Abstract
The cytoplasmic viral RNA sensors RIG-I and MDA5 are important for the production of type I interferon and other inflammatory cytokines. DDX60 is an uncharacterized DEXD/H box RNA helicase similar to Saccharomyces cerevisiae Ski2, a cofactor of RNA exosome, which is a protein complex required for the integrity of cytoplasmic RNA. Expression of DDX60 increases after viral infection, and the protein localizes at the cytoplasmic region. After viral infection, the DDX60 protein binds to endogenous RIG-I protein. The protein also binds to MDA5 and LGP2 but not to the downstream factors IPS-1 and IκB kinase ε (IKK-ε). Knockdown analysis shows that DDX60 is required for RIG-I- or MDA5-dependent type I interferon and interferon-inducible gene expression in response to viral infection. However, DDX60 is dispensable for TLR3-mediated signaling. Purified DDX60 helicase domains possess the activity to bind to viral RNA and DNA. Expression of DDX60 promotes the binding of RIG-I to double-stranded RNA. Taken together, our analyses indicate that DDX60 is a novel antiviral helicase promoting RIG-I-like receptor-mediated signaling.
INTRODUCTION
RIG-I and MDA5 are cytoplasmic viral RNA sensors belonging to the group of RIG-I-like receptors (RLRs), which includes LGP2 (57–59). RIG-I recognizes RNAs from vesicular stomatitis virus (VSV), hepatitis C virus (HCV), Sendai virus (SeV), and influenza A virus (21, 36, 37), while MDA5 recognizes RNA from picornaviruses such as encephalomyocarditis virus and poliovirus (PV) (3, 19, 21). RLRs are also involved in the recognition of cytoplasmic B-DNA. RNA polymerase III transcribes cytoplasmic AT-rich double-stranded DNA (dsDNA), and the transcribed RNA is recognized by RIG-I (1, 6). In contrast, Choi et al. have reported that RIG-I associates with dsDNA (7).
When RIG-I or MDA5 is activated by viral infection, the N-terminal caspase recruitment domains (CARDs) associate with the adaptor protein IPS-1 (also called MAVS/Cardif/VISA) on the outer mitochondrial membrane (22, 26, 42, 55). After this association occurs, IPS-1 activates TBK1 and IκB kinase ε (IKK-ε) and signals interferon (IFN) regulatory factor 3 (IRF-3)- and NF-κB-responsive genes, such as those for type I IFNs or other inflammatory cytokines (22, 23, 26, 42, 44, 55).
Both the helicase and C-terminal domain (CTD) of RIG-I bind to RNA, but it is the CTD that is responsible for the recognition of the 5′ triphosphate double-stranded structure typical of viral RNA (16, 39, 40). Recently, Rehwinkel et al. showed that the physiological ligand of RIG-I during influenza A virus or SeV infection is the full-length viral genomic single-stranded RNA (ssRNA), which possesses base-paired regions or defective interfering (DI) genomes (35). In contrast to RIG-I, MDA5 recognizes long viral double-stranded RNA (dsRNA) (21). The RNA binding activity of the MDA5 CTD is relatively weak compared with that of the RIG-I CTD, because the basic surface of the MDA5 CTD has a more extensive flat region than the RIG-I CTD (8, 45, 46). Although the RNA binding activity of the MDA5 CTD is weak, this protein plays a pivotal role in the recognition of picornavirus RNA (20, 21).
For the efficient recognition of viral RNA, RIG-I and MDA5 require protein modification and association with upstream factors. LGP2 is one of the upstream factors. LGP2 lacks an N-terminal CARD; thus, LGP2 itself cannot transmit the signal in the absence of RIG-I or MDA5 (36, 38, 49). The CTD of LGP2, which binds to the terminal region of viral double-stranded RNA, is more similar to the CTD of RIG-I than to that of MDA5 (24, 33, 45). LGP2 knockout studies have revealed that LGP2 is essential for type I IFN production by MDA5 but plays only a minor role in type I IFN production by RIG-I (38, 49). RIG-I requires modification of K63-linked polyubiquitination by TRIM25 and Riplet/REUL ubiquitin ligases for its full activation (11, 13, 30, 31). High-mobility-group box (HMGB) proteins also act as upstream factors of RLRs. Recently, Yanai and colleagues reported that HMGB1, HMGB2, and HMGB3 serve as sentinels for the nucleic acids required for both RIG-I and MDA5 recognition of viral RNA (56). Hayakawa and colleagues reported that ZAPS associates with RIG-I to promote oligomerization and ATPase activity of RIG-I (15). Another factor interacting with RLRs is DDX3, a DEXD/H box RNA helicase, which is similar to LGP2 in that it does not contain a CARD but promotes signaling by forming a complex with either RIG-I or MDA5 (32). DDX3 also plays important roles in TBK-1- and IKK-ε-mediated IRF activation, and Schröder et al. and Soulat et al. were the first to describe results showing that DDX3 is a non-RLR helicase involved in innate immune responses (41, 43).
DDX60, a DEXD/H box helicase, was annotated in a genome project, and the protein function is unknown. The protein is weakly similar to SKIV2L and SKIV2L2 and is the human homolog of Saccharomyces cerevisiae (budding yeast) Ski2, a cofactor of the RNA exosome (9, 18). The RNA exosome is a macromolecular protein complex that includes ribonucleases and helicases and controls the quality of host RNA molecules in both the nucleus and cytoplasm (17). It is composed of nine core components and several cofactor proteins (18). In budding yeast, the RNA exosome and Ski2 together exhibit antiviral activity (53, 54); similarly, the mammalian RNA exosome, together with its cofactors, shows antiviral activity against Moloney leukemia virus and Sindbis virus (5, 14). Our microarray analysis has shown that DDX60 is upregulated in human dendritic cells during infection with measles virus (MV) (our unpublished results). Thus, we expected DDX60 to be a novel antiviral protein and found that DDX60 is involved in RIG-I-like receptor-dependent antiviral pathways.
Here, we show that DDX60 is induced during viral infection and suppresses viral replication. DDX60 was found to form a complex with RLRs, promoting signaling; the results of knockdown experiments indicated that DDX60 is involved in RLR-dependent pathways. Moreover, the DDX60 helicase domain was observed to bind to viral RNA and DNA. Furthermore, DDX60 is required for type I IFN expression after DNA virus infection. These data indicate that DDX60 is a novel antiviral helicase involved in RLR-dependent pathways.
MATERIALS AND METHODS
Cell cultures.
HEK293 and Vero cells were cultured in Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal calf serum (Invitrogen), and HeLa cells were cultured in minimum Eagle's medium with 2 mM l-glutamine and 10% heat-inactivated fetal calf serum (JRH Biosciences). HEK293FT cells were maintained in Dulbecco's modified Eagle's high-glucose medium containing 10% heat-inactivated fetal calf serum (Invitrogen). RAW 264.7 cells were cultured in RPMI 1640 medium with 10% heat-inactivated fetal calf serum (Invitrogen). Mouse bone marrow-derived dendritic cells (BM-DCs) were induced as described in reference 2. PV receptor (PVR)-transgenic mice were provided by S. Koike (Tokyo Metropolitan Institute for Neuroscience).
Plasmids.
Full-length human DDX60 cDNA was obtained from HeLa cell total RNA by reverse transcription-PCR (RT-PCR). The obtained cDNA fragments were sequenced, and we confirmed by PCR that the obtained cDNA clones do not contain nucleotide mutations. The DDX60 cDNA clone was cloned into XhoI and NotI restriction sites of pEF-BOS, and a hemagglutinin (HA) tag sequence was inserted just before the stop codon. EXOSC1, EXOSC4, or EXOSC5 was amplified by RT-PCR from HeLa cell total RNA. The obtained cDNA fragment was cloned into XhoI and NotI restriction sites of pEF-BOS vector, and the FLAG tag was fused at the C-terminal end. The DDX6 cDNA carrying a full-length open reading frame (ORF) was amplified by RT-PCR using primers DDX6-F (GGC CGC TCG AGC CAC CAT GAG CAC GGC CAG AAC AGA G) and DDX6-R (GGC GGG GTA CCC CAG GTT TCT CAT CTT CTA CAG). The fragment was cloned into XhoI and NotI sites of pEF-BOS vector. For in vitro viral RNA synthesis, we amplified VSV-G region cDNA by PCR using primers VSV-G-F and VSV-G-R. The obtained cDNA fragment was cloned into pGEM-T Easy vector. The primer sequences are as follows: for VSV-G-F, ACAGGAGAATGGGTTGATTC; and for VSV-G-R, ATGCAAAGATGGATACCAAC. Vectors expressing full RIG-I or RIG-I fragments were described before (30). The plasmids expressing TLR3 or TICAM-1 are described in reference 29. The p125luc reporter plasmid was a gift from T. Taniguchi (University of Tokyo, Tokyo, Japan). Mutant DDX60 expression constructs were amplified using primers for DDX60 (amino acids [aa] 1 to 169), (aa 169 to 334), (aa 334 to 490), (aa 478 to 656), (aa 657 to 857), (aa 857 to 1054), (aa 1049 to 1256), (aa 1256 to 1409), (aa 1407 to 1543), and (aa 1543 to 1712). The primer sequences are shown in Table S1 in the supplemental material.
Phylogenetic analysis.
The amino acid sequences of the DEXD/H box domain were aligned using ClustalW software on the NIG server. The phylogenetic tree was drawn using the neighbor-joining method and GENETYX-MAC software (version 13.0.3).
Northern blotting.
A human DDX60 644-bp cDNA fragment (from the region at bp 3978 to 4621) was used for the probe for Northern blotting. The Northern blot membranes (human 12-lane multiple-tissue Northern [MTN] blot and MTN blot III) were purchased from Clontech. The probe was labeled using [α-32P]dCTP and a Rediprime II random prime labeling system (GE Healthcare). The labeled probe was hybridized to the membrane by the use of ExpressHyb hybridization solution (Clontech) at 68°C for 1 h. The membrane was washed with washing solution I (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.05% sodium dodecyl sulfate [SDS]) for 40 min and then washed with washing solution II (0.1× SSC, 0.1% SDS) for 40 min. DDX60 mRNA bands were detected with X-ray film.
RT-PCR and real-time PCR.
Total RNA was extracted with TRIzol reagent (Invitrogen), and then the samples were treated with DNase I to remove the DNA contamination. The reverse transcription reaction was carried out using a high-capacity cDNA reverse transcription kit (ABI). Quantitative PCR (qPCR) analysis was performed using Step One software version 2.0 (ABI) and SYBR Green Master Mix (ABI). The primer sequences for qPCR and PCR are described in Table S2 and S3 in the supplemental material, respectively.
Microarray analysis.
Mouse bone marrow-derived dendritic cells were stimulated with poly(I·C) or infected with MV with anti-IFN-AR antibody (Ab) (see Fig. 2A). Total RNA was extracted from the cells, and we performed microarray analysis using Affimetrix GeneChip Mouse 430.2 software (10). The data were analyzed by GeneSpring GX 11 software.
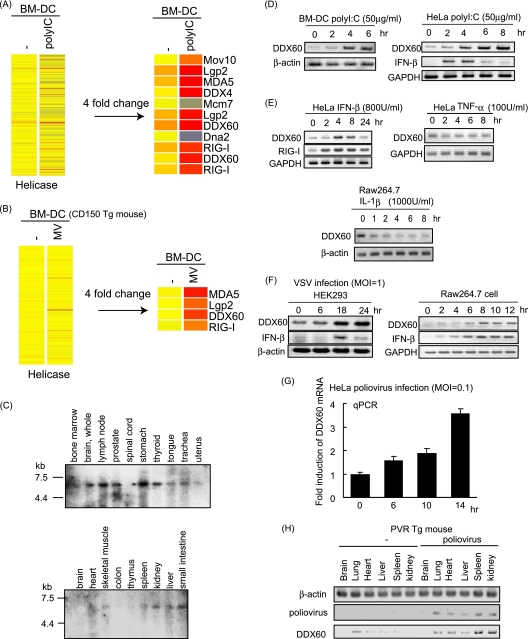
Fig. 2.
Expression of DDX60 mRNA. (A and B) Mouse BM-DCs were stimulated with poly(I·C) (A) or infected with MV in the presence of anti-IFN-AR antibody (B). Total RNA was extracted from the cells, and microarray analysis was performed. The heat maps in the left column show the expression profiles of the genes encoding the helicase domain. The heat maps in the right column show the genes encoding the helicase domain whose expression levels changed more than 4-fold. (C) Northern blot of human DDX60 mRNAs in specified tissues. Northern blots of human tissues were probed with DDX60 cDNA. (D and E) Mouse BM-DCs, HeLa cells, or RAW 264.7 cells were stimulated with 50 μg of poly(I·C)/ml (D), 800 U of IFN-β/ml (E), 100 U of TNF-α/ml (E), or 1,000 U of IL-1β/ml (E). Expression of DDX60, RIG-I, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and β-actin mRNA was examined by RT-PCR. (F and G) HEK293 cells, RAW 264.7 cells, or HeLa cells were infected with VSV at an MOI of 1 (F) or PV at an MOI of 0.1 (G). Expression of DDX60, IFN-β, β-actin, and/or GAPDH was examined by RT-PCR (F) or RT-qPCR (G). (H) PV was injected intraperitoneally (i.p.) into PVR-transgenic mice susceptible to PV. Tissue RNA extraction was performed before or 3 days after infection, and RT-PCR was carried out on these samples.
RNA interference.
RNA interference (RNAi) vectors were constructed by the insertion of oligonucleotides into the XbaI and PstI site of the pH 1 vector. The target sequence for DDX60 is 5′-CTTTACCACTTTCCTACGA-3′, the target sequence for EXOSC4 is 5′-TATAGTTCAGCGACCTT-3′, and the target sequence for EXOSC5 is 5′-GGATCCTACATCCAAGCAA-3′. HeLa cells or HEK293 cells were transfected with RNAi vector (0.5 μg) by the use of 24-well plates and FuGENE HD (Roche). After incubation for 24 h, cells were recovered and suspended in 12 ml of medium and then seeded to a 24-well plate. At 24 h after incubation, puromycin (1.0 μg/ml) was added. The medium containing puromycin was changed every 5 days, puromycin-resistant colonies were recovered, and the mRNAs of endogenous DDX60 or EXOSC4 and EXOSC5 were checked by RT-PCR. The sequences of the primers used for RT-PCR are described in Table S3 in the supplemental material. Small interfering RNA (siRNA) for DDX60 was purchased from Ambion and was transfected with Lipofectamine 2000 reagent (Invitrogen). The target sequence was 5′-GGC TAA CAA ACT TCG AAA A-3′.
Immunoprecipitation.
HEK293FT cells were transfected in 6-well plates with plasmids encoding FLAG-tagged RIG-I, MDA5, EXOSC1, EXOSC4, EXOSC5, LGP2, IKK-ε, and Ubc13- and/or HA-tagged DDX60. The plasmid amounts were normalized by the addition of empty plasmid. At 24 h after transfection, cells were lysed with lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Nonidet P-40, 30 mM NaF, 5 mM Na3VO4, 20 mM iodoacetamide, 2 mM phenylmethylsulfonyl fluoride), and then proteins were immunoprecipitated with rabbit anti-HA polyclonal antibody (Sigma) or anti-FLAG M2 monoclonal antibody (Sigma). The precipitates were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with anti-HA polyclonal or anti-FLAG M2 monoclonal antibody or monoclonal antibody to RIG-I (Alme-1) (Alexis Biochemicals).
Confocal microscopy.
HeLa and HEK293 cells were plated onto Micro Cover glass sheets (Matsunami) and poly-l-lysin-coated cover glass sheets (eBioscience), respectively, in a 24-well plate. The following day, cells were transfected with the indicated plasmids. At 24 h after transfection, cells were infected with VSV or transfected with poly(I·C) by the use of 0.5 μg/ml DEAE-dextran for 4 h and then fixed using 3% formaldehyde-phosphate-buffered saline (PBS) for 30 min and permeabilized with 0.2% Triton X-100 for 15 min. Fixed cells were blocked in 1% bovine serum albumin (BSA)-PBS for 10 min and labeled with the indicated primary Abs (5 μg/ml) for 60 min at room temperature. Alexa-conjugated secondary Abs (1:400) were used to visualize staining of the primary Abs for 30 min at room temperature, and the mixture was mounted onto glass slides by the use of PBS containing 2.3% 1,4-diazabicyclo[2.2.2]octane and 50% glycerol or Prolong Gold antifade reagent with DAPI (4′,6′diamidino-2-phenylindole; Invitrogen). Cells were visualized at a magnification of ×63 with an LSM510 Meta microscope (Zeiss). To stain the mitochondria, endoplasmic reticulum (ER), early endosome, and autophagosome, we used Mitotracker (Invitrogen), anti-calnexin polyclonal antibody (Stressgen), anti-EEA1 polyclonal antibody (ABR), and anti-LC3 polyclonal antibody (MBL), respectively.
Reporter gene analysis.
HEK293 cells were transiently transfected using 24-well plates and FuGENE HD (Roche) with expression vectors, reporter plasmids, and an internal control plasmid coding Renilla luciferase. The total amounts of plasmids were normalized with empty vector. For poly(I·C) stimulation, at 24 h after transfection, cells were stimulated with medium containing poly(I·C) (50 μg/ml) and DEAE-dextran (0.5 μg/ml) for 1 h, and then the medium was replaced with normal medium and incubation was performed for an additional 3 h. dsRNA was transfected using Lipofectamine 2000 (Invitrogen). Cells were lysed with lysis buffer (Promega) and luciferase, and Renilla luciferase activities were measured using a dual-luciferase assay kit (Promega). Relative luciferase activities were calculated by normalizing luciferase activity by control experiments in which only empty vector and reporter and internal control plasmids were transfected.
Viruses.
VSV (Indiana strain), poliovirus (Mahoney strain), herpes simplex virus 1 (HSV-1) (K strain), and SeV (HVJ strain) were amplified using Vero cells. To determine the virus titer, we performed plaque assays using Vero cells. To observe the cytopathic effect (CPE), virus-infected cells were fixed at indicated times using 10% formaldehyde-PBS for 10 min and then stained using 1% crystal violet-PBS for 5 min at room temperature.
DDX60 helicase recombinant protein.
The DDX60 helicase domain (bp 2254 to 4047) was amplified by PCR using primers DDX60 helicase-F and DDX60 helicase-R. The obtained cDNA fragment was cloned into KpnI and SalI restriction sites of pCold II DNA vector. The primer sequences were as follows: for DDX60 helicase-F, CGGGGTACCATGAGAAAAGACCCAGATCCCAG; and for DDX60 helicase-R, GACGCGTCGACTTTTCCTATTTTGGGGAATG. The DDX6 helicase domain (aa 114 to 429) was amplified by RT-PCR using primers F (GGC GGG GTA CCA TGG GCT GGG AAA AGC CAT C) and R (GGA CGC GTC GAC ACC AAA GCG ACC TGA TCT TC). The cDNA fragment was cloned into the KpnI and SalI sites of pCold II DNA vector. Expression vectors were introduced into Escherichia coli BL21-competent cells and cultured in 10 ml of LB medium with ampicillin added for 12 h at 37°C and then added to 250 ml of LB medium and cultured for 3 h at 37°C. The cells were incubated at 15°C for 30 min, and protein expression was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The cells were then cultured at 15°C for 24 h. The culture fluid was centrifuged for 10 min at 10,000 rpm and 4°C, and the E. coli cells were recovered. The cells were suspended in 5 ml of Tag binding buffer (20 mM Tris-HCl [pH 7.4], 0.5 M NaCl, 20 mM imidazol, 10% glycerol), 5 mg of lysozyme was added for 30 min on ice, and the mixture was subjected to shaking for 10 min; 500 μl of 10% Triton X-100 was then added, and the mixture was incubated for 10 min and centrifuged for 30 min at 5,000 rpm. The reaction was conducted at 4°C. The supernatant was subjected to filtration using a 0.45-μm-pore-size filter. The protein was purified using a HisTrap HP column (GE Healthcare) in accordance with the manufacturer's protocol. The protein was eluted with elution buffer (20 mM Tris-HCl [pH 7.4], 0.5 mM NaCl, 200 mM imidazol, 10% glycerol). We then collected the nonabsorbed fraction (that sample that had dropped out after the mixture was applied to the column), the wash fraction (the sample that dropped out after washing using the binding buffer was performed), and 500 μl each of elute fractions 1 to 10. The obtained samples were checked for purification by SDS-PAGE and detected by Coomassie brilliant blue (CBB) staining.
RNA synthesis.
VSV-G RNA was synthesized from plasmids by using Riboprobe combination system SP6/T7 RNA polymerase (Promega) in accordance with the manufacturer's protocol. VSV RNA was produced from PCR products by the use of VSV G-pGEM-T Easy for its template and amplified by using a 5′ primer containing the T7 promoter (TAATACGACTCACTATAGGG) and a 3′ primer containing the SP6 promoter (GATTTAGGTGACACTATAG). The dsRNA was made by mixing equal amounts of positive strand (SP6) and negative strand (T7). The obtained ssRNA or dsRNA was added to 10 U of DNase I (Promega) at 37°C for 30 min and purified by phenol-chloroform treatment.
Gel shift assay.
The components and final concentrations of the reaction solution were BSA (0.02 mg/ml), MgCl2 (10 mM), dithiothreitol (DTT; 0.1 mM), glycerol (20%), NaCl (200 mM), Tris-HCl (20 mM), and ssRNA (0.6 μg), dsRNA (0.24 μg), or dsDNA (0.1 μg); we then added DDX60 or DDX6 helicase recombinant protein. The total volume was adjusted to 20 μl by adding water. The reaction solution was incubated at 30°C for 30 min; 1% agarose gel electrophoresis was then performed at 4°C for 90 min, and we observed the RNA results by the use of ethidium bromide (EtBr).
Pulldown assay.
The RNA used for the assay was purchased from JBioS. The RNA sequences were as follows: for the sense strand, AAA CUG AAA GGG AGA AGU GAA AGU G; and for the antisense strand, CAC UUU CAC UUC UCC CUU UCA GUU U. The biotin is conjugated at the U residue at the 3′ end of antisense strand (underlined). Biotinylated dsRNAs were phosphorylated by the use of T4 polynucleotide kinase (Takara). dsRNA was incubated for 1 h at 25°C with 10 μg of protein from the cytoplasmic fractions of cells that were transfected with FLAG-tagged RIG-I and/or HA-tagged DDX60-expressing vectors. The mixture was transferred into 400 μl of lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% NP-40, 30 mM NaF, 5 mM Na3VO4, 20 mM iodoacetamide, 2 mM phenylmethylsulfonyl fluoride [PMSF]) containing 25 μl of streptavidin Sepharose beads, rocked at 4°C for 2 h, collected by centrifugation, washed three times with lysis buffer, and resuspended in SDS sample buffer.
RESULTS
Phylogenetic analysis of the DEXD/H box domain of DDX60.
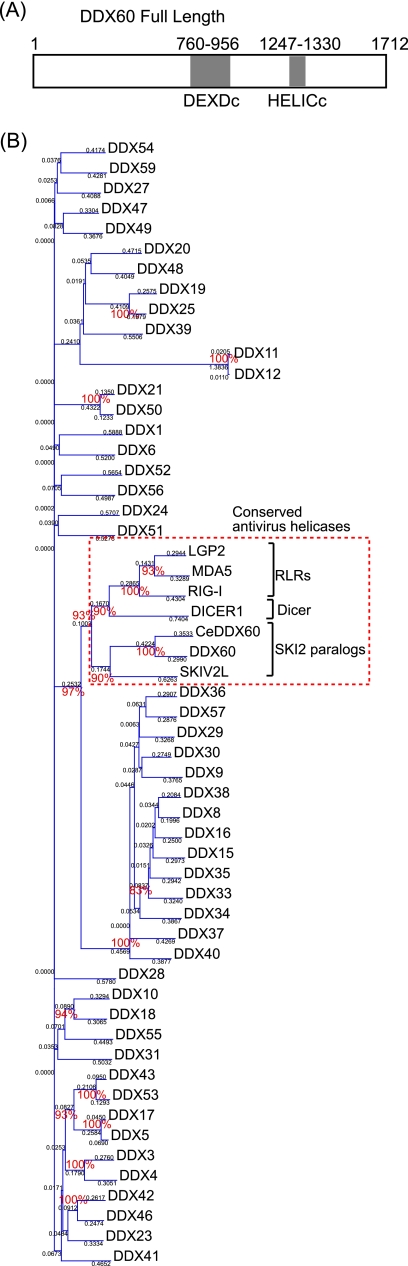
The DDX60 protein contains a DEXD/H box RNA helicase domain and long N- and C-terminal regions with no typical domains or motifs (Fig. 1A). The DEXD/H box domain is common in the human genome. Phylogenetic analysis using amino acid sequences from this domain revealed that DDX60 is clustered within a group that includes RIG-I, MDA5, DICER1, and SKIV2L (bootstrap probability, 93%) and is most closely related to SKIV2L (bootstrap probability, 90%) (Fig. 1B). The functions of RIG-I and MDA5 have been described above. Dicer is an evolutionarily conserved protein required for RNAi and is known to perform antiviral functions in Drosophila melanogaster (12, 48, 51). SKIV2L is a homolog of S. cerevisiae Ski2, an antiviral protein that acts against dsRNA virus. Thus, the DDX60 DEXD/H box domain is found to cluster into a group composed of evolutionarily conserved antiviral helicases.
Fig. 1.
The phylogenetic tree of DEXD/H box RNA helicase. (A) Schematic diagram of DDX60. DDX60 encodes a peptide of 1,712 amino acids (aa) that contains a DEXD/H box (DEXDc; aa 760 to 956) and HELICc (aa 1247 to 1330). (B) The phylogenetic tree of DEXD/H box RNA heliases. Ce, C. elegans. The bootstrap probabilities and genetic distances are shown in red and black, respectively.
Expression of DDX60 mRNA.
We carried out the microarray analysis using mouse bone marrow-derived dendritic cells (BM-DCs) and found that RLRs and DDX60 were included in the helicases whose expression had increased over 4-fold in response to viral infection (Fig. 2A and B). We investigated the expression profile of human DDX60 mRNA by Northern blot analysis and detected a single mRNA band at approximately 5.5 kb in the human brain, lymph node, prostate, stomach, thyroid, tongue, trachea, uterus, skeletal muscle, spleen, kidney, liver, and small intestine (Fig. 2C). Next, we examined the regulation of DDX60 expression. DDX60 mRNA was detected in unstimulated cells such as mouse BM-DCs, HeLa cells, HEK293 cells, and RAW 264.7 cells (Fig. 2D to F). DDX60 expression was upregulated by stimulation with poly(I·C) (Fig. 2D) or IFN-β (Fig. 2E). However, DDX60 expression was not increased by tumor necrosis factor alpha (TNF-α) or interleukin-1β (IL-1β) stimulation (Fig. 2E). Interestingly, DDX60 mRNA levels were increased by infection with VSV (Fig. 2F) or PV (Fig. 2G and H). Thus, DDX60 is an interferon-inducible gene and is upregulated during VSV or PV infection.
DDX60 associates with the components of the RNA exosome.
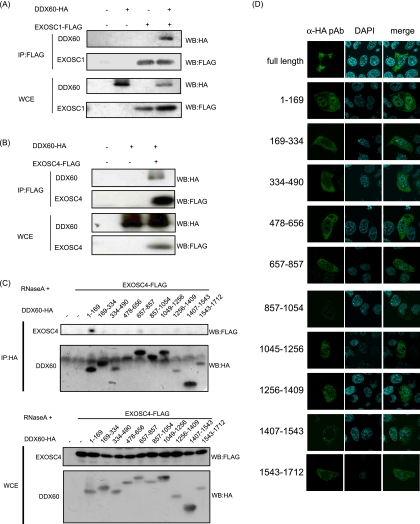
Since the sequence of DDX60 is similar to that of RNA exosome cofactor SKIV2L, we examined whether the DDX60 protein associates with the RNA exosome core components in the same manner as the cofactor proteins. The RNA exosome core components form a tight protein complex (18); thus, it is expected that one of the core components would be coimmunoprecipitated with other core components and cofactors of the RNA exosome. These core components include EXOSC1 and EXOSC4. DDX60 protein was observed to coimmunoprecipitate with EXOSC1 and EXOSC4 (Fig. 3A and B), indicating the physical interaction of DDX60 with EXOSC1 and EXOSC4. We used partial DDX60 fragments to identify the region of DDX60 that binds to the RNA exosome. EXOSC4 was coimmunoprecipitated with the N-terminal 169-aa fragment of DDX60, indicating that the RNA exosome binds to the N-terminal region of DDX60 (Fig. 3C). All fragments were localized in the cytoplasmic region (Fig. 3D).
Fig. 3.
Interactions between DDX60 and RNA exosome components. (A and B) HA-tagged DDX60 and FLAG-tagged EXOSC1 (A) and EXOSC4 (B) expression vector was transfected into HEK293FT cells. After 24 h, cell lysates were prepared and immunoprecipitation (IP) was performed with anti-FLAG antibody. The samples were analyzed by SDS-PAGE and detected by Western blotting (WB) using anti-HA or anti-FLAG antibodies. WCE, whole-cell extract (WCE). (C) HA-tagged DDX60 partial fragments and/or FLAG-tagged EXOSC4 expression vectors were transfected into HEK293FT cells, and immunoprecipitation was carried out as described for panel A in the presence of RNase A. (D) HA-tagged DDX60 fragment-expressing vectors were transfected into HeLa cells. The cells were stained with anti-HA antibody and DAPI and then observed by confocal microscopy.
Intracellular localization of DDX60.
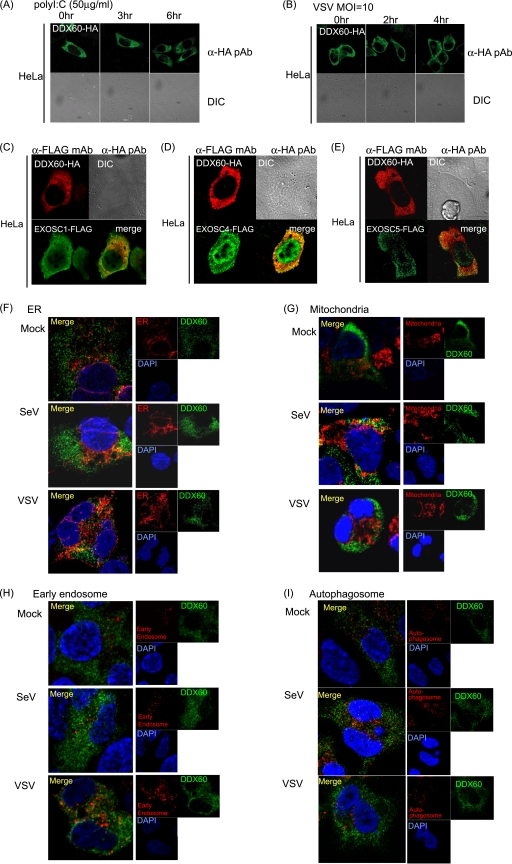
Next, we studied the intracellular localization of the DDX60 protein by the use of confocal microscopy. In resting cells, DDX60 was localized in the cytoplasmic region but not in the nucleus before and after poly(I·C) stimulation or viral infection (Fig. 4A and B). EXOSC1, EXOSC4, and EXOSC5 were localized at both the cytoplasm and nucleus. The DDX60 protein was partially colocalized with the RNA exosome components at cytoplasmic region (Fig. 4C to E). To observe the intracellular localization of DDX60 after viral infection, we used HEK293 cell clones stably expressing HA-tagged DDX60. Most DDX60 staining was not colocalized with mitochondria, the endoplasmic reticulum (ER), LC3 (autophagosome marker), and endosome markers (EEA1) (Fig. 4F to I). However, the DDX60-HA protein was partially colocalized with the ER after VSV infection (Fig. 4F) and with the mitochondria after SeV infection (Fig. 4G).
Fig. 4.
Intracellular localization of DDX60. (A and B) HA-tagged DDX60 expression vector was transfected into HeLa cells, and transfected cells were stimulated with 50 μg/ml poly(I·C) (A) or infected with VSV at an MOI of 10 (B). The cells were fixed and stained with anti-HA antibodies and observed using confocal microscopy. DIC, differential interference contrast; pAb, polyclonal antibody. (C to E) HA-tagged DDX60 was transfected into HeLa cells together with FLAG-tagged EXOSC1 (C), EXOSC4 (D), or EXOSC5 (E). Transfected cells were fixed and stained with anti-HA and anti-FLAG antibodies and observed using confocal microscopy. mAb, monoclonal antibody. (F to I) HEK293 cells stably expressing DDX60-HA were infected with VSV or SeV. DDX60-HA was stained with anti-HA antibody. The ER, mitochondria, early endosome, and autophagosome were stained with calnexin (F), Mitotracker Red (G), anti-EEA1 antibody (H), and anti-LC3 antibody (I).
Antiviral activity of DDX60 is independent of the RNA exosome.
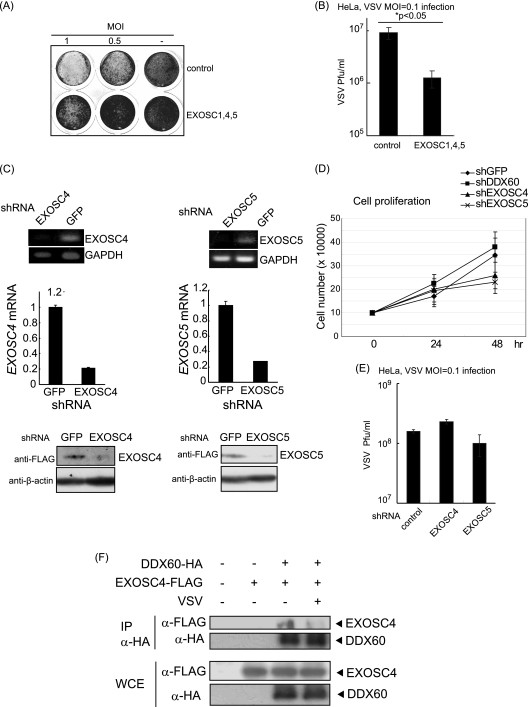
DDX60 expression was found to increase after viral infection, and the RNA exosome plays an antiviral role in lower eukaryotes such as budding yeast. Therefore, we assessed the antiviral activity of both DDX60 and the RNA exosome. Under our experimental conditions, 70% to 90% of cells expressed the proteins encoded by the transfected plasmids. Overexpression of three core components (EXOSC1, EXOSC4, and EXOSC5) suppressed the cytopathic effect (CPE) (Fig. 5A) and reduced the viral titer in the case of a low (0.1) multiplicity of infection (MOI) (Fig. 5B). Next, we performed a short hairpin RNA (shRNA) knockdown assay. We used shRNAs for EXOSC4 and EXOSC5, which were previously described as efficiently reducing expression of the target mRNA (4). We confirmed that shRNAs for EXOSC4 and EXOSC5 effectively reduced expression of their target mRNAs and the proteins (Fig. 5C). A partial decrease in levels of EXOSC4 in knockdown assays is known to reduce cell growth (47). We observed that the presence of shRNA for EXOSC4 and EXOSC5 reduced cell growth (Fig. 5D); however, knockdown of EXOSC4 or EXOSC5 showed a marginal effect on VSV replication (Fig. 5E), at least under our experimental conditions. We do not exclude the possibility that knockdown of EXOSC4 or EXOSC5 is not an efficient method by which to reduce the antivirus role of the RNA exosome. Unexpectedly, the physical interaction between EXOSC4 and DDX60 was reduced after VSV infection (Fig. 5F).
Fig. 5.
Antiviral activity of RNA exosome. (A and B) HeLa cells were transfected with expression vectors containing EXOSC1, EXOSC4, and EXOSC5, and 24 h later, the transfected cells were infected with VSV at an MOI of 1 or 0.5. One day after infection, the cells were fixed and stained with crystal violet (A). Viral titers of culture media after 76 h were measured by a plaque assay (B). (C) Expression of EXOSC4 and EXOSC5 in stable HeLa clones, which express shRNA for EXOSC4, EXOSC5, or GFP, was examined by RT-PCR (upper panel), RT-qPCR (middle panel), and Western blotting (lower panel). The amounts of EXOSC4 and EXOSC5 cDNA in each sample were normalized by dividing by the amount of GAPDH. (D and E) Cell growth rates of stable HeLa clones, which express shRNA for EXOSC4, EXOSC5, DDX60, or GFP, were determined (D). The cells were infected with VSV at an MOI of 0.1 for 48 h, and viral titers of culture media were determined by a plaque assay (E). (F) FLAG-tagged EXOSC4- and HA-tagged DDX60-expressing vectors were transfected into HEK293FT cells. After VSV or mock infection, the immunoprecipitation was performed with anti-HA antibody.
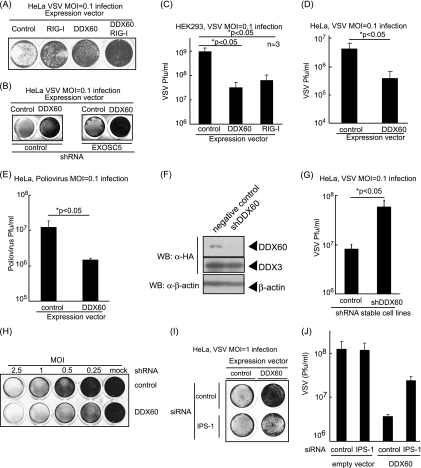
We next examined the antiviral activity of DDX60. Interestingly, DDX60 overexpression suppressed the CPE induced by VSV infection and reduced VSV replication in HEK293 and HeLa cells (Fig. 6A to D). The suppression induced by DDX60 overexpression was observed even in EXOSC5 knockdown cells (Fig. 6B). PV replication was also suppressed by overexpression of DDX60 (Fig. 6E). Next, we performed a knockdown assay using shRNA for DDX60, which reduced expression of DDX60 mRNA and protein (Fig. 6F) (see also Fig. 11 and 12). Unlike the results seen with EXOSC4 and EXOSC5, knockdown of DDX60 increased VSV replication and enhanced CPE and did not inhibit cell growth (Fig. 5D and Fig. 6G and H). Because the interaction between DDX60 and the RNA exosome core component was reduced after viral infection, we examined the molecular mechanism by which DDX60 suppresses viral replication.
Fig. 6.
Antiviral activity of DDX60. (A and B) HeLa cells (A) or the cells stably expressing shRNA for EXOSC5 or GFP (B) were transiently transfected with empty, DDX60-expressing, and/or RIG-I-expressing vectors. After 24 h, cells were infected with VSV at an MOI of 0.1 for 30 h. Cells were fixed and stained with crystal violet. (C and D) HEK293 clones stably expressing DDX60 or RIG-I (C) and HeLa cells subjected to mock treatment or transiently expressing DDX60 (D) were infected with VSV at an MOI of 0.1 for 24 h (C) or 48 h (D). Viral titers of culture media were measured by plaque assay. (E) Empty or DDX60-expressing vectors were transfected into HeLa cells, and 24 h after transfection, cells were infected with PV at an MOI of 0.1 for 26 h. Viral titers of culture media were measured by a plaque assay. (F) DDX60-expressing vector was transfected into HeLa clones stably expressing shRNA for GFP or DDX60, and the protein results were observed by Western blotting. (G) Control or DDX60 knockdown cells were infected with VSV at an MOI of 0.1 for 12 h, and viral titers of the culture media were measured by a plaque assay. (H) Control and DDX60 knockdown cells were infected with VSV at the indicated MOI for 12 h. The cells were fixed and stained with crystal violet. (I and J) Empty or DDX60-expressing vectors were transfected into HeLa cells with negative-control siRNA or siRNA for IPS-1. The cells were infected with VSV at an MOI of 1. After 24 h, cells were stained with crystal violet (I), and the viral titers of culture media were determined by a plaque assay (J).
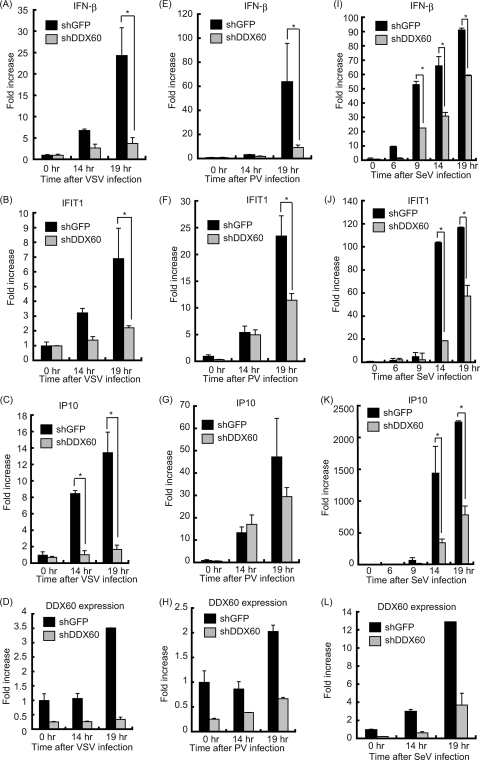
Fig. 11.
Knockdown of DDX60 decreases expression of type I IFN during viral infection in HeLa cells. (A to L) Control cells or HeLa cells from a stable cell line expressing shRNA for DDX60 were infected with VSV (A to D), PV (E to H), or SeV (I to L). Total RNA was extracted at the indicated times. RT-qPCR was performed to measure expression of IFN-β (A, E, and I), IFIT1 (B, F, and J), IP10 (C, G, and K), and DDX60 (D, H, and L). The expression level of each sample was normalized to GAPDH expression.
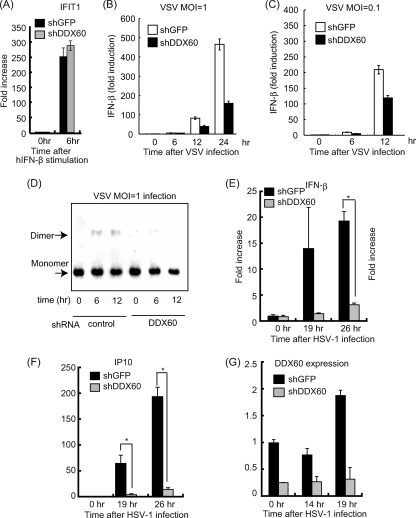
Fig. 12.
Effects of DDX60 knockdown on antiviral responses. (A to C) Control or DDX60 knockdown cells were stimulated with human IFN-β (A) or infected with VSV at an MOI of 1 (B) or 0.1 (C), and expression of IFIT1 (A) and IFN-β (B and C) mRNA was examined by RT-qPCR. (D) Control or DDX60 knockdown HEK293 cells were infected with VSV at an MOI of 1, and cell lysates were prepared at the indicated times. Lysates were analyzed by native PAGE, and Western blot analysis was performed with anti-IRF-3 antibody. (E to G) Control or DDX60 knockdown HeLa cells were infected with HSV-1, and total RNA was extracted at the indicated times. RT-qPCR was performed to examine expression of IFN-β (E), IP10 (F), and DDX60 (G).
DDX60 associates with RIG-I-like receptors.
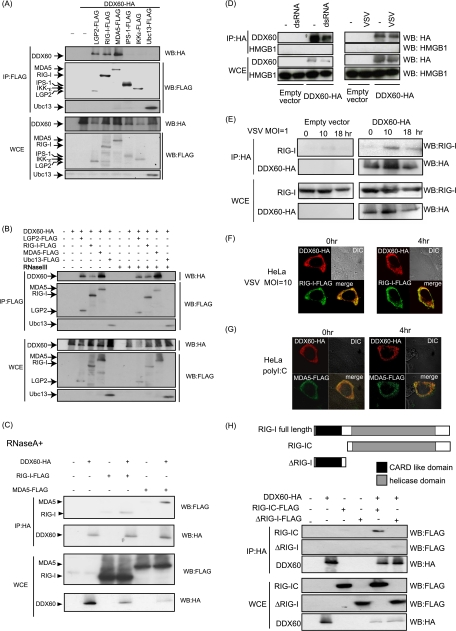
To identify the antiviral pathway in which DDX60 is involved, we used immunoprecipitation assays to search for the protein that binds to DDX60. Because the RLR-dependent pathway plays an important role in the antiviral activity of the host cell, we examined the binding of DDX60 to proteins involved in this pathway. Interestingly, DDX60 was coimmunoprecipitated with RIG-I, MDA5, and LGP2 but not with IPS-1 or IKK-ε, which are downstream factors of RIG-I and MDA5 (Fig. 7A). RNase A or RNase III treatment did not abolish the interaction between DDX60 and RIG-I or MDA5 (Fig. 7B and C), indicating that these associations are not mediated via RNA. DDX60 did not bind to HMGB1 before or after dsRNA stimulation or VSV infection (Fig. 7D). To further confirm the binding of DDX60 to RIG-I, we examined the interaction of DDX60 with endogenous RIG-I by the use of anti-RIG-I monoclonal antibody. RIG-I mRNA levels are known to increase after viral infection (59). We observed an increase in RIG-I protein levels after poly(I·C) stimulation (data not shown). However, the protein level in HEK293FT cells was not increased after VSV infection in our experiment for unknown reasons (Fig. 7E). Endogenous RIG-I was found to interact with DDX60 after VSV infection but not in its absence (Fig. 7E), indicating that interaction between DDX60 and endogenous RIG-I is dependent on viral infection, although the interaction between overexpressed RIG-I and DDX60 was independent of viral infection.
Fig. 7.
Association of DDX60 with RLRs. (A to C) Vectors expressing HA-tagged DDX60 were transfected into HEK293FT cells with FLAG-tagged LGP2, RIG-I, MDA5, IPS-1, IKK-ε, and/or Ubc13, and cell lysates were prepared. The lysates were treated with RNase III (B) or RNase A (C). Immunoprecipitation was carried out with anti-FLAG antibody, and the precipitates (IP) and 10% of whole-cell extract (WCE) were analyzed using SDS-PAGE. Proteins were stained by Western blotting using anti-HA or anti-FLAG antibody. (D and E) HEK293FT cells were transfected with empty or HA-tagged DDX60-expressing vectors, and cells were stimulated with dsRNA or infected with VSV. Cell lysates were prepared at the indicated times, and immunoprecipitation was performed with anti-HA antibody. The precipitates were analyzed using SDS-PAGE, and Western blotting was carried out using anti-HA and anti-HMGB1 (D) or anti-RIG-I (E) antibodies. (F and G) Vectors expressing HA-tagged DDX60 and FLAG-tagged RIG-I (F) or MDA5 (G) were transfected into HeLa cells. After 24 h, cells were fixed and stained with anti-HA or anti-FLAG antibody and then observed using confocal microscopy. (H) The upper panel shows a schematic diagram of RIG-I partial fragments. The lower panel shows results of an immunoprecipitation assay performed as described for panel A. DDX60 was found to bind to the RIG-IC region.
Next, we used confocal microscopy to examine the intracellular colocalization of DDX60 with RIG-I and MDA5. Consistent with the immunoprecipitation assay, confocal microscopic analysis showed that DDX60 was partially colocalized with overexpressed RIG-I and MDA5 before and after VSV infection or poly(I·C) stimulation (Fig. 7F and G). We tried to observe endogenous RIG-I by confocal microscopy; however, we could not detect endogenous RIG-I in our tests for technical reasons. We also used RIG-I partial fragments to identify the region of RIG-I that binds to DDX60 (Fig. 7H). RIG-IC, which includes a helicase domain and CTD, was coimmunoprecipitated with DDX60, while the N-terminal CARD of RIG-I was not (Fig. 7H). These data indicate that DDX60 binds to the RIG-IC fragment.
The DDX60 helicase domain binds to viral RNA.
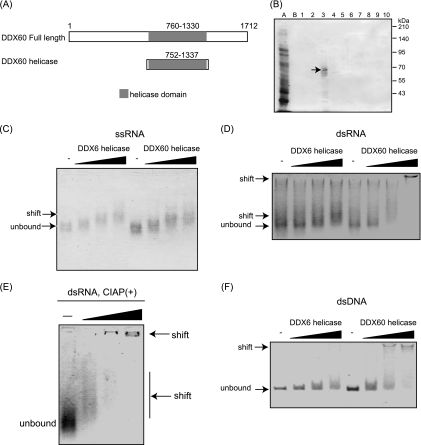
In light of the binding of DDX60 to viral RNA sensors RIG-I and MDA5, we hypothesized that the RNA helicase domain of DDX60 binds to viral RNA. To test this hypothesis, we expressed a histidine-tagged DDX60 RNA helicase domain (aa 752 to 1337) in E. coli (Fig. 8A). The protein was purified using nickel-nitrilotriacetic acid (Ni-NTA) resin, analyzed by SDS-PAGE, and stained with CBB. Protein purity was found to be greater than 90% (Fig. 8B). VSV single- and double-stranded RNA was synthesized in vitro. Binding of the DDX60 helicase domain to in vitro-synthesized viral RNA was examined using gel-shift assays. Single- or double-stranded VSV RNA mobility was found to decrease as a result of the addition of DDX60 helicase (Fig. 8C and D). DDX60 was also found to bind to dsRNA treated with alkaline phosphatase, suggesting that the presence of 5′ triphosphate is nonessential for this binding (Fig. 8E). The mobility shift of ssRNA was different from that of dsRNA; this difference might have been a result of the stoichiometry assays. Interestingly, dsDNA was also shifted in the presence of DDX60 protein (Fig. 8F). As a control we used DDX6, a DEXD/H box RNA helicase distantly related to DDX60. DDX6 also bound to ssRNA (Fig. 8C); however, DDX6 only minimally reduced the mobility of dsRNA and dsDNA compared to DDX60 (Fig. 8D and F).
Fig. 8.
Binding of the DDX60 helicase domain to viral RNA. (A) Schematic diagram showing the helicase region of DDX60 used for the following gel shift assay. (B) A His-tagged DDX60 helicase fragment was expressed in E. coli and purified using Ni-NTA resin. Purified products were analyzed by SDS-PAGE and stained with CBB. Lane A represents the nonabsorbed fraction, lane B represents the wash fraction, and lanes 1 to 10 represent the eluted fractions. The lane 3 fraction contains DDX60 protein. (C to F) Purified DDX60 and DDX6 fragments were incubated with in vitro-synthesized VSV ssRNA (C), dsRNA (D), dsRNA treated with calf intestinal alkaline phosphatase (CIAP) (E), or dsDNA (F), and the products were analyzed using agarose gel. The gel was stained with ethidium bromide.
DDX60 promotes RIG-I- or MDA5-dependent expression of type I IFN.
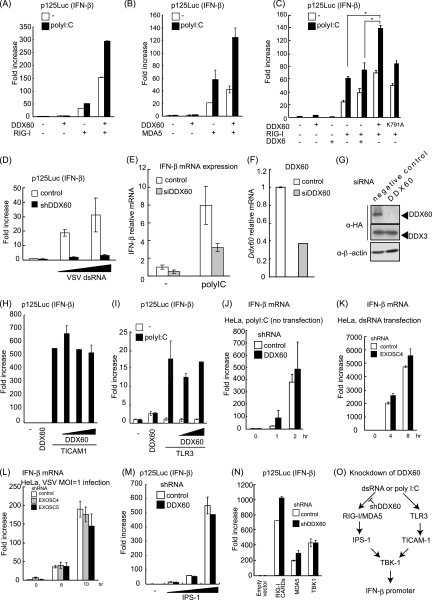
Next, we examined whether DDX60 is involved in RIG-I- or MDA5-mediated signaling. The prepared poly(I·C) solution contains various lengths of poly(I·C), both shorter and longer than 1 kbp (data not shown), in a mixture known to activate both RIG-I and MDA5 (20). Both RIG-I-mediated and MDA5-mediated IFN-β promoter activation by poly(I·C) transfection were enhanced by DDX60 expression (Fig. 9A and B). As a control we used DDX6, a helicase distantly related to DDX60. Expression of DDX6 produced neither a positive nor a negative effect on the RIG-I-dependent IFN-β promoter activation (Fig. 9C). To address the function of the DDX60 helicase domain, we introduced the mutation (K791A) on the Walker type ATP binding site, which is essential for ATPase activity of RNA helicase (50). The mutation reduced the enhancement of RIG-I-mediated IFN-β promoter activation by DDX60 (Fig. 9C). Knockdown analysis using shRNA for DDX60 showed that IFN-β promoter activation by viral dsRNA was reduced in DDX60 knockdown cells compared with control cells (Fig. 9D). To exclude the off-target effect, we also used siRNA for DDX60, whose target sequence is different from that of shRNA for DDX60. Expression of DDX60 was efficiently reduced by siRNA for DDX60, and the siRNA for DDX60 efficiently reduced IFN-β mRNA expression by poly(I·C) stimulation (Fig. 9E to G). These knockdown results are consistent with the overexpression results described above, providing further evidence that DDX60 promotes RLR-mediated IFN-β expression.
Fig. 9.
DDX60 promotes RIG-I- or MDA5-mediated signaling. (A to C) Activation of the IFN-β promoter was examined using a reporter gene assay and p125luc plasmid. Vectors expressing RIG-I (A), MDA5 (B), DDX6 (C), and the wild type (WT) or DDX60-K791A (C) were transfected into HEK293 cells together with the reporter plasmid and Renilla luciferase plasmid (internal control). After 24 h, the cells were left unstimulated or stimulated with poly(I·C) by the use of DEAE-dextran for 4 h. Cell lysates were prepared, and luciferase activity was measured. (D) Control or DDX60 knockdown HEK293 cells were transfected with the p125luc reporter, Renilla luciferase plasmid, and/or in vitro-synthesized VSV dsRNA. After 24 h, cell lysates were prepared and luciferase activity was measured. (E and F) siRNA for DDX60 or control siRNA was transfected into HEK293 cells. The cells were left unstimulated or stimulated with poly(I:C), and expression of IFN-β and DDX60 mRNA was measured by RT-qPCR. Expression values were normalized using GAPDH. (G) siRNA for DDX60 or the control was transfected into HEK293 cells together with DDX60-expressing vector. The DDX60 protein was observed by Western blotting. (H) Vectors expressing TICAM-1 and/or DDX60 were transfected into HEK293 cells together with the p125luc reporter and Renilla luciferase plasmids. After 24 h, the cell lysates were prepared and luciferase activities were measured. (I) Vectors expressing TLR3 and/or DDX60 were transfected into HEK293 cells together with the p125luc reporter and Renilla luciferase plasmids. After 24 h, the cells were left unstimulated or stimulated with poly(I:C) for 4 h, the cell lysates were prepared, and luciferase activity was measured. (J and K) HeLa cells expressing shRNA for DDX60 (J) or EXOSC4 (K) were stimulated with 50 μg/ml of poly(I·C) (no transfection) (J) or dsRNA (transfection) (K). RT-qPCR was performed to measure IFN-β mRNA expression. (L) HeLa cells expressing shRNA for GFP, EXOSC4, or EXOSC5 were infected with VSV at an MOI of 1. Levels of induction of IFN-β mRNA were calculated as described for panel J. (M and N) Empty or IPS-1-expressing vector (M) and RIG-I CARD-, MDA5-, or TBK1-expressing vector (N) were transfected into control or DDX60 knockdown HEK293 cells together with p125luc reporter and Renilla luciferase plasmids. After 24 h, cell lysates were prepared and luciferase activity was measured. (O) shRNA for DDX60 did not inhibit the signaling from TLR3 (H to J). Although DDX60 promotes RLR-dependent signaling (A to E), shRNA for DDX60 did not reduce the signaling induced by RIG-I CARD, MDA5, IPS-1, or TBK1 overexpression (M and N). These data suggest that shRNA suppresses signaling upstream of RIG-I and MDA5.
In contrast, TICAM-1- and TLR3-mediated IFN-β promoter activation was not increased by overexpression of DDX60 (Fig. 9H and I). In addition, poly(I·C) stimulation of TLR3 without transfection resulted in normal expression of IFN-β in DDX60 knockdown cells (Fig. 9J). These data suggest that DDX60 is specific to the RLR pathway. Because the knockdown of EXOSC4 or EXOSC5 did not reduce the promoter activation resulting from VSV infection or dsRNA transfection (Fig. 9K and L), the data suggest that these proteins do not play a major role in DDX60-mediated enhancement of RIG-I or MDA5 signaling, at least under our experimental conditions. We also assessed the effect of DDX60 knockdown on IFN-β promoter activation by overexpressing TBK1, IPS-1, RIG-I CARDs, or MDA5 to discover the step at which DDX60 plays a role in RIG-I-mediated signaling. All of these procedures led to autoactivation, inducing transcription from the IFN-β promoter in the absence of RIG-I or MDA5 ligands (22). Although DDX60 knockdown reduces the IFN-β promoter activation induced by dsRNA transfection, it was not found to affect this autoactivation (Fig. 9M and N). These data suggest that shRNA suppression of DDX60 occurs upstream of RIG-I and MDA5 (Fig. 9O).
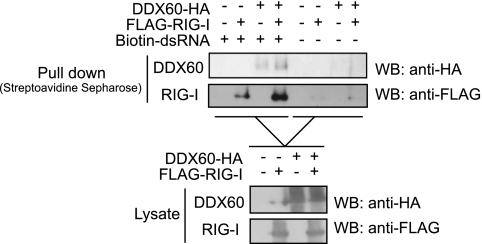
To examine the effect of DDX60 on the binding of RIG-I to dsRNA, we performed pulldown assays. The proteins were exogenously expressed in HEK293FT cells, and the proteins were recovered from cell lysates by the use of biotin-conjugated dsRNA and streptavidin Sepharose. RIG-I or DDX60 protein was recovered from cell extracts, suggesting the binding of RIG-I or DDX60 to dsRNA. Because both RIG-I and DDX60 can bind to dsRNA, the data do not imply an interaction between RIG-I and DDX60. Interestingly, coexpression of both proteins increased the levels of proteins recovered from the cell lysate (Fig. 10).
Fig. 10.
DDX60 increases the association of RIG-I to short synthetic dsRNA. DDX60- and RIG-I-expressing vectors were transfected into HEK293FT cells. At 24 h later, cell lysate was prepared. The lysate was incubated with or without biotin-conjugated dsRNA, and the dsRNA was recovered using streptavidin Sepharose beads. The recovered fraction was analyzed by Western blotting.
DDX60 is important for expression of type I IFN and IFN-inducible genes during viral infection.
We next tested whether DDX60 is involved in cytokine expression during viral infection. RIG-I recognizes VSV and SeV, and MDA5 recognizes PV (3, 21). Interestingly, knockdown of DDX60 reduced IFN-β expression in HeLa cells after VSV, PV, and SeV infection. IFIT-1 and IP10 expression after VSV and SeV infection was also reduced by DDX60 knockdown (Fig. 11). Knockdown of DDX60 caused a marginal effect on expression of IFIT1 and IP10 in response to PV infection. Some of the IFN-α gene expression induced by other sensor molecules might have been responsible for the difference. Unlike viral infection, knockdown of DDX60 did not reduce expression of IFIT1 after IFN-β stimulation (Fig. 12A). Reduction of type I IFN expression was also observed after VSV infection in HEK293 cells (Fig. 12B and C). In addition, DDX60 knockdown reduced IRF-3 dimerization after VSV infection (Fig. 12D). These data indicate that DDX60 is required for RIG-I- and MDA5-dependent type I IFN and IFN-inducible gene expression during viral infection. We also observed that suppression of CPE and viral titers in culture medium induced by DDX60 overexpression can be reduced by IPS-1 knockdown (Fig. 6I and J), confirming that the antiviral activity of DDX60 is dependent on the presence of RLRs. Because the DDX60 helicase domain was found to bind to dsDNA, we examined whether DDX60 is involved in type I IFN expression after infection with HSV-1, a DNA virus. In this case, knockdown of DDX60 reduced expression of IFN-β and IP10 after HSV-1 infection (Fig. 12E and F).
DISCUSSION
Here, we report that DDX60 is a novel antiviral factor in human cells. The amino acid sequence of DDX60 is similar to that of Ski2 homologs, which are cofactors of the RNA exosome. DDX60 interacts with core components of the RNA exosome. After viral infection, the DDX60 protein binds to endogenous RIG-I protein and is involved in RIG-I-dependent pathway. The protein also binds to MDA5 and LGP2. The DDX60 helicase domain binds to viral RNA and DNA, and coexpression of RIG-I with DDX60 increases the binding of RIG-I and DDX60 to dsRNA. Knockdown of DDX60 reduces expression of type I IFN and IFN-inducible genes after VSV, PV, SeV, and HSV-1 infections. Therefore, we concluded that DDX60 is a novel antiviral helicase involved in RLR-dependent pathways.
Schröder et al. and Soulat et al. first reported that the non-RLR helicase DDX3 plays pivotal roles in RLR-dependent pathways (41, 43). DDX3 is ubiquitously expressed in a variety of cells and exerts its positive effect as a part of TBK-1- and/or IKK-ε-containing complexes that activate IRF-3 (41). DDX3 also binds to RIG-I and IPS-1 and promotes the activation of those proteins (28, 32). Our study showed that another non-RLR helicase, DDX60, is also involved in RLR-dependent pathways. Thus, our reports and previous studies demonstrate the important roles of non-RLR helicases in RLR-mediated signaling and antiviral response.
Because DDX60 protein does not contain CARDs, which are required for the interaction with IPS-1, it seems unlikely that DDX60 directly activates IPS-1 without RLRs. The results of knockdown studies suggest that DDX60 is an upstream factor of IPS-1, and the immunoprecipitation assay results suggest that DDX60 binds to the RLR upstream factors. On the basis of these findings, we expected that DDX60 would bind to viral RNA. The gel shift assay showed that DDX60 helicase can bind to dsRNA, and the pulldown assay showed that coexpression of DDX60 with RIG-I increased the binding of RIG-I to dsRNA. Thus, we speculate that DDX60 binds to viral dsRNA and associates with RLRs during viral infection. Further study is required to reveal the precise molecular mechanism by which DDX60 activates the RLR-dependent pathway.
The DDX60 helicase domain binds to dsDNA in vitro, and DDX60 was required for type I IFN expression during infection with HSV-1, a DNA virus. In human cells, RIG-I is involved in the pathway activated by cytoplasmic B-DNA or DNA virus infection (1, 6, 7). AT-rich dsDNA is transcribed by RNA polymerase III, and these transcripts are recognized by RIG-I (1, 6). In contrast, Choi et al. reported that the RIG-I protein associates with B-DNA and activates the signaling (7). Previously, Takahasi et al. reported that purified RIG-I protein does not itself bind to dsDNA (46). Therefore, RIG-I seems to associate with B-DNA via another protein that directly binds to dsDNA. HSV-1 was reported to produce considerable amounts of dsRNA and to activate RIG-I and MDA5 (25, 34, 52). Thus, we do not exclude the possibility that DDX60 is involved in recognition not of dsDNA but of dsRNA derived from HSV-1. Although RIG-I and MDA5 are involved in the signaling induced by cytoplasmic B-DNA, IPS-1 is dispensable for the signaling (23). Further study is required to reveal the molecular mechanism by which DDX60 plays an important role in the signaling induced by cytoplasmic B-DNA.
In addition to the DDX60 gene, the human genome includes the closely related DDX60L gene, which is located 5′ upstream of DDX60 on chromosome IV. However, the mouse genome encodes only one DDX60 protein. Our phylogenetic analysis indicated that the mouse gene is a shared ancestor of human DDX60 and DDX60L genes, and a pilot study has revealed that DDX60L is expressed after viral infection (unpublished results). Therefore, DDX60L is also expected to be an antiviral protein. Considering that knockdown of DDX60 severely reduced type I IFN expression after viral infection, there seem to be functional differences between DDX60 and 60L. Further study is required to reveal the functional differences between DDX60 and 60L.
The amino acid sequence of DDX60 is weakly similar to those of the exosome cofactors SKIV2L and SKIV2L2. In immunoprecipitation experiments, DDX60 protein was found to coimmunoprecipitate with core components of the RNA exosome. However, we could not determine the physiological role of the interaction between DDX60 and RNA exosome by knockdown analysis. We examined whether or not knockdown of DDX60 and the RNA exosome delays degradation of viral genomic RNA. We found that knockdown of DDX60 or RNA exosome components does not substantially delay degradation of transfected viral RNA (unpublished results). This observation is consistent with our results showing that knockdown of the RNA exosome does not increase viral titers. However, we do not exclude the possibility that the RNA exosome is involved in antiviral responses. There exist several antiviral nucleases, such as RNase L and ISG20. Thus, it is possible that those antiviral nucleases compensate for the defect of the RNA exosome. This possibility is not surprising, because there are several redundant pathways in the innate immune system. For example, poly(I·C) is a ligand common to TLR3 and MDA5; thus, not single but double knockout is required to abolish poly(I·C)-dependent NK cell cytotoxicity (27). Type I IFNs are produced from various kinds of cells, such as fibroblasts, dendritic cells, and macrophages; thus, we do not exclude the possibility that the RNA exosome performs some roles in DDX60 antiviral activity in other cells. The RNA exosome is required to maintain the integrity of host RNA and to disrupt RNA that lacks a 5′ catabolite gene activator protein (CAP) or 3′ poly(A) tail. Thus, it is also possible that DDX60 is involved in host RNA integrity. Further study is required to reveal the physiological role of the association of DDX60 with the RNA exosome.
In single-cell organisms such as budding yeast (S. cerevisiae), Ski2 plays a major role in the antiviral response to dsRNA virus. DDX60, a homolog of Ski2, is conserved among eukaryotes. For example, DDX60 is also encoded by the Caenorhabditis elegans genome. In a pilot study, we found nematode DDX60 expression to be increased after viral infection (unpublished results), leading us to predict that DDX60 possesses antiviral activity in this species as well. Phylogenetic tree analysis has shown that antiviral helicases such as DDX60, RLRs, and Dicer are clustered into one node. Considering that budding yeast uses Ski2 helicase for its antiviral activity, Ski2-DDX60 protein might represent the most primitive antiviral helicase, which has since diverged into several distinct but similar proteins, such as Dicer and RLRs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants-in-aid from the Ministry of Education, Science and Culture of Japan, the Ministry of Health, Labor, and Welfare of Japan, Mitsubushi Foundation, and Akiyama Foundation.
We thank K. Matsumoto, H. Saito (National Research Institute for Child Health and Development), and H. Takaki for microarray data.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Ablasser A., et al. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10: 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akazawa T., et al. 2007. Antitumor NK activation induced by the Toll-like receptor 3-TICAM-1 (TRIF) pathway in myeloid dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 104: 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barral P. M., et al. 2007. MDA-5 is cleaved in poliovirus-infected cells. J. Virol. 81: 3677–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brouwer R., et al. 2001. Three novel components of the human exosome. J. Biol. Chem. 276: 6177–6184 [DOI] [PubMed] [Google Scholar]
- 5. Chen G., Guo X., Lv F., Xu Y., Gao G. 2008. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc. Natl. Acad. Sci. U. S. A. 105: 4352–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chiu Y. H., Macmillan J. B., Chen Z. J. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi M. K., et al. 2009. A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA. Proc. Natl. Acad. Sci. U. S. A. 106: 17870–17875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui S., et al. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29: 169–179 [DOI] [PubMed] [Google Scholar]
- 9. Dangel A. W., Shen L., Mendoza A. R., Wu L. C., Yu C. Y. 1995. Human helicase gene SKI2W in the HLA class III region exhibits striking structural similarities to the yeast antiviral gene SKI2 and to the human gene KIAA0052: emergence of a new gene family. Nucleic Acids Res. 23: 2120–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ebihara T., et al. 2010. Identification of a polyI:C-inducible membrane protein that participates in dendritic cell-mediated natural killer cell activation. J. Exp. Med. 207: 2675–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gack M. U., et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446: 916–920 [DOI] [PubMed] [Google Scholar]
- 12. Galiana-Arnoux D., Dostert C., Schneemann A., Hoffmann J. A., Imler J. L. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 7: 590–597 [DOI] [PubMed] [Google Scholar]
- 13. Gao D., et al. 2009. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One 4: e5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo X., Ma J., Sun J., Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. U. S. A. 104: 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hayakawa S., et al. 2011. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat. Immunol. 12: 37–44 [DOI] [PubMed] [Google Scholar]
- 16. Hornung V., et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997 [DOI] [PubMed] [Google Scholar]
- 17. Houseley J., LaCava J., Tollervey D. 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7: 529–539 [DOI] [PubMed] [Google Scholar]
- 18. Houseley J., Tollervey D. 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- 19. Kato H., et al. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23: 19–28 [DOI] [PubMed] [Google Scholar]
- 20. Kato H., et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205: 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kato H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105 [DOI] [PubMed] [Google Scholar]
- 22. Kawai T., et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6: 981–988 [DOI] [PubMed] [Google Scholar]
- 23. Kumar H., et al. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203: 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X., et al. 2009. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 284: 13881–13891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Melchjorsen J., et al. 2010. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J. Virol. 84: 11350–11358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meylan E., et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- 27. Miyake T., et al. 2009. Poly I:C-induced activation of NK cells by CD8 alpha+ dendritic cells via the IPS-1 and TRIF-dependent pathways. J. Immunol. 183: 2522–2528 [DOI] [PubMed] [Google Scholar]
- 28. Oshiumi H., et al. 2010. Hepatitis C virus core protein abrogates the DDX3 function that enhances IPS-1-mediated IFN-beta induction. PLoS One 5: e14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4: 161–167 [DOI] [PubMed] [Google Scholar]
- 30. Oshiumi H., Matsumoto M., Hatakeyama S., Seya T. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J. Biol. Chem. 284: 807–817 [DOI] [PubMed] [Google Scholar]
- 31. Oshiumi H., et al. 2010. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8: 496–509 [DOI] [PubMed] [Google Scholar]
- 32. Oshiumi H., Sakai K., Matsumoto M., Seya T. 1 February 2010, posting date DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta inducing potential. Eur. J. Immunol. doi:10.1002/eji.200940203 [DOI] [PubMed] [Google Scholar]
- 33. Pippig D. A., et al. 2009. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 37: 2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rasmussen S. B., et al. 2009. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene-like receptors, which synergize to induce type I interferon production. J. Gen. Virol. 90: 74–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rehwinkel J., et al. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140: 397–408 [DOI] [PubMed] [Google Scholar]
- 36. Saito T., et al. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104: 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saito T., Owen D. M., Jiang F., Marcotrigiano J., Gale M., Jr 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454: 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Satoh T., et al. 26 January 2010, posting date LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. doi:10.1073/pnas.0912986107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schlee M., et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmidt A., et al. 2009. 5′-Triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U. S. A. 106: 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schröder M., Baran M., Bowie A. G. 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 27: 2147–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seth R. B., Sun L., Ea C. K., Chen Z. J. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- 43. Soulat D., et al. 2008. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 27: 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun Q., et al. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24: 633–642 [DOI] [PubMed] [Google Scholar]
- 45. Takahasi K., et al. 2009. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J. Biol. Chem. 284: 17465–17474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takahasi K., et al. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29: 428–440 [DOI] [PubMed] [Google Scholar]
- 47. van Dijk E. L., Schilders G., Pruijn G. J. 2007. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA 13: 1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Rij R. P., et al. 2006. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20: 2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Venkataraman T., et al. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178: 6444–6455 [DOI] [PubMed] [Google Scholar]
- 50. Walker J. E., Saraste M., Runswick M. J., Gay N. J. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1: 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang X. H., et al. 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weber F., Wagner V., Rasmussen S. B., Hartmann R., Paludan S. R. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80: 5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wickner R. B. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol. Rev. 60: 250–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Widner W. R., Wickner R. B. 1993. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol. Cell. Biol. 13: 4331–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu L. G., et al. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- 56. Yanai H., et al. 2009. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462: 99–103 [DOI] [PubMed] [Google Scholar]
- 57. Yoneyama M., Fujita T. 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 18: 545–551 [DOI] [PubMed] [Google Scholar]
- 58. Yoneyama M., et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175: 2851–2858 [DOI] [PubMed] [Google Scholar]
- 59. Yoneyama M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5: 730–737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.