Abstract
TrbB, a periplasmic protein encoded by the conjugative plasmid F, has a predicted thioredoxin-like fold and possesses a C-X-X-C redox active site motif. TrbB may function in the conjugative process by serving as a disulfide bond isomerase, facilitating proper folding of a subset of F-plasmid-encoded proteins in the periplasm. Previous studies have demonstrated that a ΔtrbB F plasmid in Escherichia coli lacking DsbCE.coli, its native disulfide bond isomerase, experiences a 10-fold decrease in mating efficiency but have not provided direct evidence for disulfide bond isomerase activity. Here we demonstrate that trbB can partially restore transfer of a variant of the distantly related R27 plasmid when both chromosomal and plasmid genes encoding disulfide bond isomerases have been disrupted. In addition, we show that TrbB displays both disulfide bond isomerase and reductase activities on substrates not involved in the conjugative process. Unlike canonical members of the disulfide bond isomerase family, secondary structure predictions suggest that TrbB lacks both an N-terminal dimerization domain and an α-helical domain found in other disulfide bond isomerases. Phylogenetic analyses support the conclusion that TrbB belongs to a unique family of plasmid-based disulfide isomerases. Interestingly, although TrbB diverges structurally from other disulfide bond isomerases, we show that like those isomerases, TrbB relies on DsbD from E. coli for maintenance of its C-X-X-C redox active site motif.
INTRODUCTION
Bacterial conjugation is a process by which single-stranded DNA is unidirectionally transferred from donor to recipient bacterium (18, 26). Conjugative plasmid transfer can accelerate the evolution of bacteria (15), facilitate various pathogenic processes (12), and disseminate genes encoding antibiotic resistance (13), including New Delhi metallo-β-lactamase, which has recently become a global health concern (41).
The transfer (tra) region of plasmid F encodes proteins involved in plasmid transfer. A subset of Tra proteins assembles to form the pore complex that spans the inner and outer membranes of the donor, while others are thought to assist in assembly of the pore complex (25). The periplasmic protein TrbB is proposed to assist pore complex assembly through disulfide isomerase activity (2, 21, 44).
In its mature state, TrbB consists of 161 amino acids. TrbB has an N-terminal periplasmic localization signal and a predicted thioredoxin-like fold containing a C-X-X-C (C=Cys; X=any residue) redox active site motif. The variable residues in C-X-X-C motifs influence the active site redox potential and, in turn, influence function (10, 49). TrbB has a C-P-Y-C motif sequence, a combination common in DsbC- and DsbG-like families of disulfide isomerases (50). Based on the above characteristics, TrbB has been placed into the thioredoxin-like superfamily, which includes reductases, oxidases, and disulfide bond isomerases. Disulfide bond isomerases facilitate formation of correct disulfide bonds in protein targets through the direct rearrangement of erroneous disulfide bonds. Recently, it has also been demonstrated that disulfide isomerization can occur through a reduction-oxidation cycling mechanism promoted by periplasmic reductases (58).
In Gram-negative bacteria, disulfide bonds are formed as proteins are secreted from the reducing cytoplasm into the oxidizing periplasm (35). In most Gram-negative bacteria, the conserved disulfide bond-forming (Dsb) protein DsbA introduces disulfide bonds, almost always between consecutive Cys residues in the primary structure (16). This mechanism can form incorrect disulfide linkages, causing misfolding, inhibition of function, and possibly aggregation of periplasmic proteins (5, 29, 30, 48). Disulfide bond isomerases, such as DsbC from Escherichia coli (DsbCE.coli) (47, 57), can “shuffle” incorrect disulfide linkages, allowing proteins to assume their natively folded state. Proteins exhibiting disulfide bond isomerase activity have also been recently implicated in protecting single Cys residues from sulfenylation by reactive oxygen species, thereby regulating the global level of sulfenylation in the periplasm (19).
The C-X-X-C active site motif of disulfide isomerases must be reduced to be active. After an isomerase has bound its substrate, the N-terminal active site Cys residue initiates isomerization by attacking a disulfide bond in the substrate. A successful attack yields a disulfide-linked enzyme-substrate complex that is resolved after a second attack from either the C-terminal active site Cys or a free thiol in the substrate (34, 36). If the secondary attack originates from the C-terminal Cys of the isomerase, the C-X-X-C active site becomes oxidized, rendering it inactive. Disulfide isomerases rely on the inner membrane protein DsbD for redox state maintenance (51). DsbD accepts electrons from cytoplasmic thioredoxin and shuttles these electrons via a well-defined cascade through the membrane-spanning region and into the periplasm (11, 38, 39, 59). In the absence of DsbD, disulfide isomerases accumulate in an oxidized and inactive state (51). No DsbD homologs are found in the 108 open reading frames in F (GenBank sequence accession number AP001918), suggesting that TrbB may rely on the E. coli system for maintaining its reduced and hypothetically active state.
In conjugative systems, the role of disulfide isomerases has only recently been appreciated (21), even though genomic comparisons of several conjugative plasmids show a conservation of thioredoxin-like family members. In particular, TrbBs from plasmids F, R100, and pSLT are nearly identical (Fig. 1), leading to the hypothesis that TrbB acts to assist proper folding of a subset of plasmid-encoded proteins (Fig. 2). In early studies on Tra proteins, mating efficiencies were unaffected for a ΔtrbB plasmid, suggesting that TrbB was not integral to the conjugative process (23). However, later studies performed in a ΔdsbCE.coli ΔtrbB background exhibited a small but significant reduction in F mating efficiency. This reduction is fully rescued by complementation in trans with either DsbCE.coli or TrbB, but not by a TrbB mutant lacking its active site cysteines (21). These studies, however, did not directly analyze the redox activity of TrbB.
Fig. 1.
Analysis of a putative new family of disulfide isomerases. Shown is a multiple sequence alignment of TrbB-like disulfide isomerases from plasmids F, R100, and pSLT done using ClustalW. F and R100 TrbB are 90% identical in amino acid sequence while F and pSLT TrbB are 77% identical. Predicted signal sequences are italicized. The N-terminal arms predicted from results of limited proteolysis, Phyre, and PONDR are underlined. The C-X-X-C redox active sites are highlighted in bold. Sequence conservation is indicated as follows: *, fully conserved; :, strongly similar residues;., weakly similar residues.
Fig. 2.
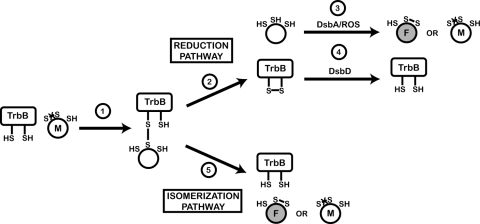
Model of TrbB mechanism in the E. coli periplasm on a substrate containing three Cys residues. Cys residue side chains are represented as -SH (reduced/thiol) or S-S (oxidized/disulfide). (1) Reduced TrbB recognizes misfolded substrate (M) and forms a disulfide-linked enzyme-substrate complex. This complex is resolved by secondary attack of a free thiol from either TrbB or the substrate protein. When the secondary attack originates from TrbB (2), the substrate is released in a fully reduced state and depends on either DsbA or a reactive oxygen species (ROS) for oxidation (3). This nonspecific oxidation can lead to either recurrence of the substrate's misfolded state (M) or a correctly folded state (F). TrbB becomes oxidized and therefore dependent on an inner membrane protein (potentially DsbD) for reactivation (4). When the secondary attack originates from the substrate protein (5), the substrate is released in an oxidized state. Depending on which free thiol from the substrate initiates the secondary attack, either the correctly folded (F) or misfolded (M) state will be achieved. In this case, TrbB remains reduced and therefore active.
Here, results of in vitro and in vivo assays demonstrate that TrbB can act as a disulfide isomerase and rule out the model of TrbB promoting disulfide isomerization through a reduction-oxidation cycling mechanism. In addition, phylogenetic analyses, secondary structure predictions, and analytical ultracentrifugation show that TrbB-like proteins are unique among thioredoxin-like family members and should be considered a distinct family of disulfide isomerases. We have also discovered that the bacterial protein DsbD is required for maintenance of the C-X-X-C redox active site in TrbB, which is responsible for its enzymatic activity. Our observations not only give insight into TrbB's in vivo activity but also illustrate how the F plasmid system depends on E. coli cellular machinery to ensure proper function.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain BL21(DE3) was purchased from Novagen. Strains JP114 and RGP475 were kindly provided by J. Bardwell (University of Michigan). Strain CH79 was constructed by replacing the dsbD gene of E. coli strain MB69 with the kanamycin cassette from plasmid pKD4 as previously described (17). pDSW204-dsbCR27 was created by PCR amplifying dsbCR27 from JC3272 (1) and cloning it into the EcoRI/KpnI sites of pDSW204 (61). pDSW204-dsbCE.coli was created by excising dsbCE.coli from pBAD33-dsbCE.coli (6, 27) by KpnI/HindIII digestion and ligating into pDSW204. pDSW204-trbB-FLAG was created by amplifying trbB from the F′ in E. coli strain DHB4 (7) and ligating into the NcoI/XbaI sites in pMER77. Plasmid pDSW204-trbB was made by replacing the native stop codon in pDSW204-trbB-FLAG through QuikChange mutagenesis. To construct pMER77, the 3×FLAG tag-containing XbaI/HindIII fragment of pNB100 (9) was cloned into the XbaI/HindIII sites of expression vector pDSW204. TrbBΔ29 was created by PCR amplifying trbB with primers that exclude codons encoding residues 21 to 50. The resulting product was digested and ligated into the EcoRV/HindIII sites of pDSW204. Plasmid pET24a-trbB lacking the N-terminal native signal peptide (−sp) was created by PCR amplifying bp 61 to 546 of trbB from the ER2738 F′ plasmid and cloning the product into the NdeI and EcoRI sites of pET24a. Additional TrbB and TrbB (−sp) mutants were created by QuikChange mutagenesis. Plasmids encoding proteins used for activity assays were transformed into various E. coli strains as specified in Tables 1 and 2. A list of primers is available upon request.
Table 1.
Bacterial strains
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| DHB4 | MC1000 [F−araD139 Δ(ara-leu)7697 ΔlacX74pho-510galE15galK16relA1spoT1rpsLthi] phoR68dphoA PvuII ΔmalF3 (F′ lacIqpro) | 7 |
| ER2738 | F′ proA+B+lacIq Δ(lacZ)M15 zzf::Tn10(TetR)/fhuA2glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5 | NEB |
| JC3272 | JC3051 gal mutant lys mutant | 1 |
| MB69 | DHB4 ΔdsbC | M. Berkmen |
| MB322 | DHB6501 with SM551 | M. Berkmen |
| MB614 | DHB4 with pBAD33-appA | M. Berkmen |
| MB637 | MB69 with pBAD33-appA, pDSW204-dsbC_E.coli | M. Berkmen |
| MB721 | MB69 with pBAD33-appA, pDSW204 | M. Berkmen |
| MB790 | JC3272 with pR27 | M. Berkmen |
| MB794 | MB69 with pBAD33-appA, pDSW204-dsbC-R27 | M. Berkmen |
| MB823 | DHB4 Δara714leu+ | M. Berkmen |
| MB838 | MB69 with pBAD33-appA | M. Berkmen |
| MB872 | MB790 with ΔdsbC-R27::cam | M. Berkmen |
| MB898 | MB790 with ΔdsbC::kan | M. Berkmen |
| MB899 | MB872 with ΔdsbC::kan | M. Berkmen |
| MB900 | MB899 with pDSW204-dsbC_R27 | M. Berkmen |
| MB904 | MB899 with pDSW204-dsbC_E.coli | M. Berkmen |
| MB1171 | MB823 with pBAD33-phoA | M. Berkmen |
| MB1174 | MB823 ΔdsbA::kan | M. Berkmen |
| MB1191 | MB1174 with pBAD33-phoA | M. Berkmen |
| JP114 | ER1821: NEB F−glnV44 e14 mutant (McrA−) rfbD1? relA1? endA1spoT1? thi-1 Δ(mcrC-mrr)114::ISO | J. Bardwell |
| RGP475 | JP114 ΔdsbD::kan | J. Bardwell |
| CH37 | MB899 with pDSW204-trbB | This study |
| CH44 | MB69 with pBAD33-appA, pDSW204-trbB_F | This study |
| CH46 | MB69 with pBAD33-appA, pDSW204-trbB_F C81S/C84S | This study |
| CH50 | MB69 with pBAD33-appA, pDSW204-trbB_F Δ29 | This study |
| CH60 | JP114 with pMER77-trbB_F-FLAG | This study |
| CH61 | RGP475 with pMER77-trbB_F-FLAG | This study |
| CH79 | MB69 ΔdsbD | This study |
| CH80 | CH79 with pBAD33-appA | This study |
| CH81 | CH79 with pBAD-appA, pDSW204-trbB_F | This study |
| CH120 | MB1191 with pDSW204-trbB | This study |
Table 2.
Plasmids
| Plasmid | Relevant genotype | Source or reference |
|---|---|---|
| pET24a | T7 promoter, Kanr | Novagen |
| pNB100 | pBAD18-flag (3×) | 9 |
| pDSW204 | Mutant lacUV5 promoter, pBR322 origin, Ampr | 61 |
| pMER77 | Based on pDSW204 with C-terminal FLAG tag sequence | This study |
| pMER78 | Based on pNB100 with DsbA signal sequence and C-terminal FLAG tag | 58 |
| pBAD33 | Arabinose promoter, pACYC origin, Camr | 27 |
| pKD4 | pANTSγ derivative, FRTa-flanked Kan cassette from pCP15 | 17 |
| pET24a-trbB (−sp) | Mutant trbB from plasmid F with signal sequence removed | This study |
| pET24a-trbB C81S/C84S | pET24a-trbB (−sp) with C81S and C84S mutations | This study |
| pET24a-dsbC_E.coli (−sp) | Mutant dsbC from E. coli with signal sequence removed | This study |
| pDSW204-dsbC_E.coli | Wild-type dsbC from E. coli with native signal sequence | This study |
| pDSW204-dsbC_R27 | Wild-type dsbC from plasmid R27 with native signal sequence | This study |
| pDSW204-trbB | Wild-type trbB from plasmid F with native signal sequence | This study |
| pDSW204-trbB C81S/C84S | Mutant trbB from plasmid F with mutations C81S and C84S in active site | This study |
| pDSW204-trbB Δ29 | Mutant trbB from plasmid F excluding residues 21 to 50 | This study |
| pDSW204-trbB-FLAG | Wild-type trbB from plasmid F with C-terminal FLAG tag in pMER77 | This study |
| pMER78-trxA | trxA with DsbA signal sequence | 58 |
| pBAD33-appA | Wild-type appA | 58 |
| pBAD33-phoA | Wild-type phoA | 8 |
FRT, FLP recombination target.
Protein purification.
TrbB (−sp), TrbB C81S/C84S (−sp), and DsbCE.coli (−sp) were all expressed in E. coli strain BL21(DE3) from pET24a constructs. Five-hundred-milliliter Luria-Burtani (LB) broth cultures with kanamycin (30 μg/ml) were inoculated and grown to an A600 of 0.5 to 0.7 at 37°C. Expression was induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG), and cells were incubated for an additional 3 h at 37°C. Cells were pelleted by centrifugation and stored at −80°C. For purification, cells were thawed and resuspended in either 50 ml of 50 mM Tris-HCl (pH 7.5) with 3 M NaCl and 1 mM EDTA (TrbB constructs) or 50 mM bis-Tris-HCl (pH 6.5) (DsbC). Cells were lysed by sonication, and cellular debris was removed by centrifugation. Purifications were performed using a Gradi-Frac system. Column fractions were analyzed by 10% Tris-Tricine SDS-PAGE gels stained with Coomassie blue (54).
TrbB (−sp) and TrbB C81S/C84S (−sp) were applied to a 5-ml Hi-Trap Butyl FF column (Amersham Biosciences) equilibrated with 50 mM Tris-HCl (pH 7.5) containing 3 M NaCl and 1 mM EDTA. Using a 3 M to 25 mM NaCl gradient, TrbB eluted as a single peak beginning at 0.6 M. Pooled fractions were dialyzed overnight at 4°C against 50 mM Tris-HCl (pH 7.5) containing 25 mM NaCl and 1 mM EDTA. The dialyzed sample was then applied to a 5-ml Hi-Trap Blue HP column (Amersham Biosciences) equilibrated with 50 mM Tris-HCl (pH 7.5) containing 25 mM NaCl and 1 mM EDTA. Protein eluted in a single peak at ∼0.2 M using a 25 mM to 1 M NaCl gradient. Pooled samples were dialyzed against 50 mM Tris-HCl (pH 7.5) containing 100 mM NaCl and 1 mM EDTA and concentrated to 15 to 40 mg/ml using Amicon Ultra 10,000-molecular-weight-cutoff (MWCO) concentrators (Millipore). The purity of the final sample was assessed with 10% Tris-Tricine SDS-PAGE gels stained with Coomassie blue.
DsbC (−sp) was purified using a 1-ml Hi-Trap Q FF anion-exchange column (Amersham Biosciences) equilibrated with 50 mM bis-Tris-HCl (pH 6.5). Protein eluted as a single peak at 0.2 M in a 0 to 1 M NaCl gradient. Pooled fractions were dialyzed against 50 mM bis-Tris-HCl (pH 6.5) with 100 mM NaCl and concentrated to 7 mg/ml using Amicon Ultra concentrators (Millipore). Sample purity was assessed with a 10% Tris-Tricine SDS-PAGE gel stained with Coomassie blue.
Limited trypsin proteolysis.
Trypsin (25 μg/ml in protein sample buffer: 50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) was added to TrbB (1.0 mg/ml in protein sample buffer) at a 1:250 trypsin-to-TrbB molar ratio. Three-microliter aliquots were removed at 1, 2, 5, 10, 20, and 30 min, and proteolysis was quenched by addition of 1 μl of 100 mM phenylmethylsulfonyl fluoride (PMSF). Samples were analyzed using a 10% Tris-Tricine SDS-PAGE gel with Coomassie blue staining. The molecular weight of the single stable fragment was determined using a Voyager DE-STR matrix-assisted laser desorption ionization-time of flight mass spectrometer (MALDI-TOF MS) (Applied Biosystems) at the Johns Hopkins University Mass Spectrometry Facility.
Mating assays.
Mating assays were performed as described previously (62) with some modifications. Briefly, strain JC3272 or MB899 containing various pDSW204 or derivative constructs was used as donor and MB322 was used as recipient (Table 1). Donor cells were grown in LB broth containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) (LB-Amp/Chlor) while recipients were grown in LB containing nalidixic acid (15 μg/ml) (LB-Nal). Cells were grown and mated at 30°C due to the temperature sensitivity of plasmid R27 (24, 56). Following mating, cells were plated on LB-Amp/Chlor to select for donors and LB agar containing nalidixic acid (34 μg/ml) and tetracycline (25 μg/ml) (LB-Nal/Tet) to select for transconjugants. Mating efficiencies are reported as the number of transconjugants per donor cell.
AppA assay for acid phosphatase activity.
Acid phosphatase activity was assayed using paranitrophenyl phosphate (pNPP) (Sigma N2765) as described previously (5). Reactions were stopped after 15 min by addition of 200 μl of 5 N NaOH. Data are presented as relative activity (percent), which was calculated by setting the mean activity of the wild-type bacteria in each experiment to 100%.
Insulin assay for reductase activity.
Disulfide reductase activity of TrbB was examined using 1 mg/ml (172 μM) insulin as substrate as previously described (31) with slight modifications. Each experiment was run in the presence of 2.5 μM assayed protein unless otherwise noted. Light scattering was monitored at 600 nm using a Tecan Infinite M200 plate reader equilibrated at 25°C. Absorbances were recorded every 60 s until an A650 of 0.15 was reached. Curves presented in this work are the means of five individual reactions.
PhoA assay to determine thiol-oxidase activity.
In vivo thiol-oxidase activity was measured by reactivation of periplasmic alkaline phosphatase PhoA in ΔdsbA E. coli. Overnight cultures grown in LB broth at 37°C were diluted 1:100 into M63 medium supplemented with 0.2% glycerol and 1% LB. Cells were grown to an A600 of 0.3, and expression of phoA from pBAD33 was induced by addition of l-arabinose to 0.2%. After 3 h of growth, 1-ml culture aliquots were centrifuged at 14,000 rpm for 1 min. Pellets were resuspended in BugBuster protein extraction reagent (Novagen) (10 μl per A600 of 0.1). Cell lysate was cleared by centrifugation at 14,000 rpm for 1 min. Four microliters of cleared lysate was added to 100 μl of 25 mM pNPP in 1.0 M Tris-HCl, pH 8.0, with 5 mM magnesium chloride and 0.5 mM zinc acetate. Hydrolysis of pNPP was measured as the change in A410 as a function of time. Absorbances were measured once every 4 min for 15 h on a Tecan Infinite M200 plate reader equilibrated at 37°C. Data were plotted as A410 versus time and fit by nonlinear regression of a straight line to calculate the rate of pNPP hydrolysis.
AMS trapping.
The in vivo redox state of FLAG-tagged TrbB was analyzed using alkylation by 4-acetamido-4′-maleimidylstilbene-2-2′-disulfonic acid (AMS; Molecular Probes) as described previously (33). A reduced control was made by incubating resuspended cells in 100 mM dithiothreitol (DTT) on ice for 1 h prior to trichloroacetic acid (TCA) precipitation. An oxidized control was prepared by performing a reaction in the absence of AMS. Following TCA precipitation and AMS incubation, thiol modification by AMS was quenched by adding 5 μl freshly prepared 2× SDS loading dye (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.001% bromophenol blue) (42). Samples were resolved using 15% Tris-glycine SDS-PAGE gels (19:1 bis-acrylamide) run at 20 mA for 2.5 h. TrbB bands were visualized by immunoblot analysis using a monoclonal anti-FLAG antibody (Sigma F3165), a goat anti-mouse IgG-IgM alkaline phosphatase (AP)-conjugated antibody (Applied Biosystems T2192), and the CDP-Star chemiluminescent substrate system (Applied Biosystems T2305).
Phylogenetic analysis of thioredoxin-like proteins from enterobacteria.
Protein sequences of thioredoxin-like proteins from enterobacteria were obtained from GenBank (http://www.ncbi.nlm/nih/gov/GenBank) using the search terms disulfide isomerase, thiol-disulfide interchange protein, and protein disulfide oxidoreductase. Protein sequence alignments were created with Mega 5.1 (http://www.megasoftware.net/index.html) using ClustalW (http://www.ebi.ac.uk/Tools/clustalw/index.html) with a pairwise alignment method. Maximum likelihood phylogenetic trees were built with Mega 5.1 using a bootstrap number of 1,000.
Analytical ultracentrifugation.
Sedimentation equilibrium analytical ultracentrifugation experiments were performed on a Beckman Optima XL-I analytical ultracentrifuge with an AN-60Ti rotor (Beckman Instruments, Fullerton, CA). Cells with 12-mm six-channel charcoal-filled Epon centerpieces and sapphire windows were used. Buffer density was calculated from its composition using SEDNTERP (43). Partial specific volume of TrbB was calculated from its amino acid composition according to the method of Cohn and Edsall (14) as implemented in SEDNTERP. Calculated partial specific volume of TrbB is 0.7389 ml/g. TrbB solutions at concentrations corresponding to A280 of 0.4, 0.7, and 1 for a 1.2-cm path length (0.375 mg/ml, 0.657 mg/ml, and 0.734 mg/ml, respectively) were centrifuged at rotor speeds of 28,000 and 31,000 rpm at 20°C. Absorbance data were collected at 280 nm. Equilibrium at each speed was assumed when successive scans taken 2 h apart were unchanged. Data were analyzed using the Origin-based data analysis software for Beckman XL-A/XL-I (MicroCal Software Inc., Northampton, MA).
RESULTS
TrbB can rescue mating deficiency of R27-ΔdsbCR27 in ΔdsbC E. coli.
F plasmid mating efficiency is reduced 10-fold when both dsbCE.coli and trbB are absent in the donor cell (21). This decrease is fully rescued by complementation in trans with either DsbCE.coli or TrbB, suggesting that TrbB can function as a disulfide isomerase, at least in the context of F plasmid transfer. To further examine TrbB's ability to act as a disulfide isomerase, we used ΔdsbCR27 R27 in a ΔdsbCE.coli host strain. DsbCR27, a protein encoded by the IncH1 plasmid R27, shares 35% identity with DsbCE.coli and little sequence identity with F TrbB and is believed to function as a disulfide isomerase (21). Similar to what was seen in the F system, individual disruption of either dsbCR27 (MB872) or dsbCE.coli (MB898) yields only minor effects (Table 3). When both genes are disrupted, however, mating efficiency falls 10,000-fold (compare MB790 and MB899; Table 3). The double mutant provides an excellent model to assess the function and specificity of DsbCE.coli, DsbCR27, and TrbB, as the range of observed transfer efficiencies, 4 orders of magnitude, allows for a clearer quantitative analysis than does the single order of magnitude assessed by studies using plasmid F (21).
Table 3.
Effect of dsbCE.coli, dsbCR27, and trbB complementation on the mating efficiency of R27 in a ΔdsbCR27 ΔdsbCE.coli double mutant background
| Donor | Genotype | Mating efficiency (no. of transconjugants/donor) | Relative efficiency |
|---|---|---|---|
| MB790 | Wild type | (1.11 ± 0.14) × 10−1 (n=6) | 1 |
| MB872 | ΔdsbCR27 | (1.93 ± 0.20) × 10−2 (n=6) | 0.2 |
| MB898 | ΔdsbCE.coli | (2.88 ± 0.89) × 10−1 (n=6) | 2.6 |
| MB899 | ΔdsbCR27 ΔdsbCE.coli | (6.34 ± 1.05) × 10−6 (n=4) | 0.00006 |
| MB904 | MB899 + dsbCE.coli | (2.90 ± 1.36) × 10−2 (n=3) | 0.26 |
| MB900 | MB899 + dsbCR27 | (6.25 ± 1.41) × 10−3 (n=4) | 0.06 |
| CH37 | MB899 + trbB | (1.76 ± 0.03) × 10−4 (n=8) | 0.002 |
| CH121 | MB899 + trxA | (1.21 ± 0.19) × 10−5 (n=7) | 0.0001 |
Complementation of a ΔdsbCE.coli ΔdsbCR27 R27 donor (MB899) with DsbCE.coli (MB904) increases mating efficiency ∼5,000-fold, to 25% of the wild-type value (Table 3). This result is the first demonstration that DsbCE.coli can successfully complement a dsbCR27 knockout in R27. Complementation with DsbCR27 (MB900) increases mating efficiency nearly as well, by ∼1,000-fold. Complementation in trans with TrbB (CH37) is less successful but still significant, causing a 30-fold increase in transfer efficiency over the ΔdsbCE.coli ΔdsbCR27 donor. Complementation with periplasmically localized thioredoxin A (TrxA) (CH121) results in only a 2% increase in mating efficiency, demonstrating that merely expressing a thioredoxin-like protein does not promote proper plasmid transfer.
Despite the comparatively modest activity of TrbB, these results suggest that TrbB can act similarly to DsbCE.coli and, in the context of R27, can be used to partially complement the disulfide isomerase-like protein, DsbCR27. The failure of TrbB to restore R27 mating to wild-type levels might be explained by an inherent partial specificity of TrbB for its F-plasmid-encoded substrates or by the fact that TrbB merely exhibits lower activity. To ensure that the partial complementation by TrbB was not due to lower concentrations of proteins in the periplasm, matings were performed in the presence of 1 mM IPTG, which causes an ∼6-fold increase in the expression level of the complementing gene. Mating efficiencies were unaffected in these experiments (data not shown), suggesting that these results are not biased by the periplasmic concentration of the proteins.
TrbB can restore AppA activity in vivo.
To assess the in vivo disulfide isomerase activity of TrbB, AppA activities were monitored in strains lacking dsbCE.coli. AppA is a periplasmic acid phosphatase that contains a disulfide bond between nonconsecutive cysteines when properly folded and therefore depends on a disulfide isomerase to assume its natively folded state. In the absence of a disulfide isomerase, a significant percentage of AppA molecules will contain an errant disulfide linkage, rendering the protein inactive (5). Functional AppA is able to dephosphorylate the substrate paranitrophenyl phosphate, and the resulting accumulation of the paranitrophenolate species can be quantitatively analyzed in solution. Complementation of a system lacking dsbC with a disulfide isomerase leads to enhanced AppA activity resulting from proper AppA folding, thus enabling an indirect analysis of disulfide isomerase activity in vivo.
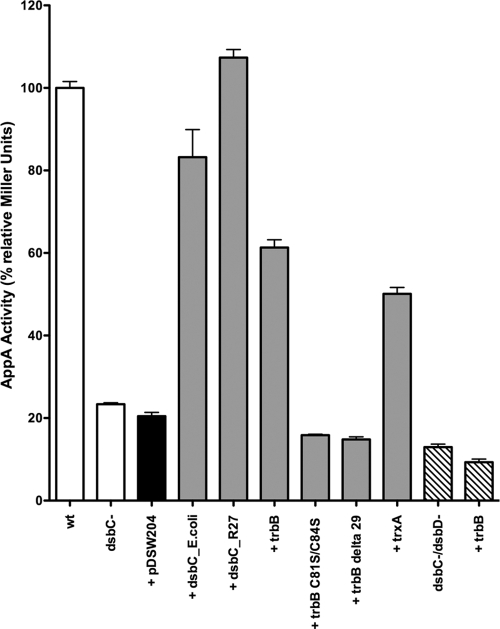
Data were normalized to the AppA activity in the E. coli strain DHB4, which contains AppA expressed from the plasmid pBAD33 under the control of its arabinose-inducible promoter (MB614) (Fig. 3). In ΔdsbCE.coli DHB4 (MB838), activity fell to ∼20% of the wild-type level, consistent with previous results (5). Complementation of MB838 with an empty vector (MB721) resulted in no significant change. Complementation with DsbCE.coli (MB637), however, increased AppA activity to 80% of wild-type levels, and complementation with TrbB (CH44) resulted in 60% of wild-type activity. The TrbB increase is consistent with mating results and further supports the hypothesis that TrbB functions as a disulfide isomerase. Complementation with DsbCR27 increases AppA function to levels higher than those in the wild-type cell, further solidifying its role as a disulfide isomerase in the R27 system. Complementation of MB838 with periplasmically localized thioredoxin A (TrxA) results in a level of AppA activity close to 50% of the wild-type level (Fig. 3, lane 9).
Fig. 3.
Examination of disulfide isomerase activity by AppA pNPP hydrolysis. AppA requires a disulfide isomerase to fold properly; therefore, pNPP hydrolysis is an indirect measure of disulfide isomerase activity. AppA activity is shown for a ΔdsbCE.coli mutant (black bar) complemented with an empty plasmid (pDSW204) or plasmids encoding disulfide isomerase DsbCE.coli, putative disulfide isomerases DsbCR27 and TrbB, TrbB N-terminal truncation mutant TrbBΔ29, TrbB active site mutant TrbB C81S/C84S, and reductase TrxA (gray bars). AppA activity in a ΔdsbCE.coli ΔdsbD double mutant strain, with or without TrbB, was also assessed (striped bars). DHB4 (wild-type [wt]) activity was set to 100%.
TrbB activity requires the active site cysteines and the N terminus.
To confirm that the disulfide isomerase activity exhibited by TrbB is due to specific redox activity, the AppA assay was repeated using an active site mutant. Cys81 and Cys84, the Cys residues in the TrbB C-X-X-C motif, were replaced with Ser. Complementing MB838 with TrbB C81S/C84S (CH46) yielded 16% of wild-type AppA activity levels, comparable to the empty vector. This result confirms that the activity of TrbB observed in the AppA assay is mediated by its C-X-X-C redox active site.
As part of a structural characterization of TrbB, a stable, N-terminal truncation mutant, TrbBΔ29, was isolated following limited trypsin proteolysis (C. W. Hemmis and J. F. Schildbach, unpublished results). The N-terminal TrbB “arm” appears to be fairly unstructured (see Fig. S2 in the supplemental material), and its removal has no effect on the overall structure of the remaining TrbB protein, based on a comparison of TrbB and TrbBΔ29 heteronuclear single-quantum coherence (HSQC) spectra (data not shown). In the AppA assay, complementation with periplasmically targeted TrbBΔ29 (CH50) results in 15% of wild-type activity (Fig. 3), equivalent to the negative control. These results indicate that the N-terminal arm is also required for function.
TrbB is not an efficient disulfide reductase in vitro.
Disulfide isomerization is the culmination of several steps, including reduction of the aberrant disulfide bond, refolding of the substrate, and oxidation to form the new disulfide linkage (58). To explore the difference in activities of TrbB and DsbCE.coli in the AppA assay, we examined their abilities to catalyze the reduction of a disulfide bond. In vitro reduction of the two disulfide bonds that link α- and β-chains of insulin causes the individual chains to aggregate, allowing relative disulfide reductase activity to be followed via increasing light scattering. Because this assay measures enzymatic activity indirectly, it provides only a qualitative assessment but nevertheless allows for comparison of the activities.
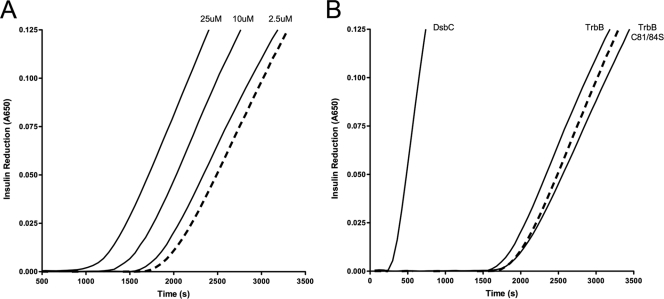
As seen in Fig. 4A, TrbB disulfide reductase activity correlates with TrbB concentration. Increased light scattering over time was observed in the negative control (dashed line) due to the presence of 1.0 μM DTT in the reaction cocktail. Addition of 2.5 μM TrbB (70:1 substrate/enzyme molar ratio) causes a slight decrease in the time required to detect light scattering, and increasing TrbB concentration to 10 and 25 μM TrbB reduces time further, indicating higher rates of disulfide reduction.
Fig. 4.
Monitoring disulfide reductase activity through insulin reduction. Comparison of rates of insulin reduction as visualized by light scattering at 600 nm. Unless otherwise noted, 2.5 μM protein was added to assay solution containing 172 μM insulin. The reaction was initiated by addition of 10 μl of 100 mM dithiothreitol, and light scattering was recorded every 60 s until an absorbance of 0.125 was reached. Residual reduction of disulfide bonds is seen in the negative control (dashed line) due to the presence of DTT in the reaction cocktail. (A) Concentration dependence of reductase activity displayed by TrbB. (B) Comparison of reductase activities of TrbB, TrbB C81S/C84S, and DsbCE.coli.
In a separate experiment, we compared the disulfide reductase activity of wild-type TrbB to that of TrbB mutant C81S/C84S and the disulfide bond isomerase DsbCE.coli (Fig. 4B). In the presence of 2.5 μM DsbCE.coli, the time required to detect light scattering was significantly reduced while the rate of increase in light scattering was enhanced. For comparison, previous work has shown that TrxA and DsbCE.coli have nearly identical insulin reduction profiles (64). As previously described, 2.5 μM TrbB causes only a small shift from the negative control. In contrast, 2.5 μM TrbB C81S/C84S produces a curve that is slightly delayed in comparison to that of the negative control, illustrating the essential role of the active site cysteines in the reductase activity of TrbB. Similar results were obtained with 2.5 μM bovine serum albumin (BSA) (data not shown), indicating that this shift is not protein specific. These results demonstrate that TrbB is clearly able to reduce disulfide bonds but that it catalyzes this reaction in insulin at levels lower than that of DsbCE.coli.
TrbB does not exhibit oxidase activity in vivo.
If TrbB is active and maintained in a reduced state as hypothesized, it should be unable to act as an oxidase in vivo on the model substrate PhoA. PhoA is a periplasmic alkaline phosphatase dependent upon the thiol-oxidase DsbA for proper folding and therefore function. Wild-type cells expressing phoA from pBAD33 catalyze hydrolysis of pNPP at a rate well above that of a negative control that includes only reaction buffer (Table 4). Removal of dsbA from the wild-type background reduces pNPP hydrolysis nearly to background levels, while complementation with TrbB increases specific pNPP hydrolysis only slightly. Neither of these rates is statistically significantly different from the background rate. Because this method is sensitive to even weak levels of oxidase activity (22), these results indicate that under physiological conditions, the redox state of the C-X-X-C active site is tightly controlled, and therefore, TrbB cannot act as a thiol-oxidase in vivo.
Table 4.
Reactivation of PhoA in the presence of TrbB
| Genotype | PhoA activity (ΔA410/h) | Normalized activity (%) |
|---|---|---|
| Wild type | (42.4 ± 1.0) × 10−4 | 100.0 |
| ΔdsbA | (9.9 ± 0.4) × 10−4 | 3.0 |
| ΔdsbA + trbB | (10.7 ± 0.2) × 10−4 | 5.4 |
| Blank | (8.9 ± 1.0) × 10−4 | 0.0 |
The C-X-X-C active site of TrbB is maintained in a reduced state by DsbD.
As redox processes of thioredoxin-like proteins are largely dictated by the redox state of their active site, we determined the in vivo redox state of the TrbB C-X-X-C active site motif using AMS alkylation studies. Chemical modification by AMS covalently links an ∼500-Da maleimide group to free thiols, causing a shift in electrophoretic mobility in the thiol-containing protein. Following AMS treatment, we observe an ∼1-kDa shift in the molecular mass of TrbB, relative to unmodified TrbB, on a 15% Tris-glycine SDS-PAGE gel (Fig. 5). This shift is identical to that seen in the reduced control that was prepared by incubation with DTT prior to AMS modification. These results indicate that TrbB is kept in the reduced state in a wild-type background (CH60). AMS alkylation was also performed in a dsbD mutant (CH61) background. DsbD is the inner membrane protein responsible for maintaining E. coli disulfide isomerases in a reduced state. In a DsbD deletion strain, TrbB is found in its oxidized state, implying that TrbB requires DsbD activity to maintain its reduced state in the periplasm.
Fig. 5.
AMS alkylation to analyze in vivo redox state of TrbB. First lane, oxidized TrbB control (ox); second lane, reduced TrbB control (red); third lane, TrbB in wild-type bacteria (wt); fourth lane, TrbB in dsbD mutant bacteria. TCA-precipitated whole-cell samples were reacted with AMS and visualized by immunoblot staining. TrbB experiences a 1-kDa increase in molecular mass when reduced in vivo.
Disulfide isomerase activity is lost in a dsbD mutant background.
In the absence of DsbD, TrbB accumulates in an oxidized state. How does this affect TrbB activity? As previously mentioned, in a ΔdsbC but dsbD+ strain (MB721), background levels of AppA activity fell to ∼20% of wild-type values (Fig. 3). When, in the ΔdsbC background, the dsbD gene is replaced with a kanamycin cassette (CH80), background levels of AppA activity fall to 13% of wild-type activity. Complementation in trans with TrbB (CH81) has little effect, with the strain showing 9% of wild-type activity (Fig. 3). Therefore, in the absence of DsbD, oxidized TrbB does not influence AppA function.
Phylogenetic analysis reveals distinct clustering of TrbB-like proteins.
As a further assessment of whether TrbB has unique characteristics relative to other disulfide isomerases, we performed a phylogenetic analysis. Protein sequences of thioredoxin-like proteins annotated as disulfide isomerases, thiol-disulfide interchange proteins, or protein disulfide oxidoreductases from enterobacteria were obtained from GenBank, yielding 3,639 “unique” sequences. Although these search terms retrieved a large number of accurately annotated protein sequences, our analysis may inadvertently exclude relevant thioredoxin-like proteins that have not been annotated or have been annotated incorrectly. This list was further refined by removing duplicates, any sequences lacking a C-X-X-C redox active site motif or an N-terminal methionine, and those sequences containing more than 300 or fewer than 60 residues. The resulting 511 sequences were randomized, separated into groups of 50, and aligned using the ClustalW algorithm. Maximum likelihood trees were then built with a bootstrap number of 1,000 in Mega 5.1 using sets of 50 or 100 aligned sequences. To limit the number of sequences used in the analysis, we used an iterative process in which sequences were removed from clusters in which a large number of protein sequences aligned with >90% identity. Using this method, the number of sequences was limited to 277. Eight known TrbB-like family members along with DsbCE.coli and DsbCR27 were then removed from this list, and the remaining 267 sequences were randomized and sorted into three groups of roughly 100 sequences. The 10 removed sequences were then added to each of these lists, and the analysis was repeated as described previously.
Each of the resulting trees (see Fig. S1 in the supplemental material) was analyzed to determine how TrbB clustered among other thioredoxin-like family members. Overall, protein sequences were found to cluster in families indicative of function, while TrbB was found to cluster separately and distinctly from other thioredoxin-like families. Clusters can be categorized as DsbA-like, DsbB-like, DsbC-like, DsbG-like, CcmG-like, or TrbB-like, with a limited number of sequences falling outside these clusters.
The distinct clustering of TrbB-like family members provides additional support for the hypothesis that TrbB belongs to a unique family of disulfide isomerases. In addition, it was found that each of the proteins clustered in the TrbB-like family is plasmid based. Those family members not found on a mobile plasmid are instead located in plasmid transfer regions that have been incorporated into the bacterial genome, presumably through a recombination event.
Secondary structure predictions of DsbCE.coli, DsbCR27, and TrbB were made using Phyre (40). Comparison of DsbCE.coli and DsbCR27 reveals high conservation (see Fig. S2 in the supplemental material), which correlates with similar clustering in the phylogenetic analyses (Fig. S1). By aligning the C-X-X-C redox active sites of DsbCE.coli, DsbCR27, and TrbB (Fig. S2), we find that the thioredoxin-like fold is similar, yet TrbB lacks an inserted alpha-helical domain present in DsbA, DsbC, and DsbG-like families (Fig. S2). In addition, the N-terminal region of TrbB is much smaller than and differs from both DsbCE.coli and DsbCR27, suggesting that it may not be a dimerization domain, as is the case in typical disulfide isomerases.
TrbB does not form homodimers.
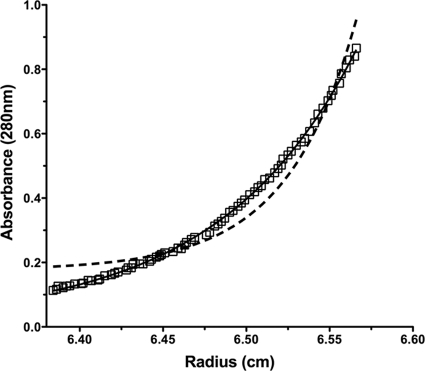
Sedimentation equilibrium ultracentrifugation was utilized to determine the oligomeric state of TrbB (−sp). Figure 6 shows representative sedimentation equilibrium data collected at 28,000 rpm. The resulting data were compared to curves generated by constraining the molecular mass to either monomeric or dimeric TrbB species. The raw experimental data align well with the curve representing an ideal monomeric species, indicating that TrbB is monomeric and does not detectably form homodimers under these conditions.
Fig. 6.
Analytical ultracentrifugation of TrbB (−sp). Sedimentation equilibrium data were collected at 28,000 and 31,000 rpm and monitored at 280 nm. Representative data collected at 28,000 rpm are shown here. Using Ultrascan software, the molecular mass was constrained to represent 100% monomeric (solid line) or dimeric (dashed line) TrbB species. The raw experimental data (open squares) fit those of the curve representing the modeled ideal TrbB monomer, indicating that TrbB exists in a monomeric state in solution.
DISCUSSION
TrbB displays disulfide isomerase activity.
Predicted members of the thioredoxin-like superfamily are conserved in F-like plasmid systems, suggesting that their role in plasmid transfer is integral. Transfer of a ΔtrbB F plasmid is unaffected under standard laboratory conditions in wild-type bacteria. However, when the trbB deletion is coupled with a DsbCE.coli knockout, mating efficiency is reduced 10-fold (21). In this work, we aimed to further elucidate TrbB function and to determine how TrbB's redox active site is maintained in the cell.
Our studies of TrbB function in plasmid transfer extend previous work by Elton et al. (21) to the IncH1 plasmid R27. We have shown here that TrbB partially restores R27 transfer efficiency in a ΔdsbCR27 ΔdsbCE.coli background (Table 3). TrbB function was also examined outside the context of plasmid transfer for the first time. Results indicate that TrbB is able to reduce disulfide bonds in insulin in vitro. TrbB can also partially restore function of the disulfide bond isomerase-dependent protein, AppA, an activity that requires both the TrbB C-X-X-C redox active site motif and its N-terminal “arm” (Fig. 3). TrbB exhibits lower activity than DsbCE.coli in these assays, possibly the result of a partial TrbB substrate specificity for its F-plasmid-encoded substrates or, alternatively, a lower catalytic efficiency of TrbB.
TrbB is not a periplasmic reductase.
Recent evidence indicates that disulfide isomerization can be achieved through reduction/oxidation cycling (58). Because TrbB is maintained in the reduced state in vivo (Fig. 5), it is possible that TrbB is a periplasmic reductase instead of a disulfide isomerase. If so, the relatively low TrbB activity in the AppA and mating assays could be attributed to an inefficient reduction/oxidation cycling mechanism. Some disulfide reductases consist of only a single thioredoxin-like domain, including the cytoplasmic protein thioredoxin (32, 37) and the periplasmic protein CcmG (DsbE) from E. coli (20). This is an intriguing observation considering the monomeric state of TrbB. The disulfide reductase activity of TrbB was analyzed using insulin from bovine pancreas as a substrate (Fig. 4). TrbB does exhibit disulfide reductase activity, but it is not significant in comparison to DsbCE.coli and, by extension, TrxA. Additional experiments utilizing periplasmically localized TrxA have shown that periplasmic expression of TrxA can stimulate disulfide isomerization through reduction-oxidation cycling (Fig. 3). TrxA, however, is unable to promote proper plasmid transfer (Table 3), indicating a distinct functional difference between TrxA and TrbB.
Disulfide bond isomerases, which typically exist as homodimers, generally contain distinct N-terminal dimerization domains (4, 46, 63). Structural alignments with homodimeric thioredoxin family members such as DsbCE.coli and DsbCR27 (see Fig. S2 in the supplemental material) suggest that TrbB-like proteins lack this dimerization domain. Analytical ultracentrifugation studies indicate that TrbB does not dimerize (Fig. 6) under the conditions tested, indicating that the N terminus is both structurally and functionally different from the N termini in DsbCE.coli-like disulfide isomerases.
Although TrbB does not have the canonical dimerization domain of a disulfide isomerase, the functional studies presented here support the conclusion that TrbB is a disulfide isomerase rather than a disulfide reductase. Our results then lead to the proposal that TrbB from plasmid F is a member of a previously unrecognized group of disulfide isomerases that also includes TrbB from plasmids R100 and pSLT.
Putative substrates.
The conservation of disulfide isomerases in conjugative plasmids suggests that at least one plasmid protein requires a disulfide isomerase to properly fold or to maintain its correct redox state. Of the 108 proteins encoded in plasmid F, only a few are predicted to be exported to the periplasm. Of these, only TraH (6 Cys residues), TraU (11 Cys residues), and TraN (22 Cys residues) contain more than 2 Cys residues. TraH, TraU, and TraN all have homologs in plasmid R27 (Table 5), consistent with their putative roles as TrbB substrates. Surprisingly, TrhU from plasmid R27, which is 32% identical to TraU from plasmid F and contains 16 Cys residues, does not have a predicted N-terminal signal sequence, suggesting that it may not be exported to the periplasm.
Table 5.
Comparison of periplasmic F plasmid proteins containing two or more cysteine residues with homologs from plasmid R27
| Plasmid F | No. of Cys residues | Plasmid R27 | No. of Cys residues | % identical | % similar |
|---|---|---|---|---|---|
| TraB | 2 | TrhB | 2 | 30 | 50 |
| TraH | 6 | TrhH | 6 | 26 | 44 |
| TraN | 22 | TrhN | 32 | 27 | 60 |
| TraU | 11 | TrhU | 16 | 32 | 51 |
Recent work by Arutyunov et al. suggests that TraH is not significantly affected in the absence of TrbB (2), but these experiments were performed in E. coli strain MC4100, which contains DsbCE.coli. Since the mating-deficient phenotype of a ΔtrbB plasmid in ΔdsbCE.coli bacteria is rescued by complementation with either TrbB or DsbCE.coli, a requirement of TraH for TrbB might be masked by the presence of DsbCE.coli. For this reason, we cannot rule out the possibility that TraH is a TrbB substrate. There are currently no data available on the remaining putative substrates. Interestingly, a yeast two-hybrid screen performed by Harris and Silverman (28) identified a putative TrbB/TraW interaction in plasmid F. TraW is a periplasmic protein essential to proper F plasmid transfer that contains only a single Cys residue. Since TraW cannot form an intramolecular disulfide linkage, it would not be a suitable substrate for disulfide bond isomerization unless it forms an intermolecular disulfide bond. Depuydt et al., however, recently discovered that in addition to isomerization, certain proteins exhibiting disulfide isomerase activity are able to protect single, unpaired Cys residues from oxidation (19). In the case of TraW and TraB, the latter an F-plasmid-encoded protein that contains 2 Cys residues (Table 5), TrbB may be required to maintain single Cys residues to preserve their functional states.
Conclusions.
The work presented here supports the hypothesis that F TrbB is a disulfide isomerase that is maintained in its reduced and functional state by DsbD. In addition to disulfide isomerization, TrbB may also be required for the maintenance of unpaired cysteine residues in certain substrates. Interestingly, we discovered that the N-terminal domain of TrbB, which is predicted to be largely unstructured when analyzed by both PONDR (45, 52, 53) and Phyre (40), is absolutely required for proper function, although it does not act as a dimerization domain under conditions tested. Previous work has suggested that the dimerization domain and a conserved α-helical linker that is missing from TrbB are required for proper disulfide isomerase function (3, 55, 60). The TrbB-like family of disulfide isomerases, which includes, among others, TrbB from plasmids F, R100, and pSLT (Fig. 1), seems to be unique both functionally and structurally and does not fit well into the definition of typical disulfide isomerases. The discovery of this family emphasizes the need for further investigation into their structures, mechanisms, and roles in the cell.
We have also shown in this work that on each of the substrates examined, TrbB exhibits lower levels of enzymatic activity than does DsbCE.coli. This difference in enzymatic activity may merely be the result of TrbB being less efficient at performing the redox chemistries required for proper disulfide isomerization. Alternatively, this difference could be explained by a partial specificity of TrbB for its substrates. The latter possibility, coupled with the predicted differences in structure and an apparent absence of dimerization in TrbB, might indicate a fundamental difference in substrate recognition between these two proteins.
Our results lay the foundation for future work investigating the mechanism by which DsbD regulates the redox active site of TrbB. Additional studies will also focus on identifying those F-plasmid-encoded substrates that require a disulfide isomerase for proper folding and function and environmental conditions that may lead to a specific requirement of TrbB for proper plasmid transfer.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported, in whole or in part, by National Institutes of Health grant GM61017 (to J.F.S.).
We specially acknowledge M. Bolisetty for invaluable help with the phylogenetic analyses.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Achtman M., Willetts N., Clark A. J. 1971. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J. Bacteriol. 106:529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arutyunov D., Arenson B., Manchak J., Frost L. S. 2010. F plasmid TraF and TraH are components of an outer membrane complex involved in conjugation. J. Bacteriol. 192:1730–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bader M. W., et al. 2001. Turning a disulfide isomerase into an oxidase: DsbC mutants that imitate DsbA. EMBO J. 20:1555–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banaszak K., Mechin I., Frost G., Rypniewski W. 2004. Structure of the reduced disulfide-bond isomerase DsbC from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 60:1747–1752 [DOI] [PubMed] [Google Scholar]
- 5. Berkmen M., Boyd D., Beckwith J. 2005. The nonconsecutive disulfide bond of Escherichia coli phytase (AppA) renders it dependent on the protein-disulfide isomerase, DsbC. J. Biol. Chem. 280:11387–11394 [DOI] [PubMed] [Google Scholar]
- 6. Bessette P. H., Aslund F., Beckwith J., Georgiou G. 1999. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. U. S. A. 96:13703–13708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyd D., Manoil C., Beckwith J. 1987. Determinants of membrane protein topology. Proc. Natl. Acad. Sci. U. S. A. 84:8525–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brandon L. D., Goldberg M. B. 2001. Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. J. Bacteriol. 183:951–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buddelmeijer N., Beckwith J. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52:1315–1327 [DOI] [PubMed] [Google Scholar]
- 10. Chivers P. T., Prehoda K. E., Raines R. T. 1997. The CXXC motif: a rheostat in the active site. Biochemistry 36:4061–4066 [DOI] [PubMed] [Google Scholar]
- 11. Cho S.-H. H., Porat A., Ye J., Beckwith J. 2007. Redox-active cysteines of a membrane electron transporter DsbD show dual compartment accessibility. EMBO J. 26:3509–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christie P. J., Vogel J. P. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen S. N., Chang A. C., Hsu L. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. U. S. A. 69:2110–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cohn E. J., Edsall J. T. 1943. Proteins, amino acids and peptides as ions and dipolar ions. Reinhold, New York, NY [Google Scholar]
- 15. Cooper T. F. 2007. Recombination speeds adaptation by reducing competition between beneficial mutations in populations of Escherichia coli. PLoS Biol. 5:e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Darby N. J., Creighton T. E. 1995. Catalytic mechanism of DsbA and its comparison with that of protein disulfide isomerase. Biochemistry 34:3576–3587 [DOI] [PubMed] [Google Scholar]
- 17. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de la Cruz F., Frost L. S., Meyer R. J., Zechner E. L. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34:18–40 [DOI] [PubMed] [Google Scholar]
- 19. Depuydt M., et al. 2009. A periplasmic reducing system protects single cysteine residues from oxidation. Science 326:1109–1111 [DOI] [PubMed] [Google Scholar]
- 20. Edeling M. A., Guddat L. W., Fabianek R. A., Thöny-Meyer L., Martin J. L. 2002. Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment. Structure 10:973–979 [DOI] [PubMed] [Google Scholar]
- 21. Elton T. C., Holland S. J., Frost L. S., Hazes B. 2005. F-like type IV secretion systems encode proteins with thioredoxin folds that are putative DsbC homologues. J. Bacteriol. 187:8267–8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eser M., Masip L., Kadokura H., Georgiou G., Beckwith J. 2009. Disulfide bond formation by exported glutaredoxin indicates glutathione's presence in the E. coli periplasm. Proc. Natl. Acad. Sci. U. S. A. 106:1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Firth N., et al. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377–2401 In Neidhardt F. C., et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 24. Forns N., Baños R. C., Balsalobre C., Juárez A., Madrid C. 2005. Temperature-dependent conjugative transfer of R27: role of chromosome- and plasmid-encoded Hha and H-NS proteins. J. Bacteriol. 187:3950–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fronzes R., Christie P. J., Waksman G. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7:703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frost L. S., Ippen-Ihler K., Skurray R. A. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harris R. L., Silverman P. M. 2004. Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J. Bacteriol. 186:5480–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hiniker A., Bardwell J. C. A. 2003. Disulfide bond isomerization in prokaryotes. Biochemistry 42:1179–1185 [DOI] [PubMed] [Google Scholar]
- 30. Hiniker A., Collet J.-F. F., Bardwell J. C. A. 2005. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J. Biol. Chem. 280:33785–33791 [DOI] [PubMed] [Google Scholar]
- 31. Holmgren A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254:9627–9632 [PubMed] [Google Scholar]
- 32. Holmgren A., Söderberg B. O., Eklund H., Brändén C. I. 1975. Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 A resolution. Proc. Natl. Acad. Sci. U. S. A. 72:2305–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Inaba K., Ito K. 2002. Paradoxical redox properties of DsbB and DsbA in the protein disulfide-introducing reaction cascade. EMBO J. 21:2646–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ito K., Inaba K. 2008. The disulfide bond formation (Dsb) system. Curr. Opin. Struct. Biol. 18:450–458 [DOI] [PubMed] [Google Scholar]
- 35. Kadokura H., Beckwith J. 2009. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell 138:1164–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kadokura H., Katzen F., Beckwith J. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72:111–135 [DOI] [PubMed] [Google Scholar]
- 37. Katti S. K., LeMaster D. M., Eklund H. 1990. Crystal structure of thioredoxin from Escherichia coli at 1.68 A resolution. J. Mol. Biol. 212:167–184 [DOI] [PubMed] [Google Scholar]
- 38. Katzen F., Beckwith J. 2000. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103:769–779 [DOI] [PubMed] [Google Scholar]
- 39. Katzen F., Beckwith J. 2003. Role and location of the unusual redox-active cysteines in the hydrophobic domain of the transmembrane electron transporter DsbD. Proc. Natl. Acad. Sci. U. S. A. 100:10471–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kelley L. A., Sternberg M. J. E. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 41. Kumarasamy K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 43. Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90–125 In Harding S. E., Rowe A. J., Horton J. C. (ed.), Analytical ultracentrifugation in biochemistry and polymer science. The Royal Society of Chemistry, Cambridge, United Kingdom [Google Scholar]
- 44. Lawley T. D., Klimke W. A., Gubbins M. J., Frost L. S. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1–15 [DOI] [PubMed] [Google Scholar]
- 45. Li X., Romero P., Rani M., Dunker A. K., Obradovic Z. 1999. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 10:30–40 [PubMed] [Google Scholar]
- 46. McCarthy A. A., et al. 2000. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat. Struct. Biol. 7:196–199 [DOI] [PubMed] [Google Scholar]
- 47. Missiakas D., Georgopoulos C., Raina S. 1994. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 13:2013–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakamoto H., Bardwell J. C. A. 2004. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim. Biophys. Acta 1694:111–119 [DOI] [PubMed] [Google Scholar]
- 49. Quan S., Schneider I., Pan J., Von Hacht A., Bardwell J. C. A. 2007. The CXXC motif is more than a redox rheostat. J. Biol. Chem. 282:28823–28833 [DOI] [PubMed] [Google Scholar]
- 50. Ren G., et al. 2009. Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J. Biol. Chem. 284:10150–10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rietsch A., Bessette P., Georgiou G., Beckwith J. 1997. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 179:6602–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Romero P. Z., Obradovic Z., Dunker A. K. 1997. Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform. Ser. Workshop Genome Inform. 8:110–124 [PubMed] [Google Scholar]
- 53. Romero P., et al. 2001. Sequence complexity of disordered protein. Proteins 42:38–48 [DOI] [PubMed] [Google Scholar]
- 54. Schägger H., von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 55. Segatori L., et al. 2006. Conserved role of the linker alpha-helix of the bacterial disulfide isomerase DsbC in the avoidance of misoxidation by DsbB. J. Biol. Chem. 281:4911–4919 [DOI] [PubMed] [Google Scholar]
- 56. Sherburne C. K., et al. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shevchik V. E., Condemine G., Robert-Baudouy J. 1994. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. EMBO J. 13:2007–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shouldice S. R., et al. 2010. In vivo oxidative protein folding can be facilitated by oxidation-reduction cycling. Mol. Microbiol. 75:13–28 [DOI] [PubMed] [Google Scholar]
- 59. Stewart E. J., Katzen F., Beckwith J. 1999. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J. 18:5963–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sun X. X., Wang C. C. 2000. The N-terminal sequence (residues 1-65) is essential for dimerization, activities, and peptide binding of Escherichia coli DsbC. J. Biol. Chem. 275:22743–22749 [DOI] [PubMed] [Google Scholar]
- 61. Weiss D. S., Chen J. C., Ghigo J. M., Boyd D., Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Williams S. L., Schildbach J. F. 2006. Examination of an inverted repeat within the F factor origin of transfer: context dependence of F TraI relaxase DNA specificity. Nucleic Acids Res. 34:426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yeh S. M., Koon N., Squire C., Metcalf P. 2007. Structures of the dimerization domains of the Escherichia coli disulfide-bond isomerase enzymes DsbC and DsbG. Acta Crystallogr. D Biol. Crystallogr. 63:465–471 [DOI] [PubMed] [Google Scholar]
- 64. Zhao Z., Peng Y., Hao S. F., Zeng Z. H., Wang C. C. 2003. Dimerization by domain hybridization bestows chaperone and isomerase activities. J. Biol. Chem. 278:43292–43298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.