Abstract
We present the complete genome sequence and proteogenomic map for Acholeplasma laidlawii PG-8A (class Mollicutes, order Acholeplasmatales, family Acholeplasmataceae). The genome of A. laidlawii is represented by a single 1,496,992-bp circular chromosome with an average G+C content of 31 mol%. This is the longest genome among the Mollicutes with a known nucleotide sequence. It contains genes of polymerase type I, SOS response, and signal transduction systems, as well as RNA regulatory elements, riboswitches, and T boxes. This demonstrates a significant capability for the regulation of gene expression and mutagenic response to stress. Acholeplasma laidlawii and phytoplasmas are the only Mollicutes known to use the universal genetic code, in which UGA is a stop codon. Within the Mollicutes group, only the sterol-nonrequiring Acholeplasma has the capacity to synthesize saturated fatty acids de novo. Proteomic data were used in the primary annotation of the genome, validating expression of many predicted proteins. We also detected posttranslational modifications of A. laidlawii proteins: phosphorylation and acylation. Seventy-four candidate phosphorylated proteins were found: 16 candidates are proteins unique to A. laidlawii, and 11 of them are surface-anchored or integral membrane proteins, which implies the presence of active signaling pathways. Among 20 acylated proteins, 14 contained palmitic chains, and six contained stearic chains. No residue of linoleic or oleic acid was observed. Acylated proteins were components of mainly sugar and inorganic ion transport systems and were surface-anchored proteins with unknown functions.
INTRODUCTION
Mollicutes are a class of microorganisms which have the smallest known genome sizes among autonomously replicating organisms, the smallest one being Mycoplasma genitalium (8). The genomic nucleotide sequence of the latter was among the first bacterial genome sequenced, in the mid-1990s (13), and was the first one artificially synthesized and cloned as a yeast artificial chromosome (15). Moreover, the first artificial chromosome transplanted to another species was the Mycoplasma mycoides genome (4). Mycoplasmas, together with the Bacillus/Clostridium group, form the Firmicutes phylum, and almost all of them have small genome sizes, absolutely or partially require externally supplied sterols, have low GC contents, and demonstrate high pheno- and genotypic variation. A distinctive feature of the Mollicutes is the absence of the cell wall, as well as pronounced metabolic dependence on external sources (culture medium, host cells, etc.) (41).
An important group of the Mollicutes is the phytoplasmas, which are phytopathogens notable for high genome plasticity caused by numerous repetitive elements. This allows them to easily shuffle adhesion and virulence factors and hence to infect a broad range of host organisms (2). While nearly 80 Mollicutes genomes have been sequenced so far, a genomic nucleotide sequence of a representative of the Acholeplasmataceae family has not yet been characterized. In contrast to the well-studied Mycoplasmataceae family, the acholeplasmas have relatively large genomes of 1.5 to 1.8 Mbp. In addition, unlike other mycoplasmas, they do not require sterols for cultivation and are able to synthesize fatty acids from precursors (47).
Acholeplasma laidlawii is the best-studied organism of this family. It was isolated from wastewaters in 1936 by Laidlaw and Elford (26) and was among the first mycoplasmas successfully cultivated on an artificial growth medium (42). One peculiarity of Acholeplasma laidlawii is the presence of NADH oxidase, a flavin mononucleotide (FMN)-containing membrane enzyme able to catalyze electron transfer from reduced NAD to oxygen, generating hydrogen peroxide and other active forms of oxygen (39). The plasmatic membrane of acholeplasmas is pigmented. Its main pigment contains neurosporene-C40, a linear carotenoid that is a precursor of lycopene and other carotenoid pigments with cyclic groups (32). A. laidlawii synthesizes neurosporene from acetate; it has a complete set of enzymes from the carotenoid synthesis pathway. A. laidlawii can infect the plant Vinca minor L., with phytopathogenic effects analogous to the phytoplasma infection (32). It has been suggested that the acholeplasmas are evolutionary ancestors of the phytoplasmas that have evolved by further degenerative evolution (2).
We report the complete genome sequence of A. laidlawii PG-8A and its annotated gene complement, which we augmented using proteomic techniques (18). We further characterize posttranslation modifications (acylation and phosphorylation), which may play a role in the cell's function.
MATERIALS AND METHODS
Acholeplasma laidlawii and DNA isolation.
A. laidlawii PG8-A was cultured in the modified Edward medium as described in reference 11. DNA was isolated by proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation.
Sequencing strategy.
Shotgun libraries were constructed as follows: 10 mg of DNA was sheared using a nebulizer (Invitrogen) to produce DNA fragments of 2 kb and 4 kb on average. The sheared DNA was loaded on a 0.7% agarose gel, and DNA fractions corresponding to 2 to 2.5 kb and 4 to 4.5 kb, respectively, were extracted from the agarose gel. Size-selected fragments were cloned into the pCR4Blunt-Topo vector using the TOPO shotgun subcloning kit (Invitrogen). They were then introduced into Escherichia coli TOP10 and sequenced with the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems). Sequence quality assessment and subsequent assembly were performed with Phred (12), LUCY (7), TIGR Assembler (56), and BAMBUS (40). To close gaps, custom primers were designed near the ends of the contigs, and PCRs were performed with chromosomal template DNA. Sequences were obtained from PCR products that spanned the gaps. The sequence coverage of both strands was 10×. For the 10× coverage of each strand, the error rate was 0.36%, and we made 45.4 reads/kb.
Genome annotation.
An initial set of open reading frames (ORFs) likely to encode proteins was identified by Artemis (46). Predicted ORFs longer than 100 codons (300 nucleotides) were searched using BLASTP (1) against the nonredundant protein database at the National Center for Biotechnology Information and then manually annotated based on protein homology. Manual annotation was performed using ad hoc software developed with Oracle Express Edition (Oracle). Orthologs were defined using the bidirectional best-hit criterion (38). Translational start codons were corrected by inspecting BLASTP alignments. TMHMM (23) and HMMTOP (59) servers were used to identify transmembrane domains. Glimmer (48) and GipsyGene (36) tools were used to identify candidate genes without known homologs. Glimmer, GipsyGene, and comparison with the phytoplasma genomes AY-WB (2) and OY-M (37) were used to identify short protein-coding genes (<100 codons). Regions of possible frameshifts and errors were identified by visual inspection for interrupted or truncated genes. Several frameshifts identified in the initial genome sequence were corrected, and the remaining ones were confirmed by resequencing. Finally, each gene was functionally classified by assigning a cluster of orthologous group (COG) number (57) using ad hoc COG classification software. rRNAs and tRNAs were identified by BLASTN (1) and tRNA-Scan-SE (27), respectively. Riboswitches and T boxes were identified by the RNA pattern software (62). The origin of replication was identified using the analysis of GC-skew by GraphDNA (58) and with a search for candidate DnaA boxes (51). The circular representation of the genome was plotted using the GenomeViz software (14). Metabolic reconstruction was done using KEGG (http://www.genome.jp/kegg/pathway.html).
SDS-PAGE of A. laidlawii.
Proteins of A. laidlawii, solubilized by boiling in sample buffer, were separated by SDS-PAGE gels consisting of 7.5% T or 16.5% T and 2.6% C (% T, gel acrylamide concentration; % C, degree of cross-linking within the polyacrylamide gel), according to the Laemmli method (25). The gels were fixed (20% C2H5OH and 10% CH3COOH) and stained with Coomassie G-250 dye.
Two-dimensional PAGE.
Prior to two-dimensional (2-D) PAGE, cells were treated with the nuclease mix and antiprotease cocktail (Amersham Bioscience). Cell pellets (10 μl) were dissolved in the buffer of 8 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonia]-1-propanesulfonate (CHAPS), 2% (wt/vol) NP-40, 1% Triton X-100, and 2% ampholytes (Bio-Rad) (pH range, 3 to 10), and 80 mM dithiothreitol (DTT). The protein concentration was determined using the Quick Start Bradford dye reagent (Bio-Rad). For gel zooming, the Rotofor liquid prefractioning system (Bio-Rad) was used according to the manufacturer's protocol. The total cell lysate in the isoelectric focusing (IEF) solution was separated by the Rotofor system into 10 fractions. Each fraction was subjected to standard two-dimensional protein separation. The first-dimension separation was performed using tube gels (20 cm by 1.5 mm) containing carrier ampholytes and applying a voltage gradient in the IEF chamber Protean II XL cell (Bio-Rad). Isoelectric focusing was performed in the following mode: 100, 200, 300, 400, 500, and 600 V for 45 min; 700 V for 10 h; and 900 V for 1 h. After the first dimension, the ejected tube gels were incubated in the equilibration buffer (125 mM Tris-HCl, 40% [wt/vol] glycerol, 3% [wt/vol] SDS, 65 mM DTT, pH 6.8) for 30 min. The tube gels were placed onto the SDS-PAGE gels consisting of 7.5% T or 16.5% T and 2.6% C, were run using a 20-by-20-cm format (Protean II Multi-Cell; Bio-Rad), and were fixed using 0.9% (wt/vol) agarose containing 0.01% (wt/vol) bromphenol blue. Electrophoresis was performed in Tris-glycine buffer under cooling in the following mode: 20 mA on glass for 20 min, 40 mA on glass for 2 h, and 35 mA on glass for 2.5 h under chamber cooling to 10°C.
Gel staining and detection of proteins.
The gels were fixed and silver stained as described previously (52). For specific phosphoprotein staining, the gels were fixed in two steps in 500 ml of the fixation solution (50% methanol and 10% acetic acid). The first fixation step was carried out for 60 min, with the second step lasting overnight. The gels were washed three times with 500 ml of double-distilled water (ddH2O), for 15 min every wash, in gentle agitation (50 rpm). Once the gels were washed, they were incubated with 500 ml Pro-Q Diamond phosphoprotein stain (Molecular Probes/Invitrogen) in the dark for 2 h and destained with 500 ml of destaining solution (20% acetonitrile, 50 mM sodium acetate, pH 4), followed by three changes, 30 min per wash, in the dark with gentle agitation. An image was acquired on the Typhoon Trio scanner (Amersham Biosciences) with a 532-nm laser excitation and a 555-nm band-pass emission filter.
For specific glycoprotein staining, the gels were fixed in two steps in 500 ml of fixation solution (50% methanol and 5% acetic acid). The first fixation step was carried out for 60 min, with the second step lasting overnight. The gels were washed three times with 500 ml of 3% glacial acetic acid in ddH2O, for 10 min every wash, in gentle agitation (50 rpm). To oxidize the carbohydrates, the gels were incubated for 1 h with 500 ml of oxidizing solution (Molecular Probes/Invitrogen). Then, the gels were washed three times with 500 ml of 3% glacial acetic acid in ddH2O, for 10 min every wash. Once the gels were washed, they were incubated with 500 ml Pro-Q Emerald 488 staining solution (Molecular Probes/Invitrogen) while gently agitating in the dark for 2.5 h and were washed three times with 500 ml of 3% glacial acetic acid in ddH2O, for 15 min every wash, in gentle agitation. An image was obtained on the Typhoon Trio scanner (Amersham Biosciences) with a 510-nm laser excitation and a 520-nm band-pass emission filter.
The image analysis was performed using the PDQest software (Bio-Rad). All spots were extracted for matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis.
Trypsin digestion and mass spectrometry.
The protein bands/spots after 1-D or 2-D PAGE were subjected to trypsin in-gel hydrolysis mainly as described in reference 21. Gel pieces of 1 mm3 were excised and washed twice with 100 μl of 0.1 M NH4HCO3 (pH 7.5) and 40% acetonitrile mixture for 30 min at 37°C and then dehydrated with 100 μl of acetonitrile and air dried. Then, they were treated with 3 μl of 12 mg/ml solution of trypsin (Promega) in 50 mM ammonium bicarbonate for 12 h at 37°C. Peptides were extracted with 6 μl of 0.5% trifluoroacetic acid water solution for 30 min.
MALDI analysis.
Aliquots (1 μl) from the sample were mixed on a steel target with 0.3 μl of 2,5-dihydroxybenzoic acid (Aldrich) solution (10 mg ml−1 in 30% acetonitrile and 0.5% trifluoroacetic acid), and the droplet was left to dry at room temperature. Mass spectra were recorded on the Ultraflex II MALDI-TOF/TOF mass spectrometer (Bruker Daltonik, Germany), equipped with the Nd laser. MH+ molecular ions were measured in the reflector mode, and the accuracy of the mass peak measurement was 0.007%.
Fragment ion spectra were generated by laser-induced dissociation slightly accelerated by low-energy collision-induced dissociation using helium as a collision gas. The accuracy of the fragment ion mass peak measurement was 1 Da. Matching of the tandem mass spectrometry (MS-MS) fragments to proteins was performed using the Biotools software (Bruker Daltonik, Germany) and the Mascot MS-MS ion search.
Protein identification was carried out with a peptide fingerprint search against the NCBI A. laidlawii protein database using the Mascot software (Matrix Science Inc.). One missed cleavage of Met oxidation and Cys-propionamide per peptide each was permitted. Protein scores greater than 44 were assumed to be significant (P < 0.05).
The LC-ESI-MS analysis.
The liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) analysis (of tryptic peptides after 1-D SDS-PAGE separation of proteins) was performed on the Agilent 1100 series LC/MSD Trap (Agilent Technologies), equipped with a Zorbax 300-SB C18 column and nano-ESI source. The elution conditions consisted of a 0.3-μl/min 20-min ablution by 5% solvent B (80% acetonitrile, 20% water, 0.1% formic acid), a 50-min gradient of 5 to 60% solvent B in solvent A, and then a 20-min gradient of 60 to 90% solvent B in solvent A (0.1% formic acid-water solution). MH1+ to MH3+ ions were detected in the range of 200 to 2,200 m/z optimized to 800. MS-MS spectra were obtained automatically for all perceptible MS signals. The accuracy of the mass peak measurement was 0.5 Da. Protein identification was carried out by an MS-MS ion search using the Mascot software as described above. Protein scores greater than 36 were assumed to be significant. To make a deeper inventory of A. laidlawii proteins, we used different variants of 2-D electrophoresis followed by one- or two-dimensional chromate mass spectrometry. The gel track was divided into approximately 50 parts, extracting both densely and slightly stained zones. Peptide extracts after the tryptic hydrolysis of complex protein mixes were separated by ion-exchange and/or by reverse-phase C18 high-performance liquid chromatography (HPLC) and sent to acquire the MS-MS spectra. The data were analyzed by Mascot as described above. Data validation, integration, and comparison with the sequenced genome were done by an ad hoc software system, allowing for screening for misannotated proteins using the MS-MS data.
Additional screening for phosphopeptides was done by the X!Tandem software package (9) with potential phospho-S/T/Y modification.
The validation of scoring by both algorithms was done by searching a meaningless database constructed by a reverse reading of the initial database.
Incorporation of exogenous 14C-labeled fatty acids into protein of A. laidlawii in vivo.
To label proteins, 74 kBq of either palmitic acid (16:0), oleic acid (18:1), stearic acid (18:0), or linoleic acid (18:2c) (Amersham Biosciense) was added per ml of growth medium. Exponentially growing cells were collected by centrifugation at 15,000 × g for 15 min at 4°C and were washed twice in buffer containing 150 mM NaCl, 50 mM Tris, and 2 mM MgCl2, pH 7.4. Proteins from these cells were resolved by 2-D PAGE and silver stained. The gels were dried between two cellophane sheets and exposed to a storage phosphor screen (Amersham Bioscience) for 2 weeks. The image was obtained on the Typhoon scanner (Amersham Biosciences).
Nucleotide sequence accession number.
The complete genomic nucleotide sequence of Acholeplasma laidlawii PG8-A has been deposited in the GenBank database under accession number CP000896.
RESULTS
General genomic features.
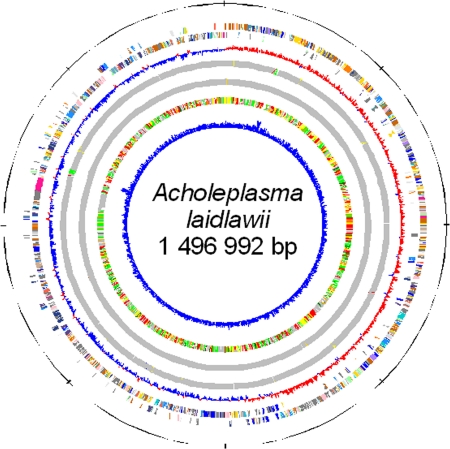
The genome of A. laidlawii is represented by a single 1,496,992-bp circular chromosome (Fig. 1). This is the longest genome among the Mollicutes with a known nucleotide sequence (before sequencing the A. laidlawii genome, the largest genome was Mycoplasma penetrans) (49). The genome contains two rRNA gene operons (16S-23S-5S), 34 tRNA genes, and 1,380 predicted ORFs. Table 1 represents the general A. laidlawii genomic features compared to genomes of phytoplasmas onion yellows phytoplasma strain M (OY-M) (37) and aster yellows phytoplasma strain witches' broom (AY-WB) (2).
Fig. 1.
Circular representation of the A. laidlawii PG-8A genome. Circles 1 and 2 (counting from the outer side), the predicted coding sequences on the plus and minus strands, respectively, colored according to COG functional categories: translation, ribosomal structure, and biogenesis (gold); RNA processing and modification (orange); transcription (dark orange); DNA replication, recombination, and repair (maroon); cell division and chromosome partitioning (yellow); defense mechanisms (light pink); signal transduction mechanisms (purple); cell envelope biogenesis, outer membrane (peach); cell motility and secretion (medium purple); intracellular trafficking, secretion, and vesicular transport (dark pink); posttranslational modification, protein turnover, chaperones (light green); energy production and conversion (lavender); carbohydrate transport and metabolism (blue); amino acid transport and metabolism (red); nucleotide transport and metabolism (green); coenzyme metabolism (light blue); lipid metabolism (cyan); inorganic ion transport and metabolism (dark purple); secondary metabolites biosynthesis, transport, and catabolism (sea green); general function prediction only (light gray); function unknown (ivory); not in COGs (dark gray). Circle 3, GC skew ([G − C]/[G + C] using 1-kb sliding window); circle 4, tRNAs (green), rRNAs (red), T boxes (yellow), and riboswitches (blue); circle 5, IS elements (gold), groups of repeated sequences (the members of the same group are shown by the same color); circle 6, best-hit organisms, compared (BLASTP search) against the nonredundant database of previously sequenced genomes: Acholeplasmatales (yellow), Mycoplasmatales (pink), Bacillales (red), Lactobacillales (orange), Clostridiales (green), Thermoanaerobacteriales (dark green), all others displayed in gray; circle 7, G+C content (using 1-kb sliding window).
Table 1.
General features of the chromosomes of A. laidlawii, Phytoplasma AY-WB, and Phytoplasma OY-M
| Feature | Value |
||
|---|---|---|---|
| A. laidlawii PG-8A | AY-WB | OY-M | |
| Size (bp) | 1,496,992 | 706,57 | 860,631 |
| G+C content (%) | 31 | 27 | 28 |
| No. of CDSs | 1,380 | 671 | 754 |
| With predicted function | 1,003 | 450 | 446 |
| Conserved hypothetical | 62 | 149 | 51 |
| Hypothetical | 315 | 72 | 257 |
| Coding density (%) | 90 | 72 | 73 |
| Avg gene size (bp) | 984 | 779 | 785 |
| No. of: | |||
| rRNA operons (16S-23S-5S) | 2 | 2 | 2 |
| tRNAs | 34 | 31 | 32 |
UGA was used as a stop codon for ORF prediction. This conforms to previous reports that acholeplasmas and phytoplasmas use UGA as a stop codon, unlike the SEM branch Mollicutes (Mycoplasmatales and Entomoplasmatales), where this codon encodes tryptophan (43). The average total content of guanine (G) and cytosine (C) in the A. laidlawii chromosome sequence is 31%. Unlike phytoplasmas, which have plasmids (four in AY, two in OY), A. laidlawii has no plasmids.
The predicted products of A. laidlawii protein-coding sequences (CDSs) were categorized according to function and compared with the Phytoplasma species and with a model firmicute, Bacillus subtilis (Table 2).
Table 2.
Functional analysis of predicted proteins among A. laidlawii, B. subtilis, and Phytoplasma sp. strains AY-WB and OY-M
| COG |
Proteins among: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
A. laidlawii PG-8A |
B. subtilis |
AY-WB |
OY-M |
||||||
| Code | Description | No. | % | No. | % | No. | % | No. | % |
| J | Translation, ribosomal structure, biogenesis | 133 | 12.25 | 151 | 3.70 | 105 | 27.27 | 106 | 22.70 |
| K | Transcription | 69 | 6.35 | 276 | 6.70 | 18 | 4.68 | 28 | 6.00 |
| L | Replication, recombination, repair | 94 | 8.66 | 134 | 3.30 | 74 | 19.22 | 114 | 24.41 |
| D | Cell cycle control, cell division, chromosome partitioning | 8 | 0.74 | 32 | 0.80 | 4 | 1.04 | 4 | 0.86 |
| V | Defense mechanisms | 46 | 4.24 | 52 | 1.30 | 5 | 1.30 | 5 | 1.07 |
| T | Signal transduction mechanisms | 32 | 2.95 | 123 | 3.00 | 5 | 1.30 | 6 | 1.28 |
| M | Cell wall/membrane/envelope biogenesis | 32 | 2.95 | 161 | 3.90 | 5 | 1.30 | 7 | 1.50 |
| N | Cell motility | 4 | 0.37 | 97 | 2.40 | 0 | 0.00 | 0 | 0.00 |
| U | Intracellular trafficking, secretion, vesicular transport | 10 | 0.92 | 42 | 0.93 | 5 | 1.30 | 5 | 1.07 |
| O | Posttranslational modification, protein turnover, chaperones | 43 | 3.96 | 87 | 2.10 | 16 | 4.16 | 21 | 4.50 |
| C | Energy production and conversion | 54 | 4.97 | 163 | 4.00 | 13 | 3.38 | 14 | 3.00 |
| G | Carbohydrate transport and metabolism | 87 | 8.01 | 274 | 6.70 | 14 | 3.64 | 15 | 3.21 |
| E | Amino acid transport and metabolism | 76 | 7.00 | 294 | 7.10 | 19 | 4.94 | 31 | 6.64 |
| F | Nucleotide transport and metabolism | 37 | 3.41 | 81 | 2.00 | 19 | 4.94 | 23 | 4.93 |
| H | Coenzyme transport and metabolism | 26 | 2.39 | 109 | 2.70 | 6 | 1.56 | 9 | 1.93 |
| I | Lipid transport and metabolism | 41 | 3.78 | 85 | 2.10 | 8 | 2.08 | 9 | 1.93 |
| P | Inorganic ion transport and metabolism | 54 | 4.97 | 148 | 3.60 | 19 | 4.94 | 20 | 4.28 |
| Q | Secondary metabolite biosynthesis, transport, and catabolism | 12 | 1.10 | 129 | 3.10 | 2 | 0.52 | 2 | 0.43 |
| R | General function prediction only | 137 | 12.62 | 335 | 8.10 | 35 | 9.09 | 35 | 7.49 |
| S | Function unknown | 91 | 8.38 | 233 | 5.70 | 13 | 3.38 | 13 | 2.78 |
| (−) | No COG | 385 | 1,200 | 307 | 311 | ||||
As seen in Table 2, A. laidlawii has 133 gene products (12.25% of CDSs) involved in translation and 69 gene products (6.35% of CDSs) involved in transcription. Of all A. laidlawii CDSs, 385 were not categorized in the COG database. Of the latter, 282 (71%) were annotated as hypothetical proteins, as they had no homologs with known function, and 47 (12%) were characterized as integral membrane proteins.
OriC structure.
The A. laidlawii chromosome does not demonstrate a distinct GC-skew inversion, which in bacteria often corresponds to the origin of replication (oriC) (6). Another typical feature of oriC regions is the rnpA-rpmH-dnaA-dnaN-recF-gyrB locus (34). The presence of the recF gene distinguishes A. laidlawii from other Mollicutes with sequenced genomes.
As in many bacterial chromosomes, in the A. laidlawii chromosome, the direction of transcription changes between the rpmH and dnaA genes. This region contains nine candidate DnaA boxes: three TTATCCACA (one inverted), two TTtTCCACA (one inverted), one TTATtCACA, one TTATCaACA (inverted), one TTccCCACA, and one TTgcCACA (nonconsensus nucleotides are set in lowercase letters). The dnaA boxes are present near the replication origin, as demonstrated in M. genitalium, Mycoplasma pneumoniae, and Ureaplasma urealyticum (10, 13).
Mobile elements.
The A. laidlawii genome contains transposase genes of the types IS3 (ACL_0571 and ACL_0939, both containing in-frame stop codons), IS4 (ACL_0778), IS10 (ACL_0003), and IS150 (ACL_0782), a partial gene similar to the transposase N-terminal domain (ACL_0779), and XerD-like integrase genes (ACL_1160 and ACL_0584).
Regulation of transcription and translation.
A. laidlawii has three genes that encode σ factors from the ECF subfamily, compared to 14 in B. subtilis (3). On the other hand, other sequenced Mycoplasmatales, including M. penetrans, whose genome length is comparable to that of A. laidlawii, have only σ70. However, aster yellow phytoplasma has one chromosomal rpoD gene that encodes a standard σ70 factor of 465 amino acids and several copies of the sigF gene localized in potential mobile units (PMUs) and PMU-like loci (2).
A. laidlawii has a variety of transcription factors from the LacI (five), MarR (five), TetR (three), PadR (three), RpiR (one), and XRE (one) families. It also has two membrane-bound DNA-binding proteins, a TmrB-like factor, an iron-dependent repressor, and the heat shock repressor HrcA. In addition, it has three two-component signal transduction systems, one being incomplete (ACL_0010 and ACL_0011 with a CheY-like domain regulator, ACL_1298 and ACL_1297 with an OmpR family regulator, and ACL_1421, a LytR/AlgR-like regulator). The only other two-component signal transduction system observed in the Mollicutes are in M. penetrans (49), and they are not related to the A. laidlawii ones.
A. laidlawii is a second Mollicutes that has regulatory RNA structures, riboswitches, and T boxes. Riboswitches are structures that, upon binding small ligands, lead either to premature termination of transcription or to the inhibition of translation initiation (30, 63). Earlier guanine riboswitches were found in Mesoplasma florum (22). A. laidlawii has four predicted types of the riboswitches: a flavin mononucleotide (FMN)-dependent riboswitch, a thiamine pyrophosphate (TPP)-responsive riboswitch, a purine riboswitch, and a yybP-ykoY element (Table 3). One more RNA-based regulatory system in A. laidlawii is T boxes, a system of transcription termination control widely used by Gram-positive bacteria for the regulation of expression of aminoacyl-tRNA synthetase genes and other amino acid-related genes. The genome contains 19 T boxes upstream of genes encoding aminoacyl-tRNA synthetases, transporters (ABC type), and enzymes (tryptophan synthase, beta subunit) (Table 4). By comparison, no T boxes were found in the genomes of phytoplasmas.
Table 3.
Riboswitches predicted in A. laidlawii genome
| Riboswitch | Locus tag | Riboswitch |
Regulated gene |
||||
|---|---|---|---|---|---|---|---|
| Start position | End position | Start position | End position | Product | COG | ||
| FMN riboswitch | ACL_0401 | 421358 | 421470 | 421553 | 422281 | Integral membrane protein | COG3601 |
| TPP riboswitch | ACL_0834 | 869174 | 869264 | 868478 | 869104 | Putative proton-coupled thiamine transporter | COG3859 |
| Purine riboswitch | ACL_1370 | 1432561 | 1432632 | 1431225 | 1432475 | GTP-binding protein, HflX subfamily | COG2262 |
| yybP-ykoY element | ACL_0803 | 839365 | 839452 | 839473 | 842247 | Cation transporting ATPase | COG0474 |
Table 4.
Predicted T boxes in the A. laidlawii genome
| Locus tag | Gene's start position | Gene's end position | Product |
|---|---|---|---|
| ACL_0009 | 13187 | 14458 | Seryl-tRNA synthetase |
| ACL_0132 | 115135 | 116685 | Methionyl-tRNA synthetase |
| ACL_0249 | 248035 | 249048 | Phenylalanyl-tRNA synthetase alpha chain |
| ACL_0250 | 249048 | 251402 | Phenylalanyl-tRNA synthetase beta chain |
| ACL_0354 | 372064 | 373992 | Threonyl-tRNA synthetase |
| ACL_0540 | 575203 | 575763 | Acetyltransferase, GNAT family |
| ACL_0541 | 575735 | 578314 | Valyl-tRNA synthetase |
| ACL_0649 | 676648 | 677457 | ABC-type transport system, substrate-binding component |
| ACL_0650 | 677459 | 678136 | ABC-type transport system, permease component |
| ACL_0651 | 678126 | 678875 | ABC-type transport system, ATP-binding component |
| ACL_0702 | 732914 | 735604 | Isoleucine amino-acyl tRNA synthetase |
| ACL_0777 | 813255 | 814430 | Tryptophan synthase, beta subunit |
| ACL_0825 | 859075 | 859623 | Putative acetyltransferase, GNAT family |
| ACL_0824 | 857816 | 859078 | Histidyl-tRNA synthetase |
| ACL_0823 | 856110 | 857813 | Aspartyl-tRNA synthetase |
| ACL_0906 | 934989 | 937544 | Alanyl-tRNA synthetase |
| ACL_1182 | 1235584 | 1236588 | Tryptophanyl-tRNA synthetase |
| ACL_1185 | 1239231 | 1240463 | Tyrosyl-tRNA synthetase |
| ACL_1427 | 1490339 | 1491730 | Asparaginyl-tRNA synthetase |
Known mycoplasmal genomes contain 19 aminoacyl-tRNA synthetase genes, with glutaminyl-tRNA synthetase missing. This is also typical for B. subtilis and other Gram-positive bacteria where tRNAGlu is first charged with Gln by glutamyl-tRNA synthetase and the charged Gln is subsequently converted to Glu by glutamyl-tRNA aminotransferase. However, the A. laidlawii genome, like both phytoplasmas (AY and OY) and Clostridium spp. (35), does contain a glutaminyl-tRNA synthetase gene (ACL_1352) and lacks glutamyl-tRNA aminotransferase.
Metabolism.
It is well known that many Mollicutes have rather limited biosynthetic capabilities (13, 49, 60, 64). These are limited mainly to energy acquisition, with synthetic pathways being considerably reduced or absent. They lack the full di- and tricarbon acid cycles, possess minimal capabilities for amino acid synthesis, and lack de novo purine and pyrimidine synthesis. The metabolism of A. laidlawii is more complex.
Carbohydrate metabolism.
Like other Mollicutes, A. laidlawii has no di- and tricarbon acid cycles, and its only source of ATP, as in other fermenting Mollicutes (13, 49, 60, 64), is the glycolysis pathway completely represented in the genome. Unlike the Phytoplasma spp. (2, 37), A. laidlawii is able to ferment pyruvate into O-lactate and, through transformation of pyruvate into acetyl coenzyme A (acetyl-CoA), acetic acid. But unlike phytoplasmas, it is able to form acetyl-CoA, which is required for the carotenoid synthesis, from pyruvate.
The most significant distinctive feature of the sugar metabolism of A. laidlawii is the presence of the complete pentosophosphate pathway, absent in all other Mollicutes sequenced so far. The Entner-Doudoroff pathway is represented by both oxidative and nonoxidative branches. Apparently, the hexamonophosphate bypass in the glucose metabolism yields a high demand in reduced NADH required for the carotenoids and fatty acid biosynthesis pathways.
In addition to glucose as a source for carbon, A. laidlawii, like many other mycoplasmas, can catabolize d-fructo-1-phosphate, phosphor derivatives of mannose, N-acetylglucosamine, N-acetylmannosamine, and several other sugars and amino sugars (17). It also has glycogen-phosphorylase and alpha-amylase, allowing the bacterium to degrade starch to glucose-6-phosphate and metabolize cleavage products in the glycolysis. In other Mollicutes genomes, this metabolic pathway was not found, and the capability of Mollicutes to use starch as a carbon source was not known (17).
As reported before, some representatives of the Acholeplasma spp. are able to use not only glucose and fructose but also galactose as a starting point in the carbohydrate metabolism (53). Indeed, the genome contains UDP-glucose-4-epimerase (isomerizing UDP-galactose to UDP-glucose) and UDP-glucose-pyrophosphorylase (transforming UDP-glucose to glucose-1-phosphate, incoming into glycolysis). These metabolic pathways were not previously found in genome sequences of the Phytoplasma spp. (2, 37) and were present only in the genomes of two mycoplasmas with sequenced genomes: M. pneumoniae (10) and M. mycoides (64).
Biosynthesis and degradation of amino acids.
Unlike other Mollicutes, A. laidlawii has several enzymes for partial or complete de novo synthesis of some amino acids. In particular, it has all genes forming the pathway of the phenylalanine and tyrosine biosynthesis from phosphoenolpyruvate via chorismate and prephenate. In addition, A. laidlawii has several genes for the tryptophan biosynthesis from indole-3-glycerolphosphate via indole.
The genome contains NAD+ synthase and glutamine-dependent glutamate-dehydrogenase, providing for the synthesis of NAD+ and its reduction required in the biosynthetic processes involving NADH with formation of ammonia and 2-oxyglutarate.
A. laidlawii has several enzymes of the methionine metabolism, as do other Mollicutes (31). Among them is the metK1 gene for encoding S-adenosylmethionine (SAM) synthase, providing for the biosynthesis of SAM from l-methionine. It also has partial pathways of lysine biosynthesis from aspartic acid, l-ornithine from N-2-acetylornithine, and several other amino acids.
Cofactor and vitamin metabolism.
A. laidlawii gets most vitamins from the medium, again similar to other Mollicutes (31). Minor differences between A. laidlawii and other Mollicutes are the genes encoding several enzymes for the vitamin and cofactor synthesis, mainly the NAD+ synthesis from nicotinamide, the one-carbon pool synthesis by folate, and the coferment A biosynthesis from 4-phosphopantetheine. It also has flavin adenine dinucleotide (FAD)-synthase/riboflavin kinase, providing for the FAD synthesis from flavin mononucleotide. But, in general, A. laidlawii mainly takes up vitamins and cofactors in the same manner as other Mollicutes with smaller genome sizes do.
Carotenoid biosynthesis.
An important distinguishing feature of the A. laidlawii metabolism is its ability to synthesize carotenoids de novo from acetyl-CoA and acetoacetyl-CoA incoming from glycolysis (33). The A. laidlawii genome contains almost all enzymes of this pathway, including 3-hydroxy-3-methylglutaryl-CoA synthase, with one exception. So far, no gene of farnesyltranstransferase, providing for the biosynthesis of trans-geranylgeranyl diphosphate from farnesyl-diphosphate and isopentenyl diphosphate (the latter being the first intermediate product in the chain of neurosporene and lycopene), has been identified. All other enzymes of this biosynthetic pathway (trans-geranylgeranyl diphosphate-lycopene) (53) are encoded in the genome. It is likely that the farnesyltranstransferase activity is encoded by a unique A. laidlawii gene, having no known functionally characterized homologs in the sequenced genomes. An alternative is that one of the other enzymes has a broad specificity.
Further, A. laidlawii has enzymes of the undecaprenyl phosphate biosynthesis pathway from farnesyl diphosphate, including two reactions from the terpenoid synthesis pathway (farnesyl-diphosphate-trans,trans,cis-geranylgeranyl-diphosphate–di-trans,poly-cis–undecaprenyl diphosphate) and one reaction from the peptidoglycan biosynthesis pathway (poly-cis-undecaprenyl diphosphate-undecaprenyl phosphate). The presence of this pathway was unexpected, as it was known that eubacteria rarely use undecaprenyl phosphate in the cell wall (5).
Fatty acid biosynthesis.
The composition of the A. laidlawii cell membrane differs from that of other Mollicutes (33). The main components of its cytoplasmic membrane are glycolipids and Acholeplasma-specific lipoglycans, whereas other mycoplasmas have cholesterol as a major membrane component (33). Earlier reports showed that most Mollicutes do not have fatty acid synthesis pathways (11, 53), while the activity of enzymes from this metabolic pathway had been observed in A. laidlawii (33).
The functional annotation of the A. laidlawii genome identified enzymes from the fatty acids biosynthesis pathway, except for acyl-ACP-dehydrogenase, catalyzing the dehydration of enoyl-acyl-acyl-carrier protein derivatives with a carbon chain length of 4 to 16 and a reduction of NAD+ to NADH. Apparently, this function is performed by an unidentified protein. This metabolic pathway was never observed in the Mollicutes.
Glycerolipid, glycerophospholipid, and sphingolipid biosynthesis.
Only two enzymes from the glycerolipid biosynthesis pathways were identified, acetol kinase for ATP-dependent phosphorylation of glycerin to phosphoglycerin and 1,2-diacylglycerol 3-glycosyltransferase for carrying glycosyl residue from UDP-glucose to 1,2-diacylglycerol. These enzymes have not been observed in the Phytoplasma spp., and they are not connected to other metabolic pathways in A. laidlawii (2, 37). Hence, their presence does not allow for a proper glycerolipid biosynthesis. 1,2-Diacylglycerol-3-phosphate, a product of the reaction catalyzed by 1-acylglycerol-3-phosphate O-acyltransferase, is one of the initial substrates in the cardiolipin and phosphatidyl glycerophosphate synthesis. These biosynthetic pathways are complete in the A. laidlawii genome, while they have not been described in the Phytoplasma spp. (2, 37).
In addition, the A. laidlawii genome has a partial biosynthesis pathway for choline and glycerol-3-phosphate, which is absent in the Phytoplasma spp. and other Mollicutes (2, 10, 13, 37, 49, 60, 64).
The sphingolipid biosynthesis in A. laidlawii is represented by two copies of sphingosine kinase: phosphorylating sphingosine to sphingosine-1-phosphate. Sphingosine-1-phosphate is one of the cytoplasmic membrane components in A. laidlawii.
Nucleotide metabolism.
Mollicutes are unable to synthesize nucleotides de novo (31). The metabolism of purines and pyrimidines in A. laidlawii is similar to that of other Mollicutes, but there are several differences in the interconversion and degradation of nucleotides and nucleosides. In particular, the genome contains NADP+ oxidoreductase and ribonucleoside-triphosphate reductase, which is not found in the Phytoplasma genomes (2, 37). The genome contains genes encoding purine-nucleoside phosphorylase (transforming desoxyuridine to uracil), dCMP deaminase (converting dCMP to dUMP), cytidine deaminase (catalyzing transformation of deoxycytidine to deoxyuridine and cytidine to uridine), purine-nucleoside phosphorylase (cleaving purine nucleosides to purine and ribose or desoxyribose), and several other enzymes of the nucleotide metabolism.
Comparative genome analysis of A. laidlawii.
The A. laidlawii genome is the largest among all known Mollicutes genomes (138,359 bp longer then the genome of M. penetrans).
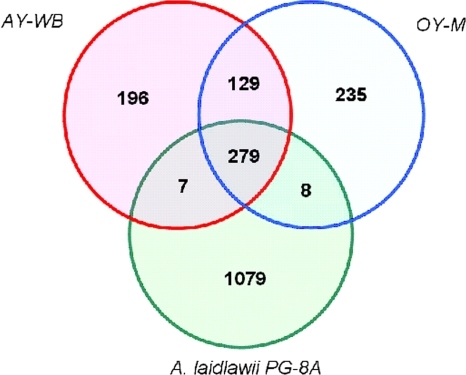
The Venn diagram in Fig. 2 shows that the overlap between the genomes of A. laidlawii and two closely related Phytoplasma species is not large: only 279 genes are common to all three genomes, while more than a thousand genes are specific for A. laidlawii. On the other hand, 560 genes, mainly hypothetical ones, are specific to the Phytoplasma genomes, and the majority of them again are genome specific.
Fig. 2.
Venn diagram of orthologous clusters in the A. laidlawii PG-8A, AY-WB, and OY-M genomes.
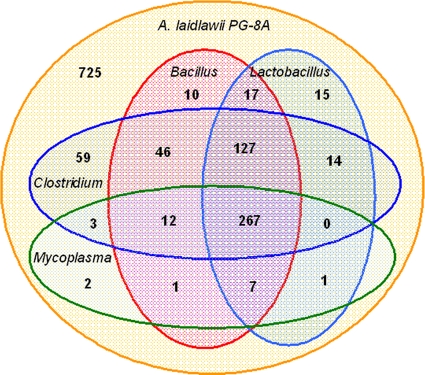
At a larger scale, we compared the A. laidlawii genome to all Bacillales, Clostridiales, Lactobacillales, and Mollicutes genomes (Fig. 3). Again, the number of A. laidlawii-specific genes is rather large, as only true orthologs and not paralogs were considered (see Materials and Methods, “Genome annotation”). A more interesting observation is the virtual absence of the Mollicutes signature: only two genes are common to A. laidlawii and the Mycoplasma spp. (ACL_0737 and ACL_0738, encoding hypothetical proteins). The reductive character of the Mycoplasma spp. is seen in a large number of genes common to A. laidlawii and the Bacillales and Clostridiales but absent from the Mycoplasma genomes. The Firmicutes core (the set of genes common to all three lineages, that is, the Mollicutes as represented by A. laidlawii, the Bacillales, and the Clostridiales) contains 387 genes. Accepting genes present in A. laidlawii and one of the latter lineages adds 197 genes (note that this does account for genes present in the Bacillales and Clostridiales but not in A. laidlawii, and thus the ancestral Firmicutes genome should be larger than 584 genes).
Fig. 3.
Venn diagram of orthologous rows of genes from of all Bacillus, Lactobacillus, Clostridium, and Mycoplasma sequenced genomes and A. laidlawii PG-8A. Bacillus, Lactobacillus, Clostridium, and Mycoplasmas indicate that a gene is present in, respectively, at least one Bacillus sp., Lactobacillus sp., Clostridium sp., and Mycoplasma sp. genome. This diagram takes into account only true orthologs, not homologs, hence the large number of acholeplasma-specific genes.
Proteomic profiling of A. laidlawii.
A. laidlawii is a universalist, adapting to various media and conditions. It is the only mycoplasma capable of living outside a host organism. It survives and reproduces in animals, plants, and wastewaters. Such a broad spectrum of environments requires regulatory switches and intersecting metabolic pathways, allowing for the fast and effective adaptation to changes in nutrition fluxes from the environment. The growth of A. laidlawii in an optimal medium should lead to a reduced functionality, requiring a minimal set of protein products.
We employed a combination of several proteome analysis methods to obtain the saturated proteome of A. laidlawii. It allowed us to identify not only major proteins but also a substantial number of low-copy-number proteins.
The application of 2-D gel electrophoresis with preliminary zooming and subsequent tryptic hydrolysis of separate protein spots in the gel led to the identification of 237 individual proteins (see Fig. S1 in the supplemental material). Most of them were major proteins, such as elements of the translation elongation system and glycolysis. Further, the 2-D map resolved a considerable number of membrane transport systems. Twenty proteins produced a series of more than three spots. Some of them were sequences of isoforms, similar in mass and different in pI (for instance, Tex and TpiA). Others proteins were different in both dimensions, like the Tig protein represented by three spots, with a monotonically decreasing mass and pI. This distribution was observed in each experiment, which is likely explained by intracellular protein proteolysis.
Even with liquid prefractionation and other methods of zooming, 2-D electrophoresis cannot provide a satisfactory resolution to identify low-copy-number proteins. To obtain a more complete inventory of the A. laidlawii proteome, various approaches to 1-D electrophoresis were applied (low-molecular-mass electrophoresis, electrophoresis of total lysate fractions separated by protein hydrophobicity) followed by one- or two-dimensional chromatography-mass spectrometry. The application of these complex approaches allowed us to identify 562 additional proteins that were absent in the 2-D map. Hence, the total number of identified A. laidlawii proteins reached 803.
This is equivalent to 58% of all annotated A. laidlawii proteins (Table 5). To characterize the functional distribution of proteins, COG (cluster of orthologous group) categories were used. Proteins were classified in 20 categories of functional activity (http://www.ncbi.nlm.nih.gov/COG/) (Table 5).
Table 5.
Number of proteins found in the proteome and genome in each of COG group
| Function of COG group | No. of proteins in: |
|
|---|---|---|
| Proteome | Genome | |
| Amino acid transport and metabolism | 55 | 70 |
| Carbohydrate transport and metabolism | 58 | 79 |
| Cell cycle control, cell division, chromosome partitioning | 6 | 7 |
| Cell motility | 1 | 2 |
| Cell wall/membrane/envelope biogenesis | 23 | 30 |
| Coenzyme transport and metabolism | 13 | 20 |
| Defense mechanisms | 28 | 44 |
| Energy production and conversion | 39 | 53 |
| Unknown | 169 | 504 |
| General function prediction only | 73 | 114 |
| Inorganic ion transport and metabolism | 23 | 45 |
| Intracellular trafficking, secretion, and vesicular transport | 5 | 9 |
| Lipid transport and metabolism | 29 | 40 |
| Nucleotide transport and metabolism | 31 | 37 |
| Posttranslational modification, protein turnover, chaperones | 36 | 41 |
| Replication, recombination, and repair | 65 | 98 |
| Secondary metabolites biosynthesis, transport, and catabolism | 5 | 6 |
| Signal transduction mechanisms | 20 | 26 |
| Transcription | 29 | 56 |
| Translation, ribosomal structure, and biogenesis | 95 | 111 |
As in most microorganisms, the functions of approximately one-third of proteins are unknown. The fraction of identified proteins among them is relatively low. Among proteins with a known function, the highest percentage of proteomic identification (91%) was observed in the “nucleotide transport and metabolism” group. The fact that not all proteins of the transcription and translation systems were found in the proteome could be due to the dispensability of the active regulation as well as deactivation of most reparative functions in a rich medium with optimal culture growth conditions, where these proteins are not needed.
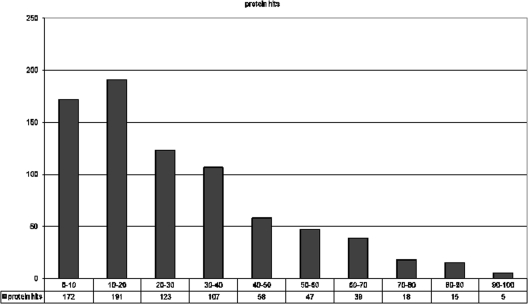
We observed peptides with lengths ranging from 6 to 40 amino acids (aa). For 803 detected proteins, the theoretical number of tryptic peptides is 12,254, out of which 3,078 peptides were experimentally observed (the total number of observed peptides is 4,999). We analyzed the reproducibility of the experiments using two-dimensional chromatography and observed no new peptide hits in gel zooming experiments. Thus, it seems that more than half of tryptic peptides could not be observed due to their physical-chemical parameters, such as solubility, hydrophobicity, and ionization ability. The average number of peptides per protein was 6.3. The protein coverage in total ORFs was 17.5%. Most proteins (191) had coverage ranging from 10 to 20% (Fig. 4).
Fig. 4.
Fraction of proteins with different percentages of amino acid sequence coverage. Each bar corresponds to a bin of the protein coverage (the first bar is for the coverage from 0 to 10%), and the height of a bar corresponds to the fraction of proteins in the bin.
N-terminal peptides were used to correct the start codon annotation of several genes (see Table S2 in the supplemental material). In the A. laidlawii genome, 55 genes have N-terminal codons different from AUG (for comparison, in E. coli this number is 320 [54], and in B. subtilis, 121 [44]). Overall, in A. laidlawii, N-terminal peptides comprise less than 3% of all possible tryptic peptides produced by theoretical trypsin digestion. Experimentally, we identified 56 N-terminal peptides with the score exceeding the threshold. This is less than 1% of all observed peptides, with conditions set so as to observe more than one peptide in the same band of a one-dimensional gel. To search for the potential misannotation of N-terminal amino acids, we examined the spectra for peptides with both start sites differing up to 10 aa from annotated ones and nontryptic cleavage at the N terminus. Setting the same conditions as for true peptides (scoring threshold and presence of two identified peptides per protein), we found 69 peptides whose genes, hence, are candidates for change in the start codon annotation.
In the optimal conditions, A. laidlawii expresses about 60% of all annotated proteins. The fraction is lower (37%) for highly hydrophobic membrane proteins and proteins of small mass and size, while it was 75% for membrane-associated proteins and 60% for cytoplasmic proteins. An additional analysis of RNA expression for missing proteins identified 3 to 5% of transcripts from a variety of categories. Most proteins not seen in the proteomic analysis but having expressed mRNA are hydrophobic (such as permease components of ABC transporters) or small (ribosomal proteins with mass less than 10 kDa) or belong to the low-copy-number stress response group.
Phosphorylation.
We characterized the phosphoproteome of the CHAPS-soluble A. laidlawii protein fraction. We applied the two-dimensional protein separation method followed by staining with the fluorescent dye Pro-Q Diamond for the identification of phosphorylated proteins and with Sypro Ruby for staining all separated proteins. The obtained gels were scanned in the Typhoon Trio scanner. Next, we performed a computer overlay of the images from the two channels using the ImageQuant software to identify proteins containing phosphoric acid groups. The MALDI mass spectrometry identified nine phosphorylated proteins, constituting 0.6% of the total A. laidlawii proteome (Table 6). However, this number may not be final since many proteins, which may be phosphorylated, are not expressed in an amount sufficient for identification by the available methods. Indeed, two-dimensional electrophoresis as a detection method has substantial limitations in protein pI and solubility. In particular, most membrane proteins do not enter the separation area. All proteins observed in the phosphorylated form have a calculated pI of less than 6. For comparison, currently E. coli has 79 identified phosphoproteins comprising 2% of its annotated proteins (29), and B. subtilis has 78 (1.9%) (30). At that, A. laidlawii has 4 candidate kinases, E. coli has 35, and B. subtilis has 44. Hence, there is some correlation between the number of protein kinases and the fraction of phosphorylated proteins.
Table 6.
Posttranslational protein modification in A. laidlawii
| Protein | Description | Scoreb | Mr | pI | Functiona |
|---|---|---|---|---|---|
| Phosphorylated | |||||
| GI:161986262 | speB putative agmatinase | 77 | 32,649 | 5.51 | Amino acid transport and metabolism |
| GI:161986348 | DHH domain protein | 89 | 35,604 | 5.46 | General function prediction only |
| GI:161985360 | Hypothetical surface-anchored protein | 46 | 38,667 | 4.34 | General function prediction only |
| GI:161985444 | ABC-type transport system, ligand-binding component | 102 | 40,993 | 4.03 | Amino acid transport and metabolism |
| GI:161985091 | rpoA DNA-directed RNA polymerase, alpha subunit | 185 | 36,967 | 4.83 | Transcription |
| GI:161985374 | eno enolase | 94 | 46,488 | 4.97 | Carbohydrate transport and metabolism |
| GI:161985165 | Translation elongation factor EF-Tu | 115 | 42,824 | 5.21 | Translation, ribosomal structure, and biogenesis |
| GI:161985165 | Translation elongation factor EF-Tu | 49 | 42,824 | 5.21 | Translation, ribosomal structure, and biogenesis |
| GI:161985998 | glmM phosphoglucosamine mutase | 41 | 48,072 | 5.84 | Carbohydrate transport and metabolism |
| GI:161985175 | dps starvation-inducible DNA-binding protein, ferritin like | 69 | 16,521 | 5.04 | Inorganic ion transport and metabolism |
| Acylated | |||||
| GI:161985444 | potD spermidine/putrescine ABC transport system, ligand-binding component | 76 | 40,993 | 4.03 | Amino acid transport and metabolism |
| GI:161985628 | ABC transport system, ligand-binding component | 55 | 48,485 | 4.64 | Carbohydrate transport and metabolism |
| GI:161985628 | ABC-type transport system, ligand-binding component | 44 | 48,485 | 4.64 | Carbohydrate transport and metabolism |
| GI:161986352 | ABC-type transport system, substrate-binding component | 316 | 54,628 | 5 | Carbohydrate transport and metabolism |
| GI:161986352 | ABC-type transport system, substrate-binding component | 316 | 54,628 | 5 | Carbohydrate transport and metabolism |
| GI:161985047 | apbE thiamine biosynthesis lipoprotein | 100 | 40,561 | 4.58 | Coenzyme metabolism |
| GI:161986256 | pdhC dihydrolipoamide acetyltransferase | 89 | 57,225 | 5 | Energy production and conversion |
| GI:161985360 | Hypothetical surface-anchored protein | 145 | 38,667 | 4.34 | General function prediction only |
| GI:161985360 | Hypothetical surface-anchored protein | 64 | 38,667 | 4.34 | General function prediction only |
| GI:161986349 | Hypothetical surface-anchored protein | 251 | 44,832 | 4.74 | General function prediction only |
| GI:161986349 | Hypothetical surface-anchored protein | 251 | 44,832 | 4.74 | General function prediction only |
| GI:161985964 | ABC transporter, periplasmic | 114 | 29,998 | 4.36 | Inorganic ion transport and metabolism |
| GI:161985964 | ABC-type transport system, substrate-binding component | 118 | 29,998 | 4.36 | Inorganic ion transport and metabolism |
| GI:161986249 | pstS ABC-type transport system, substrate-binding component | 153 | 33,583 | 4.96 | Inorganic ion transport and metabolism |
| GI:161985246 | ABC-type transport system, substrate-binding component | 132 | 56,866 | 4.69 | NA |
| GI:161985029 | Hypothetical protein | 239 | 50,917 | 4.7 | NA |
| GI:161985692 | Hypothetical protein | 166 | 61,139 | 4.55 | NA |
| GI:161985639 | Hypothetical surface-anchored protein | 88 | 35,318 | 4.57 | NA |
| GI:161985714 | Peptidyl-prolyl cis-trans isomerase, cyclophilin type | 107 | 24,252 | 4.78 | Posttranslation modification, protein turnover, chaperones |
| GI:161985164 | Translation elongation factor EF-G | 346 | 76,287 | 5.29 | Translation, ribosomal structure, and biogenesis |
NA, not applicable.
Score acquired by fragment ion spectra of peptides belonging to designated proteins after identification by the Mascot algorithm.
Phosphorylation was additionally analyzed based on the MS-MS spectra from ESI-Trap, and 74 candidate phosphorylated proteins were found to have a sufficient score (see Table S1 in the supplemental material). A comparison with known phosphoproteomes of E. coli (29), B. subtilis (30), and Mycoplasma spp. (55) confirmed 11 peptides as having phosphorylated orthologs at least in one of the species. Notably, 16 candidates are proteins unique to A. laidlawii, and 11 of them are surface-anchored or integral membrane proteins, which implies the presence of active signaling pathways. Out of nine phosphoproteins identified by MALDI, four are found in the ESI experiments. The list of phosphorylated proteins obtained by both techniques is rich in proteases, kinases, transferases, and nucleases, again suggesting active regulation by phosphorylation.
Acylation.
For the identification of acylated proteins, 14C-labeled palmitic, stearic, linoleic, and oleic fatty acids were introduced into the incubation medium during the A. laidlawii culture growth, as described previously (20). The standard procedure of two-dimensional protein separation was applied, and silver-stained gels were dyed for radioactivity buildup in the Storage phosphor screen. One month later, the screen was scanned on the Typhoon Trio scanner. Among 20 acylated proteins, 14 contained palmitic chains, and six contained stearic chains (Table 6). No residues of linoleic or oleic acid were observed. Acylated proteins were components of mainly sugar and inorganic ion transport systems or were surface-anchored proteins with unknown functions. The prevalence of the palmitic acid acylation agrees with earlier reports on fatty acid representation in various Mycoplasma species (65). All these modified proteins except two (translation elongation factor EF-G and dihydrolipoamide acetyltransferase) have a cysteine in the N-terminal region in a position favorable for fatty-acid modification, which corresponds to the acylation mechanism proposed for bacteria (65). For several proteins, MALDI/MS-MS analyses yielded a more precise acylation pattern because the N-acylated peptide mass allowed for the determination of the modification pattern. It has been suggested earlier the Mollicutes are characterized by diacylation (45). However, we determined that A. laidlawii has three acyl residues (50).
DISCUSSION
The biological properties of A. laidlawii are rather different from those of already sequenced mycoplasmas. Its genome of 1,496,992 bp is the longest sequenced genome of the Mollicutes. M. penetrans, whose genome is only 1,500 bp shorter (49), differs from A. laidlawii by having a substantially lower number of regulatory and structural elements. We found genes of polymerase type I, SOS response, and signal transduction systems, as well as RNA regulatory elements, riboswitches, and T boxes. This demonstrates a significant capability in the regulation of gene expression and mutagenic response to stress. We believe that these profound differences in the genome and molecular machinery organization between the acholeplasmas and mycoplasmas indicate that the acholeplasmas form a unique branch of evolution, either as a side trend in the evolution of parasitism or as an intermediate in the genome reduction and decrease in adaptive mechanisms and specialization.
The proteomic mapping of the A. laidlawii genome identified 803 (58%) proteins synthesized in optimal growth conditions. In a model species, Mycoplasma pneumoniae, 70% of the annotated proteins were expressed in the studied conditions, and this fraction of the proteome is sufficient for sustaining the work of all cellular systems (24). The presence of multimeric protein complexes containing components of different systems implies not only an active exchange of the complexes' components but the probable multifunctionality of the proteins comprising them (24). In M. mobile, a Mollicutes species with a genome size less than 800 kb, the expression of 88% of genes was shown (19). In A. laidlawii, likely due to the presence of signal transduction systems and RNA regulatory elements, the fraction of expressed genes is lower, below 70%. We suppose that the remaining 30% of genes, for which the protein products were not observed, are either adaptive genes expressed in specific conditions or recent pseudogenes with an intact reading frame but with disrupted promoters. In Mycoplasma pneumoniae, a considerable number of noncoding transcripts was found (16), which could explain the absence of products of various genes in the A. laidlawii proteome.
One of the major difficulties in the proteogenomic analysis was determining the accuracy of methods used for the peptide identification. To validate the techniques used, we analyzed several proteins involved in the replication and transcription, the rationale being that polymerase III is represented in prokaryotes by 10 to 20 molecules per cell. We identified all subunits of polymerase III, thus demonstrating the sensitivity of our techniques toward low-copy-number proteins. The analysis of proteins involved in the transcription and translation yielded a distinct regular pattern: in optimal growth conditions, the constitutive elements of these systems are present and SOS-induced proteins are absent.
The proteogenomic profiling of A. laidlawii characterized a representative of the Mollicutes which occupies an intermediate position between the Clostridia/Bacillus and the Mycoplasmataceae. Being mainly free-living, some Mollicutes representatives also parasitize a wide spectrum of hosts. A. laidlawii retains a significant number of genes used for adaptive response to environmental challenges.
Modern data on genomes and functional proteomes of mycoplasmas, as well as other organisms, make one doubt that the minimal set of genes sufficient for sustaining the main life functions in a cell may be derived based solely on genome analyses, not taking into account the variability of gene products present in cells in different functional states. In the mycoplasma species, the number of gene products present in cells varies from 65 to 80% of the total ORFs, although the size of the genome may vary from 570,000 bp, as in M. genitalium, to 800,000 to 1,500,000 bp. Therefore, the precise specification of the minimal genome requires taking into account experimental data, including growth conditions, protein posttranslational modifications, protein interactions, presence of metabolites in the medium, etc.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Ilya Borovok, Tel Aviv University, for helpful comments on annotation of RNR genes.
This study was partially supported by state contract no. 2.740.11.0101, the Russian Academy of Sciences via the “Cellular and Molecular Biology” and “Basic Science for Medicine” programs, and the Russian Foundation of Basic Research via grants 09-04-92745, 09-04-01299, and 10-07-00610.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Bai X., et al. 2006. Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188:3682–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bairoch A., Apweiler R. 2000. The SWISSPROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28:45–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benders G. A., et al. 2010. Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 38:2558–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bugg T. D., Brandish P. E. 1994. From peptidoglycan to glycoproteins: common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol. Lett. 119:255–262 [DOI] [PubMed] [Google Scholar]
- 6. Chambaud I., et al. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chou H. H., Holmes M. H. 2001. DNA sequence quality trimming and vector removal. Bioinformatics 17:1093–1104 [DOI] [PubMed] [Google Scholar]
- 8. Colman S. D., Hu P. C., Litaker W., Bott K. F. 1990. A physical map of the Mycoplasma genitalium genome. Mol. Microbiol. 4:683–687 [DOI] [PubMed] [Google Scholar]
- 9. Craig R., Beavis R. C. 2003. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 17:2310–2316 [DOI] [PubMed] [Google Scholar]
- 10. Dandekar T., et al. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edwards J. C., Chapman D., Cramp W. A. 1983. Radiation studies of Acholeplasma laidlawii: the role of membrane composition. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 44:405–412 [DOI] [PubMed] [Google Scholar]
- 12. Ewing B., Hillier L., Wendl M. C., Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 13. Fraser C. M., et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397–403 [DOI] [PubMed] [Google Scholar]
- 14. Ghai R., Hain T., Chakraborty T. 2004. GenomeViz: visualizing microbial genomes. BMC Bioinformatics 5:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gibson D. G., et al. 2008. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319:1215–1220 [DOI] [PubMed] [Google Scholar]
- 16. Guell M., et al. 2009. Transcriptome complexity in a genome-reduced bacterium. Science 326:1268–1271 [DOI] [PubMed] [Google Scholar]
- 17. Halbedel S., Hames C., Stulke J. 2007. Regulation of carbon metabolism in the mollicutes and its relation to virulence. J. Mol. Microbiol. Biotechnol. 12:147–154 [DOI] [PubMed] [Google Scholar]
- 18. Jaffe J. D., Berg H. C., Church G. M. 2004. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4:59–77 [DOI] [PubMed] [Google Scholar]
- 19. Jaffe J. D., et al. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jan G., Fontenelle C., Le Henaff M., Wroblewski H. 1995. Acylation and immunological properties of Mycoplasma gallisepticum membrane proteins. Res. Microbiol. 146:739–750 [DOI] [PubMed] [Google Scholar]
- 21. Jensen O. N., Wilm M., Shevchenko A., Mann M. 1999. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol. Biol. 112:513–530 [DOI] [PubMed] [Google Scholar]
- 22. Kim J. N., Roth A., Breaker R. R. 2007. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc. Natl. Acad. Sci. U. S. A. 104:16092–16097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 24. Kuhner S., et al. 2009. Proteome organization in a genome-reduced bacterium. Science 326:1235–1240 [DOI] [PubMed] [Google Scholar]
- 25. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 26. Laidlaw P. P., Elford W. J. 1936. A new group of filterable organisms. Proc. R. Soc. Lond. Ser. B Biol. Sci. 120:292–303 [Google Scholar]
- 27. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of tRNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reference deleted.
- 29. Macek B., et al. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6:697–707 [DOI] [PubMed] [Google Scholar]
- 30. Mandal M., Breaker R. R. 2004. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 5:451–463 [DOI] [PubMed] [Google Scholar]
- 31. Maniloff J., Morowitz H. J. 1972. Cell biology of the mycoplasmas. Bacteriol. Rev. 36:263–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCoy R. E., et al. 1989. Plant diseases associated with mycoplasma-like organisms, p. 545–560.In Whitcomb R., Tully J. G. (ed.), The mycoplasmas. Academic Press Inc., San Diego, CA. [Google Scholar]
- 33. McElhaney R. N. 1984. The structure and function of the Acholeplasma laidlawii plasma membrane. Biochim. Biophys. Acta 779:1–42 [DOI] [PubMed] [Google Scholar]
- 34. McLean M. J., Wolfe K. H., Devine K. M. 1998. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 47:691–696 [DOI] [PubMed] [Google Scholar]
- 35. Myers G. S., et al. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neverov A. D., Gelfand M., Mironov A. A. 2003. GipsyGene: a statistics-based gene recognizer for fungal genomes. Biophysics (Moscow) 48:71–75 [Google Scholar]
- 37. Oshima K., et al. 2004. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 36:27–29 [DOI] [PubMed] [Google Scholar]
- 38. Overbeek R., Fonstein M., D'Souza M., Pusch G. D., Maltsev N. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. U. S. A. 96:2896–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pollack J. D., Tryon V. V., Beaman K. D. 1983. The metabolic pathways of Acholeplasma and Mycoplasma: an overview. Yale J. Biol. Med. 56:709–716 [PMC free article] [PubMed] [Google Scholar]
- 40. Pop M., Kosack D. S., Salzberg S. L. 2004. Hierarchical scaffolding with Bambus. Genome Res. 14:149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Razin S. 1978. The mycoplasmas. Microbiol. Rev. 42:414–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Razin S. 1962. Nucleic acid precursor requirements of Mycoplasma laidlawii. J. Gen. Microbiol. 28:243–250 [DOI] [PubMed] [Google Scholar]
- 43. Razin S., Yogev D., Naot Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rocha E. P., Danchin A., Viari A. 1999. Translation in Bacillus subtilis: roles and trends of initiation and termination, insights from a genome analysis. Nucleic Acids Res. 27:3567–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rottem S. 2002. Sterols and acylated proteins in mycoplasmas. Biochem. Biophys. Res. Commun. 292:1289–1292 [DOI] [PubMed] [Google Scholar]
- 46. Rutherford K., et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 47. Saito Y., Silvius J. R., McElhaney N. 1977. Membrane lipid biosynthesis in Acholeplasma laidlawii B: de novo biosynthesis of saturated fatty acids by growing cells. J. Bacteriol. 132:497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salzberg S. L., Delcher A. L., Kasif S., White O. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sasaki Y., et al. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30:5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Serebryakova M. V., et al. 2011. The acylation state of surface lipoproteins of mollicute Acholeplasma laidlawii. J. Biol. Chem. 286:22769–22776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sernova N. V., Gelfand M. S. 2008. Identification of replication origins in prokaryotic genomes. Brief. Bioinform. 9:376–391 [DOI] [PubMed] [Google Scholar]
- 52. Shevchenko A., Wilm M., Vorm O., Mann M. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850–858 [DOI] [PubMed] [Google Scholar]
- 53. Smith P. F. 1984. Lipoglycans from mycoplasmas. Crit. Rev. Microbiol. 11:157–186 [DOI] [PubMed] [Google Scholar]
- 54. Stormo G. D., Schneider T. D., Gold L. M. 1982. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 10:2971–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Su H. C., Hutchison C. A., III, Giddings M. C. 2007. Mapping phosphoproteins in Mycoplasma genitalium and Mycoplasma pneumoniae. BMC Microbiol. 7:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sutton G., White O., Adams M., Kerlavage A. 1995. TIGR Assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci. Technol. 1:9–19 [Google Scholar]
- 57. Tatusov R. L., Koonin E. V., Lipman D. J. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 58. Thomas J. M., Horspool D., Brown G., Tcherepanov V., Upton C. 2007. GraphDNA: a Java program for graphical display of DNA composition analyses. BMC Bioinformatics 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tusnady G. E., Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850 [DOI] [PubMed] [Google Scholar]
- 60. Vasconcelos A. T., et al. 2005. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 187:5568–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reference deleted.
- 62. Vitreschak A. G., Gelfand M. A. M. S. 2001. RNApattern program: searching for RNA secondary structure by the pattern rule, p. 623–625. Abstr. 3rd Int. Conf. Complex Syst. NECSI, Samara, Russia. [Google Scholar]
- 63. Vitreschak A. G., Rodionov D. A., Mironov A. A., Gelfand M. S. 2004. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 20:44–50 [DOI] [PubMed] [Google Scholar]
- 64. Westberg J., et al. 2004. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP). Genome Res. 14:221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Worliczek H. L., Kampfer P., Rosengarten R., Tindall B. J., Busse H. J. 2007. Polar lipid and fatty acid profiles—re-vitalizing old approaches as a modern tool for the classification of mycoplasmas? Syst. Appl. Microbiol. 30:355–370 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.