Abstract
The plant pathogen Pseudomonas syringae pv. tomato DC3000 (DC3000) is found in a wide variety of environments and must monitor and respond to various environmental signals such as the availability of iron, an essential element for bacterial growth. An important regulator of iron homeostasis is Fur (ferric uptake regulator), and here we present the first study of the Fur regulon in DC3000. Using chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq), 312 chromosomal regions were highly enriched by coimmunoprecipitation with a C-terminally tagged Fur protein. Integration of these data with previous microarray and global transcriptome analyses allowed us to expand the putative DC3000 Fur regulon to include genes both repressed and activated in the presence of bioavailable iron. Using nonradioactive DNase I footprinting, we confirmed Fur binding in 41 regions, including upstream of 11 iron-repressed genes and the iron-activated genes encoding two bacterioferritins (PSPTO_0653 and PSPTO_4160), a ParA protein (PSPTO_0855), and a two-component system (TCS) (PSPTO_3382 to PSPTO_3380).
INTRODUCTION
The Gram-negative bacterium Pseudomonas syringae pv. tomato DC3000 (DC3000), a widely used model organism for the study of bacterial plant-pathogen interactions, is the causal agent of bacterial speck on tomato and is also a pathogen of the model plant Arabidopsis thaliana (4). All pathogens must closely monitor and respond to various environmental signals to secure nutrients from and defend against their hosts. One important environmental signal is iron, which is an essential element for bacterial growth due to the fact that it is a cofactor in many proteins involved in major biological processes such as metabolism and electron transport. While iron is the fourth most abundant element in the earth's crust, it is found mostly in insoluble iron (III) oxide forms and is therefore not readily available to bacteria. For this reason many organisms, including P. syringae, rely on siderophores to chelate and solubilize the available iron. However, because excess iron can generate toxic hydroxyl radicals through Fenton chemistry in the presence of oxygen, bacteria must also regulate intracellular iron concentrations carefully to avoid damage or death.
The primary regulator of iron homeostasis in bacteria is Fur (ferric uptake regulator). This DNA binding protein has been identified in many bacterial species where it regulates expression of iron uptake and storage genes and is also involved in virulence and protection against oxidative stress (reviewed in reference 5). Fur typically functions as a repressor by binding to DNA in the presence of iron and preventing gene transcription. Under low-iron conditions the affinity of Fur for these binding sites is greatly reduced, resulting in expression of the genes that it negatively regulates. The role of Fur has been well studied in the opportunistic human pathogen Pseudomonas aeruginosa, which must respond to the low-iron conditions encountered in the host to ensure its survival. The first crystal structure of Fur was determined using protein isolated from P. aeruginosa PAO1 (36), and many studies have led to the elucidation of the Fur regulon in this species (31, 32, 35). Fur is an essential protein in PAO1 and, like in many other organisms, represses genes involved in iron acquisition (some of which contribute to virulence) such as siderophores and heme uptake systems. It also indirectly regulates (via PvdS) the expression of extracellular proteinases, which degrade the host iron binding proteins, and the virulence factor exotoxin A, which is a potent extracellular toxin (reviewed in reference 11). In addition, PAO1 Fur represses the expression of other regulators such as 10 alternative extracytoplasmic function (ECF) sigma factors and two regulatory noncoding RNAs (ncRNAs) (prrF1 and prrF2). The prrF1 and prrF2 ncRNAs are functionally similar to ryhB in Escherichia coli and act by causing the degradation of mRNAs encoding iron-containing proteins (33, 41).
While the classical repression model represents the most familiar mechanism by which Fur controls gene expression, it is now understood that Fur can exert regulation in other ways. For example, Fur-dependent activation of genes can occur indirectly when Fur blocks the binding of a second repressor (e.g., Fur inhibits H-NS binding upstream of fntA in E. coli [29]). In at least one organism (Neisseria meningitidis), Fur appears to directly activate the expression of genes that have iron centers (13). In Helicobacter pylori, the iron-depleted (apo) form of Fur represses a subset of genes under low-iron conditions (12, 14, 17, 28). Although it is not yet known whether these mechanisms are broadly shared among bacteria, they provide evidence that the regulatory role of Fur may be unexpectedly subtle.
Little is known about the regulation by Fur and its role in pathogenicity in P. syringae. In the pathovar tabaci (causal agent of wildfire disease in tobacco), Fur regulates the swarming motility and production of tabtoxin, siderophores, and the quorum-sensing molecule N-acyl homoserine lactone (6). The tabaci fur mutant is also less virulent on tobacco leaves. In DC3000, we determined the regulon of the iron-responsive extracytoplasmic function (ECF) sigma factor, PvdS, which regulates expression of genes responsible for production of the siderophore pyoverdine (39), and used microarrays to identify genes that were differentially expressed in response to iron bioavailability (3). The microarray analysis revealed a wide variety of genes that are repressed in an iron-dependent manner, many of which are downstream of putative Fur motifs, suggesting that they are members of the Fur regulon in DC3000. However, these experiments did not provide direct evidence for DNA binding by Fur. In this study, we use high-throughput methods to identify genomic sites bound to Fur in the presence of iron. Chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq) has been extensively used to study nucleic acid-protein interactions in eukaryotes. However, this technology has only recently been applied to the study of DNA binding proteins in bacteria (24, 27). Integration of our results with our previously published microarray and transcriptome data sets provides stronger evidence for regulation by Fur and suggests that Fur function in P. syringae involves activation as well as repression.
MATERIALS AND METHODS
Creation of a strain containing a FLAG-tagged Fur.
Integration of the FLAG epitope tag at the 3′ end of fur was achieved using a pK18mobsacB (37)-based construct as follows: A 1.053-kb region containing the fur gene and upstream sequence was amplified using oligomers oSWC2099 and oSWC02101 (all primer sequences are listed in Table S1 in the supplemental material). Primer oSWC02101 inserts the FLAG sequence in front of the fur stop codon. A 0.939-kb region downstream of the fur gene was amplified using primers oSWC02102 and oSWC02100. oSWC02102 contains a sequence complementary to oSWC02101. These fragments were amplified using the Expand high-fidelity PCR system (Roche). The PCR products were gel purified using the Zymoclean gel DNA recovery kit (Zymo Research) and were joined using the Expand high-fidelity PCR system (Roche) with primers oSWC02099 and oSWC02100 and the up- and downstream fragments as the PCR template. The product (including the fur gene with a FLAG epitope at the 3′ end, up- and downstream sequences, and flanking XbaI sites) was cloned into pK18MobsacB. This suicide plasmid was introduced into DC3000 by electroporation, and integration events were selected on modified Luria medium (LM) (10 g Bacto tryptone, 6 g yeast extract, 0.6 g NaCl, 0.4 g MgSO4·7H2O, and 1.5 g K2HPO4 per liter) with 50 μg/ml kanamycin. Two independent colonies (BBPS29#1 and #2) were chosen and subjected to counterselection on 10% sucrose to select for the loss of the sacB gene. Kanamycin-sensitive colonies were then screened for the presence of the FLAG tag by PCR with primers oSWC02099 and oSWC02103 (a primer specific to the FLAG sequence) using Premix Taq (Ex Taq version 2.0) (Takara), and three colonies were selected (BBPS30#1 and #2 and BBPS31) for further experiments. The fur-FLAG and flanking areas in these strains were sequenced to confirm that no other mutations had been introduced in these regions during strain construction.
Growth of the Fur-FLAG and WT strains in bioreactors.
BBPS30#1, BBPS30#2, and BBPS31 along with three independent wild-type (WT) DC3000 cultures were prepared using a Sixfors bioreactor system as described previously (3). Briefly, strains were grown in 400 ml mannitol-glutamate (MG) medium (10 g/liter mannitol, 2 g/liter l-glutamic acid, 0.5 g/liter KH2PO4, 0.2 g/liter NaCl, 0.2 g/liter MgSO4, final pH of 7). Cultures were adjusted to 50 μM iron citrate (+ Iron) or 50 μM Na citrate (− Iron) at an optical density at 600 nm (OD600) of 0.35 to 0.4 and grown for an additional 30 min prior to harvest. These conditions correspond to those used to study the global transcriptional response of DC3000 to the addition of iron (3). One-milliliter samples were used to confirm the presence of the FLAG-tagged Fur protein by Western blotting (data not shown). One hundred milliliters from each culture was subjected to cross-linking for chromatin immunoprecipitation (ChIP) as described below.
ChIP-seq to enrich for Fur binding sites.
The 100-ml samples removed from the bioreactor after exposure for 30 min to iron citrate or sodium citrate were treated with 1% formaldehyde at room temperature (RT) with gentle shaking for 20 min. Cross-linking was quenched by the addition of 0.36 M glycine, followed by incubation at RT with gentle agitation for 5 min. The cells were harvested by centrifugation at 5,000 × g, and the pellet washed twice with an equal volume of ice-cold Tris-buffered saline buffer (TBS; 10 mM Tris HCl [pH 7.4], 150 mM NaCl, and 2.7 mM KCl). The cell pellet was stored at −80°C until required. Cells were lysed by incubation at 37°C for 10 min in 1 ml CelLytic B lysis reagent (Sigma), 10 μl Longlife lysozyme (G-Biosciences), and 1 mM phenylmethylsulfonyl fluoride (PMSF) followed by sonication six times for 30 s at 15% power (using a Fisher Scientific 550 sonic dismembrator with a microtip). This had previously been determined to shear the DNA to an average size of approximately 400 bp. Insoluble material was removed by centrifugation at 16,000 × g for 10 min at 4°C. One hundred microliters of the supernatant was retained as the lysate control. The remaining supernatant was applied to 40 μl of anti-FLAG M2 affinity gel (Sigma), which had been washed twice with TBS as described by the manufacturer. The supernatant-anti-FLAG suspension was incubated at 4°C for 2 h with gentle agitation. The resin was collected by centrifugation at 5,000 × g for 30 s at 4°C and the supernatant carefully removed and discarded. The resin was resuspended in 500 μl of ice-cold TBS, transferred to a Spin-X centrifuge tube filter (Sigma), and collected by centrifugation at 3,000 × g for 30 s. The resin was then washed two times with ice-cold TBS. The bound material was eluted from the resin by incubation with 100 μl TBS containing 300 ng/μl FLAG peptide (Sigma) for 30 min at 4°C with gentle agitation. The Spin-X column was transferred to a new tube and the elutant (IP sample) collected by centrifugation at 5,000 × g for 30 s. The cross-links in 20 μl of the lysate or 100 μl of the IP sample were reversed by the addition of 2 mg/ml pronase (protease from Streptomyces griseus [Sigma]) followed by incubation at 42°C for 2 h and then heating to 65°C for 6 h. DNA was purified using the QIAquick PCR purification kit (Qiagen) and eluted with 30 μl of Milli-Q H2O.
Purified DNA from the + Iron samples was subjected to library preparation and high-throughput sequencing using the Illumina Solexa platform at the Cornell University Life Sciences Core Laboratories Center (http://cores.lifesciences.cornell.edu/). The number of raw Illumina reads for each sample is shown in the second column of Table S2 in the supplemental material. Reads were not obtained for one of the three WT samples. Reads were aligned to the P. syringae pv. tomato DC3000 genome (NCBI accession numbers: NC_004578.1, NC_004632.1, and NC_004633.1) and to controls consisting of primer and adapter sequences by using SOAPaligner/soap2 version 2.20 (26). Reads that aligned to more than one location or to the artificial sequence were discarded. The number of uniquely aligned reads is shown in the third column of Table S2.
qPCR.
Quantitative PCR (qPCR) was performed to confirm enrichment of predicted Fur targets upstream of known or predicted Fur targets pvdS (PSPTO_2133; primers oSWC01393/oSWC01394) and PSPTO_1209 (primers oSWC1395/oSWC1396) before submitting DNA from the + Iron sample for sequencing. qPCR was also performed on the + Iron and − Iron samples to test for iron-dependent enrichment of additional targets identified through ChIP-seq (see Table S1 in the supplemental material for primers). qPCR was performed with 10 ng of purified DNA from the lysate or IP samples as the template using IQ SYBR green Supermix (Bio-Rad) on a iQ5 multicolor real-time PCR detection system (Bio-Rad). Enrichment was determined relative to regions within the gap1 (primers oSWC00381/oSWC00382) and gyrA genes (primers oSWC00379/oSWC00380).
Profile generation.
Two sets of Artemis-loadable user plots (profiles) were generated from the reads that aligned with the DC3000 chromosome as described in reference 21. Sinister profiles are histograms of the number of aligned reads whose 5′ end aligns at each position of the genome, with coincident locations on the positive and negative strands being counted separately. Naive profiles are histograms of the number of aligned reads that overlap each genomic position. Sinister profiles were used to construct an input data set for peak calling. Naive profiles are used in some figures as noted. All profiles are included in File S1 in the supplemental material.
Peak calling.
ChIP-seq data corresponding to the main DC3000 chromosome (NC_004578.1) were analyzed using CSDeconv (27) to detect regions that had been enriched by immunoprecipitation (“peaks”). The log-likelihood threshold for the initial phase of enriched region estimation was set to 200, and the penalty term for estimating multiple binding sites within an enriched region was set to 20,000, resulting in a total of 322 called binding sites within the set of 300 enriched regions. To identify conserved genomic motifs, 80 nucleotides of flanking sequence on either side of a called binding site (161 nucleotides total) were extracted. Enriched regions containing multiple binding sites were merged if the flanking sequence windows around those sites overlapped. This procedure resulted in 312 regions that were subjected to further analysis (listed in File S2 in the supplemental material). A GFF-formatted file describing the genomic locations of these regions is provided in File S3 in the supplemental material.
We can estimate a false-discovery rate (FDR) as part of the peak-calling procedure using CSDeconv (described in reference 27) by swapping treatment and control samples and rerunning CSDeconv to obtain enriched regions and binding sites for the swapped samples. Using this method with the parameters described previously, no enriched regions were found in the swapped sample as compared to 300 for the actual sample, placing an upper bound on the FDR for enriched region identification at an FDR of less than 1/300 (0.0033). Because no enriched regions were identified in the swapped sample, the second phase of binding site estimation could not be carried out; therefore, we can only place an upper bound for the second phase at an FDR of less than 1/312 (0.0032).
Motif identification.
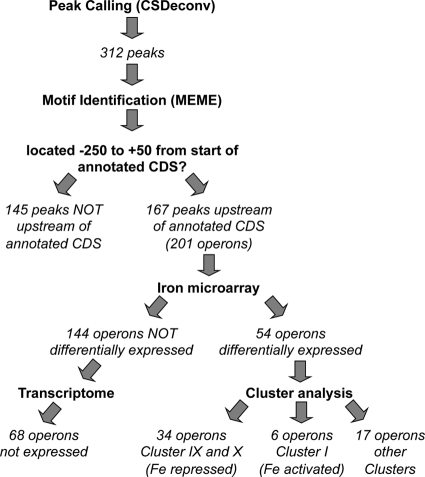
Putative Fur-DNA binding motifs were generated using MEME (version 4.4, http://meme.sdsc.edu/meme4_4_0/intro.html), with input sequences derived from ChIP-seq peak calling (described above) or via DNase I footprinting (described below). A preliminary search for motifs between 10 and 30 nucleotides in width (based on the expected size of the dimeric Fur box of 15 to 21 nucleotides), at a frequency of zero or one times per enriched region revealed a motif (P = 1.2e-166) of 19 nucleotides which was found in each of the 312 input regions (see Fig. 3A). A second MEME analysis in which we required a palindromic (inverted repeat) structure but where other parameters were unchanged produced a motif (P = 1.6e-84) of 15 nucleotides wide also found in each of the 312 input regions (see Fig. 3B). Sites were sorted into one of two subsets depending on whether they overlapped genomic intervals spanning 250 nucleotides upstream of an annotated coding sequence (CDS) to 50 nucleotides downstream. The 167 sites satisfying this test are referred to as “upstream of annotated CDS” in our flowchart (see Fig. 2). Operon predictions for DC3000 from VIMSS (Virtual Institute for Microbial Stress and Survival; http://microbesonline.org), along with a small number of manual curations (20, 39), were used to identify operons under putative regulatory control by Fur.
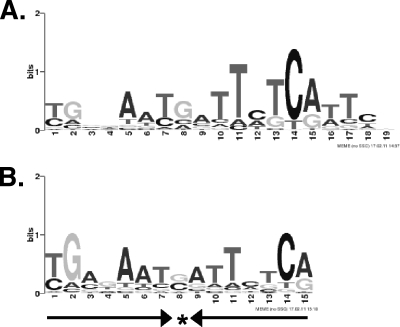
Fig. 3.
Sequence logos representing motifs generated by MEME and identified in all 312 ChIP-seq peaks. (A) MEME motif generated using a target motif size between 10 and 30 bp. (B) MEME was rerun to search for a motif between 10 and 30 bp but constrained to identify a palindromic motif. Here, the motif consists of a 7-1-7 inverted repeat, as indicated by the arrows.
Fig. 2.
Workflow of the analysis performed on the ChIP-seq data. A computational method, CSDeconv, was applied to the data, and 312 peaks were identified for further analysis by integration with our previously published iron microarray and DC3000 transcriptome data sets.
Overproduction and purification of P. syringae pv. tomato DC3000 Fur.
The fur gene from P. syringae pv. tomato DC3000 was amplified by PCR with primers oSWC02036 and oSWC01249 from genomic DNA by using Expand high-fidelity PCR system (Roche) (see Table S1 in the supplemental material for primer sequences). The PCR product was gel purified using the Zymoclean gel DNA recovery kit (Zymo Research), digested with NcoI and BamHI (New England BioLabs), and cloned into pET-16b (Novagen) digested with the same restriction enzymes to create pBB54. This plasmid places the fur gene under the control of the T7 promoter. Plasmid pBB54 was transformed into One Shot BL21(DE3)pLysS chemically competent E. coli (Invitrogen). One liter of Luria-Bertani (LB) medium was inoculated from an overnight culture of the transformant and grown at 37°C until an OD600 of 0.6 was reached. IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) was added to induce protein expression, the culture was incubated for an additional 2 hours, and 800 ml of cells was harvested by centrifugation and frozen at −80°C. Cells were thawed, resuspended in 10 ml buffer A (50 mM Tris-HCl [pH 8], 150 nM NaCl, 2 mM dithiothreitol [DTT], and 10% vol/vol glycerol), lysed by sonication, and centrifuged at 10,000 × g for 10 min. Fur protein was purified from the soluble extract using fast-protein liquid chromatography (FPLC) by sequential application to mono-Q ion-exchange and Superdex-200 size exclusion columns. Fractions were analyzed for the presence of Fur by using SDS-PAGE, and those containing Fur were stored at −80°C for later use.
EMSAs.
PCR fragments containing predicted Fur binding sites upstream of pvdS (PSPTO_2133), prrF2 (PSPTO_5657), and PSPTO_0070 or a control region with no Fur binding site (upstream of PSPTO_1900) were purified and labeled with [γ-32P]ATP by using T4 polynucleotide kinase (NEB) for 1 h at 37°C (see Table S1 in the supplemental material for primers). In addition, eight intragenic Fur binding sites were also amplified and labeled as described above (see Table S1 in the supplemental material for primers). Labeled probes were purified using a QIAquick PCR purification column (Qiagen) and eluted in 50 μl Milli-Q H2O. Electrophoretic mobility shift assays (EMSAs) were performed as described by de Lorenzo et al. (15). Briefly, increasing concentrations of Fur protein (0 to 50 nM) were added to the labeled probes in binding buffer (10 mM bis-Tris [pH 7.5], 5 μg/ml salmon sperm DNA, 5% [vol/vol] glycerol, 1 mM MgCl2, 100 μM MnCl2, 40 mM KCl, 100 μg/ml bovine serum albumin) and incubated for 10 min at 37°C. Specific competition assays were performed by adding excess unlabeled probe to the binding reaction and as a negative control the unlabeled PSPTO_1900 probe was added instead (see Fig. S1 in the supplemental material). The reaction mixtures were loaded onto 5% polyacrylamide gels polymerized in 20 mM bis-Tris (pH 7.5) buffer containing 100 μM MnCl2 and were subjected to electrophoresis for 1.5 h at 100 V. The gels were dried, exposed to a PhosphorImager screen, and imaged on a Storm 840 PhosphorImager scanner (Molecular Dynamics).
Nonradioactive DNase I footprinting.
To examine additional candidate Fur binding sites identified by ChIP-seq, we used fluorescently labeled primers to visualize standard DNase I protection assays by using an Applied BioSystems 3730xl DNA analyzer (42). DNA fragments (∼300 bp in size) containing predicted Fur binding sites were amplified using a primer labeled with the 6-carboxyfluorescein (6-FAM) fluorescent dye (Integrated DNA Technologies). If necessary, PCR products were purified using the QIAquick PCR purification kit (Qiagen). Binding reaction mixtures (50-μl total volume) consisted of 100 ng of labeled DNA probe and either 0 nM, 17.5 nM, 25 nM, or 50 nM Fur in binding buffer (10 mM bis-Tris [pH 7.0], 2.5 μg/ml salmon sperm DNA, 100 μM MnCl2, 100 μg/ml bovine serum albumin, 2 mM MgCl2, 100 mM KCl, and 1 mM CaCl2). These were incubated at 37°C for 10 min, transferred to room temperature, and allowed to cool for 2 min. DNase I (0.1 U or 0.2 U; New England BioLabs) was added to the reaction mixtures and digestion was allowed to proceed for 2 min at room temperature. The reaction was stopped by the addition of 50 μl of DNase I stop solution (20 mM EDTA [pH 8], 1% SDS, and 200 nM NaCl). The DNA was purified using the QIAquick 96 PCR purification kit (Qiagen) and eluted in 60 μl H2O. Ten microliters of each purified fragment was mixed with 0.2 μl of the LIZ 500 size standard and 9.8 μl of HiDi formamide (Applied Biosystems) and submitted to the Cornell University Life Sciences Core Laboratories Center (CLC) for fragment analysis on an Applied BioSystems 3730xl DNA analyzer. The results were visualized using Peak Scanner Software v1.0 from Applied Biosystems. Protein binding sites were indicated by the disappearance of peaks. The position of the binding site was estimated by comparison of the size of the peaks with the sequence of the amplified target.
5′ RACE.
Transcriptional start sites (TSS) were determined using Invitrogen's 5′ RACE system for rapid amplification of cDNA ends (version 2.0) as recommended by the manufacturer. RNA was isolated from cells grown as described above and used as the template. Oligonucleotides used for reverse transcription and PCR are listed in Table S1 in the supplemental material. Products were separated by agarose gel electrophoresis to assess purity and product size. If necessary, products were excised and gel eluted (Qiagen). The 5′ RACE products were sequenced by the Cornell University Life Sciences Core Laboratories Center (CLC). The sequencing results were interpreted by pairwise alignments of the 5′ RACE product sequence with the DC3000 genomic sequence.
RESULTS
ChIP-seq of DC3000 Fur-FLAG binding sites.
We performed ChIP using three independently isolated DC3000 strains containing the fur-FLAG integrated at the native locus. Our goal was to facilitate comparisons of the ChIP-seq data with other data sets already available (3, 21); therefore, strains were grown under the same conditions in a minimal medium (mannitol-glutamate [MG]) by using a bioreactor system. Since Fur binds DNA in the presence of iron, the medium was supplemented with 50 μM ferric citrate 30 min before harvesting the cells.
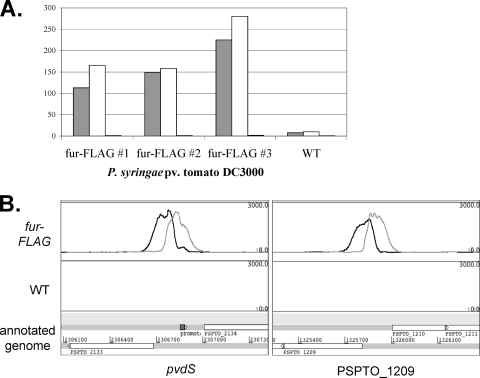
Quantitative PCR of the DNA collected by immunoprecipitation confirmed enrichment of predicted Fur targets pvdS and PSPTO_1209 (Fig. 1A). These genes encode FecI-type ECF sigma factors, which are known or hypothesized to be involved in the regulation of iron transport systems (34, 39). Both genes are a short distance downstream from predicted Fur binding sites (3) and homologs of both pvdS and PSPTO_1209 in P. aeruginosa PAO1 (pvdS and PA3899, respectively) are Fur regulated (31). The pvdS and PSPTO_1209 upstream regions were enriched 100- to 300-fold in the DNA isolated from ChIP with the fur-FLAG strains, while little enrichment was observed when ChIP was performed on the wild-type DC3000 (Fig. 1A). For comparison, a region within the gyrA coding sequence was not enriched after ChIP in either strain (Fig. 1A).
Fig. 1.
(A) qPCR performed on ChIP samples showing enrichment of the pvdS (gray) and PSPTO_1209 (white) promoter regions relative to DNA sequences within the gap1 gene after immunoprecipitation with Fur-FLAG-tagged strains, but not with WT strains. No enrichment of DNA within the gyrA gene was observed (black bars). (B) Naive profiles of ChIP-seq data displayed above the annotated genome by using Artemis. Enrichment of DNA sequences (black and gray peaks) upstream of the pvdS and PSPTO_1209 genes was observed when ChIP-seq was performed with FLAG-tagged Fur (top panel) but not with the wild-type DC3000 strain (lower panel). The black line represents sequences corresponding to the positive strand, while the gray line shows sequences corresponding to the negative strand. Annotated CDSs (open arrows) are shown on the genome below the profiles.
After massively parallel sequencing was performed on these samples, sequences were aligned to the DC3000 genome and DNA enrichment by ChIP was visualized as peaks containing larger numbers of sequence reads relative to other areas of the genome (see File S1 in the supplemental material). Representative regions displaying predicted Fur binding upstream of pvdS and PSPTO_1209 are shown in Fig. 1B. A total of 312 enriched regions were computationally identified and extracted for further characterization (see Materials and Methods; enriched regions are listed in File S2 in the supplemental material).
Motif identification from ChIP-seq data.
Figure 2 shows an overview of the analysis of the ChIP-seq data. Using MEME we identified a 19-bp motif that was found in each of the 312 regions enriched after ChIP-seq (Fig. 3A). Inspection of the alignment reveals similarity to a Fur-DNA consensus binding sequence (GATAATGATAATCATTATC) that we reported previously (3), and to Fur-DNA motifs in E. coli, P. aeruginosa, and Bacillus subtilis (1, 8). The sequence logo shown in Fig. 3A is offset by two nucleotides with respect to the classical consensus sequence and does not exhibit the inverted repeat structure implied by dimeric Fur binding. Various models of Fur-DNA binding sites have been proposed, including a single 9-1-9 inverted repeat (8), a head-to-head-to-tail 6-mer repeat (18, 19), and a minimal 7-1-7 repeat that can be paired with a second 7-1-7 repeat offset by 6 bp (1). We therefore conducted a second MEME analysis in which we required a palindromic (inverted repeat) structure but with other parameters unchanged. The most informative motif was 15 nucleotides wide and was found in each of the 312 input regions (Fig. 3B). This motif corresponds to the 7-1-7 inverted repeat proposed to be the minimal Fur binding sequence in B. subtilis (1). In 209 out of 312 cases, the aligned sequence associated with the palindromic motif has the same start site as the corresponding sequence in the nonpalindromic motif. In the remaining cases, motifs either overlapped (39 instances) or did not align (64 instances).
Given the large number of aligned sequences in our data set, the repetitive nature of the Fur-DNA binding sequence (which allows shifted versions of sequences to match models), and the fact that we see less sequence conservation in Fur-DNA binding than has been observed with much smaller sequence alignments in other organisms, we cannot discriminate between these models. However, since the 7-1-7 inverted repeat we identified sometimes corresponds to one end of the larger, nonpalindromic motif (Fig. 3A), we believe this larger motif may be a mixture of binding sites associated with single Fur-dimer binding sites (15 bp) and double, offset binding sites (21 bp), as proposed in reference 1. String searches in the putative binding regions (data not shown) suggest possible candidates for double 7-1-7 binding sites offset by 6 bp, but we did not further characterize Fur binding to these regions.
Confirmation of Fur binding.
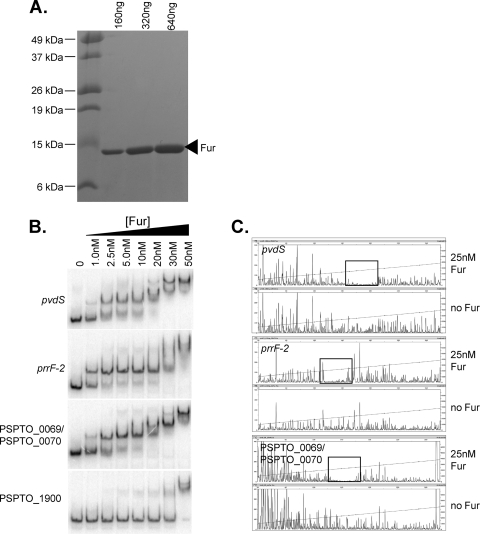
To test Fur binding in vitro, we purified native (untagged) DC3000 Fur protein and confirmed binding to three sites by EMSA (Fig. 4). One was located upstream of the FecI-type ECF sigma factor pvdS, the second was upstream of the ncRNA prrF2, and the third lay between the divergent PSPTO_0070 gene and the PSPTO_0069 operon (encoding a TonB-dependent transport system). The EMSA was performed in the presence of Mn2+ which, like Fe2+, promotes DNA binding of Fur but is stable in the presence of oxygen. Fur binding to all three targets was evident at 1 nM Fur, with nearly complete binding observed at concentrations between 10 to 20 nM Fur (Fig. 4B). For pvdS and PSPTO_0070 a second shift was observed at 30 nM, suggesting that at higher concentrations additional dimers of Fur may bind to lower affinity overlapping binding sites (as has been observed, for example, upstream of pvdS in P. aeruginosa [30] and in B. subtilis [1]). No specific binding was observed to a control of the same size containing a promoter region of PSPTO_1900 where no enrichment had been detected by the ChIP-seq procedure (Fig. 4B).
Fig. 4.
(A) Coomassie-stained SDS-PAGE gel showing DC3000 Fur after purification. (B) EMSA of Fur targets, regions upstream of pvdS and prrF2, and region between PSPTO_0069 and PSPTO_0070. As a negative control, Fur binding to the PSPTO_1900 promoter (not predicted to be bound by Fur) was also assessed. Unbound probes are shown in the first lane and reduced mobility is observed with increasing Fur concentrations for all three Fur targets. Competition assays with specific unlabeled probe were performed and resulted in a loss of band shift (see Fig. S1 in the supplemental material). (C) Fragment analysis from nonradioactive footprinting assays. In each case, the lower panel shows the fragments obtained after DNase I digestion of the probe alone, while in the top panel the probes were incubated with 25 nM Fur prior to DNase I digestion. Protection by Fur can be seen in the boxed regions.
To test Fur binding at a larger number of putative Fur binding sites, we performed nonradioactive DNase I footprinting assays. Fur binding sites were confirmed for all three of the promoter regions tested above (Fig. 4C), situated approximately in the centers of the ChIP-seq peaks and overlapping with the predicted MEME motifs. Interestingly, the area of protection within the pvdS and PSPTO_0070 promoters is extended with increasing concentrations of Fur providing more evidence of a second Fur binding site as suggested above (data not shown). The same procedure was used to confirm binding at 41 of 49 candidate Fur binding sites (Table 1). The corresponding ChIP-seq peaks lay both in expected regions (i.e., upstream of annotated genes) and in unexpected positions (e.g., within CDS). Using the 41 regions as input to MEME, we recovered a palindromic motif that closely resembles the Fur binding site shown in Fig. 3B. The eight negative results may represent regions where additional factors are required for Fur to bind or where Fur binds at concentrations higher than those used in the footprinting assay.
Table 1.
Results of nonradioactive DNase I footprint analysis
| Peak no. | Operon associated with peaka | Footprint coordinatesb | Size of footprint (bp)e | MEMEc coordinates | Overlaps MEME?d,e |
|---|---|---|---|---|---|
| Peaks not within 250 bp upstream and 50 bp downstream of annotated CDS | |||||
| 9 | Within dadA | 121232-121269 | 37 | 121244-121262 | Y |
| 21 | Within gshA | 354705-354745 | 40 | 354725-354743 | Y |
| 27 | Within betI | 485199-485243 | 44 | 485221-485239 | Y |
| 86 | Within PSPTO_1490 | 1640222-1640252 | 30 | 1640229-1640247 | Y |
| 106 | Within PSPTO_1979 | None | NA | 2164987-2165006 | NA |
| 127_1 | In divergent region between PSPTO_2310 (rmf) and PSPTO_2311 | 2557826-2557861 | 35 | 2557841-2557859 | Y |
| 152 | Upstream of PSPTO_2853 | 3204073-3204113 | 40 | 3204087-3204105 | Y |
| 272 | Within PSPTO_5056 (pseudogene) | 5758546-5758602 | 56 | 5758556-5758574 | Y |
| 290 | Within PSPTO_5375 | 6107927-6108040 | 113 | 6108017-6108035 | Y |
| Peaks associated with operons not differentially expressed in microarray study | |||||
| 10 | PSPTO_0104 | None | NA | 123901-123919 | NA |
| 20_0 | PSPTO_0312 and PSPTO_0313 | None | NA | 339930-339948 | NA |
| 28 | PSPTO_0444 | 489078-489133 | 55 | 489093-489111 | Y |
| 40 | PSPTO_0786-PSPTO_0785 and PSPTO_0787 | 853530-853584 | 54 | 853533-853551 | Y |
| 55 | PSPTO_1007 and PSPTO_1008 | 1100729-1100782 | 53 | 1100758-1100776 | Y |
| 70 | PSPTO_1202 and PSPTO_1203 | 1316485-1316528 | 43 | 1316493-1316511 | Y |
| 71 | PSPTO_1205 and PSPTO_1206 | 1318701-1318743 | 42 | 1318714-1318732 | Y |
| 87 | PSPTO_1493-PSPTO_1499 | None | NA | 1648402-1648420 | NA |
| 116 | PSPTO_2190 | 2410934-2410972 | 38 | 2410941-2410959 | Y |
| 133 | PSPTO_2358 and PSPTO_2359 | None | NA | 2613640-2613658 | NA |
| 139 | PSPTO_2507 and PSPTO_2508 | 2769686-2769738 | 52 | 2769765-2769782 | N |
| 161 | PSPTO_3038-PSPTO_3037 | 3418638-3418687 | 49 | 3418653-3418671 | Y |
| 183 | PSPTO_3440-PSPTO_3443 | None | NA | 3881499-3881517 | NA |
| 198 | PSPTO_3566 (csrA-3) | 4024812-4024856 | 44 | 4024829-4024847 | Y |
| 225 | PSPTO_4125-PSPTO_4124 and PSPTO_4126-PSPTO_4127 | 4649830-4649864 | 34 | 4649842-4649860 | Y |
| 231 | PSPTO_4183 | 4713576-4713612 | 36 | 4713580-4713598 | Y |
| 261 | PSPTO_4785-4786-4787 | 5425443-5425469 | 26 | 5425453-5425471 | Y |
| 288_0 | PSPTO_5311 | 6040507-6040541 | 34 | 6040521-6040539 | Y |
| 292 | PSPTO_5454 | None | NA | 6206258-6206276 | NA |
| 294 | PSPTO_5484-PSPTO_5483 | None | NA | 6249391-6249409 | NA |
| Peaks associated with iron-repressed operons | |||||
| 7 | PSPTO_0069-PSPTO_0067 and PSPTO_0070 | 92586-92622 | 36 | 92588-92606 | Y |
| 20_1 | PSPTO_0314-PSPTO_0315 | 340795-340834 | 39 | 340806-340824 | Y |
| 38 | PSPTO_0764 | 812007-812067 | 60 | 812044-812062 | Y |
| 72 | PSPTO_1209-PSPTO_1208 | 1325860-1325900 | 40 | 1325867-1325885 | Y |
| 77 | PSPTO_1286-PSPTO_1282 | 1415155-1415203 | 48 | 1415164-1415182 | Y |
| 112 | PSPTO_2133 (pvdS) | 2306760-2306788 | 28 | 2306761-2306779 | Y |
| 144 | PSPTO_2605 and PSPTO_2606 | 2896461-2896513 | 52 | 2896472-2896490 | Y |
| 169_0 | PSPTO_5657 (prrF2) | 3549460-3549494 | 34 | 3549465-3549483 | Y |
| 169_1 | PSPTO_3157 | 3549762-3549800 | 38 | 3549775-3549793 | Y |
| 200 | PSPTO_3596-PSPTO_3599 | 4056165-4056202 | 37 | 4056179-4056197 | Y |
| 230_0 | PSPTO_4159 | 4686544-4686583 | 39 | 4686559-4686577 | Y |
| Peaks associated with iron-activated operons | |||||
| 34 | PSPTO_0653 | 705294-705353 | 59 | 705315-705333 | Y |
| 43 | PSPTO_0855 (parA) | 925964-926017 | 53 | 925979-925997 | Y |
| 181 | PSPTO_3382-PSPTO_3380 | 3824356-3824392 | 36 | 3824383-3824401 | Partial |
| 218 | PSPTO_3959 and PSPTO_3960 | 4468082-4468112 | 30 | 4468090-4468108 | Y |
| 230_1 | PSPTO_4160 | 4686906-4686961 | 55 | 4686948-4686966 | Partial |
| 260 | PSPTO_4766 | 5404787-5404816 | 29 | 5404794-5404812 | Y |
| Peaks associated with other differentially expressed operons | |||||
| 105 | PSPTO_1975 | 2158865-2158905 | 40 | 2158881-2158899 | Y |
| 221 | PSPTO_3983 | 4490146-4490179 | 33 | 4490154-4490172 | Y |
| 285 | PSPTO_5294 | 6020046-6020067 | 21 | 6020051-6020069 | Y |
Where peaks are not upstream of annotated CDS, the location of the peak is described. Where peaks are in a region of divergent transcription, both operons are listed separated by “and.”
Coordinates refer to position on the DC3000 genome (AE01653).
MEME motif coordinates correspond to the nonpalindromic search (Fig. 3A).
Y, footprint overlaps completely with predicted motif from MEME; N, footprint does not overlap with MEME motif; Partial, footprint and MEME motif only partially overlap.
NA, not applicable since no footprint was identified.
Integration of ChIP-seq data with previously published data sets.
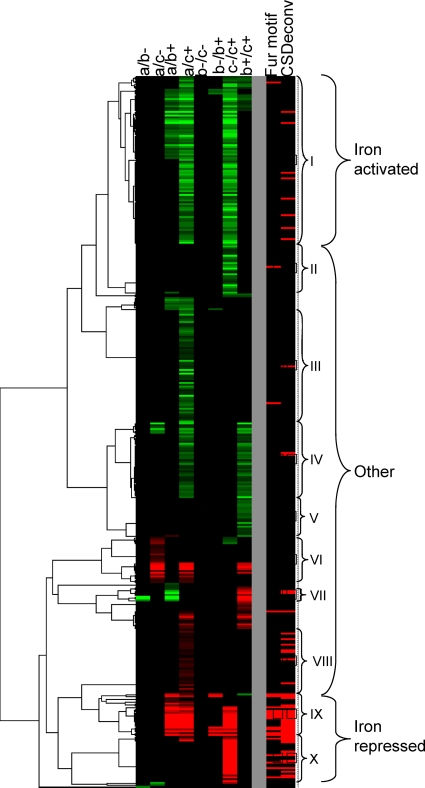
Approximately half of the regions enriched by immunoprecipitation (167/312) contain a predicted Fur binding site located between 250 bp upstream and 50 bp downstream of the annotated translation start site of a gene or presumptive operon, where transcriptional regulators typically bind in prokaryotes. Since some sites are flanked by divergently transcribed operons, Fur binding within the 167 regions could potentially control the expression of 201 operons. To assess these predictions, we compared them with results obtained from microarray experiments in which we measured the transcriptional response of DC3000 to changes in available iron (3). Of the 201 operons, 54 were differentially expressed (Table 2) and 34 appear in clusters that had been predicted to contain genes belonging to the Fur regulon based on the presence of a putative upstream “Fur box” (Fig. 5, clusters IX and X).
Table 2.
Iron-repressed Fur-regulated genes
| Operona | Footprintb | Predicted gene function |
|---|---|---|
| FecI-like sigma factors | ||
| PSPTO_1209-PSPTO_1208 | Y | RNA polymerase sigma-70 family protein/regulatory protein, putative |
| PSPTO_1286-PSPTO_1285 | Y | RNA polymerase sigma-70 family protein/regulatory protein, putative |
| PSPTO_2133 (pvdS) | Y | RNA polymerase sigma-70 family protein |
| TonB system and TonB-dependent receptors | ||
| PSPTO_0069-PSPTO_0068-PSPTO_0067 | Y | TonB system transport protein ExbB/TonB system transport protein ExbD/tonB protein |
| PSPTO_3294 | TonB-dependent siderophore receptor, putative | |
| PSPTO_3462 | TonB-dependent siderophore receptor | |
| PSPTO_3574 | TonB-dependent siderophore receptor, putative | |
| PSPTO_2152-PSPTO_2151 | TonB-dependent siderophore receptor, putative/TonB-dependent siderophore receptor, putative | |
| PSPTO_2605 | Y | TonB-dependent siderophore receptor, putative |
| PSPTO_3692 | TonB-dependent siderophore receptor | |
| PSPTO_1284-PSPTO_1283/PSPTO_1282 | TonB-dependent outer membrane heme receptor/heme oxygenase, putative/hypothetical protein | |
| Other iron transport systems | ||
| PSPTO_0763-PSPTO_0762-PSPTO_0761-PSPTO_0760 | Y | Iron(III) dicitrate transport system, periplasmic iron-binding protein FecB/iron(III) dicitrate transport system, permease protein FecC/iron(III) dicitrate transport system, permease protein FecD/iron(III) dicitrate transport system, ATP-binding protein FecE |
| PSPTO_0314-PSPTO_0315 | Y | Iron ABC transporter, periplasmic iron-binding protein/iron ABC transporter, permease protein |
| Noncoding RNAs | ||
| PSPTO_5657 (prrF2) | Y | ncRNA prrF2 |
| PSPTO_5658 | ncRNA cobalamin-4 | |
| PSPTO_5649 (prrF1) | ncRNA prrF1c | |
| Other | ||
| PSPTO_3157 | Y | σ70/FecR hybrid |
| PSPTO_2146 | Pyoverdine biosynthesis regulatory gene, putative | |
| PSPTO_2134-PSPTO_2135-PSPTO_2136-PSPTO_2137 | Y | Pyoverdine synthetase, thioesterase component/pyoverdine chromophore precursor synthetase/2,4-diaminobutyrate 4-transaminase/MbtH-like protein |
| PSPTO_2145-PSPTO_2144-PSPTO_2143-PSPTO_2142-PSPTO_2141-PSPTO_2140-PSPTO_2139-PSPTO_2138 | Iron-regulated membrane protein, putative/hypothetical protein/hypothetical protein/hypothetical protein/cation ABC transporter, periplasmic cation-binding protein/cation ABC transporter, ATP-binding protein/cation ABC transporter, permease protein/ABC transporter, periplasmic substrate-binding protein, putative | |
| PSPTO_4159 | Y | Bacterioferritin-associated ferredoxin |
| PSPTO_4366 | Iron-regulated protein A, putative | |
| PSPTO_4923 | Azurin | |
| PSPTO_1456-PSPTO_1457 | Multicopper oxidase/cytidine/deoxycytidylate deaminase family protein | |
| PSPTO_0426-PSPTO_0425-PSPTO_0424 | Peptidase, M16 family/hypothetical protein/methyltransferase, putative | |
| PSPTO_3122-PSPTO_3121-PSPTO_3120-PSPTO_3119 | 6-Phosphogluconate dehydrogenase/glucose-6-phosphate 1-dehydrogenase/Cof-like hydrolase family protein/glycosyl hydrolase, family 15 | |
| PSPTO_1915-PSPTO_1916 | Bacterial transferase, hexapeptide repeat protein/3-oxoacyl-(acyl carrier protein) synthase III, putative | |
| PSPTO_5310 | Glutamine synthetase | |
| PSPTO_4381 | Hypothetical protein | |
| PSPTO_4580 | Hypothetical protein | |
| PSPTO_4462-PSPTO_4461-PSPTO_4460-PSPTO_4459 | Hypothetical protein/fumarate hydratase, class II/hypothetical protein/superoxide dismutase, Mn | |
| PSPTO_3596-PSPTO_3597-PSPTO_3598-PSPTO_3599 | Y | Hypothetical protein/hypothetical protein/dyp-type peroxidase family protein/hypothetical protein |
| PSPTO_1210-PSPTO_1211-PSPTO_1212 | Y | Hypothetical protein/hypothetical protein/hypothetical protein |
| PSPTO_4368-PSPTO_4369 | Lipoprotein, putative/lipoprotein, putative | |
| PSPTO_4367-PSPTO_4368-PSPTO_4369 | Lipoprotein, putative/lipoprotein, putative/lipoprotein, putative |
Operon predictions determined by VIMSS (see Materials and Methods).
Footprint detected by nonradioactive DNase I footprint assay (indicated with “Y”); blank cells indicate that targets were not tested.
Not present on microarray; iron-dependent differential expression determined independently (data not shown).
Fig. 5.
Clustering of transcripts based on differential expression at time points taken before (a) and 30 min (b) or 4 h (c) after the addition of sodium citrate (−) or ferric citrate (+) to DC3000 grown in MG medium (adapted from reference 3). Roman numerals show cluster groups that appeared to be regulated in a similar manner (see reference 3 for discussion of these clusters). The panel to the right shows whether these genes are associated with a computationally derived Fur motif (3) or with a peak identified by CSDeconv from the ChIP-seq data (this study).
Genes located in cluster IX include TonB-dependent siderophore receptors (PSPTO_3294, PSPTO_3574, PSPTO_2152, and PSPTO_2151), components of the TonB transport system (PSPTO_0069 operon), and regulatory RNAs (ncRNA prrF2 and the cobalamin-4 riboswitch upstream of the PSPTO_3256-PSPTO_3258 putative iron ABC transport system). Except for two genes (PSPTO_4136 and PSPTO_4145, encoding an amino acid ABC transporter periplasmic amino acid-binding protein and CapB cold shock protein, respectively), all genes in cluster IX are associated with ChIP-seq peaks (Fig. 5).
Cluster X contains genes involved in the production of the siderophores yersiniabactin and pyoverdine (3). Several genes in this group are regulated by the FecI-type ECF sigma PvdS but appear to be regulated by Fur as well (as indicated by the presence of a Fur ChIP-seq peak) (Fig. 5). Also in this group are two other FecI-type sigma factors (PSPTO_1209 and PSPTO_1286) as well as three TonB-dependent siderophore receptors (PSPTO_3462, PSPTO_2605, and PSPTO_3692) and a TonB-dependent outer membrane heme receptor (PSPTO_1284). Genes not associated with Fur ChIP-seq peaks in this cluster are either involved in yersiniabactin production or regulated by PvdS without direct Fur involvement.
Many (144/201) operons that were associated with Fur binding were not identified as differentially regulated in our microarray experiments (Fig. 2), and 68 were not detectably expressed in our global transcriptome analysis of DC3000 (21) by using cells grown in iron-limited medium. It is possible that some of these are differentially expressed but did not meet the threshold for statistical significance. Alternatively, other factors not present under these conditions may be required for expression of these genes even though they are regulated by Fur. In fact it has been noted that in P. aeruginosa the growth medium (rich medium versus minimal medium) has an effect on which groups of genes are most active under high-iron conditions (32). We also know that several of the genes not expressed during growth in our medium are controlled by alternative sigma factors not active under these conditions (unpublished results). For example, expression of the FecI-like sigma factor PSPTO_1203 is induced by the presence of hyroxamate siderophores not produced by DC3000 (E. Markel et al., submitted for publication). At least one confirmed target of PSPTO_1203, the TonB-dependent siderophore receptor PSPTO_1206, is also preceded by a Fur binding site but was not differentially expressed in our microarray studies. Other Fur-regulated but not differentially expressed genes include the two remaining FecI-like sigma factors (PSPTO_0444 and PSPTO_1203), five TonB-dependent receptors, two type III effectors, genes involved in alginate biosynthesis, and several ABC transporters, as well as a number of putative regulators (see Table S3 and File S2 in the supplemental material).
Fur-regulated regulators.
DC3000 Fur binds upstream of a variety of putative regulators (Table 2; also see Table S3 in the supplemental material), including a RNA-binding protein and several ncRNAs and FecI-type sigma factors. CsrA-3 (PSPTO_3566) belongs to the RsmA/CrsA family of proteins that bind to target RNAs and block interaction of the ribosome with the ribosome binding site, thereby preventing initiation of translation eventually leading to destabilization and degradation of the transcript (2). We confirm that Fur binds upstream of the prrF1 and prrF2 ncRNAs, which are homologous to Fur-regulated ncRNAs in P. aeruginosa (21), and that Fur also regulates a third ncRNA, the cobalamin-4 riboswitch located upstream of the PSPTO_3256-PSPTO_3258 operon. All five FecI-type ECF sigma factors in DC3000 are immediately downstream of Fur binding sites, but only three (pvdS, PSPTO_1209, and PSPTO_1286) were differentially expressed in our microarray experiments. The two remaining sigma factors may be repressed by additional proteins or require activators that are unavailable under the conditions of our experiments.
Fur-regulated iron-activated genes.
Eight genes from six Fur peak-associated operons are found in cluster I (Table 3), and Fur binding was confirmed by footprinting at all six peaks. Cluster I contains genes that are induced rather than repressed in the presence of iron. Four of the six peak-associated operons encode bacterioferritins (PSPTO_0653 and PSPTO_4160), a ParA family protein (PSPTO_0855), and an operon containing a two-component system (TCS) (PSPTO_3382-3379). Although Fur-dependent gene activation can occur indirectly, mediated by a Fur-repressed ncRNA, the confirmation of Fur binding sites in close proximity to these genes suggests that Fur plays a direct role in their regulation.
Table 3.
Iron-activated genes associated with Fur binding
| Peak no. | Gene/operon associated with peaka | Predicted function of gene(s)a |
|---|---|---|
| 34 | PSPTO_0653 | Bacterioferritin |
| 43 | PSPTO_0855-PSPTO_0856 | ParA family protein/hypothetical protein |
| 181 | PSPTO_3382-PSPTO_3381-PSPTO_3380-PSPTO_3379 | Lipoprotein, putative/DNA-binding response regulator/sensor histidine kinase/methyl-accepting chemotaxis protein |
| 218 | PSPTO_3959b | Quinolinate synthetase complex, subunit A |
| 230_1 | PSPTO_4160 | Bacterioferritin |
| 260 | PSPTO_4766 | Hypothetical proteinc |
For operons, genes are separated by dashes, and functions of genes are separated by slashes.
Peak located in region with divergent PSPTO_3960 transcription.
The C-terminal residues of this open reading frame (ORF) are encoded by an adjacent insertion sequence.
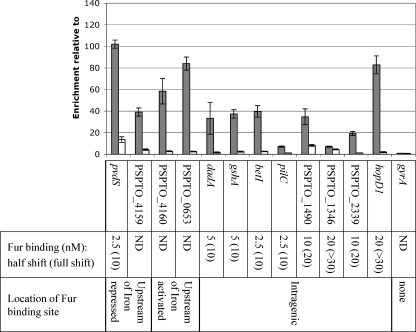
In H. pylori, activation of genes in an iron-dependent manner was found to be the result of repression by apo-Fur under low-iron conditions (12, 14, 17). To investigate this possibility, we repeated ChIP using samples supplemented with either ferric citrate (+ Iron) or sodium citrate (− Iron). If apo-Fur were binding to the promoter elements, we would expect to see enrichment of these regions under low-iron conditions. However, we found that while regions upstream of PSPTO_4160 and PSPTO_0653 were enriched in the + Iron sample, there was little enrichment after ChIP of the − Iron sample (see Fig. 7). Since we are unable to make a fur mutant in DC3000, we cannot confirm that apo-Fur is not involved; however, these data suggest that iron-bound Fur is binding upstream of these genes.
Fig. 7.
qPCR performed on ChIP samples from cultures grown with iron citrate (+ Iron; gray) or sodium citrate (− Iron; white) for 30 min. Enrichment of selected Fur binding regions is calculated relative to DNA sequences within the gap1 gene after immunoprecipitation with Fur-FLAG-tagged strains. No enrichment is observed of DNA within the gyrA gene. Shown below are the estimated Fur concentrations (nM) at which half shift and complete shift of a radiolabeled probe was observed after EMSA (see Fig. S2 in the supplemental material). For comparison, EMSA of other intergenic probes can be seen in Fig. 4B. Nonspecific shift of a PSPTO_1900 probe was observed only at concentrations of 50 nM Fur and higher (Fig. 4B). ND, not done.
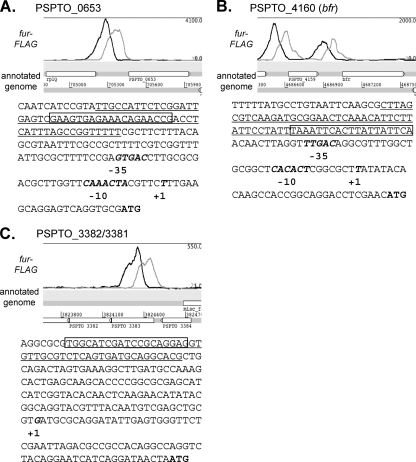
Using 5′ RACE, the transcriptional start sites (TSS) for the two bacterioferritins (PSPTO_0653 and PSPTO_4160) were mapped using RNA isolated from cells grown under high-iron conditions (Fig. 6). Putative RpoD (σ70) promoter sequences can be identified upstream of the start sites, and the positions of the Fur binding sequences are upstream (75 bp and 31 bp, respectively) of the promoter sequences. This location is consistent with the role of Fur as an activator of these genes (Fig. 6) and is reminiscent of the arrangement of other promoters thought to be regulated positively by Fur (13). A TSS was also mapped upstream of PSPTO_3382, within PSPTO_3383 and 112 bp downstream of the Fur footprint (Fig. 6). However, we were unable to identify a promoter in this region. PSPTO_3382 encodes a putative lipoprotein with a PepSY peptidase domain, and PSPTO_3381 and PSPTO_3380 are the response regulator and histidine kinase of a predicted TCS that when deleted results in a decrease in virulence on Arabidopsis (data not shown).
Fig. 6.
Fur binds upstream of two iron-activated bacterioferritin genes, PSPTO_0653 (A) and PSPTO_4160 (B), and upstream of PSPTO_3382/3381/3380, an operon containing a TCS (C). For each gene, an Artemis screen shot shows the naive profile of the ChIP-seq data indicating the region of enrichment after immunoprecipitation. Below is the sequence of the region upstream of the start (ATG) of each gene, showing the transcriptional start site (+1; mapped by 5′ RACE) and the position of a putative RpoD promoter sequence (−10 and −35). The underlined sequence corresponds to the region protected by Fur in a nonradioactive footprinting assay, while the boxed region is the predicted MEME motif generated from the ChIP-seq peaks (corresponding to the motif in Fig. 3A).
Fur binding in unexpected locations.
Over 45% (145/312) of ChIP-seq peaks do not fall into regions expected to be associated with regulatory activity (from 250 bp upstream to 50 bp downstream of an annotated CDS start codon). Some unexpected peaks (29) appear in intergenic regions, many flanked by convergently transcribed genes (see Table S4 in the supplemental material). Several regions are long enough to encode unannotated genes or ncRNAs. Five appear to contain genes for unannotated hypothetical proteins based on predictions using Easy Gene version 1.2 (PSPTO_1097-PSPTO_1098, PSPTO_3216-PSPTO_3217, PSPTO_3615-PSPTO_3616, PSPTO_3948-PSPTO_3949, and PSPTO_4755-PSPTO_4756) (see Table S4 in the supplemental material). Transcriptional activity under low-iron conditions can be detected for three of these CDSs by using global transcription profiles (21). Two of the unannotated CDSs (located between PSPTO_1097 and PSPTO_1098 and PSPTO_3216 and PSPTO_3217) share 87 to 92% identity with hypothetical proteins from Pseudomonas syringae pv. aesculi but do not appear to be present in the genomes of the other pseudomonads. The large intergenic regions were also evaluated to determine if they were associated with known or predicted ncRNAs, and Fur peaks were found upstream of the ncRNAs prrF1 and prrF2. In addition, a peak was found between two divergently transcribed genes (PSPTO_2310 and PSPTO_2311) in a region where a ncRNA (rmf) is predicted upstream of PSPTO_2310 (40). Transcriptional activity has been detected in this region (21) and is consistent with the presence of the ncRNA.
In four cases, peaks were located adjacent to regions containing insertion sequences. It is possible that the insertions have separated the Fur binding sites from their original neighbors. For example, one predicted Fur binding site is located upstream of PSPTO_2167, a transposase. The surrounding region (without the transposase) is conserved in the P. syringae strains 1448a and B728a. It is possible that Fur may have regulated the PSPTO_2168-2170 operon and that the Tn insertion separated the Fur binding site from this set of genes in DC3000.
Finally, in two cases the peaks are located well upstream of the closest potential regulatory targets, PSPTO_2853 and PSPTO_3365. These regions do not appear to contain possible open reading frames (ORFs) or predicted ncRNAs. Comparison with the transcriptional profile shows that transcription begins close to the predicted peak/Fur binding sites, suggesting that these genes are preceded by long 5′ untranslated regions (UTRs). Fur binding was confirmed by DNase I footprinting upstream of PSPTO_2853 (TonB-dependent receptor). PSPTO_3365 is the first gene in the nuoA-nuoN gene cluster, encoding subunits of the NADH dehydrogenase of the aerobic respiratory chain, and in Neisseria, these genes are induced in response to iron (13, 23). In Neisseria meningitidis, Fur was shown to bind to and activate expression from a promoter upstream of nuoA (13).
The majority of the Fur binding sites in this group (116; 80%) lie within coding sequences. Fur binding was confirmed by footprint analysis in six of these cases (Table 1). To further investigate intragenic binding sites, we selected eight regions (four of which had been footprinted) and performed qPCR on DNA isolated after ChIP on samples with and without iron added. We found that in all except one case (PSPTO_1346) enrichment was iron dependent (Fig. 7). In most cases, levels of enrichment under high-iron conditions were also comparable to those of intergenic Fur binding sites. We used EMSA to assess whether Fur bound at these intragenic sites with a relative affinity lower than that at intergenic sites, as has been shown for one site in N. gonorrhoeae (23). We found that while half of the sites displayed lower binding affinities, binding at the remaining sites was comparable to that at pvdS, prrF2, and PSPTO_0070 (Fig. 7).
Fur binding at intragenic sites is difficult to interpret in terms of current models for regulation by Fur. One possibility is that Fur binding results in altered transcription patterns in these regions in response to iron. To test this hypothesis, quantitative reverse transcription-PCR (qRT-PCR) was performed on targeted regions upstream and downstream of the Fur binding sites within these genes by using RNA isolated from cells grown in high- or low-iron conditions. Gene transcription was detected under both growth conditions, but we were unable to demonstrate any difference in RNA levels, either by comparing transcript levels between conditions or by comparing levels upstream and downstream of the Fur binding sites (data not shown).
DISCUSSION
This study is the first to characterize the Fur regulon in DC3000, an important model organism for bacterium-plant interactions, and is one of only a handful of studies to apply ChIP-seq to bacterial regulators. We have observed Fur binding to genomic targets in vivo in the presence of iron citrate and have integrated this information with data from prior experiments in which we investigated regulation by iron. The synthesis significantly expands our understanding of the Fur regulon and highlights the complexity of Fur function.
While the Fur regulon has been well characterized in the closely related human pathogen P. aeruginosa, where iron is sequestered from the pathogen by the mammalian host, the situation is very different in the P. syringae host environment. P. syringae is likely to experience iron limitation on the surface of the leaf, where it is in competition with other epiphytes producing siderophores with binding affinities higher than those produced by DC3000 (16); however, once DC3000 has entered the apoplast it will likely be presented with an environment richer in iron (22). We expect that the role that Fur plays in virulence will be shown to be different between P. aeruginosa and DC3000. Unfortunately, we have been unable to construct a fur mutant in DC3000 and so are unable to assess the role of this protein in virulence. Therefore, this elucidation of the DC3000 Fur regulon provides targets for further study. For example, we have identified several additional regulators controlled by Fur extending the role of Fur in response to iron levels in DC3000. In addition, the Fur-regulated iron-activated genes such as the PSPTO_3381 and PSPTO_3380 TCSs may play an important role in virulence once the pathogen enters the iron-rich environment of the leaf apoplast.
Fur has been studied in many bacteria, but in most cases global regulation is investigated by microarray studies, and often, additional Fur binding sites are determined computationally. Our previous microarray study identified several genes that appeared to be Fur regulated, but in the absence of a fur mutant we were unable to confirm these predictions. Our ChIP-seq results highlight that at least in DC3000 Fur binding is more extensive than we had suspected and that our previous computational predictions missed many bona fide binding sites. In addition, since the ChIP-seq method detects direct Fur binding, we identified many binding sites upstream of genes that were not expressed and therefore would not be identified in our earlier transcription-based studies, even though cells had been grown in low-iron conditions (when Fur repression is relieved).
As expected, the DC3000 Fur regulon (consisting of genes involved in iron uptake, storage, and homeostasis) resembles that identified in other bacteria. Nonetheless, DC3000 offers several interesting differences. For example, DC3000 (like P. aeruginosa) has an increased number of TonB-dependent receptors, allowing the bacteria competitive fitness under iron-limited conditions by scavenging an array of siderophores produced by other microorganisms (10, 25). However, unlike P. aeruginosa where most (28/32) of the receptors are indirectly regulated by Fur via FecI-type ECF sigma factors or other regulators (9), in DC3000 we find that more than half (14/25) are associated with Fur binding (Table 2; also see Table S3 in the supplemental material). In addition, we find that many of the TonB-dependent transducers are both controlled by Fur and expressed via a Fur-regulated FecI-like sigma factor (B. Swingle, unpublished results).
We were also surprised to find 37% of the DC3000 Fur binding sites within annotated CDSs. While Fur binding within CDSs has been reported before (23), this is the first study to extensively identify intragenic binding sites in vivo. We confirmed Fur binding (by footprinting and/or EMSA) at 10 of these sites and in seven cases binding appears to occur predominately under high-iron conditions (Fig. 7). A recent study in N. gonorrhea showed that Fur bound within one gene with lower affinity than at a binding site upstream of a second gene, leading to the hypothesis that the binding affinity at intragenic Fur binding sites is lower than that at intergenic sites (23). However, in DC3000 the binding affinity of Fur at some intragenic sites is comparable to that observed for intergenic sites (e.g., betI versus pvdS) (Fig. 7), and thus we see no obvious general differences between the two classes. The role Fur plays at these sites remains puzzling. One obvious model is that Fur acts as a repressor and prevents transcription downstream of the binding site, resulting in different levels of expression upstream and downstream of the Fur complex. However, our test of this hypothesis at Fur binding sites in eight genes yielded negative results. We also explored the possibility that Fur controls expression on the noncoding strands of these genes. If so, we would expect to detect antisense transcription under conditions when Fur does not bind. Our previous RNA sequencing experiments, conducted using cells grown in low-iron conditions, failed to reveal any convincing examples where expression does not correspond to the annotated genome in the region of these Fur binding sites. We cannot exclude the possibility that Fur binding at these sites may play a nonregulatory role in the cell (e.g., on organization of the bacterial chromatin). Further studies are clearly required to determine the role of Fur in these cases.
Interestingly we also found Fur associated with genes that are activated in the presence of iron. The Fur-regulated iron-activated genes PSPTO_0653 and PSPTO_4160 encode bacterioferritins homologous to BfrA and BfrB of P. aeruginosa PAO1. These proteins oxidize Fe2+ and store Fe3+ to protect the cell from toxic reactive oxygen species and reserve iron for use under iron-limited conditions (38). In P. aeruginosa PAO1, expression of bfrB is positively regulated by Fur and Fe2+ and is independent of the prrF ncRNAs (41). The authors suggested that this positive regulation may be due to direct activation by Fur, an undetected ncRNA, or a Fur-repressed repressor. Bacterioferritin homologs bfrα and bfrβ from Pseudomonas putida KT2440 are also activated by iron in a Fur-dependent manner (7) but since no Fur motif was identified, the authors noted that positive regulation by Fur is likely to be indirect. Here we confirm that at least in DC3000 Fur does bind upstream of the bacterioferritin genes, despite the absence of an easily recognizable Fur box consensus sequence (Fig. 6), suggesting that Fur directly regulates expression of these genes.
There are three ways in which Fur binding has been shown to result in increased expression under high-iron conditions. First, binding of iron-bound Fur may block binding of a second repressor. For example, in E. coli Fur was found to activate ftnA expression by blocking histone-like nucleoid-associated protein (H-NS)-mediated repression (29). As such, Fur still acts in the classical manner by binding DNA in the presence of iron and blocking the action of another protein. Second, binding of iron-bound Fur directly activates expression of the downstream gene. In N. meningitidis Delany and coworkers (13) showed that the binding of Fur upstream of three genes encoding iron-containing respiratory proteins (pan1, norB, and nuoA) was essential for activation of these genes both in vitro and in vivo. Third, binding of apo-Fur represses expression of genes under low-iron conditions. To date, direct evidence for this type of binding has been demonstrated only in H. pylori, where apo-Fur represses expression of sodB and pfr (14, 17). We found that enrichment of the PSPTO_4160 and PSPTO_0653 upstream regions was significantly reduced under low-iron conditions, suggesting that apo-Fur binding is not occurring at these sites. However, further investigation will be required to determine whether Fur binding activates expression of the DC3000 genes or merely blocks binding of a second repressor.
Supplementary Material
Acknowledgements
J.D.H. and A.G. were supported by NIH grant GM059323.
We thank Desmond Lun for assistance with the use of the CSDeconv peak calling program.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Baichoo N., Helmann J. D. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brencic A., et al. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73:434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bronstein P. A., et al. 2008. Global transcriptional responses of Pseudomonas syringae DC3000 to changes in iron bioavailability in vitro. BMC Microbiol. 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buell C. R., et al. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 100:10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carpenter B. M., Whitmire J. M., Merrell D. S. 2009. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect. Immun. 77:2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cha J. Y., Lee J. S., Oh J. I., Choi J. W., Baik H. S. 2008. Functional analysis of the role of Fur in the virulence of Pseudomonas syringae pv. tabaci 11528: Fur controls expression of genes involved in quorum-sensing. Biochem. Biophys. Res. Commun. 366:281–287 [DOI] [PubMed] [Google Scholar]
- 7. Chen S., Bleam W. F., Hickey W. J. 2010. Molecular analysis of two bacterioferritin genes, bfrα and bfrβ, in the model rhizobacterium Pseudomonas putida KT2440. Appl. Environ. Microbiol. 76:5335–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Z., et al. 2007. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res. 35:6762–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86:1637–1645 [DOI] [PubMed] [Google Scholar]
- 10. Cornelis P., Bodilis J. 2009. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ. Microbiol. Reports 1:256–262 [DOI] [PubMed] [Google Scholar]
- 11. Cornelis P., Matthijs S., Van Oeffelen L. 2009. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22:15–22 [DOI] [PubMed] [Google Scholar]
- 12. Delany I., Pacheco A. B., Spohn G., Rappuoli R., Scarlato V. 2001. Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein. J. Bacteriol. 183:4932–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delany I., Rappuoli R., Scarlato V. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52:1081–1090 [DOI] [PubMed] [Google Scholar]
- 14. Delany I., Spohn G., Rappuoli R., Scarlato V. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297–1309 [DOI] [PubMed] [Google Scholar]
- 15. De Lorenzo V., Giovannini F., Herrero M., Neilands J. B. 1988. Metal ion regulation of gene expression: Fur repressor-operator interaction at the promoter region of the aerobactin system of pCoIV-K30. J. Mol. Biology. 203:875–884 [DOI] [PubMed] [Google Scholar]
- 16. Dulla G. F., Krasileva K. V., Lindow S. E. 2010. Interference of quorum sensing in Pseudomonas syringae by bacterial epiphytes that limit iron availability. Environ. Microbiol. 12:1762–1774 [DOI] [PubMed] [Google Scholar]
- 17. Ernst F. D., et al. 2005. Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. J. Bacteriol. 187:3687–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Escolar L., Perez-Martin J., de Lorenzo V. 1998. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283:537–547 [DOI] [PubMed] [Google Scholar]
- 19. Escolar L., Perez-Martin J., de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ferreira A. O., et al. 2006. Whole-genome expression profiling defines the HrpL regulon of Pseudomonas syringae pv. tomato DC3000, allows de novo reconstruction of the Hrp cis clement, and identifies novel coregulated genes. Mol. Plant Microbe Interact. 19:1167–1179 [DOI] [PubMed] [Google Scholar]
- 21. Filiatrault M. J., et al. 2010. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J. Bacteriol. 192:2359–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hernandez-Morales A., et al. 2009. Transcriptional profile of Pseudomonas syringae pv. phaseolicola NPS3121 in response to tissue extracts from a susceptible Phaseolus vulgaris L. cultivar. BMC Microbiol. 9:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jackson L. A., et al. 2010. Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon. J. Bacteriol. 192:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kahramanoglou C., et al. 2011. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 39:2073–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koebnik R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13:343–347 [DOI] [PubMed] [Google Scholar]
- 26. Li R., et al. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967 [DOI] [PubMed] [Google Scholar]
- 27. Lun D. S., Sherrid A., Weiner B., Sherman D. R., Galagan J. E. 2009. A blind deconvolution approach to high-resolution mapping of transcription factor binding sites from ChIP-seq data. Genome Biol. 10:R142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miles S., Carpenter B. M., Gancz H., Merrell D. S. 2010. Helicobacter pylori apo-Fur regulation appears unconserved across species. J. Microbiol. 48:378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nandal A., et al. 2010. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe(2+)-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol. Microbiol. 75:637–657 [DOI] [PubMed] [Google Scholar]
- 30. Ochsner U. A., Vasil A. I., Vasil M. L. 1995. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J. Bacteriol. 177:7194–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ochsner U. A., Vasil M. L. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. U. S. A. 93:4409–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ochsner U. A., Wilderman P. J., Vasil A. I., Vasil M. L. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277–1287 [DOI] [PubMed] [Google Scholar]
- 33. Oglesby A. G., et al. 2008. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J. Biol. Chem. 283:15558–15567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oguiza J. A., Kiil K., Ussery D. W. 2005. Extracytoplasmic function sigma factors in Pseudomonas syringae. Trends Microbiol. 13:565–568 [DOI] [PubMed] [Google Scholar]
- 35. Palma M., Worgall S., Quadri L. E. 2003. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch. Microbiol. 180:374–379 [DOI] [PubMed] [Google Scholar]
- 36. Pohl E., et al. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903–915 [DOI] [PubMed] [Google Scholar]
- 37. Schafer A., et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 38. Smith J. L. 2004. The physiological role of ferritin-like compounds in bacteria. Crit. Rev. Microbiol. 30:173–185 [DOI] [PubMed] [Google Scholar]
- 39. Swingle B., et al. 2008. Characterization of the PvdS-regulated promoter motif in Pseudomonas syringae pv. tomato DC3000 reveals regulon members and insights regarding PvdS function in other pseudomonads. Mol. Microbiol. 68:871–889 [DOI] [PubMed] [Google Scholar]
- 40. Weinberg Z., et al. 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 11:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilderman P. J., et al. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101:9792–9797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zianni M., Tessanne K., Merighi M., Laguna R., Tabita F. R. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17:103–113 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.