surface of the culture and the OD600 was measured.
Biofilm formation.
Biofilms produced in LB and high-glucose DMEM were observed and quantified by fixing and staining with crystal violet using standard methods (36). Briefly, 10 μl of overnight culture was added to 1 ml of test medium in a 24-well plate. Plates were incubated without shaking or with rocking at 37°C. At various time points, culture medium was aspirated and each well was washed three times with phosphate-buffered saline (PBS), fixed for 10 min with 75% ethanol, and then allowed to dry. To quantify the biofilm, crystal violet was eluted with 1 ml of a 3:1 solution of ethanol-acetone. The OD570 of the eluted crystal violet was measured. Data were analyzed by an unpaired Student t test.
HEp-2 adherence.
The HEp-2 adherence assay originally described by Cravioto et al. (6) was used with modifications for delineating aggregative adherence (42). HEp-2 cells were cultured overnight in 8-well chamber slides to 50% confluence, and bacterial strains were cultured overnight in LB. Following the overnight incubation, the HEp-2 monolayer was washed three times with PBS. The growth medium was replaced by DMEM containing 1% mannose, and the wells were infected with 10 μl of overnight culture of the appropriate bacterial strain. Chamber slides were incubated at 37°C in 5% CO2. After 3 h, the culture medium was removed and each well was washed three times with PBS. To fix cells, 70% methanol was added for 20 min. Cells were stained by the addition of a 1:10 dilution of Giemsa in PBS for 20 min. Multiple fields of each slide were examined by light microscopy at 1,000× magnification with oil immersion.
Quantitative adherence assay.
To quantify the difference in the amount of adherence between strains, a quantitative adherence assay was performed as described by Torres et al. (40) with some modifications (1). HEp-2 cells were cultured overnight in 24-well plates to 50% confluence, and bacterial strains were cultured overnight in LB. Following the overnight incubation, the HEp-2 monolayer was washed three times with PBS. The growth medium was replaced by DMEM containing 1% mannose, and the wells were infected with 10 μl of overnight culture of the appropriate bacterial strain. The 24-well plates were incubated at 37°C in 5% CO2. After 3 h, the culture medium was removed and each well was washed three times with PBS to remove nonadherent bacteria. Eukaryotic cells were lysed with a solution of 0.1% Triton X-100 in PBS, and the plate was incubated at room temperature for 15 min. Serial dilutions of lysate from each well and from the initial inoculum were prepared in PBS and plated on LB agar plates, with selective antibiotics where appropriate. After overnight incubation at 37°C, the colonies arising were counted and the proportion of the inoculum that was adherent was computed.
Invasion assay.
A gentamicin protection assay was used to quantify invasiveness of bacterial strains as previously described by Kihlström (22) with modifications. HEp-2 cells were cultured overnight in 24-well plates to 50% confluence, and bacterial strains were cultured overnight in LB. Following the overnight incubation, the HEp-2 monolayer was washed three times with phosphate-buffered saline with calcium and magnesium (PBS-CM; Invitrogen). The growth medium was replaced by DMEM with 10% fetal bovine serum (FBS), and the wells were infected with 10 μl of overnight culture of each bacterial strain. The 24-well plates were incubated at 37°C with 5% CO2.. After 3 h, the culture medium was removed and each well was washed three times with PBS-CM and then once with PBS containing 100 mg/ml of gentamicin. The medium was replaced with DMEM with 10 mg/ml gentamicin, and the 24-well plate was incubated for an additional hour. The medium was then removed, and the monolayer was washed three times with PBS-CM. Triton X-100 (1%) in PBS was added to lyse the monolayer, and the lysates as well as the inoculum were diluted in PBS and plated out onto MacConkey agar plates for viable counts. After overnight incubation of the agar plates at 37°C, the colonies were counted, and results are expressed as the percentage of invasive (gentamicin-protected) bacteria relative to the inoculum for each strain.
Construction of an EAEC phylogenetic tree based on MLST data.
Multilocus sequence typing (MLST) data from 150 EAEC strains generated previously were used to construct a phylogenetic tree using ClonalFrame, a Bayesian method of constructing evolutionary histories that takes both mutation and recombination into account (8, 35). ClonalFrame version 1.1 was downloaded from http://www.xavierdidelot.xtreemhost.com/clonalframe.htm, and MLST allele sequences and profiles are publicly available from http://mlst.ucc.ie/mlst/dbs/Ecoli/. A 75% consensus tree was created from four independent runs of the Markov chain.
Nucleotide sequence accession number.
The sequence of hra2 and its immediate flank from EAEC strain 60A has been submitted to GenBank under accession number JF808724.
RESULTS
Genes hra1 and tia are common among intestinal colonizers and are similar to a third agglutinin gene.
The first 125 nucleotides of tia from ETEC strain H10407 (ORF ETEC3907) are 96% identical to those of hra1 from EAEC strain 042 (ORF EC042_3176), while the 200-bp sequences at the 3′ ends are 83% identical. In contrast, the central gene regions lack significant similarity. We designed primers agglF and agglR, complementary to the conserved 5′ and 3′ ends of the hra1 and tia genes, and performed restriction analysis of the amplified intervening regions, digesting with EcoRI and HincII, each of which produced restriction fragment length polymorphisms in tia and hra1 (Fig. 1). This PCR-RFLP protocol was used to screen 288 independent fecal E. coli isolates. Eighty-four (29.2%) of the strains produced the predicted 614-bp amplicon (Table 2). Agglutinin-positive strains, that is, strains producing an amplicon, were seen in all categories of fecal E. coli screened but were most prevalent among EAEC strains (Table 2). RFLP analysis demonstrated that 21 strains carried only the tia gene, 53 carried only the hra1 gene, and 9 isolates possessed both genes (Table 2). Thus, hra1 predominated overall, particularly among EAEC strains. Of 31 E. coli strains from healthy individuals, all of which lacked virulence genes defining any of the five E. coli pathotypes, four harbored hra1 and three more carried both hra1 and tia (Table 2). Hypothesizing that these genes might be present in other commensals, we screened 23 Providencia fecal isolates obtained from healthy individuals and, as shown in Table 2, identified hra1 in two of them.
Fig. 1.
Detection and identification of agglutinin genes in EAEC strains by PCR-RFLP. (A) EcoRI digests. (B) HincII digests. Lanes (A and B): 1 to 3, 5, and 8 to 10, strains containing hra1; 4, strain 60A (hra2); 11, strain D32 (tia); 6 and 7, agglutinin-negative strains.
Table 2.
Detection of agglutinin genes in fecal E. coli and Providencia isolates
| Strains or species | No. of isolates with: |
|||||
|---|---|---|---|---|---|---|
| No agglutinin gene | hra1 alone | tia alone | hra1 and tia | hra2 | At least one agglutinin genea | |
| Enteroaggregative E. coli | 95 | 38 | 12 | 4 | 1 | 55 (36.7) |
| Enterotoxigenic E. coli | 21 | 3 | 3 | 1 | 0 | 7 (25.0) |
| Diffusely adherent E. coli | 16 | 3 | 5 | 0 | 0 | 8 (33.3) |
| Enteropathogenic E. coli | 30 | 1 | 1 | 0 | 0 | 2 (6.3) |
| Enterohemorrhagic E. coli | 18 | 4 | 0 | 1 | 0 | 5 (21.7) |
| Commensal E. coli | 24 | 4 | 0 | 3 | 0 | 7 (22.6) |
| Providencia spp. | 21 | 2 | 0 | 0 | 0 | 2 (8.7) |
| Total | 231 | 55 | 21 | 9 | 1 | 86 (27.1) |
Numbers in parentheses are percentages.
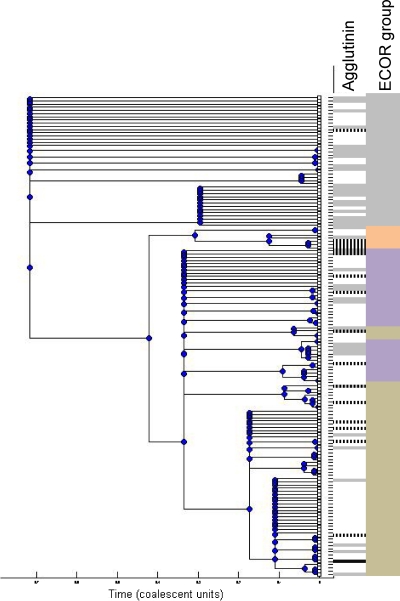
Examination of the distribution of hra1 and tia genes among EAEC strains that have been subjected to multilocus sequence typing revealed that the hra1 and tia genes are distributed throughout the tree (Fig. 2). EAEC strains belonging to the ECOR D lineage carried hra1 significantly more commonly than other lineages (P = 0.0003, chi-square test), but the genes were more randomly distributed in other lineages, suggesting that EAEC strains outside the ECOR D lineage may have acquired the genes very recently in evolutionary time. As shown in Fig. 2, ECOR group A EAEC strains were least likely to harbor an agglutinin gene. tia, alone or with hra1, was most commonly encountered outside ECOR group D. EAEC strain 101-1, an outbreak isolate belonging to sequence type complex 12, possessed the tia gene, as did isolates most closely related to this strain (19, 20, 35).
Fig. 2.
Seventy-five percent consensus ClonalFrame tree for MLST data from 151 EAEC strains, incorporating recombination as well as mutation. The column to the immediate right of the tree indicates whether each strain harbored no agglutinin gene (white), hra1 alone (gray), tia alone (hatched black on white), both hra1 and tia (hatched black on gray), or hra2 (black). Principal E. coli subclades corresponding to the four major groups originally defined by multilocus enzyme electrophoresis (MLEE), A, B1, D, and B2, are marked to the far right of the tree in brown, purple, gray, and orange, respectively.
One amplicon from EAEC strain 60A produced a restriction fragment length polymorphism profile that was distinct from those of hra1 and tia (Fig. 1). We sequenced the amplicon and determined that it comprised a novel gene, which we have designated hra2. hra2 was not identified in any of the other 310 strains screened. By chromosomal walking, we cloned the 3′ end of the hra2 gene and most of a downstream gene predicted to encode a regulatory protein. We used the partial sequence of hra2 from EAEC strain 60A to search the GenBank nucleotide database. A routine BLAST-N search revealed that the available sequence was identical to an open reading frame annotated as a heat-resistant agglutinin ORF in the in-process Salmonella enterica serovar Heidelberg strain SL476 genome (ORF SeHA_C4728). Furthermore, an open reading frame designated deoR (SeHA_C4729), situated immediately downstream of hra1, was identical to the predicted regulator gene we identified in 60A. We speculated that this gene cluster was conserved in the two species and therefore designed primers for the upstream region of hra2 in S. Heidelberg strain SL476 and successfully used these to clone 1,511 bp of sequence representing the 747-bp full-length hra2 gene as well as 341 bp of upstream and 423 bp of downstream flanking sequences. We determined that hra2 lies between a P4-like integrase gene and a putative regulator gene in EAEC strain 60A as well as S. Heidelberg SL476.
hra2 from EAEC strain 60A is 99% identical to the agglutinin gene found in S. enterica serovar Heidelberg strain SL476, with only 3 base mismatches. The flanking 5′ and 3′ sequences of both agglutinin genes are also nearly identical. Additionally, Hra2 shares 72% maximum identity with the Tia invasin determinant in ETEC at the amino acid level. Hra2 from EAEC strain 60A showed less homology to Hra1 from EAEC strain 042 and the porcine pathogenic E. coli O9:H10:K99 strain, with only 69% shared identity across full coverage (see Fig. S1 in the supplemental material). Hra2 is 67% similar to Hek from NMEC strain RS218, but only at 89% coverage.
The hra2 ORF has two potential start codons, both of which are in the same reading frame and lie within the predicted signal sequence, predicting a full-length, unprocessed protein of 248 or 245 residues. Based on PredictProtein software (http://www.predictprotein.org/), the first 25 amino acids signify a prototypical signal peptide cleavage sequence required for targeting proteins to the outer membrane in Gram-negative organisms.
Hra2 is not an autoagglutinin like Hra1.
We placed the hra2 gene under the control of the arabinose promoter by cloning it into the expression vector pBAD102. Protein expression and outer membrane localization following induction with arabinose were confirmed by SDS-PAGE and Coomassie staining (data not shown). Using this construct, we assessed whether Hra2 expression was capable of conferring bacterial autoaggregation on E. coli K-12 strain TOP10 and compared these results to the autoagglutination phenotypes due to expression of Hra1, Tia, or empty vector controls. Expression of Hra1, but not Tia, was sufficient to confer autoagglutination on E. coli TOP10. E. coli TOP10 expressing Hra2 demonstrated a phenotype of weak to no autoagglutination, similar to that seen with Tia (see Fig. S2 in the supplemental material). We used lambda Red-mediated mutagenesis to construct an in-frame deletion of hra2, replacing the gene with an aphA-3 cassette. The 60A hra2 mutant was not deficient in autoaggregation (data not shown). We measured biofilm formation on polystyrene surfaces using the crystal violet assay. Although 60A formed strong biofilms, we found no significant difference in biofilm formation between the wild-type strain and its isogenic mutant (see Fig. S3 in the supplemental material). Furthermore, unlike hra1 (1), hra2 was not sufficient to confer biofilm formation. Thus, we conclude that, unlike its homolog Hra1, Hra2 is not an autoagglutinin and does not contribute to the exceptional biofilm formation demonstrated by strain 60A.
Hra2 is not an invasin like Tia.
Hra2 is slightly more similar to the Tia invasin than to the Hra1 autoagglutinin (72 versus 67%), and the similarity includes the predicted surface-exposed region required for invasion (13). The similarity to Tia and the absence of autoagglutinating activity led us to hypothesize that Hra2 might act as an invasin in 60A. However, consistent with previous studies suggesting that most EAEC strains do not invade significantly, 60A is 4-fold less invasive than the moderately invasive tia-positive ETEC strain H10407 (see Fig. S4 in the supplemental material). Moreover, there were no significant differences in invasion between 60A and its hra2 mutant (see Fig. S4 in the supplemental material) (P = 0.077). Thus, we conclude that Hra2 is not an invasin.
Hra2 contributes to epithelial cell adherence, but not aggregative adherence, of EAEC strain 60A.
Both hra1 and tia are sufficient to confer epithelial cell adherence on laboratory E. coli strains (1, 13). As shown in Fig. 3, hra2 expression is also sufficient to confer HEp-2 adherence on E. coli TOP10 (Fig. 3E and F). However, the pattern of adherence conferred by hra2, unlike that conferred by the hra1 gene from EAEC strain 042, which we have reported previously (1), was not aggregative. Instead, as illustrated in Fig. 3, hra2 conferred a diffuse pattern of adherence.
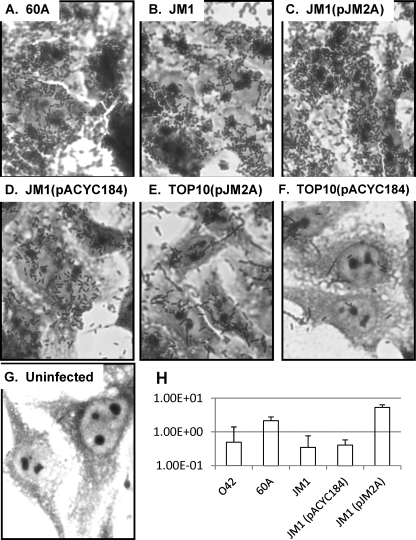
Fig. 3.
Adherence to epithelial cells in the HEp-2 adherence assay. Light microscopy photomicrographs were taken at a magnification of ×1,000. (A) 60A, demonstrating aggregative adherence; (B) JM1, the 60A hra2 mutant; (C) JM1 complemented with hra2 clone pJM2A; (D) JM1 carrying the pACYC184 vector; (E) pJM2A in TOP10; (F) nonadherent TOP10 carrying vector pACYC184; (G) uninfected HEp-2 cells; (H) % of inoculum adherent (y axis) for EAEC strains 042, 60A, JM1, JM1(pJM2a), and JM1(pACYC184). Adherence of strains JM1 and JM1(pACYC184) was significantly less than that of wild-type strain 60A (P < 0.005).
EAEC strains 60A and 042 both demonstrate aggregative adherence, but an 042 ahra1 mutant loses its aggregative stacked-brick conformation, producing a diffuse pattern of adherence (1). As shown in Fig. 3A, 60A adheres proficiently and in an aggregative pattern. The 60A hra2 mutant, JM1, remains adherent, but there is a slight change of pattern, with more diffuse adherent bacteria surrounding localized microcolonies. These small clusters or microcolonies were not evident in the largely two-dimensional stacked-brick orientation of wild-type 60A but were seen in the less adherent mutant. Thus, like hra1, the hra2 gene appears to moderate the adherence pattern in its cognate EAEC host strain. Complementing the mutation with the hra2 gene in trans, but not the pACYC184 vector control, obliterated the appearance of microcolonies and restored the wild-type pattern and degree of adherence.
Given the slight change in adherence pattern, a quantitative adherence assay was necessary to determine whether the apparently less-dense adherence seen with hra2 mutant JM1 compared to that with parent 60A was due to a reduction in the number of adherent bacteria. The quantitative adherence assay we used measured viable counts of adherent bacteria and corrected for inoculum size. Wild-type 60A-infected monolayers contained quantitatively more adherent 60A bacteria than seen for EAEC strain 042. The 60A hra2 mutant was significantly less adherent to cultured HEp-2 cells than the wild-type strain (P < 0.005), and this phenotype could be complemented in trans (Fig. 3H).
DISCUSSION
Nonstructural bacterial adhesins and other colonization factors are important contributors to pathogenesis and commensalism. One-third to two-thirds of extraintestinal E. coli isolates carry the hra1 (hek) gene, which is believed to contribute to virulence (9, 39). In this study, we sought hra1 and tia among a wide variety of enteric E. coli strains and found that these genes are surprisingly widely disseminated. Agglutinin genes are common among commensal E. coli and Providencia isolates. Cooke et al. (5) found that the hek gene, detected with primers that are specific for hra1 (hek) but that would not amplify tia, was significantly more commonly detected in community-acquired and health care (non-hospital-)-associated bloodstream E. coli isolates than in nosocomial isolates. It is probable that this gene is or was disseminated horizontally among enteric organisms irrespective of pathogenicity.
Although present in some commensals, agglutinin genes are especially common in aggregative and diffusely adherent E. coli, providing one explanation for why the repertoire of known adhesins does not always correlate with colonization-associated phenotypes in these categories (27). Moreover, the presence of hra1 or tia in some commensals illustrates that these are colonization rather than virulence genes, even though their presence may enhance the pathogenicity of select pathogenic lineages. One such lineage is ECOR D EAEC strains, including EAEC strain 042, which we have previously shown uses Hra1 as an accessory adhesin (1, 35). The predominance of hra1 among this subgroup of EAEC, compared to other EAEC as well as non-EAEC intestinal colonizers, strongly suggests that it does provide a selective advantage to strains like 042.
Hra1 and Tia are homologous proteins that confer different properties associated with colonization on E. coli strains (1, 12, 13, 25). Among EAEC strains, we have additionally identified a novel member of this agglutinin family, which we have named Hra2 (heat-resistant agglutinin 2). Hra2 was detected only in a single ECOR A EAEC strain, 60A, and is also present in an S. Heidelberg genome. The hra2 gene product shares 64% identify with Hra1 and 68% identity with Tia, with most of the similarity lying within parts of the proteins predicted to be embedded within the outer membrane. As the four predicted surface-exposed loops vary among the three proteins (see Fig. S1 in the supplemental material), it is significant that Hra2, unlike Hra1, lacks autoagglutination- and biofilm-conferring properties and is also not an invasin like Tia. Hra2 is not essential for epithelial cell adherence by strain 60A but was found to contribute quantitatively and qualitatively to this phenotype. The hra2 gene was also sufficient to confer diffuse adherence to HEp-2 cells on a laboratory E. coli strain. We are yet to identify the hra2 gene in any other E. coli strain. However, we note that a virtually identical gene is present in the S. Heidelberg strain SL476 genome and is therefore not a unique variant of Hra1 or Tia. Although the strains used in this study include isolates from different countries in Africa, Europe, Asia, and the Americas, it is possible that other isolates obtained at a similar time from Mexico may also harbor hra2, which warrants further investigation using other strain collections.
Upstream of hra2 in 60A and S. Heidelberg strain SL476 is a P4 integrase-like gene. This could suggest that hra2 is part of a genetic element that was acquired horizontally in the recent evolutionary past. Srinivasan et al. (39) made a similar suggestion with respect to hra1 when they drew attention to the variable, plastic organization of genes flanking that gene in uropathogenic E. coli. It is probable that the agglutinin genes are (or were recently) mobile, which could account for their wide distribution among E. coli isolates of different pathotypes and phylogenetic lineages. Additionally, horizontal transfer into organisms that have adapted differently to host selection may account for evolution of different functions among members of very similar gene families, an example of how horizontal gene transfer can shape protein family expansion (41).
S. Heidelberg is among the more virulent nontyphoidal Salmonella serovars, and its isolates possess a wide variety of adhesins and other colonization factors (2). The potential role that hra2 might play in S. Heidelberg virulence and its distribution among this and other Salmonella serovars remain to be studied.
In summary, we have found that Hra1, Tia, and Hra2 constitute a family of small, integral outer membrane proteins that share considerable sequence similarity but differ functionally. More-distant homologs of the genes that encode these three agglutinins are present in the genomes of other colonizing bacteria (see Fig. S5 in the supplemental material). These include sapA from Salmonella Typhimurium, which is present in an operon required for antimicrobial peptide resistance (37), an S. Typhimurium adhesin-invasin gene known as pagN (16, 23), and gene T2544 from the Salmonella Typhi CT18 genome, which is annotated as encoding a putative outer membrane protein (Poma). The agglutinin genes show the most similarity in regions predicted to be embedded within the outer membrane and vary considerably in their predicted surface-exposed loops (see Fig. S1 in the supplemental material). This means that they are an effective module for host-specific adaptation, immune evasion, and other types of in vivo positive selection. Ghosh et al. (15) recently demonstrated that Poma/T2544 is an adhesin and that convalescent-patient sera contained anti-T544 IgG. Moreover, antiserum against this protein enhanced macrophage-mediated clearance and contained antibodies that are protective in mice. The E. coli agglutinin proteins are more similar to each other than the Salmonella genes, and all the agglutinins share similarity with the Opa proteins from Neisseria spp. (see Fig. S5 in the supplemental material).
The agglutinin family is widely disseminated among pathogenic and commensal intestinal colonizers. With the characterization of Hra2 in this study, a spectrum of phenotypes conferred by agglutinins has been identified. Hra1, the first member of the family reported in the literature, is an autoagglutinin and adhesin (1, 25). Tia is an adhesin and invasin (12, 13), while Hra2, newly reported in this study, is an adhesin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NSF RUI awards 0516591 and 0948460 to I.N.O. J.C.D., R.S.L., and S.B. were supported by undergraduate research awards from the Howard Hughes Medical Institute through Haverford College. R.S.L. was an Arnold and Mabel Beckman Foundation-supported undergraduate research scholar, and J.M. and S.A. received summer research support from the Loewey-Santer-Finger Fund at Haverford College. I.N.O. was a Branco Weiss Fellow of the Society in Science and a Fellow of the Wissenshaftskolleg zu Berlin.
We are grateful to Rachel Song and Jason Park for technical assistance and Kenan Murphy for pKM200.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Bhargava S., et al. 2009. The heat-resistant agglutinin 1 is an accessory enteroaggregative Escherichia coli colonization factor. J. Bacteriol. 191:4934–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bronowski C., Winstanley C. 2009. Identification and distribution of accessory genome DNA sequences from an invasive African isolate of Salmonella Heidelberg. FEMS Microbiol. Lett. 298:29–36 [DOI] [PubMed] [Google Scholar]
- 3. Chart H., Spencer J., Smith H. R., Rowe B. 1997. Identification of entero-aggregative Escherichia coli based on surface properties. J. Appl. Microbiol. 83:712–717 [DOI] [PubMed] [Google Scholar]
- 4. Chaudhuri R. R., et al. 2010. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One 5:e8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cooke N. M., Smith S. G., Kelleher M., Rogers T. R. 2010. Major differences exist in frequencies of virulence factors and multidrug resistance between community and nosocomial Escherichia coli bloodstream isolates. J. Clin. Microbiol. 48:1099–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cravioto A., Gross R., Scotland S., Rowe B. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional enteropathogenic serotypes. Curr. Microbiol. 3:95–99 [Google Scholar]
- 7. Czeczulin J., Whittam T., Henderson I., Navarro-Garcia F., Nataro J. 1999. Phylogenetic analysis of virulence genes in enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Didelot X., Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dobrindt U., et al. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365–6372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Evans D. G., Silver R. P., Evans D. J., Jr., Chase D. G., Gorbach S. L. 1975. Plasmid-controlled colonization factor associated with virulence in Esherichia coli enterotoxigenic for humans. Infect. Immun. 12:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fagan R. P., Lambert M. A., Smith S. G. 2008. The Hek outer membrane protein of Escherichia coli strain RS218 binds to proteoglycan and utilizes a single extracellular loop for adherence, invasion and autoaggregation. Infect. Immun. 76:1135–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fleckenstein J. M., Holland J. T., Hasty D. L. 2002. Interaction of an outer membrane protein of enterotoxigenic Escherichia coli with cell surface heparan sulfate proteoglycans. Infect. Immun. 70:1530–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fleckenstein J. M., Kopecko D. J., Warren R. L., Elsinghorst E. A. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64:2256–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujiyama R., et al. 2008. The shf gene of a Shigella flexneri homologue on the virulent plasmid pAA2 of enteroaggregative Escherichia coli 042 is required for firm biofilm formation. Curr. Microbiol. 56:474–480 [DOI] [PubMed] [Google Scholar]
- 15. Ghosh S., et al. 2011. An adhesion protein of Salmonella enterica serovar Typhi is required for pathogenesis and potential target for vaccine development. Proc. Natl. Acad. Sci. U. S. A. 108:3348–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gunn J. S., Belden W. J., Miller S. I. 1998. Identification of PhoP-PhoQ activated genes within a duplicated region of the Salmonella typhimurium chromosome. Microb. Pathog. 25:77–90 [DOI] [PubMed] [Google Scholar]
- 17. Hasman H., Chakraborty T., Klemm P. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 181:4834–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang D. B., et al. 2006. Enteroaggregative Escherichia coli is a cause of acute diarrheal illness: a meta-analysis. Clin. Infect. Dis. 43:556–563 [DOI] [PubMed] [Google Scholar]
- 19. Hwang J., Mattei L. M., VanArendonk L. G., Meneely P. M., Okeke I. N. 2010. A pathoadaptive deletion in an enteroaggregative Escherichia coli outbreak strain enhances virulence in a Caenorhabditis elegans model. Infect. Immun. 78:4068–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Itoh Y., Nagano I., Kunishima M., Ezaki T. 1997. Laboratory investigation of enteroaggregative Escherichia coli O untypeable:H10 associated with a massive outbreak of gastrointestinal illness. J. Clin. Microbiol. 35:2546–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Joo L. M., Macfarlane-Smith L. R., Okeke I. N. 2007. Error-prone DNA repair system in enteroaggregative Escherichia coli identified by subtractive hybridization. J. Bacteriol. 189:3793–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kihlstrom E. 1977. Infection of HeLa cells with Salmonella typhimurium 395 MS and MR10 bacteria. Infect. Immun. 17:290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lambert M. A., Smith S. G. 2008. The PagN protein of Salmonella enterica serovar Typhimurium is an adhesin and invasin. BMC Microbiol. 8:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levine M. M., et al. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119–1122 [DOI] [PubMed] [Google Scholar]
- 25. Lutwyche P., Rupps R., Cavanagh J., Warren R. A., Brooks D. E. 1994. Cloning, sequencing, and viscometric adhesion analysis of heat-resistant agglutinin 1, an integral membrane hemagglutinin from Escherichia coli O9:H10:K99. Infect. Immun. 62:5020–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ménard R., Sansonetti P. J., Parsot C. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mohamed J. A., et al. 2007. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J. Clin. Microbiol. 45:121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monteiro-Neto V., Bando S. Y., Moreira-Filho C. A., Giron J. A. 2003. Characterization of an outer membrane protein associated with haemagglutination and adhesive properties of enteroaggregative Escherichia coli O111:H12. Cell. Microbiol. 5:533–547 [DOI] [PubMed] [Google Scholar]
- 29. Murphy K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murphy K. C., Campellone K. G. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nataro J., Scaletsky I., Kaper J., Levine M., Trabulsi L. 1985. Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect. Immun. 48:378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okeke I. N., Lamikanra A., Steinruck H., Kaper J. B. 2000. Characterization of Escherichia coli strains from cases of childhood diarrhea in provincial southwestern Nigeria. J. Clin. Microbiol. 38:7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okeke I. N., Nataro J. P. 2001. Enteroaggregative Escherichia coli. Lancet Infect. Dis. 1:304–313 [DOI] [PubMed] [Google Scholar]
- 34. Okeke I. N., Scaletsky I. C., Soars E. H., Macfarlane L. R., Torres A. G. 2004. Molecular epidemiology of the iron utilization genes of enteroaggregative Escherichia coli. J. Clin. Microbiol. 42:36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okeke I. N., et al. 2010. Multi-locus sequence typing of enteroaggregative Escherichia coli isolates from Nigerian children uncovers multiple lineages. PLoS One 5:e14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Toole G. A., Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 37. Parra-Lopez C., Baer M. T., Groisman E. A. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12:4053–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Srinivasan U., Foxman B., Marrs C. F. 2003. Identification of a gene encoding heat-resistant agglutinin in Escherichia coli as a putative virulence factor in urinary tract infection. J. Clin. Microbiol. 41:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torres A. G., et al. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Treangen T. J., Rocha E. P. 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7:e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vial P. A., Mathewson J. J., DuPont H. L., Guers L., Levine M. M. 1990. Comparison of two assay methods for patterns of adherence to HEp-2 cells of Escherichia coli from patients with diarrhea. J. Clin. Microbiol. 28:882–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vial P. A., et al. 1988. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J. Infect. Dis. 158:70–79 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.