Abstract
Cyclic AMP (cAMP) signaling and the placental transcription factor glial cell missing 1 (GCM1) regulate expression of syncytin-1 and -2 fusogenic proteins, which are critical for syncytiotrophoblast formation by trophoblast fusion. We recently revealed a cAMP/protein kinase A (PKA)/CBP signaling pathway that activates GCM1 by coordinating GCM1 phosphorylation and acetylation. In contrast, GCM1 activity is downregulated by sumoylation of Lys156. How GCM1 sumoylation is regulated was unknown. Here, we identify a novel PKA-independent cAMP signaling pathway as the critical regulator of GCM1 sumoylation. We show that Epac1 and Rap1, in response to cAMP, activate CaMKI to phosphorylate Ser47 in GCM1. This phosphorylation facilitates the interaction between GCM1 and the desumoylating enzyme SENP1 and thereby leads to GCM1 desumoylation and activation. Using RNA interference (RNAi), we further demonstrate that 8-(4-chlorophenylthio)-2′-O-Me-cAMP-AM (8-CPT-AM), an Epac activator, stimulates syncytin-1 and -2 gene expression and cell fusion of placental BeWo cells in a GCM1-dependent manner. Importantly, the cell fusion defect in GCM1-knockdown BeWo cells can be reversed and enhanced by the RNAi-resistant phosphomimetic GCM1(S47D) mutant. Our study has identified a novel cAMP/Epac1/CaMKI/GCM1 signaling cascade that stimulates trophoblast fusion through promoting GCM1 phosphorylation and desumoylation.
INTRODUCTION
The syncytiotrophoblast in human placental villus is a multinucleated cell layer that mediates gas and nutrient transport between mother and fetus. Differentiation of the syncytiotrophoblast involves cell fusion of subjacent mononucleated cytotrophoblast cells. As the syncytiotrophoblast layer undergoes apoptosis and sheds into circulation, replenishment with a new syncytiotrophoblast layer is required to maintain its structural and functional integrity. In this regard, cyclic AMP (cAMP) is a key player for syncytiotrophoblast differentiation by stimulating placental cell fusion and syncytin gene expression (4, 19, 24, 33). Stimulation of placental cell fusion by cAMP is suppressed by the protein kinase A (PKA) inhibitor H89, indicating that PKA is a critical downstream effector of cAMP signaling in the regulation of placental cell fusion (19).
The placental transcription factor GCM1 (also known as GCMa) controls trophoblast fusion via transcriptional activation of two membrane fusogenic proteins, syncytin-1 and -2 (22, 38). An RNA interference (RNAi) study by Baczyk et al. (2) has shown that GCM1 is required for the cAMP stimulant forskolin to promote placental BeWo cell fusion. Indeed, GCM1 activity can be regulated by multiple posttranslational modifications, which may be critical to fine-tune its activity in the regulation of trophoblast fusion and syncytiotrophoblast differentiation. GCM1 is a labile protein subject to ubiquitination via the F-box protein FBW2 when the Ser322 residue in GCM1 is phosphorylated by glycogen synthase kinase 3β (GSK-3β) (7, 31, 36). Our recent studies have demonstrated that PKA enhances the interaction between GCM1 and the dual-specificity phosphatase DUSP23, which facilitates Ser322 dephosphorylation and subsequently GCM1 acetylation by CBP (3, 23). As a result, GCM1 is stabilized to transactivate syncytin gene expression. These findings reveal an underlying mechanism for stimulation of placental cell fusion and differentiation by the cAMP/PKA signaling pathway.
Sumoylation is a reversible protein modification involving the formation of an isopeptide bond between the C-terminal glycine of small ubiquitin-like modifier (SUMO) and the ε-amino group of a lysine residue in the target protein (13, 16). Desumoylation is carried out by several sentrin/SUMO-specific proteases (SENPs) that constitute a family of cysteine proteases with either an intact or a split catalytic domain at their C terminus. Six mammalian SENPs (SENP1, -2, -3, -5, -6, and -7) have been identified, exhibiting differential activities toward SUMO1 to SUMO3 and functioning at different cellular locations (20, 34). For example, SENP1 and SENP2 localize in nuclear speckled foci and/or nuclear envelope and catalyze the deconjugation of SUMO1 to SUMO3. SENP3 and SENP5 localize at nucleoli and preferentially catalyze the deconjugation of SUMO2 and SUMO3. Interestingly, transgenic studies have demonstrated that ablation of the mouse SENP1 gene results in decreased maternal and fetal blood spaces and increased trophoblast population in the labyrinthine layer of the placenta (35). In addition, ablation of the mouse SENP2 gene results in abnormal differentiation of placental trophoblast lineages and defective cardiac development (8, 17). At the cellular and molecular levels, SENP1 has been shown to stabilize HIF1α during hypoxia by preventing sumoylation-enhanced HIF1α ubiquitination and degradation (6), whereas SENP2 can desumoylate a subunit of the polycomb repressive complex 1 to relieve the transcriptional repressive effect of the complex on Gata4 and Gata6 genes (17).
SUMO modification of proteins has a wide spectrum of functional consequences, including modulation of transcriptional activity, mediation of nuclear import, recruitment of transcriptional regulators in nuclear domains, protection from ubiquitination, and regulation of mitosis (13, 16). We have previously shown that sumoylation of Lys156 on GCM1 impairs its DNA-binding activity, suggesting that sumoylation negatively regulates GCM1 activity (9). Nevertheless, whether GCM1 sumoylation can be reversed by desumoylating enzymes is not known.
Multiple effectors, including PKA, cyclic nucleotide-gated ion channels, and Epac (exchange protein directly activated by cAMP) proteins, have been identified as critical mediators of cAMP signaling in many cellular activities (14). Interestingly, Epac1 and Epac2 proteins were identified as cAMP-binding proteins with guanine nucleotide exchange factor (GEF) activities for the small GTPases, Rap1 and Rap2 (14, 18). The N-terminal region of Epac proteins contains one or two cAMP-binding domains, which have an autoinhibitory effect on their catalytic GEF domain at the C-terminal region. Upon cAMP binding, Epac proteins undergo a conformational change, which relieves the autoinhibition and then activates Rap proteins to regulate numerous cellular actions. For instance, Epac and Rap proteins can activate CaMKII through phospholipase Cε and protein kinase Cε in order to regulate Ca2+ handling in cardiac myocytes (25–27).
Given that multiple effectors can be involved in the transduction of cAMP signaling, we reasoned that GCM1 activity may be regulated through different effectors in the cAMP signaling cascade that controls trophoblast cell fusion and differentiation. We found here that cAMP-dependent activation of Epac1 and Rap1 but not PKA is able to activate CaMKI to mediate Ser47 (S47) phosphorylation in GCM1. This phosphorylation enhances the interaction of GCM1 and SENP1 and consequently promotes GCM1 desumoylation and activation. RNA interference experiments further revealed that activation of Epac by the Epac activator 8-(4-chlorophenylthio)-2′-O-Me-cAMP-AM (8-CPT-AM) stimulates syncytin gene expression and placental BeWo cell fusion in a GCM1-dependent manner. Correspondingly, cell fusion defect in GCM1-knockdown BeWo cells was reversed and enhanced by the RNA interference (RNAi)-resistant phosphomimetic GCM1(S47D) mutant. Therefore, GCM1 Ser47 phosphorylation and Lys156 desumoylation underlie the stimulation of placental cell fusion by the cAMP/Epac1/CaMKI signaling pathway. Our results reveal a second pathway of cAMP signaling that coordinates GCM1 phosphorylation and desumoylation to increase GCM1 activity for trophoblast fusion.
MATERIALS AND METHODS
Plasmid constructs.
The pHA-GCM1 and pGal4-GCM1-FLAG expression plasmids and the p(GBS)4E1BLuc reporter plasmid have been described previously (3). The pHA-GCM1 expression plasmids harboring a serine- or threonine-to-alanine mutation in the CaMKI phosphorylation sites, Ser47, Thr174, Ser177, Ser178, and Ser269, were constructed by two-step PCRs with designated primer sets. Silent mutations were introduced into the region of the GCM1 sequence targeted by GCM1 short hairpin RNA (shRNA) (see below) in order to construct RNAi-resistant pHA-GCM1 expression plasmids for wild-type HA-GCM1 and S47A and S47D mutants. The pCACaMKI-FLAG (where CA stands for “constitutively active”) and pCAHA-CaMKI expression plasmids were constructed in a pEF1 expression vector (Invitrogen, Carlsbad, CA) to encode constitutively active forms of CaMKI (30) with a C-terminal triple FLAG and an N-terminal triple hemagglutinin (HA) tag, respectively. Likewise, the pDNCaMKI-FLAG and pCAEpac1-FLAG expression plasmids were constructed to encode a dominant negative form of CaMKI (32) with a C-terminal triple FLAG tag and a constitutively active form of Epac1 (12) with a C-terminal triple FLAG tag, respectively.
Cell culture, transfection, and lentivirus transduction.
293T, JAR, and BeWo cells were obtained from the American Type Culture Collection (Manassas, VA). Villous cytotrophoblast cells from term placentas were prepared and cultured as previously described (7). For transient expression, cells were transfected with the indicated reporter and expression plasmids using the Lipofectamine 2000 reagent (Invitrogen). Luciferase assays were performed as previously described (5). BeWo and JAR cells were infected with recombinant lentivirus strains harboring empty, CAHA-CaMKI, and CAEpac1-FLAG expression cassettes. The infected cells were subjected to antibiotic selection using 100 g/ml of puromycin, and the puromycin-resistant clones were pooled for studies of interaction of GCM1 and SENP1, GCM1 sumoylation, and Ser47 phosphorylation. For RNA interference, pLKO.1-Puro shRNA expression plasmids harboring a scramble sequence (5′-CCTAAGGTTAAGTCGCCCTCG-3′, Addgene plasmid 1864) and sequences for GCM1 (5′-CCTCAGCAGAACTCACTAAAT-3′), CaMKI (5′-GCTGGATGCTGTGAAATACCT-3′), Epac1 (5′-GCAGGACTTCAACCGTATCAT-3′), and Rap1 (5′-GCTCTGACAGTTCAGTTTGTT-3′) were acquired from the National RNAi Core Facility of Taiwan.
GCM1 sumoylation and desumoylation.
GCM1 sumoylation and desumoylation were studied as previously described (9). In brief, 293T cells were transfected with different combinations of pHA-GCM1, pEGFP-SUMO1, pEGFP-SUMO1-AA, pCACaMKI-FLAG, pSENP1-FLAG, and pSENP1-FLAGC603S for 48 h. Cells were then harvested in lysis buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 10% glycerol, 0.5% NP-40, 1 mM dithiothreitol (DTT), 5 mM NaF, 5 mM Na3VO4, 10 mM N-ethylmaleimide (NEM), 10 mM iodoacetamide (IAA), and a protease inhibitor cocktail (Sigma, St. Louis, MO). Detection of sumoylated GCM1 was performed by consecutive immunoprecipitation and immunoblotting with hemagglutinin (HA) monoclonal antibody (MAb) (Sigma). For in vitro desumoylation of GCM1, recombinant GCM1-FLAG prepared from the baculovirus insect cell expression system (38) was first incubated with recombinant SAE1/SAE2 (Boston Biochem, Cambridge, MA), His-SUMO1 (Boston Biochem), and Ubc9 proteins at 37°C for 2.5 h. A portion of the reaction mixture was then incubated with glutathione S-transferase (GST)-SENP1 or GST-SENP1C603S at 37°C for 40 min, followed by immunoblotting with FLAG MAb (Sigma).
Regulation of interaction between GCM1 and SENP1.
To study the interaction of GCM1 and SENP1 in vivo, 293T cells were transfected with different combinations of pHA-GCM1, pHA-GCM1K156R, pSENP1-FLAG, pSENP1C603S-FLAG, and pCAHA-CaMKI for 48 h. Cells were then harvested in lysis buffer for consecutive immunoprecipitation and immunoblotting with HA and FLAG MAbs. To study the interaction of endogenous GCM1 and SENP1 and regulation of endogenous GCM1 sumoylation, mock JAR and BeWo cells and CAHA-CaMKI-expressing JAR and BeWo cells were subjected to consecutive immunoprecipitation and immunoblotting using different combinations of GCM1, SENP1 (Santa Cruz Biotechnology, Santa Cruz, CA), and SUMO1 (Invitrogen) antibodies (Abs). Immunoblot band intensities were quantified using ImageJ densitometry software (http://rsbweb.nih.gov/ij/).
Mapping the SENP1-interacting domain in GCM1.
To map the GCM1 domain that interacts with SENP1, pulldown assays were performed. In brief, 293T cells were transfected with pGal4-FLAG, full-length pGal4-GCM1-FLAG, or the indicated deletion mutant pGal4-GCM1-FLAG plasmid. At 48 h posttransfection, cells were harvested in the lysis buffer. The cell lysate was incubated with GST or GST-SENP1 prebound to glutathione-conjugated agarose beads in lysis buffer at 4°C for 4 h before washing three times with lysis buffer. The proteins pulled down were analyzed by immunoblotting with FLAG MAb.
CaMKI-mediated GCM1 phosphorylation and mass spectrometry.
In vitro phosphorylation of GCM1 by CaMKI was performed by incubation of maltose binding protein (MBP)-GCM1 with [γ-32P]ATP and CACaMKI-FLAG, which was immunopurified from 293T cells transfected with pCACaMKI-FLAG, at 30°C for 40 min. The reaction was analyzed by SDS-PAGE and autoradiography. To identify the CaMKI sites in GCM1, the above-described reaction was scaled up with nonradioactive ATP, and the phosphorylated MBP-GCM1 was subjected to LTQ Orbitrap mass spectrometry analysis (Common Mass Spectrometry Facilities, Academia Sinica). In vivo phosphorylation of Ser47 in GCM1 was studied in mock BeWo cells and BeWo cells stably expressing CAHA-CaMKI or CAEpac1-FLAG by immunoprecipitation with Gal4 Ab (Santa Cruz Biotechnology) or phosphor-Ser47-specific Ab (p-Ser47-GCM1 Ab), followed by immunoblotting with GCM1 Ab. In a separate experiment, primary cytotrophoblast cells were infected with recombinant lentivirus strains harboring empty, CAHA-CaMKI, and CAEpac1-FLAG expression cassettes and subjected to the above-mentioned analysis. The p-Ser47-GCM1 Ab was raised against chemically synthesized phosphopeptide AKHIYSS(PO3)EDKNAQ in rabbits. A commercial antibody against phosphor-Thr177 in CaMKI (Santa Cruz Biotechnology) was used for analysis of CaMKI activation by Epac1 and Rap1.
ChIP assay.
BeWo cells were treated with or without 50 μM 8-CPT-AM for 24 h before being subjected to chromatin immunoprecipitation (ChIP) assay using normal rabbit serum or GCM1 Ab. In a separate experiment, mock or CAHA-CaMKI-expressing BeWo cells were directly subjected to ChIP assay using the same antiserum and Ab. ChIP and PCR conditions and primer sequences for a specific region containing the proximal GCM1-binding sequence in the syncytin-1 promoter have been described previously (38).
Immunofluorescence microscopy and cell-cell fusion analysis.
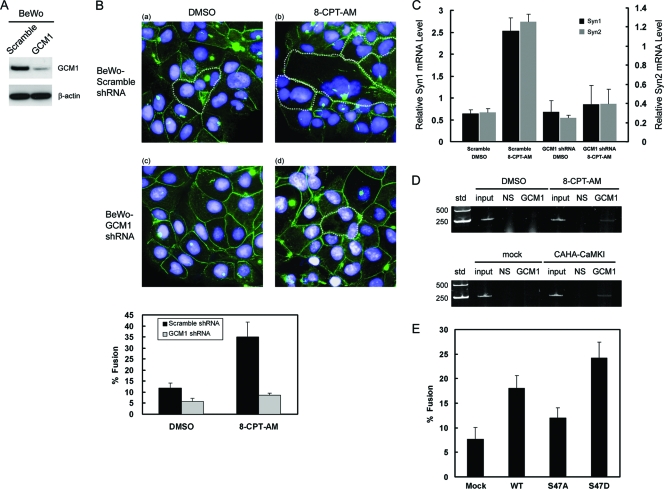
For colocalization analysis of GCM1 and SENP, 293T cells were transfected with pHA-GCM1 and the indicated expression plasmid encoding a green fluorescent protein (GFP)-SENP fusion protein. After 48 h posttransfection, cells were fixed and stained with HA MAb and then rhodamine-labeled anti-mouse IgG Ab. Nuclei were stained by DAPI (4′,6-diamidino-2-phenylindole). Immunofluorescence was examined under a Zeiss laser scanning confocal microscope (LSM510). To study the effect of 8-(4-chlorophenylthio)-2′-O-Me-cAMP-AM (abbreviated 8-CPT-AM in this study), which is an Epac activator, on GCM1-regulated placental cell fusion, BeWo cells stably expressing scramble or GCM1 shRNA were treated with or without 50 μM 8-CPT-AM. After 24 h, cells were fixed and subjected to immunostaining with anti-desmosomal protein MAb (Sigma) and then Cy2-labeled anti-mouse IgG Ab (Jackson ImmunoResearch Laboratories, West Grove, PA). Cell-cell fusions were examined under an Olympus microscope (Tokyo, Japan) equipped with a cooled charge-coupled-device camera (DP50). Four microscopic fields per sample were randomly selected for examination in each of three independent experiments. Quantification of cell-cell fusion was calculated as the ratio of the number of nuclei in the syncytia to the total number of nuclei counted in a randomly selected field. Images were prepared for presentation using Adobe Photoshop version 7.0.
Quantitative real-time PCR.
BeWo cells stably expressing scramble or GCM1 shRNA were treated with or without 8-CPT-AM. After 24 h, cells were harvested for RNA isolation using RNeasy reagents (Qiagen, Hilden, Germany) and then transcribed into cDNA using SuperScript III reagents (Invitrogen) with an oligo(dT)20 primer. Quantification of the transcript levels of GCM1 target genes was performed in the LightCycler system (Roche, Basel, Switzerland) using a commercial SYBR green reaction reagent (Qiagen) and specific primer sets. The sequences of primer sets were 5′-TGGAACAACTTCAGCACAGA-3′ and 5′-GCCATTCAAACAACGATAGG-3′ for syncytin-1, 5′-CGACTCAGTGTAAACAGCCA-3′ and 5′-CCACAGAAGCAAGACAAAGAAAAT-3′ for syncytin-2, and 5′-AACTCCATCATGAAGTGTGACG-3′ and 5′-GATCCACATCTGCTGGAAGG-3′ for β-actin.
RESULTS
CaMKI is downstream of cAMP signaling in regulation of GCM1 activity.
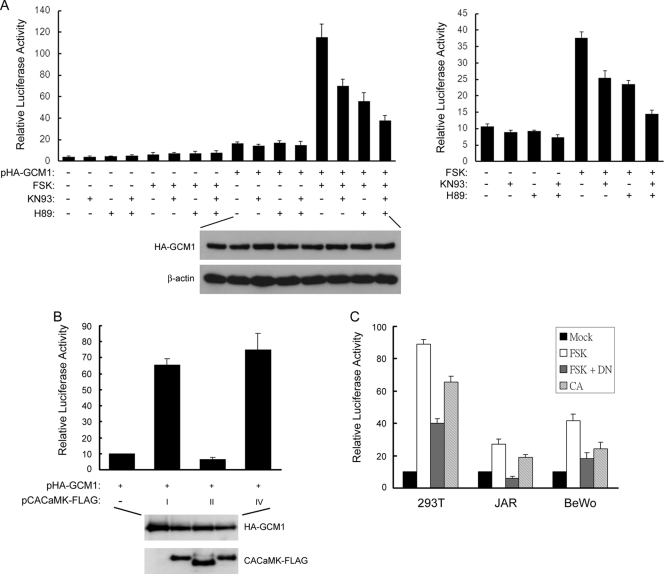
To study signaling pathways regulating GCM1 activity, we tested the effects of a variety of kinase inhibitors, including PD98059, SB203580, H89, and KN93, on GCM1 activity stimulated by forskolin in transient expression experiments. In parallel, we also tested the effects of p38, Jun N-terminal protein (JNK), extracellular signal-regulated (ERK), and CaMK kinases on GCM1 activity. These screening studies suggested that CaMK may regulate GCM1 activity (3) (data not shown). We then investigated whether CaMK plays an important role in the regulation of GCM1 activity by cAMP signaling. To this end, 293T cells were transfected with p(GBS)4E1BLuc and pHAGCM1, followed by treatment with or without forskolin, the CaMK inhibitor KN93, and the PKA inhibitor H89. In accordance with our previous study (3), forskolin stimulated the transcriptional activity of GCM1, which was counteracted by H89. Interestingly, the stimulatory effect of forskolin was also counteracted by KN93 (Fig. 1 A, left). Moreover, the stimulatory effect of forskolin was further suppressed when cells were treated with both H89 and KN93 (Fig. 1A, left), suggesting that both PKA and CaMK are involved in the stimulation of GCM1 activity by forskolin. This observation was unlikely due to differential expression of HA-GCM1, as similar levels of HA-GCM1 protein were detected among different transfection groups (Fig. 1A, left). In addition, placental BeWo cells were transfected with p(GBS)4E1BLuc and treated with or without forskolin, KN93, and H89. As shown in the right panel of Fig. 1A, forskolin stimulated the transcriptional activity of endogenous GCM1, which was also suppressed by either KN93 or H89. Likewise, cotreatment with H89 and KN93 further suppressed the stimulatory effect of forskolin (Fig. 1A, right). In a complementary approach, constitutively active CaMKI (CACaMKI), -II, or -IV was coexpressed with GCM1 in 293T cells in transient expression experiments. Indeed, both CACaMKI-FLAG and CACaMKIV-FLAG were able to upregulate the transcriptional activity of GCM1 (Fig. 1B). Because CaMKIV is barely expressed in placenta based on its expressed sequence tag (EST) profile in the UniGene database (http://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.591269), we concentrated on CaMKI for the rest of this study. Accordingly, we further demonstrated that dominant negative (DN) CaMKI is able to suppress the stimulatory effect of forskolin on GCM1-mediated transcriptional activation in 293T, JAR, and BeWo cells (Fig. 1C). Taken together, these results suggest that CaMKI is involved in the stimulation of GCM1 activity by cAMP signaling.
Fig. 1.
CaMKI is involved in the regulation of GCM1 activity by cAMP. (A) Stimulation of GCM1 transcriptional activity by forskolin (FSK) is suppressed by KN93. 293T cells (left) were transfected with p(GBS)4E1BLuc and pHA-GCM1, whereas BeWo cells (right) were transfected with p(GBS)4E1BLuc only. At 24 h posttransfection, cells were treated with or without 50 μM FSK, 6 μM KN93, and 3 μM H89 for an additional 24 h. Cells were then harvested for a luciferase reporter assay. The protein levels of HA-GCM1 and β-actin in transfected 293T cells were analyzed by immunoblotting with HA and β-actin MAbs, respectively. (B) Stimulation of GCM1 transcriptional activity by CaMKI and -IV. 293T cells were transfected with p(GBS)4E1BLuc and pHA-GCM1 plus pCACaMKI-FLAG, pCACaMKII-FLAG, or pCACaMKIV-FLAG for 48 h and then harvested for a luciferase reporter assay. The protein levels of HA-GCM1 and CACaMK-FLAG in transfected 293T cells were analyzed by immunoblotting with HA and FLAG MAbs, respectively. (C) Dominant negative CaMKI impedes the stimulation of GCM1 activity by FSK. 293T cells were transfected with p(GBS)4E1BLuc and pHA-GCM1 plus pCACaMKI-FLAG (CA) or pDNCaMKI-FLAG (DN). JAR and BeWo cells were transfected with the above-mentioned plasmids, except pHA-GCM1. At 24 h posttransfection, cells were treated with or without 50 μM FSK for an additional 24 h. Cells were then harvested for a luciferase reporter assay. Mean values and the standard deviations (SD) obtained from three independent experiments are presented in panels A to C.
CaMKI facilitates GCM1 desumoylation by enhancing the interaction of GCM1 and SENP1.
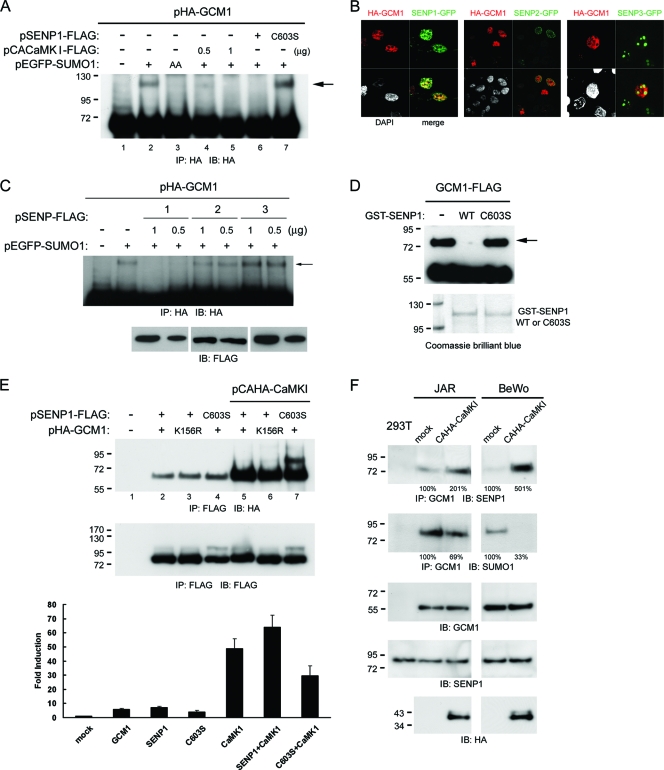
We next investigated how CaMKI regulates GCM1 activity. Because cAMP signaling, acting by PKA and CBP, stabilizes GCM1, we first tested whether CaMKI regulates the half-life of GCM1 and did not observe any significant effect of CaMKI on GCM1 stability (data not shown). Because GCM1 activity is negatively regulated by sumoylation on Lys156 (9), we then tested whether CaMKI regulates GCM1 sumoylation. In a transient expression study, sumoylation of HA-GCM1 was detected in 293T cells coexpressing EGFP-SUMO1 but not the mutant EGFP-SUMO1AA (Fig. 2A, compare lanes 2 and 3). Interestingly, the level of sumoylated HA-GCM1 was decreased when CACaMKI-FLAG was coexpressed (Fig. 2A, compare lanes 2, 4, and 5). As a control, coexpression of SENP1-FLAG but not the enzyme-dead mutant SENP1C603S-FLAG abolished sumoylation of HA-GCM1 (Fig. 2A, compare lanes 6 and 7).
Fig. 2.
Regulation of GCM1 desumoylation by CaMKI. (A) CaMKI suppresses GCM1 sumoylation. 293T cells were transfected with different combinations of pHA-GCM1, pEGFP-SUMO1, pEGFP-SUMO1AA, pSENP1-FLAG, pSENP1C603S-FLAG, and pCACaMKI-FLAG for 48 h. Cells were harvested for consecutive immunoprecipitation (IP) and immunoblotting (IB) with HA MAb. The arrow indicates HA-GCM1 conjugated with EGFP-SUMO1. (B) Nuclear colocalization of GCM1 and SENP1. 293T cells were transfected with pHA-GCM1 and the indicated expression plasmid encoding a GFP-SENP fusion protein for immunofluorescence microscopy as described in Materials and Methods. (C) In vivo desumoylation of GCM1 by SENP1. 293T cells were transfected with different combinations of pHA-GCM1, pEGFP-SUMO1, and the indicated pSENP-FLAG expression plasmid encoding SENP1, -2, or -3 for 48 h. Cells were harvested for analysis of HA-GCM1 sumoylation as described for panel A. (D) In vitro desumoylation of GCM1 by SENP1. One microgram of GCM1-FLAG was subjected to in vitro sumoylation reaction as described in Materials and Methods, followed by incubation with 0.1 μg of GST-SENP1 or GST-SENP1(C603S) and then immunoblotting with FLAG MAb. The arrow indicates GCM1-FLAG conjugated with His-SUMO1. (E) Interaction of GCM1 and SENP1 is stimulated by CaMKI. 293T cells were transfected with different combinations of pHA-GCM1, pHA-GCM1(K156R), pSENP1-FLAG, pSENP1(C603S)-FLAG, and pCAHA-CaMKI for 48 h. Cells were harvested and subjected to immunoprecipitation and immunoblotting with FLAG and HA MAbs, respectively. In a separate experiment, 293T cells were transfected with p(GBS)4E1BLuc plus different combinations of pHA-GCM1, pSENP1-FLAG, pSENP1(C603S)-FLAG, and pCAHA-CaMKI for 48 h. Cells were then harvested for a luciferase reporter assay. Fold induction was calculated relative to the luciferase expression observed for p(GBS)4E1BLuc alone. Mean values and the SD obtained from three independent experiments are presented. (F) CaMKI enhances the interaction of GCM1 and SENP1 and promotes GCM1 desumoylation in placental cells. Mock and CAHA-CaMKI-expressing JAR and BeWo cells were subjected to immunoprecipitation and immunoblotting with the indicated GCM1, SENP1, and SUMO1 Abs. The number underneath each band in the immunoblot indicates the relative intensity of the corresponding band.
Since CaMKI prevents GCM1 sumoylation, we tested whether SENP is involved in this process. We focused on SENP1 because immunofluorescence microscopy showed us that GCM1 colocalizes with SENP1, but not with SENP2 and -3, in the nucleus (Fig. 2B). Correspondingly, we demonstrated that the level of sumoylated HA-GCM1 was significantly decreased when SENP1, but not SENP2 and -3, was coexpressed in a transient expression study (Fig. 2C). To demonstrate that GCM1 is an SENP1 substrate, we performed an in vitro de-sumoylation assay by incubating sumoylated recombinant GCM1-FLAG with GST-SENP1 or GST-SENP1C603S. As shown in Fig. 2D, GST-SENP1, but not GST-SENP1C603S, was able to cleave SUMO1 from GCM1-FLAG. Because GCM1 sumoylation is decreased in the presence of CaMKI, we tested the possibility that CaMKI may regulate the interaction between GCM1 and SENP1 and thereby promote GCM1 desumoylation. Indeed, interaction between SENP1-FLAG and HA-GCM1 was detected in 293T cells by coimmunoprecipitation analysis (Fig. 2E, lane 2). Interestingly, coexpression of an N-terminally HA-tagged CACaMKI (CAHA-CaMKI) further enhanced the interaction between SENP1-FLAG and HA-GCM1 (Fig. 2E, lanes 2 and 5). As a control, the dominant negative (DN) CaMKI decreased the interaction between SENP1-FLAG and HA-GCM1 (data not shown). Of note, CAHA-CaMKI also enhanced the interaction between SENP1-FLAG and the mutant HA-GCM1K156R, which harbors a lysine-to-arginine mutation on Lys156 (Fig. 2E, lanes 3 and 6), and the interaction between SENP1C603S-FLAG and HA-GCM1 (Fig. 2E, lanes 4 and 7). This enhancement effect also enabled us to detect an interaction between SENP1C603S-FLAG and sumoylated HA-GCM1 (Fig. 2E, the upper band in lane 7). Therefore, CaMKI enhances the interaction between GCM1 and SENP1 in a manner that does not require GCM1 sumoylation or the enzyme activity of SENP1. Because CaMKI downregulates GCM1 sumoylation via SENP1, we tested whether SENP1 affects CaMKI-stimulated GCM1 activity on p(GBS)4E1BLuc in transient expression experiments. As shown in the bottom panel of Fig. 2E, CaMKI stimulated the transcriptional activity of GCM1, which was enhanced by SENP1 and suppressed by SENP1C603S.
To study whether CaMKI regulates the interaction of GCM1 and SENP1 and thereby GCM1 desumoylation in placental cells, JAR and BeWo cells stably expressing CAHA-CaMKI were established and subjected to coimmunoprecipitation analysis. As shown in the top panel of Fig. 2F, an interaction between endogenous GCM1 and SENP1 was detected in the parental JAR and BeWo cells, which was further enhanced upon expressing CAHA-CaMKI. As expected, no interaction between GCM1 and SENP1 was detected in 293T cells, which do not express GCM1 (Fig. 2F). Importantly, in comparison with the parental cells, the level of sumoylated GCM1 was significantly decreased in JAR and BeWo cells expressing CAHA-CaMKI (Fig. 2F). Taken together, these results suggest that CaMKI promotes GCM1 desumoylation by enhancing the interaction between GCM1 and SENP1 in placental cells.
CaMKI mediates Ser47 phosphorylation in GCM1.
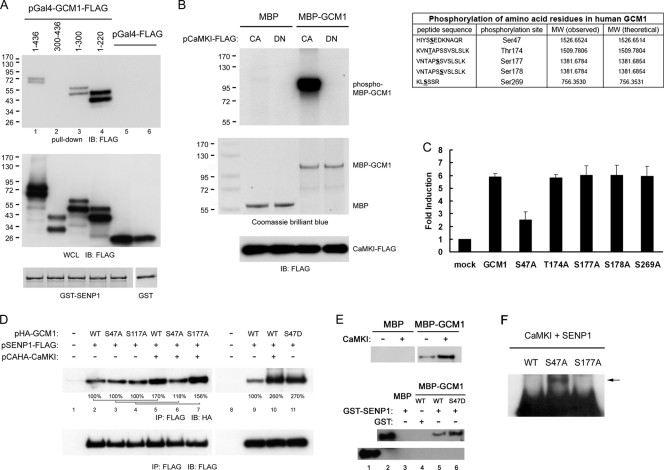
We next mapped the interaction domain in GCM1 for SENP1 by pulldown analysis. GST or GST-SENP1 was incubated with cell lysates prepared from 293T cells expressing Gal4-FLAG or different Gal4-GCM1-FLAG proteins harboring full-length or truncated GCM1. As shown in Fig. 3A, Gal4-GCM1-FLAG(1-436), -(1-300), and -(1-220) were pulled down by GST-SENP1 (lanes 1, 3, and 4), suggesting that the N-terminal region of amino acids 1 to 220 is the SENP1-interacting domain in GCM1. We further studied GCM1 phosphorylation by CaMKI and identified the CaMKI phosphorylation sites in GCM1. In vitro kinase assays were performed by incubation of MBP-GCM1, [γ-32P]ATP, and CACaMKI-FLAG or DNCaMKI-FLAG immunopurified from 293T cells transfected with pCACaMKI-FLAG or pDNCaMKI-FLAG. As shown in Fig. 3B, GCM1 was phosphorylated by CACaMKI-FLAG but not by DNCaMKI-FLAG, indicating that GCM1 is a CaMKI substrate. The CaMKI phosphorylation sites in GCM1 were further identified by mass spectrometry as Ser47, Thr174, Ser177, Ser178, and Ser269 (Fig. 3B). Correspondingly, MBP-GCM1(1-167) and -(167-349), MBP fusion proteins harboring the identified phosphorylation sites, were phosphorylated by CACaMKI-FLAG, whereas MBP-GCM1(349-436), which does not contain any of the identified phosphorylation sites, was not phosphorylated by CACaMKI-FLAG (data not shown).
Fig. 3.
CaMKI mediates Ser47 phosphorylation in GCM1 to enhance the interaction between GCM1 and SENP1. (A) Mapping of SENP1-interacting domain in GCM1. Matrix preloaded with 1 μg of GST or GST-SENP1 was incubated with cell lysates prepared from 293T cells transiently expressing Gal4-FLAG or full-length or truncated Gal4-GCM1-FLAG in pulldown analysis, followed by immunoblotting with FLAG MAb. Coomassie brilliant blue staining of GST and GST-SENP1 fusion proteins is presented in the bottom panel. (B) Identification of CaMKI phosphorylation sites in GCM1. One microgram of MBP or MBP-GCM1 was incubated with [γ-32P]ATP and immunopurified CACaMKI-FLAG or DNCaMKI-FLAG from 293T cells transfected with pCACaMKI-FLAG or pDNCaMKI-FLAG, followed by SDS-PAGE and autoradiography. In a separate experiment, 4 μg of MBP-GCM1 was incubated with ATP and immunopurified CACaMKI-FLAG for in vitro kinase reaction. Phosphorylated MBP-GCM1 was then subjected to mass spectrometry analysis to identify the CaMKI phosphorylation sites in GCM1 as underlined in the “peptide sequence” column of the table. (C) Ser47 is critical for stimulation of GCM1 activity by CaMKI. 293T cells were transfected with p(GBS)4E1BLuc and pCACaMKI-FLAG plus pHA-GCM1 encoding wild-type HA-GCM1 or HA-GCM1 mutants harboring a serine- or threonine-to-alanine mutation in the CaMKI phosphorylation sites. Fold induction was calculated relative to the luciferase expression observed for p(GBS)4E1BLuc alone. Mean values and the SD obtained from three independent experiments are presented. (D) Ser47 phosphorylation enhances the interaction of GCM1 and SENP1. 293T cells were transfected with pCAHA-CaMKI and pSENP1-FLAG plus pHA-GCM1 encoding either wild-type HA-GCM1 or S47A, S177A, or S47D mutant HA-GCM1, followed by immunoprecipitation and immunoblotting with FLAG and HA MAbs. (E) In vitro interaction of GCM1 and SENP1. Matrix preloaded with 0.5 μg of GST or GST-SENP1 was incubated with 1 μg of mock- or CaMKI-treated MBP or MBP-GCM1 in pulldown analysis, followed by immunoblotting with MBP MAb (top). In a separate experiment, wild-type MBP-GCM1 and MBP-GCM1(S47D) were used as prey proteins in pulldown analyses (bottom). The number underneath each band in the immunoblot indicates the relative intensity of the corresponding band. (F) Ser47 is critical for regulation of GCM1 desumoylation by CaMKI. 293T cells were transfected with the indicated wild-type or mutant pHA-GCM1 expression plasmid, pEGFP-SUMO1, pCACaMKI-FLAG, and pSENP1-FLAG, followed by consecutive immunoprecipitation and immunoblotting with HA MAb. The arrow indicates the sumoylated HA-GCM1(S47A).
Subsequently, we studied the functional role of each CaMKI phosphorylation site by site-directed mutagenesis in transient expression experiments. 293T cells were transfected with p(GBS)4E1BLuc, pCACaMKI-FLAG, and wild-type or mutant pHAGCM1 harboring a serine- or threonine-to-alanine mutation on the individual CaMKI phosphorylation site. As shown in Fig. 3C, the stimulatory effect of CACaMKI-FLAG on GCM1 transcriptional activity was significantly decreased in cells expressing the HA-GCM1(S47A) mutant, suggesting that Ser47 is a critical CaMKI phosphorylation site for CaMKI to stimulate GCM1 activity. We further tested whether Ser47 is required for CaMKI to enhance the interaction of GCM1 and SENP1 by coimmunoprecipitation analysis. Indeed, CaMKI enhanced the interaction between SENP1 and wild-type GCM1 but not that of the GCM1(S47A) mutant (Fig. 3D, lanes 5 and 6). As a control, CaMKI still enhanced the interaction between SENP1 and the GCM1(S177A) mutant (Fig. 3D, lanes 4 and 7). Therefore, Ser47, but not Ser177, is a phosphorylation site critical for CaMKI to regulate the interaction of GCM1 and SENP1. We further studied the in vitro interaction of GCM1 and SENP1 in pulldown assays by incubating GST-SENP1 with MBP-GCM1 pretreated with or without CaMKI. Compared with the mock-treated MBP-GCM1, CaMKI-treated MBP-GCM1 bound strongly to SENP1 (Fig. 3E, top). Moreover, changing Ser47 into aspartic acid (S47D) mimicked the enhancement effect of CaMKI on the interaction between GCM1 and SENP1 in vivo (Fig. 3D, lanes 10 and 11) and in vitro (Fig. 3E, lanes 5 and 6). Correspondingly, an in vivo sumoylation assay indicated that the GCM1(S47A) mutant is more resistant to desumoylation by SENP1 than the wild-type GCM1 and the GCM1(S177A) mutant (Fig. 3F). Taken together, these results suggest that CaMKI mediates Ser47 phosphorylation to enhance the interaction of GCM1 and SENP1 and thereby GCM1 desumoylation.
Epac activates CaMKI to regulate the interaction of GCM1 and SENP1.
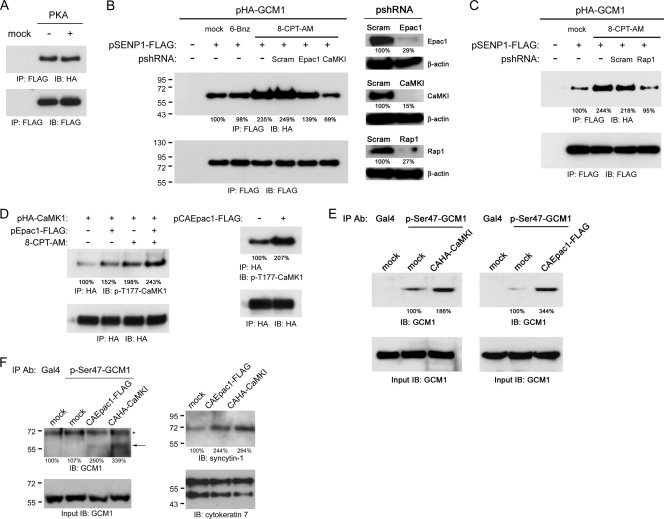
We were curious about how cAMP signaling activates CaMKI to regulate the interaction of GCM1 and SENP1. As PKA is a key effector of the cAMP signaling pathway, we first tested whether PKA is upstream of CaMKI in promoting the interaction of GCM1 and SENP1. As shown in Fig. 4A, expression of the catalytic subunit of PKA did not affect the interaction of HA-GCM1 and SENP1-FLAG in 293T cells by coimmunoprecipitation analysis. We then tested whether Epac, an exchange protein directly activated by cAMP (14), regulates the interaction of GCM1 and SENP1. To this end, 293T cells transiently expressing HA-GCM1 and SENP1-FLAG were treated with or without the Epac activator 8-CPT-AM, followed by coimmunoprecipitation analysis. Interestingly, treatment with 8-CPT-AM, but not with the PKA activator 6-Bnz, stimulated the interaction between HA-GCM1 and SENP1-FLAG (Fig. 4B). Importantly, the stimulatory effect of 8-CPT-AM on the interaction between HA-GCM1 and SENP1-FLAG was abolished when Epac1 or CaMKI was knocked down by RNA interference (Fig. 4B). Therefore, Epac proteins are involved in the CaMKI regulation of the GCM1 and SENP1 interaction.
Fig. 4.
Epac1 regulates the interaction of GCM1 and SENP1. (A) Interaction of GCM1 and SENP1 is independent of PKA. 293T cells were transfected with pHA-GCM1 and pSENP1-FLAG plus or minus an expression plasmid encoding the catalytic subunit of PKA. At 48 h posttransfection, cells were harvested for consecutive immunoprecipitation and immunoblotting with FLAG and HA MAbs. (B) Interaction of GCM1 and SENP1 is enhanced by Epac1. 293T cells were transfected with different combinations of pHA-GCM1, pSENP1-FLAG, and an expression plasmid for scramble, Epac1, or CaMKI shRNA. At 24 h posttransfection, cells were treated with or without 50 μM 6-Bnz or 8-CPT-AM for an additional 24 h. Cells were then harvested for consecutive immunoprecipitation and immunoblotting with FLAG and HA MAbs. The knockdown efficiency of Epac1, CaMKI, and Rap1 genes was analyzed by immunoblotting with the indicated Abs. (C) Rap1 knockdown impairs the interaction of GCM1 and SENP1 stimulated by Eapc1. 293T cells were transfected with pHA-GCM1, pSENP1-FLAG, and an expression plasmid for scramble or Rap1 pshRNA. At 24 h posttransfection, cells were treated with or without 50 μM 8-CPT-AM for an additional 24 h. Cells were then harvested for consecutive immunoprecipitation and immunoblotting with FLAG and HA MAbs. (D) Activation of CaMKI by Epac1. 293T cells were transfected with pHA-CaMKI and pEpac1-FLAG or pCAEpac1-FLAG. At 24 h posttransfection, cells were treated with or without 50 μM 8-CPT-AM for an additional 24 h. Cells were then harvested for consecutive immunoprecipitation and immunoblotting with HA MAb and p-T177-CaMKI Ab. (E) Stimulation of Ser47 phosphorylation by Epac1 and CaMKI in placental cells. Mock BeWo cells and BeWo cells stably expressing CAHA-CaMKI or CAEpac1-FLAG were subjected to immunoprecipitation with Gal4 or p-Ser47-GCM1 Ab, followed by immunoblotting with GCM1 Ab. (F) Regulation of Ser47 phosphorylation and syncytin-1 expression by Epac1 and CaMKI in placental cells. Primary cytotrophoblast cells were infected with recombinant lentivirus strains harboring empty, CAHA-CaMKI, and CAEpac1-FLAG expression cassettes. At 72 h postinfection, cells were harvested for immunoprecipitation and immunoblotting with the indicated Abs. The arrow and asterisk indicate the positions of the Ser47-phosphorylated GCM1 and a nonspecific protein, respectively. The number underneath each band in the immunoblot indicates the relative intensity of the corresponding band in panels B to F.
Because Epac1 regulates Rap protein activity (14, 18), we tested whether Rap1 is also involved in regulation of the interaction between GCM1 and SENP1. Activation of Rap1 by 8-CTP-AM and Epac1 was verified by pulldown assays using GST fusion protein containing the Rap-binding domain of RalGDS (data not shown). Interestingly, Rap1 knockdown also suppressed the interaction between HA-GCM1 and SENP1-FLAG stimulated by 8-CPT-AM (Fig. 4C). Because Thr177 phosphorylation is required for CaMKI activity (30), we further demonstrated that both 8-CPT-AM and Epac1 stimulate Thr177 phosphorylation in CaMKI (Fig. 4D, left). Likewise, a constitutively active form of Epac1 (CAEpac1) also stimulated CaMKI activity, in terms of Thr177 phosphorylation (Fig. 4D, right). Therefore, these results indicate that Epac1 and Rap1 activate CaMKI to stimulate the interaction of GCM1 and SENP1.
We next studied whether Ser47 phosphorylation in GCM1 is stimulated by CaMKI and Epac1 in placental cells. To this end, an antibody against phosphorylated Ser47 in GCM1 (p-Ser47-GCM1) was generated. Parental BeWo cells and BeWo cells stably expressing CAHA-CaMKI or CAEpac1-FLAG were immunoprecipitated using Gal4 or p-Ser47-GCM1 Ab, followed by immunoblotting using GCM1 Ab. As shown in Fig. 4E, Ser47 phosphorylation in GCM1 was specifically detected in mock BeWo cells using p-Ser47-GCM1 Ab but not Gal4 Ab. Importantly, a higher level of Ser47-phosphorylated GCM1 was detected in the CAHA-CaMKI- and CAEpac1-FLAG-expressing BeWo cells than in the mock BeWo cells (Fig. 4E). Stimulation of Ser47 phosphorylation in GCM1 by CAHA-CaMKI and CAEpac1-FLAG was also observed in purified cytotrophoblast cells from term placentas (Fig. 4F, left). In line with the increased GCM1 activity, the protein level of syncytin-1 in the purified cytotrophoblast cells was increased in the presence of CAHA-CaMKI or CAEpac1-FLAG (Fig. 4F, right). Taken together, these results suggest that cAMP signaling activates CaMKI through Epac1 and Rap1 to facilitate Ser47 phosphorylation and thereby the interaction between GCM1 and SENP1.
Epac regulates placental cell fusion via GCM1.
To look for further evidence that Epac1 regulates the function of GCM1, we examined the effect of 8-CPT-AM on GCM1-dependent cell-cell fusion of BeWo cells. BeWo cells expressing scramble or GCM1 shRNA (Fig. 5A) were treated with or without 8-CPT-AM, followed by immunostaining with an anti-desmosomal protein MAb. Increased cell-cell fusion events were observed in the 8-CPT-AM-treated BeWo cells expressing scramble shRNA compared with that in the mock-treated cells (compare panels a and b in Fig. 5B). Interestingly, the stimulatory effect of 8-CPT-AM on cell-cell fusion was compromised when GCM1 was knocked down in BeWo cells expressing GCM1 shRNA (compare panels b and d in Fig. 5B). We also studied the effect of 8-CPT-AM on expression of GCM1 target gene products, syncytin-1 and -2, by quantitative real-time PCR. As shown in Fig. 5C, treatment with 8-CPT-AM raised the transcript levels of syncytin-1 and -2 in the BeWo cells expressing scramble shRNA but not in the BeWo cells expressing GCM1 shRNA. Therefore, stimulation of syncytin-1 and -2 gene expression by 8-CPT-AM is also GCM1 dependent. Correspondingly, ChIP analysis indicated that association of GCM1 with the syncytin-1 promoter region is enhanced in BeWo cells treated with 8-CPT-AM and in BeWo cells expressing CAHA-CaMKI (Fig. 5D). Because Ser47 is critical for CaMKI to regulate GCM1 activity, we introduced RNAi-resistant wild-type HA-GCM1 and HA-GCM1(S47A) and HA-GCM1(S47D) mutants into GCM1-knockdown BeWo cells for cell-cell fusion analysis. Quantitatively, cell-cell fusion was reversed by wild-type HA-GCM1, which was further enhanced by the phosphomimetic HA-GCM1(S47D) mutant (Fig. 5E). Taken together, these results suggest that stimulation of placental cell fusion by cAMP can be accomplished by activation of a cAMP/Epac1/CaMKI/GCM1 signaling pathway.
Fig. 5.
Regulation of placental cell fusion by Epac1. (A) Knockdown of GCM1 in BeWo placental cells. BeWo cells stably expressing scramble or GCM1 shRNA were immunoblotted with GCM1 Ab and β-actin MAb, respectively. (B) GCM1 is required for stimulation of placental cell fusion by 8-CPT-AM. BeWo cells stably expressing scramble or GCM1 shRNA were treated with or without 50 μM 8-CPT-AM for 24 h, followed by immunofluorescence microscopy with a MAb against anti-desmosomal protein (green). Nuclei were stained by DAPI (blue). Syncytial margins are marked with a stippled line. Quantification of cell fusion was performed as described in Materials and Methods. Mean values and the SD from three independent experiments are presented. (C) Regulation of GCM1 target genes by 8-CPT-AM. BeWo cells expressing scramble or GCM1 shRNA were treated with or without 50 μM 8-CPT-AM for 24 h. Cells were harvested for quantitative real-time PCR analysis using designated primer sets for syncytin-1 (Syn1) and -2 (Syn2). Mean values and the SD from three independent experiments are presented. (D) Association of GCM1 with syncytin-1 promoter is stimulated by Epac1 and CaMKI. BeWo cells were treated with or without 50 μM 8-CPT-AM for 24 h, followed by ChIP analysis using normal rabbit serum (NS) or GCM1 Ab (top). In a separate experiment, mock and CAHA-CaMKI-expressing BeWo cells were subjected to ChIP analysis using the same serum and Ab (bottom). (E) Stimulation of BeWo cell fusion by GCM1(S47D). GCM1-knockdown BeWo cells were transduced with lentivirus harboring an expression cassette for RNAi-resistant wild-type HA-GCM1 or the phosphomimetic mutant HA-GCM1(S47D). Cells were subjected to cell-cell fusion analysis as described for panel A. Mean values and the SD of three independent experiments are presented.
DISCUSSION
Physiological cell-cell fusion is required for fertilization of egg and sperm and differentiation of myotubes, macrophages, and the syncytiotrophoblast. Interestingly, the multinucleated syncytiotrophoblast layer in a placental villus undergoes apoptosis and sheds into the maternal circulation, which underscores the need for a renewal mechanism to replenish the layer during pregnancy. How is fusion of mononuclear cytotrophoblasts into the overlying syncytiotrophoblast layer regulated? Previous studies have demonstrated that cAMP stimulates cell-cell fusion of cultured placental cells and expression of placental fusogenic proteins (19, 24, 33). Moreover, GCM1 is a critical regulator of placental cell fusion because (i) GCM1 knockout impairs syncytiotrophoblast differentiation in mouse (1), (ii) GCM1 knockdown decreases the cell-cell fusion of BeWo cells stimulated by forskolin (2), and (iii) GCM1 regulates the expression of placental fusogenic proteins (22, 38). These observations indicate that cAMP signaling may be crucial for regulation of GCM1 activity to properly maintain a functional syncytiotrophoblast layer in the outer surface of a placental villus.
Indeed, GCM1 transcription is stimulated by forskolin (21) and regulated by transcription factors such as CREB-1, TORC1, and OASIS (29). Recently, Delidaki et al. (11) have further demonstrated that p38 mitogen-activated protein kinase (MAPK) is involved in forskolin-stimulated transcription of GCM1, syncytin-1, -2, MFSD2, and OASIS. Because the syncytin-1 gene, the syncytin-2 gene, and its receptor the MFSD2 gene are GCM1 target genes (22, 38), the aforementioned stimulation of syncytin-1, -2, and MFSD2 gene expression by forskolin might be attributed to elevated GCM1 transcription.
Here, we further identified a novel cAMP signaling pathway that stimulates GCM1 activity and placental cell fusion via Epac1 but not PKA. Specifically, we demonstrated that cAMP activates Epac1, leading to activation of CaMKI, which mediates Ser47 phosphorylation in GCM1 to enhance the interaction between GCM1 and SENP1. Consequently, GCM1 is desumoylated to relieve the inhibition of its DNA-binding activity by sumoylation, thereby elevating GCM1 activity to stimulate placental cell fusion. Several lines of evidence support this scenario. First, the CaMKI inhibitor KN93 inhibits the stimulatory effect of cAMP stimulant forskolin on GCM1 transcriptional activity. Second, GCM1 steady-state sumoylation levels are lowered by a constitutively active CaMKI, which also stimulates the interaction of GCM1 and SENP1 in placental BeWo and JAR cells. Third, the interaction between GCM1 and SENP1 is not affected by the catalytic subunit of PKA or the PKA activator 6-Bnz. Instead, the interaction is significantly stimulated by the Epac activator 8-CPT-AM. Fourth, phosphorylation of Ser47 in GCM1 by CaMKI enhances the interaction between GCM1 and SENP1. Moreover, introduction of the RNAi-resistant phosphomimetic mutant GCM1(S47D) into GCM1-knockdown BeWo cells not only reversed cell fusion defect but further enhanced cell-cell fusion. A recent report published during the course of this study showed that activation of Epac proteins by 8-CPT-AM promotes cell-cell fusion of BeWo cells (37). Although Epac1 and Rap1 were implicated as downstream effectors of 8-CPT-AM in that report, the mechanism underlying Epac-stimulated cell fusion was not clear. Therefore, the results of our present study reveal more of the underpinnings of placental cell fusion stimulated by the Epac pathway of cAMP signaling.
Structural studies suggested that sumoylation of Lys156 may impose some steric interference at the DNA-binding surfaces of GCM1 and thereby inhibit the DNA-binding activity of GCM1 (9, 10). Ser47 is located in the turn linking the first strand of β-sheet and a following α-helix and is most likely on the outer surface of the GCM1 protein (10). It is feasible to speculate that Ser47 is accessible to CaMKI for phosphorylation, and once phosphorylated, it facilitates the recruitment of SENP1 to desumoylate Lys156. How Ser47 phosphorylation affects the interaction between GCM1 and SENP1 is currently not known. Whether it involves a conformational change or something else is an intriguing question that warrants further investigation.
The present study indicates that Epac1 and Rap1 are involved in activation of CaMKI for Ser47 phosphorylation in GCM1. Because phospholipase Cε and protein kinase Cε are required for Epac and Rap proteins to activate CaMKII in Ca2+ handling in cardiac myocytes (25–27), we also tested whether phospholipase C and protein kinase C are involved in CaMKI-mediated Ser47 phosphorylation in the GCM1-knockdown BeWo cells. Indeed, Ser47 phosphorylation in the RNAi-resistant HA-GCM1 stimulated by 8-CPT-AM was suppressed by KN93 and the Rap inhibitor GGTI298 but was not affected by the phospholipase C inhibitor U73122 or the protein kinase C inhibitor BISI (data not shown). These observations suggest that neither phospholipase C nor protein kinase C is likely to be involved in CaMKI activation by Epac. Therefore, just how CaMKI is activated by Epac1 and Rap1 in the placenta remains elusive.
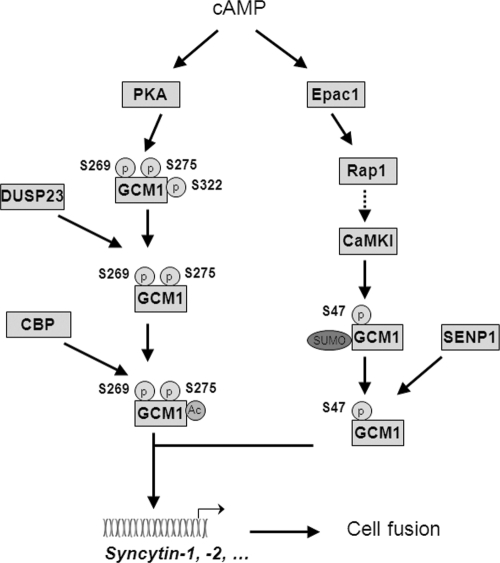
cAMP signaling can be transduced by PKA, Epac proteins, and cyclic nucleotide-gated ion channels for many cellular activities. Interestingly, Epac may synergize with PKA to promote proliferation of thyroid PCCL3 cells in response to cAMP elevation by thyroid-stimulating hormone (TSH) and to accelerate adipocyte differentiation of 3T3-L1 cells treated with forskolin or IBMX (3-isobutyl-1-methylxanthine) in conjunction with dexamethasone and insulin (15, 28). Our previous studies have demonstrated that GCM1 activity is positively regulated by different posttranslational modifications, which can be coordinated in the cAMP/PKA signaling pathway (3, 23). In this scenario, PKA phosphorylates Ser269 and Ser275 in GCM1 and enhances the recruitment of DUSP23 for Ser322 dephosphorylation, which prevents GCM1 degradation and facilitates the interaction of GCM1 and CBP (Fig. 6). As a result, CBP mediates GCM1 acetylation to further stabilize GCM1, and the CBP-GCM1 complex transactivates syncytin expression to promote placental cell fusion. Of note, we believe that p38 MAPK is not involved in the cAMP/PKA signaling pathway that upregulates GCM1 activity at the posttranslational level, because the p38 MAPK inhibitor SB 203580 does not significantly affect the transcriptional activity of GCM1 stimulated by forskolin in transient expression experiments (3). Together with the present study that demonstrates upregulation of GCM1 activity by the cAMP/Epac1/CaMKI signaling pathway, we now believe that cAMP signaling initiates two separate signaling cascades to increase GCM1 stability by protection of GCM1 from ubiquitination and to maintain the DNA-binding activity of GCM1 by enhancement of GCM1 desumoylation (Fig. 6). Importantly, both events converge to stimulate GCM1 activity in placental cells and promote placental cell fusion and differentiation.
Fig. 6.
Regulation of GCM1 activity and placental cell fusion by cAMP. GCM1 regulates expression of syncytin-1 and -2 fusogenic proteins to promote placental cell fusion. cAMP may activate PKA to mediate Ser269 and Ser275 phosphorylation in GCM1, which further enhances the interaction of GCM1 and DUSP23, leading to Ser322 dephosphorylation by DUSP23. This prevents FBW2-mediated ubiquitination and facilitates GCM1 acetylation and stabilization by CBP. In addition, cAMP may activate Epac1 and Rap1 to upregulate CaMKI activity, which mediates Ser47 phosphorylation. This enhances the interaction of GCM1 and SENP1, leading to GCM1 desumoylation by SENP1 and restoration of the DNA-binding activity of GCM1. Therefore, cAMP activates both PKA- and Epac1-dependent pathways to increase GCM1 activity to promote placental cell fusion.
ACKNOWLEDGMENTS
This work was supported by grants (to H.C.) from the National Science Council (grant no. 100-2311-B-001-010) and Academia Sinica, Taiwan.
We thank the peptide synthesis laboratory of the Institute of Biological Chemistry, Academia Sinica.
Footnotes
Published ahead of print on 26 July 2011.
REFERENCES
- 1. Anson-Cartwright L., et al. 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25:311–314 [DOI] [PubMed] [Google Scholar]
- 2. Baczyk D., et al. 2009. Glial cell missing-1 transcription factor is required for the differentiation of the human trophoblast. Cell Death Differ. 16:719–727 [DOI] [PubMed] [Google Scholar]
- 3. Chang C. W., Chuang H. C., Yu C., Yao T. P., Chen H. 2005. Stimulation of GCMa transcriptional activity by cyclic AMP/protein kinase A signaling is attributed to CBP-mediated acetylation of GCMa. Mol. Cell. Biol. 25:8401–8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen C. P., et al. 2008. Functional characterization of the human placental fusogenic membrane protein syncytin 2. Biol. Reprod. 79:815–823 [DOI] [PubMed] [Google Scholar]
- 5. Chen H., Chong Y., Liu C. L. 2000. Active intracellular domain of Notch enhances transcriptional activation of CCAAT/enhancer binding protein beta on a rat pregnancy-specific glycoprotein gene. Biochemistry 39:1675–1682 [DOI] [PubMed] [Google Scholar]
- 6. Cheng J., Kang X., Zhang S., Yeh E. T. 2007. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 131:584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiang M. H., et al. 2009. Mechanism of hypoxia-induced GCM1 degradation: implications for the pathogenesis of preeclampsia. J. Biol. Chem. 284:17411–17419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chiu S. Y., Asai N., Costantini F., Hsu W. 2008. SUMO-specific protease 2 is essential for modulating p53-Mdm2 in development of trophoblast stem cell niches and lineages. PLoS Biol. 6:e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chou C. C., et al. 2007. Small ubiquitin-like modifier modification regulates the DNA binding activity of glial cell missing Drosophila homolog a. J. Biol. Chem. 282:27239–27249 [DOI] [PubMed] [Google Scholar]
- 10. Cohen S. X., et al. 2003. Structure of the GCM domain-DNA complex: a DNA-binding domain with a novel fold and mode of target site recognition. EMBO J. 22:1835–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delidaki M., Gu M., Hein A., Vatish M., Grammatopoulos D. K. 2011. Interplay of cAMP and MAPK pathways in hCG secretion and fusogenic gene expression in a trophoblast cell line. Mol. Cell. Endocrinol. 332:213–220 [DOI] [PubMed] [Google Scholar]
- 12. de Rooij J., et al. 2000. Mechanism of regulation of the Epac family of cAMP-dependent RapGEFs. J. Biol. Chem. 275:20829–20836 [DOI] [PubMed] [Google Scholar]
- 13. Gareau J. R., Lima C. D. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11:861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gloerich M., Bos J. L. 2010. Epac: defining a new mechanism for cAMP action. Annu. Rev. Pharmacol. Toxicol. 50:355–375 [DOI] [PubMed] [Google Scholar]
- 15. Hochbaum D., Hong K., Barila G., Ribeiro-Neto F., Altschuler D. L. 2008. Epac, in synergy with cAMP-dependent protein kinase (PKA), is required for cAMP-mediated mitogenesis. J. Biol. Chem. 283:4464–4468 [DOI] [PubMed] [Google Scholar]
- 16. Johnson E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355–382 [DOI] [PubMed] [Google Scholar]
- 17. Kang X., et al. 2010. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38:191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kawasaki H., et al. 1998. A family of cAMP-binding proteins that directly activate Rap1. Science 282:2275–2279 [DOI] [PubMed] [Google Scholar]
- 19. Keryer G., Alsat E., Tasken K., Evain-Brion D. 1998. Cyclic AMP-dependent protein kinases and human trophoblast cell differentiation in vitro. J. Cell Sci. 111:995–1004 [DOI] [PubMed] [Google Scholar]
- 20. Kim J. H., Baek S. H. 2009. Emerging roles of desumoylating enzymes. Biochim. Biophys. Acta 1792:155–162 [DOI] [PubMed] [Google Scholar]
- 21. Knerr I., et al. 2005. Stimulation of GCMa and syncytin via cAMP mediated PKA signaling in human trophoblastic cells under normoxic and hypoxic conditions. FEBS Lett. 579:3991–3998 [DOI] [PubMed] [Google Scholar]
- 22. Liang C. Y., et al. 2010. GCM1 regulation of the expression of syncytin 2 and its cognate receptor MFSD2A in human placenta. Biol. Reprod. 83:387–395 [DOI] [PubMed] [Google Scholar]
- 23. Lin F. Y., et al. 2011. Dual-specificity phosphatase 23 mediates GCM1 dephosphorylation and activation. Nucleic Acids Res. 39:848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mi S., et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789 [DOI] [PubMed] [Google Scholar]
- 25. Oestreich E. A., et al. 2009. Epac and phospholipase C(epsilon) regulate Ca2+ release in the heart by activation of protein kinase C(epsilon) and calcium-calmodulin kinase II. J. Biol. Chem. 284:1514–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oestreich E. A., et al. 2007. Epac-mediated activation of phospholipase C(epsilon) plays a critical role in beta-adrenergic receptor-dependent enhancement of Ca2+ mobilization in cardiac myocytes. J. Biol. Chem. 282:5488–5495 [DOI] [PubMed] [Google Scholar]
- 27. Pereira L., et al. 2007. The cAMP binding protein Epac modulates Ca2+ sparks by a Ca2+/calmodulin kinase signalling pathway in rat cardiac myocytes. J. Physiol. 583:685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petersen R. K., et al. 2008. Cyclic AMP (cAMP)-mediated stimulation of adipocyte differentiation requires the synergistic action of Epac- and cAMP-dependent protein kinase-dependent processes. Mol. Cell. Biol. 28:3804–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schubert S. W., et al. 2008. bZIP-type transcription factors CREB and OASIS bind and stimulate the promoter of the mammalian transcription factor GCMa/Gcm1 in trophoblast cells. Nucleic Acids Res. 36:3834–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stedman D. R., Uboha N. V., Stedman T. T., Nairn A. C., Picciotto M. R. 2004. Cytoplasmic localization of calcium/calmodulin-dependent protein kinase I-alpha depends on a nuclear export signal in its regulatory domain. FEBS Lett. 566:275–280 [DOI] [PubMed] [Google Scholar]
- 31. Tuerk E. E., Schreiber J., Wegner M. 2000. Protein stability and domain topology determine the transcriptional activity of the mammalian glial cells missing homolog, GCMb. J. Biol. Chem. 275:4774–4782 [DOI] [PubMed] [Google Scholar]
- 32. Wayman G. A., et al. 2004. Regulation of axonal extension and growth cone motility by calmodulin-dependent protein kinase I. J. Neurosci. 24:3786–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wice B., Menton D., Geuze H., Schwartz A. L. 1990. Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp. Cell Res. 186:306–316 [DOI] [PubMed] [Google Scholar]
- 34. Wilkinson K. A., Henley J. M. 2010. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428:133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamaguchi T., et al. 2005. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol. Cell. Biol. 25:5171–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang C. S., et al. 2005. FBW2 targets GCMa to the ubiquitin-proteasome degradation system. J. Biol. Chem. 280:10083–10090 [DOI] [PubMed] [Google Scholar]
- 37. Yoshie M., et al. 2010. Possible role of the exchange protein directly activated by cyclic AMP (Epac) in the cyclic AMP-dependent functional differentiation and syncytialization of human placental BeWo cells. Hum. Reprod. 25:2229–2238 [DOI] [PubMed] [Google Scholar]
- 38. Yu C., et al. 2002. GCMa regulates the syncytin-mediated trophoblastic fusion. J. Biol. Chem. 277:50062–50068 [DOI] [PubMed] [Google Scholar]