Abstract
The corepressor SMRT mediates repression by thyroid hormone receptor (TR) as well as other nuclear hormone receptors and transcription factors. Here we report the isolation of a novel SMRT-containing complex from HeLa cells. This complex contains transducin β-like protein 1 (TBL1), whose gene is mutated in human sensorineural deafness. It also contains HDAC3, a histone deacetylase not previously thought to interact with SMRT. TBL1 displays structural and functional similarities to Tup1 and Groucho corepressors, sharing their ability to interact with histone H3. In vivo, TBL1 is bridged to HDAC3 through SMRT and can potentiate repression by TR. Intriguingly, loss-of-function TRβ mutations cause deafness in mice and humans. These results define a new TR corepressor complex with a physical link to histone structure and a potential biological link to deafness.
Keywords: SMRT, N-CoR, TBL1, HDAC, corepressor, repression
SMRT (silencing mediator for retinoid and thyroid receptors) and N-CoR (nuclear receptor corepressor) mediate the repression function of unliganded nuclear hormone receptors (Chen and Evans 1995; Horlein et al. 1995) and other classes of transcriptional repressors such as the leukemogenic fusion protein PLZF (Hong et al. 1997; Grignani et al. 1998; He et al. 1998; Lin et al. 1998), Notch-binding protein CBF-1 (Kao et al. 1998), and homeodomain proteins including Rpx2, Pit-1, and Pbx (Xu et al. 1998; Asahara et al. 1999) (for review, see Glass and Rosenfeld 2000; Hu and Lazar 2000). SMRT and N-CoR are thought to mediate their effects through chromatin modification, which plays a role in determining whether target genes reside in a condensed or relaxed state.
Relaxed chromatin structure is generally associated with gene activation and can be facilitated by acetylation of histone tails through recruitment of histone acetyltransferases (HATs) to the promoter region. This phenomenon can be reversed effectively by the recruitment of histone deacetylases (HDACs), which may serve to return chromatin to its condensed state through deacetylation of histone tails, thus silencing transcription (Pazin and Kadonaga 1997). Biochemical studies of HDAC-containing complexes have resulted in the isolation of Mi2/NuRD (Wade et al. 1998; Xue et al. 1998; Zhang et al. 1998a) and mSin3 (Hassig et al. 1997; Zhang et al. 1997; Laherty et al. 1998) complexes, which are distinct, yet share molecules such as Rb-associated proteins in addition to HDAC1/2.
The prevailing model for SMRT/N-CoR function invokes the recruitment of HDAC1 via a direct interaction with mSin3 (for review, see Knoepfler and Eisenman 1999). However, although mSin3 and HDAC1 copurify in large multiprotein complexes, N-CoR or SMRT were not identified as tightly binding components (Zhang et al. 1998b, 1999; Huang et al. 2000). It has been suggested that repression by SMRT/N-CoR can be mediated by mechanisms other than HDAC recruitment, and N-CoR and SMRT have been shown to interact with such basal components such as TFIIB, TAFII32, and TAFII70 (Muscat et al. 1998; Wong and Privalsky 1998).
Another major class of corepressor is typified by the Drosophila Groucho proteins (for review, see Fisher and Caudy 1998) and yeast Tup1 (for review, see Wahi et al. 1998). These proteins contain carboxy-terminal WD40-repeat domains and share the ability to interact with histone H3. However, there are a number of major differences between Groucho and Tup1, including relatively poor sequence conservation in the WD40 and repression domains, suggesting that these proteins may be analogous rather than homologous (Fisher and Caudy 1998). Functional interactions have been described between HDAC activity and both Groucho (Chen et al. 1999) and Tup1 (Edmondson et al. 1998). However, this class of corepressor has not been implicated previously in the mechanism of repression by SMRT or N-CoR.
To better define the mechanism of repression by SMRT, we purified a SMRT complex from HeLa cells. Two SMRT-associated polypeptides (SMAPs), neither of which has been thought previously to play a role in the function of SMRT, were identified by ion-trap mass specrometry. The first SMAP identified is HDAC3, a class I HDAC whose unpredicted presence in the core SMRT complex in the absence of HDAC1 and other HDACs suggests that SMRT–HDAC interactions could be both constitutive and transient. The second SMAP is related to yeast Tup1 and Drosophila Groucho corepressors both structurally and functionally. The unexpected composition of the core SMRT complex provides a new framework for understanding the mechanism of repression by nuclear receptors and other transcriptional repressors.
Results
Purification of the core SMRT complex
We have purified SMRT from HeLa cells to determine the polypeptide composition of endogenous SMRT-containing complexes. Five monoclonal antibodies that recognized SMRT but not N-CoR were developed (Fig. 1a). These were directed against the carboxy-terminal NR-interaction domain to avoid disruption of protein complexes involving the amino-terminal repression domains of SMRT. The monoclonal antibodies were pooled and used as an affinity matrix to purify SMRT from fractionated HeLa nuclear extract (Fig. 1b,c). Affinity purification of the SMRT complex from HeLa nuclear extract yielded a similar polypeptide composition (data not shown). Two putative SMAPs reproducibly coeluted from the SMRT affinity matrix along with a band that migrated at the predicted molecular mass of SMRT (polypeptides SMAP270, SMAP55, and SMAP45 in Fig. 1c, lane 2). These polypeptides were isolated and subjected to mass spectrometry sequence analysis (see Materials and Methods). SMAP270 sequences were identical to SMRT (Fig. 1d), indicating that the purification was specific.
Figure 1.
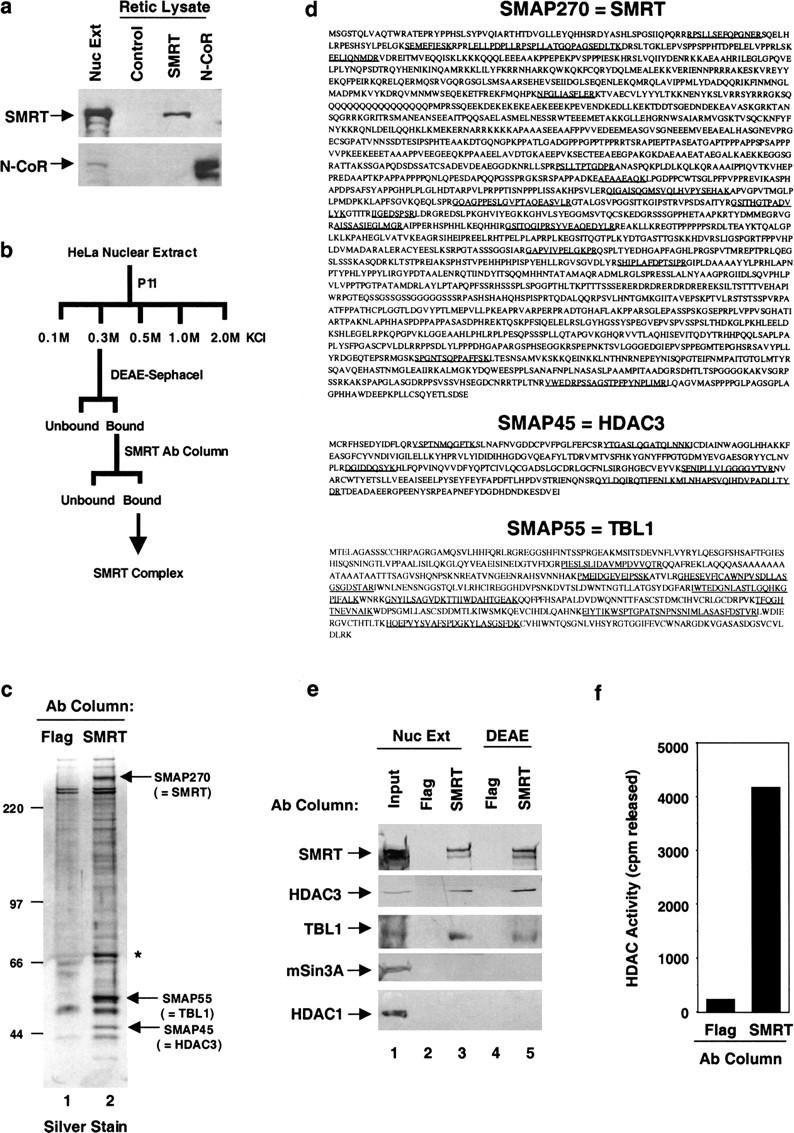
Purification, identification, and verification of SMRT complex components. (a) Pooled SMRT carboxy-terminal monoclonal antibodies are not cross-reactive with N-CoR. In vitro-translated SMRT, N-CoR, or unprogrammed rabbit reticulocyte lysate (RRL) control were subjected to immunoblot analysis with five pooled SMRT monoclonal antibodies or an anti-N-CoR monoclonal antibody. (b) Strategy to obtain SMRT-associated polypeptides. (c) SDS-PAGE and silver-staining analysis of SMRT complex purified as in b from HeLa nuclear extract. SMRT complex components are indicated by arrows. Asterisk denotes a band that was not reproducibly observed in eluates from the SMRT column. (d) Identity of SMRT complex subunits. Peptide sequences obtained by microsequencing are underlined. (e) Immunoblot analysis of SMRT complexes purified independently. Components isolated from nuclear extract or as in b were verified by immunoblot with anti-SMRT, mouse anti-TBL1, anti-HDAC3, anti-Sin3A, and anti-HDAC1. (f) HDAC assay of SMRT purified as in b.
HDAC3 and TBL1 are components of the core SMRT complex
SMAP45 was identified as HDAC3 (Yang et al. 1997; Emiliani et al. 1998; Grozinger et al. 1999) (Fig. 1d). This is consistent with the current model of corepressor function, although this is the first time that this class I HDAC has been shown to be associated with SMRT. SMAP55 was determined to be a transducin β-like 1 (TBL1) protein (Fig. 1d). The TBL1 gene is ubiquitously expressed, and has been found to be deleted in patients with sensorineural deafness associated with an X-linked syndrome of ocular ablinism (Bassi et al. 1999). TBL1 is a WD40-repeat-containing protein that is highly related to ebi (Dong et al. 1999), a regulator of the epidermal growth receptor signaling pathway in Drosophila. Similar WD40 repeats have been identified in the yeast Tup1 (Wahi et al. 1998), and Drosophila Groucho (Paroush et al. 1994) corepressors. Immunoblot analysis of SMRT affinity-purified preparations either directly from nuclear extract or following P11 and DEAE–Sephacel fractionation (Fig. 1e) confirmed copurification of HDAC3 and TBL1 with SMRT (Fig. 1e, cf. lanes 3 and 5 with 2 and 4). Also, immunopurified SMRT was associated with HDAC enzymatic activity (Fig. 1f). Notably, neither mSin3 or HDAC1 were detectable in these purified SMRT preparations (Fig. 1e.). We cannot exclude the possibility that antibody binding may have disrupted protein interactions that may be too weak to withstand biochemical purification. Nevertheless, mSin3 was also not found in immunoprecipitates of epitope-tagged TBL1 (data not shown).
Copurification of SMRT, HDAC3, and TBL1
We next confirmed the association of SMRT, HDAC3, and TBL1 without using affinity purification. SMRT was purified from HeLa cells following the chromatographic scheme presented in Figure 2a. Immunoblot analysis of the last chromatographic step revealed the coelution of SMRT, HDAC3, and TBL1 prior to the peak protein elution from a gel-filtration column, at an apparent molecular mass of 1–2 megadaltons (Fig. 2b,c). These results strongly suggest that SMRT, HDAC3, and TBL1 are present in the same high molecular weight complex.
Figure 2.
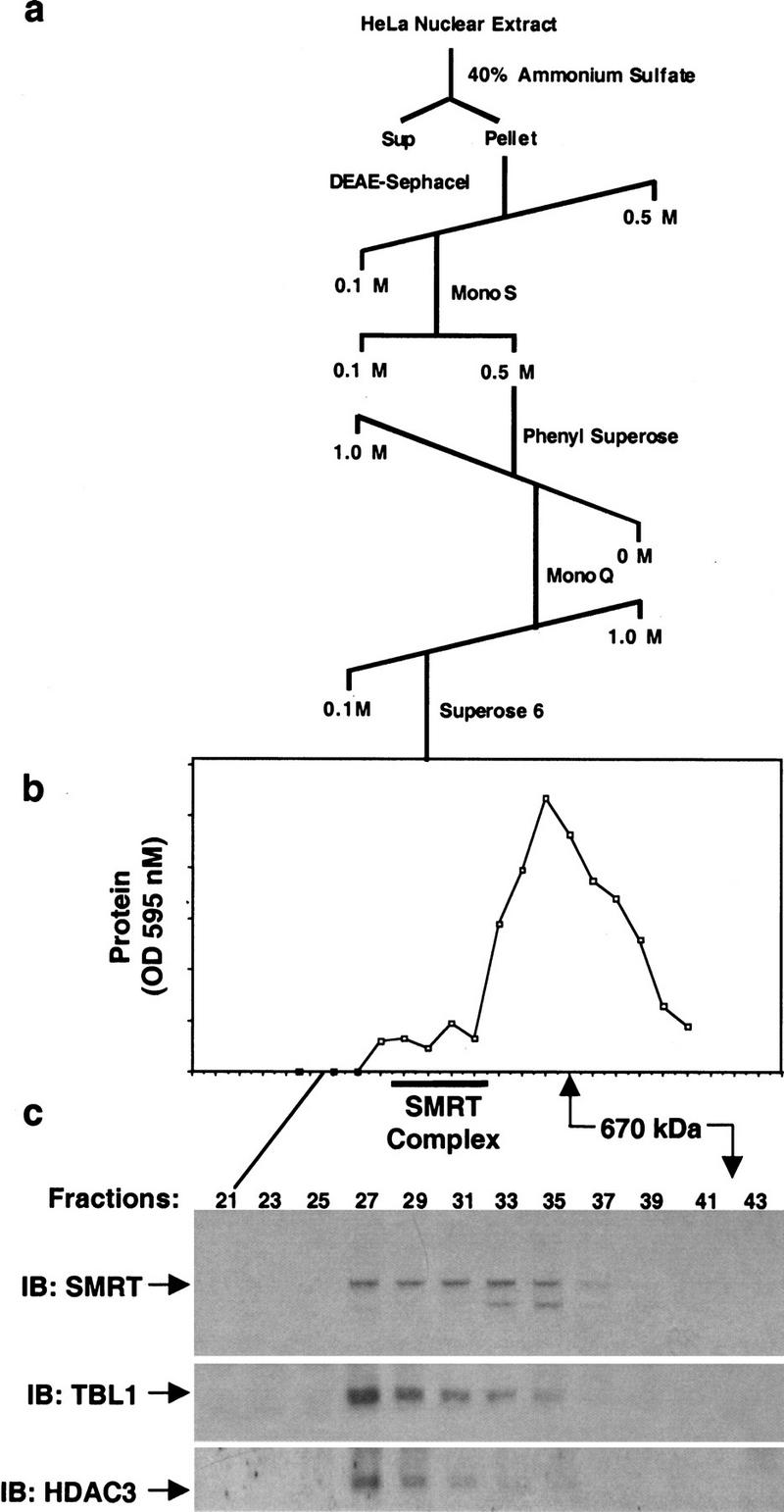
Copurification of SMRT, HDAC3, and TBL1. (a) Multistep purification scheme. (b) Protein elution from Superose 6 column. (c) Immunoblot analysis of Superose 6 column fractions. Fractions were assayed by immunoblot with anti-SMRT, rabbit anti-TBL1, and anti-HDAC3.
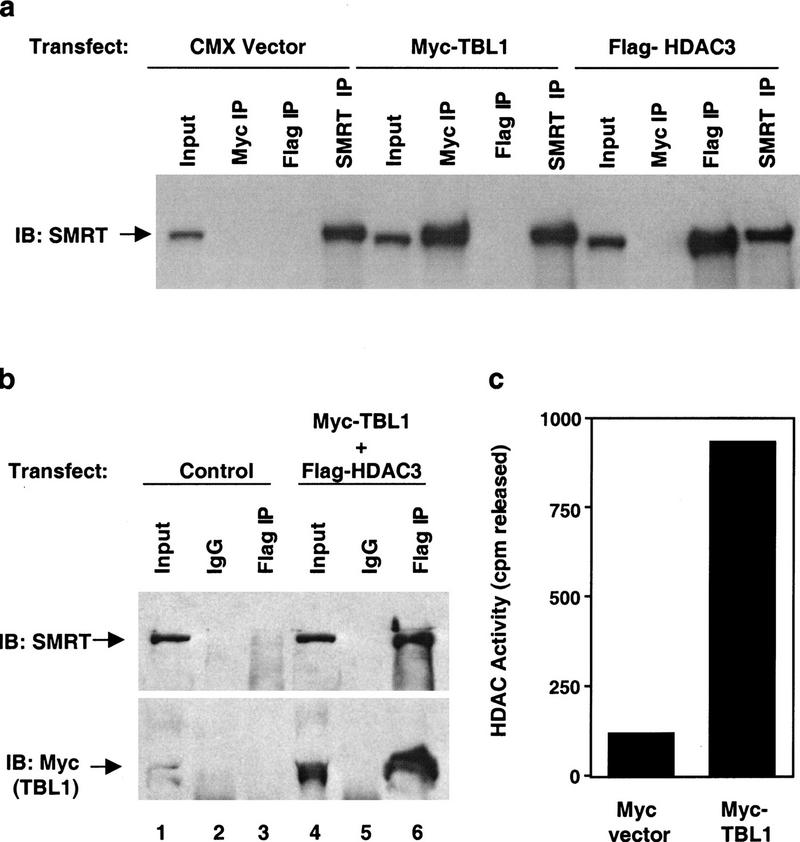
In vivo association of SMRT with myc–TBL1 and Flag–HDAC3
The ability of endogenous SMRT to interact with myc–TBL1 and Flag–HDAC3 in vivo was confirmed further by immunoprecipitation of the transfected cells with anti-myc, anti-Flag, or anti-SMRT antibodies followed by immunoblot analysis for SMRT (Fig. 3a). We also show that cotransfected myc–TBL1 and Flag–HDAC3 form a complex with one another and with endogenous SMRT (Fig. 3b, cf. lanes 3 and 6). Consistent with this novel link of TBL1 to HDAC, immunoprecipitates of myc–TBL1 from transfected cells displayed HDAC enzymatic activity (Fig. 3c). These data further demonstrate the functional association of SMRT, TBL1, and HDAC3 in vivo.
Figure 3.
In vivo characterization of the SMRT–TBL1–HDAC3 complex. (a) TBL1 and HDAC3 can each associate with SMRT. Extracts of 293T cells transfected with either Myc–TBL1 or Flag–HDAC3 were immunoprecipitated with anti-myc or anti-flag prior to immunoblot with SMRT monoclonal antibody. All inputs represent 1% of each total immunoprecipitation. (b) HDAC3 interacts with both TBL1 and SMRT in vivo. Extracts of 293T cells transfected with Myc–TBL1 and Flag–HDAC3 were immunoprecipitated with anti-flag prior to immunoblot with SMRT or myc polyclonal antibody. (c) TBL1 associates with histone deacetylase enzymatic activity. 293T cells were transfected with either myc-TBL1 or empty vector alone and subjected to immunoprecipitation with anti-myc followed by assay for deacetylase activity.
SMRT serves as a platform for HDAC3 and TBL1
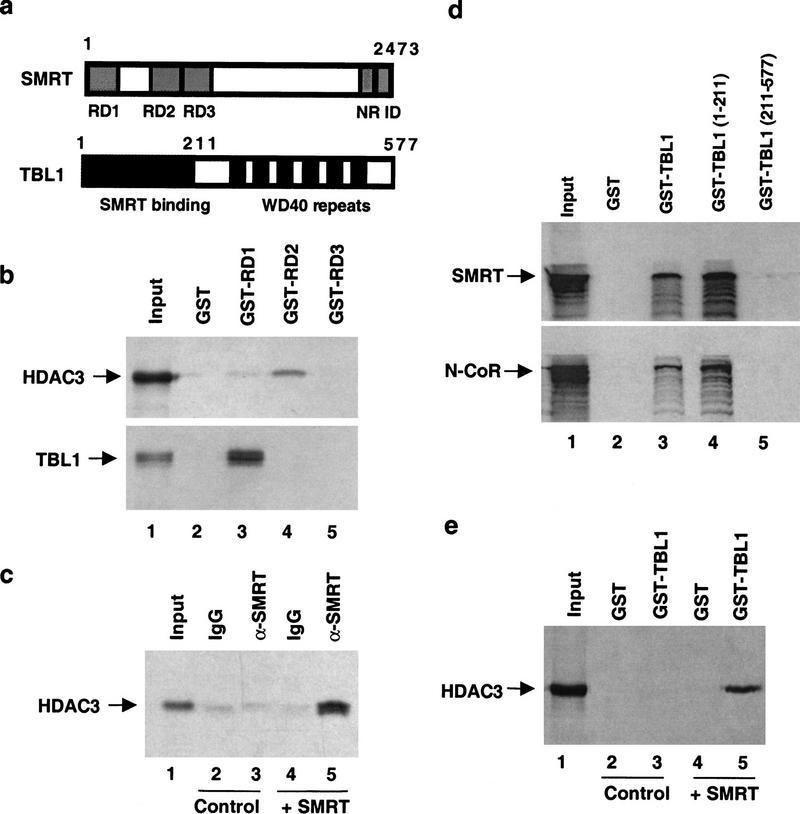
The molecular interactions among SMRT, TBL1, and HDAC3 were analyzed in vitro. SMRT contains multiple repression domains (Fig. 4a). HDAC3 interacted directly with RD2, and to a lesser extent with RD1 (Fig. 4b, top, lane 4). N-CoR RD2 interacted strongly with HDAC3 (data not shown). HDAC3 interacted with full-length SMRT in vitro (Fig. 4c, cf. lanes 3 and 5). TBL1 was found to interact directly with SMRT via interaction with RD1 (Fig. 4b, bottom, lane 3). Full-length N-CoR as well as SMRT interacted with TBL1, and this interaction required the amino terminus of TBL1, which does not include the WD40 repeats (Fig. 4d). Interestingly, HDAC3 did not interact directly with TBL1 (Fig. 4e). However, addition of SMRT to the mixture of HDAC3 and TBL1 allowed formation of a stable TBL1–SMRT–HDAC3 complex in vitro (Fig. 4e, lane 5), suggesting that SMRT mediates the association of TBL1 and HDAC3 in a single complex in vivo.
Figure 4.
SMRT–TBL1–HDAC3 interactions in vitro. (a) Schematic representation of the SMRT corepressor indicating repression domains RD1, RD2, and RD3 and nuclear hormone receptor interaction domains ID1 and ID2. (b) GST-pulldown assay using GST fusions of SMRT RD1 [amino acids 1–303 of the full-length SMRT protein (Ordentlich et al. 1999)], SMRT RD2 (amino acids 763–1028), and SMRT RD3 (amino acids 1043–1514) with 35S-labeled HDAC3 and TBL1. (c) Full-length SMRT interacts with HDAC3 in vitro. 35S-labeled HDAC3 was incubated with control (unprogrammed) RRL (lanes 2,3) or unlabeled full-length SMRT translated in RRL (lanes 4,5) and subjected to immunoprecipitation with control IgG or SMRT monoclonal antibody pool. Input lane shows 1% of total. (d) The amino terminus of TBL1 interacts with N-CoR and SMRT. GST-pulldown assay using GST fusions of TBL1 full-length(1–577), amino-terminus (1–211), and carboxy-terminal WD40-repeat domain (211–577) with 35S-labeled SMRT and N-CoR. (e) The SMRT–TBL1–HDAC3 interaction can be recapitulated in vitro. GST-pulldown assay using GST fusion to full-length TBL1 with 35S-labeled HDAC3 in the presence of control RRL (lanes 2,3) or unlabeled SMRT translated in RRL (lanes 4,5).
A functional role for TBL1 in SMRT/N-CoR-mediated repression by TR
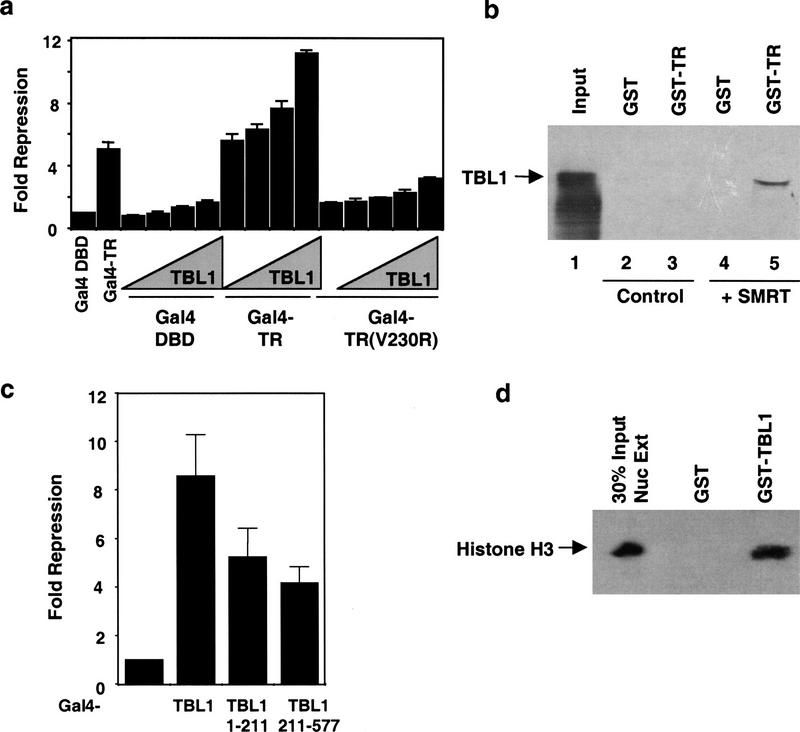
We next addressed the role of TBL1 in repression by thyroid hormone receptor (TR). Ectopic expression of TBL1 markedly potentiated repression by TR (Fig. 5a), consistent with a functional role for TBL1 as a downstream effector of TR-dependent repression. The ability of TBL1 to potentiate repression of TR was not observed with a TR mutant (V230R) that does not bind SMRT or N-CoR (Hu and Lazar 1999; Fig. 5a). This indicated that SMRT/N-CoR are required to recruit TBL1 to target genes that are repressed by unliganded TR. TBL1 does not bind directly to TR, but interaction with SMRT recruits TBL1 to TR (Fig. 5b, cf. lanes 3 and 5). Because TBL1 binds to SMRT independently of HDAC3, the potentiation of repression suggested that TBL1 may contribute an autonomous repression function to SMRT. This was verified when TBL1 or truncations thereof were fused to the Gal4 DNA-binding domain (Fig. 5c). Both amino- and carboxy-terminal TBL1 polypeptides demonstrated repression activity (Fig. 5c). The amino-terminal activity may be due to recruitment of endogenous SMRT, however, the carboxyl terminus does not interact with SMRT and thus may repress transcription directly (Fig. 5c).
Figure 5.
Role of TBL1 in repression. (a) TBL1 potentiates repression by thyroid hormone receptor. A Gal4–TK-luciferase reporter was cotransfected with either Gal4 DBD, Gal4–TR, or Gal4–TR(V230R) and increasing amounts (black ramps representing 0.25–1.5 μg of DNA) of pCMX–HA–TBL1 into HeLa cells. Fold repression was measured as relative to Gal4 DBD alone and the results of duplicate samples are shown. (b) SMRT recruits TBL1 to TR. GST-pulldown assay using GST fusion to TRβ ligand-binding domain with 35S-labeled TBL1 in the presence of control RRL (lanes 2,3) or unlabeled SMRT translated in RRL (lanes 4,5). (c) TBL1 contains an autonomous repression domain. Gal4–TBL1(1–577), Gal4–TBL1(1–211), and Gal4–TBL1(211–577) were transfected with the Gal4–TK-luciferase reporter and fold repression was measured relative to Gal4 DBD alone. (d) GST–TBL1 interacts with histone H3. HeLa Nuclear extract was incubated with either GST alone or GST–TBL1 and interacting histone H3 visualized by immunoblot.
TBL1 is a histone-binding protein like Tup1 and Groucho
TBL1 is similar to Tup1 and Groucho corepressors in its WD40-repeat domain. The sequences of all three corepressors diverge outside of the WD40 repeats. Tup1 homologs in different yeast strains are highly variant outside of the WD40 repeats, but maintain functional similarity (Mukai et al. 1999). With this in mind, we sought to determine whether TBL1 could mimic Tup1 and Groucho in their ability to interact with histone H3. Immunoblot analysis demonstrated that purified TBL1 was able to precipitate histone H3 from HeLa nuclear extract (Fig. 5d). Together, these findings show that TBL1 is likely to contribute a Tup1/Groucho-like repression function that complements the HDAC component(s) of the SMRT repression complex. Interestingly, the Drosophila TBL1 ortholog ebi down-regulates Tramtrack expression (Dong et al. 1999), consistent with a possible repression function.
Discussion
We have purified a core repression complex composed of SMRT, HDAC3, and TBL1. The existence of this complex has been verified with both endogenous and transfected components. The recapitulation of these interactions in vitro as well as their stability through multiple conventional purification steps demonstrate the stable association of SMRT with HDAC3 and TBL1.
Recently, SMRT as well as N-CoR were reported to be associated with the mSin3 multiprotein complex (Alland et al. 1997; Heinzel et al. 1997; Laherty et al. 1997; Nagy et al. 1997). However, the nuclear hormone receptor corepressors were not identified by multiple groups that isolated mSin3 and/or HDAC1 complexes. Using stringent washing conditions (0.1% Tween 20, 0.5 m KCl) for the purification of the SMRT complex from HeLa cells, we were unable to identify any components of the mSin3 complex as an integral part of the SMRT complex. This may be due to a weaker association of SMRT with the mSin3 complex. Similarly, HDAC4 was not detected in the affinity-purified SMRT complex presented here (data not shown). Although class II HDAC 4, HDAC 5, and HDAC 7 interact with SMRT (Huang et al. 2000; Kao et al. 2000), we found previously that only a small fraction of endogenous N-CoR was associated with endogenous HDAC4 despite a strong in vitro interaction (Huang et al. 2000).
The core SMRT complex eluted from a gel-filtration column with an apparent molecular mass of 1–2 mD, which is more than the simple sum of all core components. This raises the possibility that the SMRT complex may contain other components that were removed by our stringent affinity purification. However, HDAC1 did not coelute with the SMRT–TBL1–HDAC3 complex on the Superose 6 column (data not shown). Another potential explanation for the high molecular mass of the SMRT–TBL1–HDAC3 complex may be the formation of oligomers by the components of the core complex. The presence of coiled-coil motifs in SMRT as well as the findings that TBL1-related Tup1 and Groucho corepressors can form tetramers (Varanasi et al. 1996; Chen et al. 1998) are consistent with this latter possibility. Also, we have observed oligomerization of TBL1 in vitro (data not shown).
TBL1 has analogous domains to Tup1 and Groucho corepressors. Although the sequence identity wanes outside of the WD40 repeats, TBL1 is actually more homologous to Saccharomyces cerevisiae Tup1 than is transducin-like enhancer protein 1 (TLE1), a human homolog of Groucho (Stifani et al. 1992). TBL1, Tup1, and Groucho all possess intrinsic repression activity and can contact histone H3. Although the mechanism of repression by this class of WD40-repeat proteins is not known, it has been suggested that Tup1 can inhibit TBP binding (Kuras and Struhl 1999) or act through interactions with RNA Pol II-associated Srb proteins (Carlson 1997; Redd et al. 1997). Recently, Tup1 and Groucho have been implicated in repression pathways involving HDACs. Groucho is known to interact with Rpd3 (Chen et al. 1999), and although it has not been shown to interact with a deacetylase, loss of HDAC activities compromises repression by Tup1 in vivo (Edmondson et al. 1998). Furthermore, HDACs are also involved in repression by the polycomb group protein EED (van der Vlag and Otte 1999), which has a WD40 structure similar to TBL1. Thus, the core SMRT complex adds to an emerging theme of WD40–HDAC combinations mediating transcriptional repression.
The SMRT–TBL1–HDAC3 complex has three potentially distinct mechanisms of conveying a repression signal when recruited to a target gene, for example, by TR (1) enzymatic activity of HDAC3, (2) Tup1/Groucho-like functions mediated by TBL1, and (3) interactions with general transcription factors via SMRT (Fig. 6). The interactions between TBL1 and histones may serve to bring substrate to the HDACs, or could have an HDAC-independent role in repression. The ability of TBL1 to potentiate repression by TR is intriguing, as deafness associated with loss-of-function TBL1 mutation in humans (Bassi et al. 1999) phenocopies loss-of-function mutations of TRβ in humans (Refetoff et al. 1967) as well as in mice (Forrest et al. 1996). In contrast, human TRβ mutants that lose activation, but retain repression function, cause thyroid hormone resistance syndromes, but not deafness (Forrest 1996).
Figure 6.
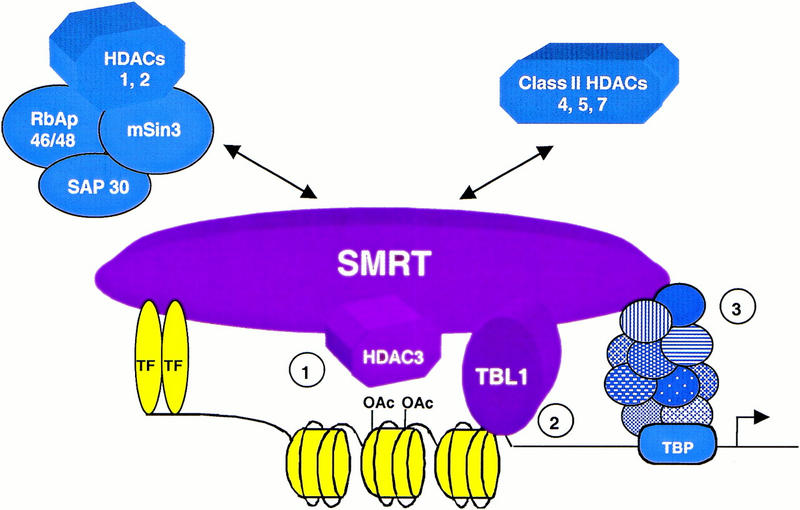
Role of the core SMRT complex in repression. A model for SMRT-mediated repression indicating the core SMRT–HDAC3–TBL1 complex, with potential repression mechanisms including (1) HDAC activity via HDAC3, (2) Tup1/Groucho-like functions mediated by TBL1, and (3) interactions between SMRT and general transcription factors. These repression functions may be augmented by additional interactions with the mSin3/HDAC1 complex, as well as HDAC4, HDAC5, and HDAC7.
The core SMRT–TBL1–HDAC3 complex is likely to be recruited to DNA elements wherever SMRT is utilized and the components are available. As a mechanism to augment or fine tune repression, the complex is also likely to functionally interact with other known SMRT-binding factors such as the mSin3/HDAC1 complex and various class II HDACs (Huang et al. 2000; Kao et al. 2000). These supplemental interactions could be regulated in the context of varying promoters, protein availability, or post-translational modifications. HDAC4 availability is regulated during differentiation by a nuclear cytoplasmic-shuttling mechanism (Miska et al. 1999). Functional differences among the HDACs, together with the specific properties of SMRT complexes built on various promoters are likely to determine the magnitude and specificity of transcriptional repression.
Materials and methods
Antibody production and screening
Mouse monoclonal antibodies were raised against GST–SMRT amino acids 2004–2310 and screened by ELISA and Western assays. The mouse N-CoR monoclonal antibody will be described elsewhere. Polyclonal mouse and rabbit serum was raised against GST–TBL1 amino acids 1–211. HDAC1 and HDAC3 specific antisera were generated by immunizing rabbits with peptides corresponding to the predicted carboxy-terminal domain of each protein coupled to keyhole limpet hemocyanin (amino acids 467–482, EEKPEAKGVKEEVKLA for HDAC1, and amino acids 413–428, FYDGDHDNDKESDVEI for HDAC3, respectively). Other antibodies used were anti-mSin3A and anti-myc rabbit polyclonal antibodies (Santa Cruz), monoclonal anti-flag antibody (Research Diagnostics), and anti-histone H3 (Upstate Biotechnology).
Affinity purification of SMRT from HeLa nuclear extract
Affinity purification of SMRT-associated polypeptides was performed using a pool of five SMRT monoclonal antibodies. Antibodies were coupled to 0.5 ml of protein A–agarose beads (GIBCO BRL) as described (Harlow and Lane 1988). The resin was incubated for 6 hr at 4°C with 35 mg of HeLa nuclear extract precleared with 10 ml of protein A beads. Binding was carried out in NEBB buffer (250 mm NaCl, 30 mm Tris-HCl at pH 7.6, 10% glycerol, 0.1 mm EDTA, 5 mm β-ME, 0.2 mm PMSF, 0.1% NP-40), then the support was washed successively with NEBB buffer containing 500 mm NaCl, 0.1% NP-40; 500 mm NaCl, 0.5% NP-40; 1.0 m NaCl, 0.1% NP-40; and 20 mm NaCl, 1.0% NP-40. The washed beads were then eluted with 3 column volumes of 100 mm glycine (pH 3.0).
In addition, HeLa nuclear extract (1.0 gram) was applied to a P11 column (Whatman) pre-equilibrated with 20 mm Tris-HCl (pH 7.9), 10% glycerol, 0.2 mm EDTA, 10 mm β-ME, 0.2 mm PMSF (Buffer BC) containing 100 mm KCl (BC100), then step eluted with BC300, BC500, BC1000, and BC2000. SMRT was followed by immunoblot analysis. The SMRT-containing BC300 fraction was dialyzed into BC100 and loaded onto a 40-ml DEAE–Sephacel column (Whatman) pre-equilibrated with BC100. Bound material was step eluted with BC350, dialysed against Buffer D (150 mm KCl, 20 mm HEPES at pH 7.9, 0.25 mm EDTA, 10% glycerol, 0.1% Tween 20), and chromatographed on the SMRT affinity resin as above except that the column was washed consecutively with Buffer D containing 150 mm, 300 mm, and 500 mm KCl before elution. Eluates were separated by 8% SDS-PAGE and bands were excised.
Conventional purification of SMRT
The procedure for chromatographic separation of SMRT is outlined in Figure 2a. A total of 1.1 gram of HeLa nuclear extract was precipitated with saturated ammonium sulfate (final concentration of 40%). Following centrifugation at 15,000 RPM for 20 min, the pellet (0.3 gram) was resuspended in BC100 and loaded onto a 40-ml DEAE–Sephacel column (Pharmacia). Following a 10 column volume linear gradient from BC100 to BC500, a 51.5-mg peak eluting at ∼200 mm KCl was dialyzed into BC100 and separated on a Mono S support (Pharmacia). The Mono S was step eluted with BC500 to yield a 6.0-mg protein peak that was then dialyzed into HB1M buffer (1.0 m KCl, 20 mm HEPES at pH 7.8, 10% glycerol, 0.5 mm EDTA, 4 mm DTT, 0.5 mm PMSF) and loaded onto a 1.0-ml phenyl superose column (Pharmacia). A 15-column volume gradient from HB1M to HB0M (HB1M containing no salt) was applied, which yielded a 3.15-mg protein peak at ∼800 mm KCl. This peak was dialyzed into BC75 and loaded onto a 1.0-ml Mono Q column (Pharmacia). A 10 column volume linear gradient was applied from BC75 to BC500 to yield a 2.1-mg protein peak at ∼250 mm KCl. Fractions were pooled and concentrated to 1.0 ml on a ultrafree cellulose concentration device (Millipore). A total of 0.7 ml (1.5 mg) was dialyzed into BC350 and separated on a Superose 6 10/30 gel filtration column (Pharmacia) with an isocratic gradient. Fractions from the Superose 6 column were subjected to immunoblot analysis and the SMRT-containing peak was found to contain ∼32 μg of protein.
Mass spectrometric peptide sequencing
Excised bands were subjected to in-gel reduction, carboxyamidomethylation, and tryptic digestion (Promega). Multiple peptide sequences were determined in a single run by microcapillary reverse-phase chromatography, (a custom New Objective 50-micron column terminating in a nanospray 15-micron tip), directly coupled to a Finnigan LCQ quadrupole ion trap mass spectrometer. The ion trap was programmed to acquire successive sets of three scan modes consisting of the following: full scan MS over alternating ranges of 395–800 m/z or 800–1300 m/z, followed by two data dependent scans on the most abundant ion in those full scans. These data-dependent scans allowed automatic acquisition of a high-resolution (zoom) scan to determine charge state and exact mass and MS/MS spectra for peptide sequence information. MS/MS spectra were acquired with relative collision energy of 30%, isolation width of 2.5 D and dynamic exclusion of ions from repeat analysis. Interpretation of the resulting MS/MS spectra of the peptides was facilitated by programs developed in the Harvard Microchemistry Facility (Chittum et al. 1998) and by database correlation with the algorithm SEQUEST (Eng et al. 1994).
Plasmid constructs
TBL1, TBL1(1–211), and TBL(211–577) were produced by PCR amplification of TBL1 cDNA, then cloned into pGEX4T (Pharmacia) to produce GST-fusion proteins or pCMX–Gal4–DBD (Zhang et al. 1997) for Gal4 fusions. Myc–TBL1 was produced by cloning the TBL1 cDNA into pcDNA3.1myc/his (Invitrogen). pCMX–HA–TBL1 was produced by cloning the TBL1 PCR product into pCMX–HA containing the HA epitope and a nuclear localization sequence (Huang et al. 2000). GST–SMRT constructs were produced by PCR amplification of the corresponding SMRT sequences followed by subcloning into pGEX4T. Flag–HDAC3 was subcloned into the pcDNA3.1 vector by standard techniques. Gal4–TR constructs have been described previously (Hu and Lazar 1999).
Transfection and reporter assays
HeLa and 293T cells were transfected with Fugene 6 (Boehringer Mannheim) according to manufacturer's instructions. After 48–52 hr of transfection, cells were washed with PBS and harvested for luciferase and β-galactosidase assays. In luciferase assays, the Gal4 UASX5 TK-luciferase reporter contains five copies of the Gal4 17mer binding site. Light units were normalized to a cotransfected β-galactosidase expression plasmid. Fold repression is relative to the Gal4–DBD, and results of duplicate samples are plotted.
Histone interaction
Purification of histones from nuclear extract was performed by incubating 500 μg of HeLa nuclear extract with 1 μg of GST fusion protein in PBS containing 10% glycerol and protease inhibitors (Boehringer Mannheim) at 4°C overnight. Beads were washed three times in binding buffer and bound material was analyzed by Western blot.
Immunoprecipitations
Cells were washed in PBS and lysed with lysis buffer A (150 mm NaCl, 40 mm Tris-HCl at pH 7.6, 10% glycerol, 0.3% NP-40) and protease inhibitors for 20 min and pelleted. Supernatants were pre-cleared with protein A–agarose, then allowed to bind either anti-SMRT, anti-myc (Santa Cruz), or anti-flag (Sigma) agarose beads for 6–8 hr at 4°C. Pellets were washed three times in lysis buffer A and three times in Lysis buffer A containing 300 mm NaCl, 0.1% NP-40 and subjected to immunoblot or HDAC assay. Immunoprecipitations of RRL-translated proteins were performed in buffer D.
Other
Cell Culture for 293T and HeLa cells, HDAC assays, and GST-pulldown assays were performed as described (Huang et al. 2000).
Acknowledgments
We thank Xiao Hu and Eric Huang for helpful discussions, Kara Punt and the Wistar Hybridoma facility for antibodies, Kerry Pierce and Dan Kirby for mass spectrometry, Peter Ordentlich and Ron Evans for full-length SMRT cDNA, Michael Rosenfeld for N-CoR expression vector, and Leo Tsuda and Larry Zipursky for providing human TBL1 cDNA. This work was supported by grants from the National Institute of Diabetes, Digestive, and Kidney Diseases of the NIH to M.A.L. R.S is a V Foundation scholar and a recipient of a W.W. Smith Charitable Trust award.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lazar@mail.med.upenn.edu; FAX (215) 898-5408.
References
- Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Asahara H, Dutta S, Kao HY, Evans RM, Montminy M. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol Cell Biol. 1999;19:8219–8225. doi: 10.1128/mcb.19.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi MT, Ramesar RS, Caciotti B, Winship IM, DeGrandi A, Riboni M, Townes PL, Beighton P, Ballabio A, Borsani G. X-linked late-onset sensorineural deafness caused by a deletion involving OA1 and a novel gene containing WD-40 repeats. Am J Hum Gen. 1999;64:1604–1616. doi: 10.1086/302408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. Genetics of transcriptional regulation in yeast: Connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- Chen G, Nguyen PH, Courey AJ. A role for Groucho tetramerization in transcription repression. Mol Cell Biol. 1998;18:7259–7268. doi: 10.1128/mcb.18.12.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes & Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, Hatfield DL. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- Dong X, Tsuda L, Zavitz KH, Lin M, Li S, Carthew RW, Zipursky SL. ebi regulates epidermal growth factor receptor signaling pathways in Drosophila. Genes & Dev. 1999;13:954–965. doi: 10.1101/gad.13.8.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Zhang W, Watson A, Xu W, Bone JR, Yu Y, Stillman D, Roth SY. In vivo functions of histone acetylation/deacetylation in Tup1p repression and Gcnp activation. Cold Spring Harb Symp Quant Biol. 1998;63:459–468. doi: 10.1101/sqb.1998.63.459. [DOI] [PubMed] [Google Scholar]
- Emiliani S, Fischle W, VanLint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McOormick AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acids sequences in a protein database. Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Caudy M. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes & Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- Forrest D. Deafness and goiter: Molecular genetic considerations. J Clin Endocrinol Metab. 1996;81:2764–2767. doi: 10.1210/jcem.81.8.8768825. [DOI] [PubMed] [Google Scholar]
- Forrest D, Erway LC, Ng L, Altschuler R, Curran T. Thyroid hormone receptor beta is essential for development of auditory function. Nat Genet. 1996;13:354–357. doi: 10.1038/ng0796-354. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes & Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Grignani F, DeMatteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, et al. Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP. Distinct interactions of PML-RARα and PLZF-RARα with co-repressors determine differential responses to RA in APL. Nat Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen T-M, Soderstrom M, Laherty CD, Torchia J, Yuang W-M, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hong SH, David G, Wong CW, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. The CoRNR motif contols the recruitment of corepressors to nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- ————— Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- Huang EY, Zhang J, Miska EA, Guenther MG, Kouzarides T, Lazar MA. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes & Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Kao H-Y, Ordentlich P, Koyano-Nakagawa K, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes & Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes & Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Billin AN, Lavinsky RM, Yochum GS, Bush AC, Sun JM, Mullen TM, Davie JR, Rose DW, Glass CK, et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- Laherty CE, Yang W-M, Sun J-M, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai Y, Matsuo E, Roth SY, Harashima S. Conservation of histone binding and transcriptional repressor functions in a Schizosaccharomyces pombe Tup1p homolog. Mol Cell Biol. 1999;19:8461–8468. doi: 10.1128/mcb.19.12.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat GE, Burke LJ, Downes M. The corepressor N-CoR and its variants RIP13a and RIP13Δ1 directly interact with the basal transcription factors TFIIB, TAFII32, and TAFII70. Nucleic Acids Res. 1998;26:2899–2907. doi: 10.1093/nar/26.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao H-Y, Chakvarkti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Ordentlich P, Downes M, Xie W, Genin A, Spinner NB, Evans RM. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Redd MJ, Arnaud MB, Johnson AD. A complex composed of tup1 and ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- Refetoff S, DeWind T, DeGroot LJ. Familial syndrome combining deafmutism, stippled epiphyses, goiter, and abnormally high PBI: Possible target organ refractoriness to thyroid hormone. J Clin Endocrinol Metab. 1967;27:279–294. doi: 10.1210/jcem-27-2-279. [DOI] [PubMed] [Google Scholar]
- Stifani S, Blaumueller C, Redhead NJ, Hill RE, Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- Varanasi US, Klis M, Mikesell PB, Trumbly RJ. The Cyc8 (Ssn6)-Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol. 1996;16:6707–6714. doi: 10.1128/mcb.16.12.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Wahi M, Komachi K, Johnson AD. Gene regulation by the yeast Ssn6-Tup1 corepressor. Cold Spring Harb Symp Quant Biol. 1998;63:447–457. doi: 10.1101/sqb.1998.63.447. [DOI] [PubMed] [Google Scholar]
- Wong C-W, Privalsky ML. Transcriptional repression by the SMRT-mSin3 corepressor: Multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol Cell Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Lavinsky RM, Dasen JS, Flynn SE, McInerney EM, Mullen TM, Heinzel T, Szeto D, Korzus E, Durokawa R, et al. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yang W-M, Yao Y-L, Sun J-M, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucelosome remodeling activities. Cell. 1998a;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun ZW, Iratni R, Erdjument BH, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998b;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]