Abstract
The cellular abundance of topoisomerase IIα (TOP2A) critically maintains DNA topology after replication and determines the efficacy of TOP2 inhibitors in chemotherapy. Here, we report that the RNA-binding protein HuR, commonly overexpressed in cancers, binds to the TOP2A 3′-untranslated region (3′UTR) and increases TOP2A translation. Reducing HuR levels triggered the recruitment of TOP2A transcripts to RNA-induced silencing complex (RISC) components and to cytoplasmic processing bodies. Using a novel MS2-tagged RNA precipitation method, we identified microRNA miR-548c-3p as a mediator of these effects and further uncovered that the interaction of miR-548c-3p with the TOP2A 3′UTR repressed TOP2A translation by antagonizing the action of HuR. Lowering TOP2A by silencing HuR or by overexpressing miR-548c-3p selectively decreased DNA damage after treatment with the chemotherapeutic agent doxorubicin. In sum, HuR enhances TOP2A translation by competing with miR-548c-3p; their combined actions control TOP2A expression levels and determine the effectiveness of doxorubicin.
INTRODUCTION
Mammalian cells express different subsets of proteins throughout the division cycle. Proteins showing altered abundance include those that drive cell cycle progression (e.g., cyclins) and those required for the cellular response to the different metabolic requirements of each cell cycle phase. Among the latter group is topoisomerase IIα (TOP2A), an enzyme that helps to maintain proper DNA topology by introducing double-strand breaks to relieve the tension created by processes like DNA replication (12, 38). Expression of TOP2A peaks during G2 and mitosis, unlike expression of the related protein TOP2B, whose abundance is constant throughout the cell division cycle (19, 39). This pattern of expression supports a role for TOP2A in relaxing the positive supercoiling that develops as the replication fork advances during the S phase and in mitotic events, such as chromosome decatenation, and kinetochore and centromere function (28, 31, 33). TOP2A is also important in chemotherapy; a growing body of literature indicates that the effectiveness of several anticancer drugs depends on TOP2A levels (29).
Since transcription by RNA polymerase II is repressed during mitosis (30), posttranscriptional processes are particularly important for controlling protein abundance in mitotic cells. The expression of TOP2A peaks in mitotic cells (19, 39); thus, the underlying mechanisms regulating TOP2A expression are critical. In mammalian cells, TOP2A function has been linked to its posttranslational modification (sumoylation, phosphorylation) and its interaction with other proteins (reviewed in reference 28). However, the transcriptional and posttranscriptional mechanisms that control TOP2A expression are virtually unknown. The posttranscriptional gene regulation (e.g., changes in mRNA splicing, transport, storage, stability, and translation) is typically controlled by the interaction of cis-acting elements in the regulated mRNA with trans-binding regulatory factors. These factors are of two main types, RNA-binding proteins (RBPs) and noncoding RNAs (principally microRNAs), and they potently modulate mRNA turnover and translation.
The ubiquitous member of the Hu/elav RBP family HuR is a well-studied regulator of the stability and translation of bound transcripts, with which it interacts through its three RNA recognition motifs (RRMs) (9, 20, 22, 24). HuR has been implicated in carcinogenesis, differentiation, and the stress and immune responses (1, 20, 26, 35). MicroRNAs (∼22 nucleotides long) typically repress gene expression posttranscriptionally by reducing the translation and/or stability of target mRNAs (13). Their action is linked to the recruitment of the RNA-induced silencing complex (RISC) to target mRNAs that associate with partial complementarity with the microRNAs (7). MicroRNAs have also been linked to physiologic and pathological events, including responses to damaging and immune signals, development, and carcinogenesis (5, 23, 36).
Here, we present evidence that TOP2A expression is potently regulated at the level of translation. Heightened TOP2A expression is associated with enhanced binding of HuR to TOP2A mRNA, in competition with binding of miR-548c-3p to the TOP2A mRNA, whose interaction with TOP2A mRNA led to its recruitment to processing bodies (PBs), cytoplasmic foci specialized in mRNA decay and translational repression. The antagonistic influence of HuR and miR-548c-3p upon TOP2A expression selectively affected the extent of DNA damage after treatment with TOP2A inhibitors. Our results underscore the usefulness of chemotherapeutic strategies that include modulating TOP2A translation.
MATERIALS AND METHODS
Cell culture, treatment, and transfection.
HeLa cells were cultured in Dulbecco's modified essential medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS) and antibiotics. Lipofectamine-2000 (Invitrogen) was used to transfect cells with small RNAs and plasmids. Small RNAs used (at 100 nM) to silence HuR were AATCTTAAGTTTCGTAAGTTA (HuR U1) and TTCCTTTAAGATATATATTAA (HuR U2), the control small interfering RNA (Ctrl siRNA) was AATTCTCCGAACGTGTCACGT (Qiagen), and the TOP2A siRNA was from Santa Cruz Biotech. Plasmid DNAs were transfected at 50 ng/ml [pEGFP, pEGFP-TOP2A(3′), pEGFP-TOP2A(3′mut), pEGFP-TOP2A(3′)ΔHuR] or at 1 to 2 μg/ml [pFlag, pHuR-Flag, pMS2, pMS2-TOP2A(3′), pMS2-YFP]. Treatment with nocodazole (100 ng/ml) lasted 16 h. Double thymidine block and flow cytometry were performed as described previously (21). TOP2A 3′-untranslated region (3′UTR) reporter constructs were made by inserting the TOP2A 3′UTR into pEGFP-C1 or pMS2. I. E. Gallouzi kindly provided pHuR-Flag; pMS2 and pMS2-YFP plasmids were described previously (25).
Microscopy.
Fluorescence microscopy was performed as described previously (25). Briefly, cells were fixed with 2% formaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 5% bovine serum albumin (BSA). After incubation with a primary antibody recognizing DCP1a (Abcam), an Alexa 568-conjugated secondary antibody (Invitrogen) was used to detect primary antibody-antigen complexes (red). Yellow fluorescent protein (YFP) fluorescence was green. Images were acquired using an Axio Observer microscope (Zeiss) with AxioVision 4.7 Zeiss image processing software or with LSM 510 Meta (Zeiss). Confocal microscopy images were acquired with Z-sectioning mode with 15 slices and 0.4 μm spacing and merged using maximum intensity.
Protein and RNA analysis.
Whole-cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer resolved in 4 to 12% Tris-glycine gels (Invitrogen) and transferred onto Immobilon-P membranes (Millipore). Incubations with primary mouse monoclonal antibodies recognizing TOP2A, HuR, green fluorescent protein (GFP), α-tubulin (Santa Cruz Biotech), or β-actin (Abcam) or a rabbit polyclonal antibody recognizing γ-H2AX (Santa Cruz) were followed by incubations with secondary antibodies (Amersham) and detection using enhanced luminescence (Amersham) or SuperSignal West Femto maximum-sensitivity substrate (Thermo Fisher). Loading controls (α-tubulin or β-actin) were chosen in each case to avoid detecting overlapping protein signals on the same blot.
After reverse transcription (RT) of total cellular RNA using random hexamers and SSII reverse transcriptase (Invitrogen), real-time quantitative PCR (qPCR) analysis was performed using SYBR green PCR master mix (Kapa Biosystems) and the following gene-specific primers (forward and reverse, respectively): GCGAGTGTGCTGGTCACTAA and ACAATTGGCCGCTAAACTTG to detect TOP2A mRNA, TGCACCACCAACTGCTTAGC and GGCATGGACTGTGGTCATGAG to detect GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA, and TGACCGCAGAGTCTTTTCCCT and TGGGTTGGTCATGCTCACTA to detect EGFP (enhanced GFP) mRNA. Mature miR-548c-3p, miR-143, miR-355, miR-410, miR-495, miR-544, and miR-548e microRNAs, as well as U6 snRNA, were quantified using a QuantiMir detection assay (System Biosciences). miR-548c-3p miRNA was from Applied Biosystems.
Detection of RNPs: biotin pulldown and RNP IP analysis.
Immunoprecipitation of native RNP complexes (RNP IP analysis) was performed as described previously (3), using primary antibodies recognizing HuR or YFP or control IgG (Santa Cruz Biotech); RNA in the IP samples was further analyzed by RT-qPCR using the primers listed above.
Biotin pulldown analysis was carried out as described previously (3). Briefly, cDNA was used as a template for PCR amplifications to prepare biotinylated transcripts spanning the TOP2A mRNA. To synthesize each template, the forward primers contained the T7 RNA polymerase promoter sequence (T7) CCAAGCTTCTAATACGACTCACTATAGGGAGA. The following primer pairs (forward and reverse) were used to synthesize TOP2A 3′UTR fragments: (T7)AATGTGAGGCGATTATTTTAAGTA and GCAGAGAAGAAAACAATGCCCAT for fragment A, (T7)CTGTCTAAATAGTGACCATCTC and AAAGGAGGAAGAGTGACACTT for fragment B, (T7)CAGTTTGATTTAAAAGTGTCACTC and CCTCTGATGATTTGAGAAGATGA for fragment C, (T7)GCTCATGTTCTTCATCTTCTCA and AATGTTGTCCCCGAGTCTTCTG for fragment D, and (T7)GAGGACTGGATTGCAGAAGAC and TTTATTAAAGTACAAAATTGTTGGAAT for fragment E. The TOP2A coding region (CR) biotinylated RNA was synthesized using the primer pair (T7)CAGCAAATGTGGGTTTACGATGA and GGTGATACATGTATAATCTTCTCCA. The biotinylated GAPDH 3′UTR was synthesized using the primer pair (T7)CCTCAACGACCACTTTGTCA and GGTTGAGCACAGGGTACTTTATT. Whole-cell lysates (100 μg per sample) were incubated with 3 μg of purified biotinylated transcripts (1 h, 25°C); the complexes were isolated with streptavidin-coupled Dynabeads (Invitrogen). Proteins present in the pulldown material were analyzed by Western blotting.
Translation assays.
Polyribosome fractionation assays were carried out as explained previously (4, 25). In short, cells were incubated with cycloheximide (100 μg/ml, 15 min; Sigma), and cytoplasmic lysates (500 μl) were fractionated by centrifugation through 15 to 60% linear sucrose gradients and divided into 10 fractions for RT-qPCR analysis to determine the distribution of TOP2A mRNA and GAPDH mRNA.
Nascent translation of TOP2A was studied by two methods (4, 15). In one, HeLa cells were incubated with 1 mCi l-[35S]methionine and l-[35S]cysteine (NEN/Perkin Elmer) per 60-mm plate for 15 min; after lysis, IP reactions were carried out (16 h, 4°C) using IgG1 (BD Pharmingen), anti-GAPDH, or anti-TOP2A antibodies, and reactions were resolved by SDS-PAGE. Reaction products were transferred to polyvinylidene difluoride (PVDF) filters and visualized using a PhosphorImager (Molecular Dynamics). In the other, HeLa cells were incubated for 1 h in methionine- and cysteine-free medium containing l-azidohomoalanine (AHA; Invitrogen) that was incorporated into nascent polypeptide chains; lysates were then labeled using alkyne-biotin and a Click-iT protein reaction buffer kit (Invitrogen), following the manufacturer's instructions. Biotinylated proteins were isolated using streptavidin-coupled Dynabeads (Roche), and TOP2A and GAPDH were detected by Western blot analysis.
RESULTS
HuR associates with the TOP2A 3′UTR and increases TOP2A translation.
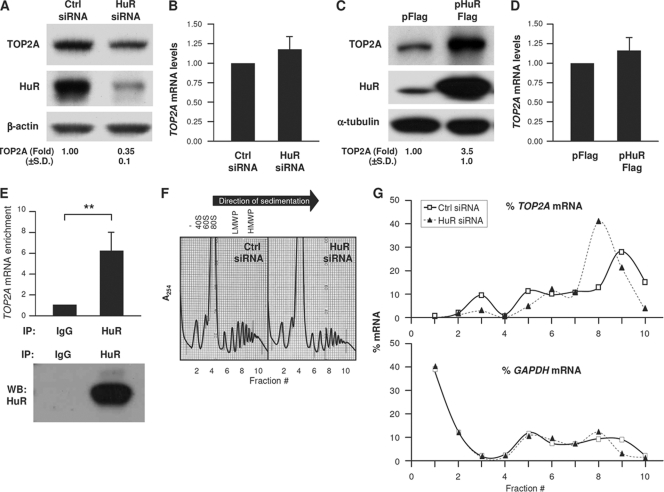
A survey of HuR-regulated gene expression suggested that TOP2A mRNA was a likely target of HuR (27). Reducing HuR levels by transfection of siRNA potently lowered HuR levels (by <90%) and reduced TOP2A to one-third of its original abundance, while TOP2A mRNA levels remained unchanged (Fig. 1A and B). Conversely, HuR overexpression increased the levels of TOP2A protein but not TOP2A mRNA (Fig. 1C and D). HuR similarly promoted TOP2A expression in other carcinoma cells (not shown). As HuR modulates the translation of some target mRNAs (2), we tested if it associated with TOP2A mRNA. Immunoprecipitation (IP) of HuR ribonucleoprotein (RNP) complexes was followed by RNA isolation from the IP materials and RT-qPCR amplification of TOP2A mRNA; control reactions included IP with IgG and measurement of the abundant housekeeping GAPDH mRNA, a nonspecific transcript present in all RNP IP samples that was used to normalize sample input. This analysis revealed that TOP2A mRNA was enriched ∼6-fold in HuR IP samples (Fig. 1E).
Fig. 1.
HuR associates with TOP2A mRNA and promotes TOP2A translation. (A and B) Forty-eight hours after transfection of HeLa cells with Ctrl siRNA or siRNA targeting HuR (HuR siRNA), Western blot analysis of TOP2A, HuR, and loading control β-actin was performed (A), and the levels of TOP2A mRNA relative to GAPDH mRNA were calculated by RT-qPCR analysis (B). (C and D) Twenty-four hours after transfection of cells with a control (pFlag) or HuR overexpression (pHuR-Flag) plasmid, the levels of TOP2A, HuR, and loading control β-tubulin were assessed by Western blot analysis (C) and TOP2A mRNA/GAPDH mRNA by RT-qPCR analysis (D). (E) RNP IP analysis of the interaction of TOP2A mRNA with HuR using anti-HuR and mouse IgG antibodies. (Top) TOP2A mRNA was detected by RT-qPCR and normalized to GAPDH mRNA levels (see Materials and Methods). (Bottom) Western blot (WB) analysis of HuR in the IP samples. (F and G) Lysates prepared from cells that were transfected as described for panel A were fractionated through sucrose gradients (F), and the relative distribution (%) of TOP2A mRNA and control GAPDH mRNA (G) was studied by RT-qPCR analysis of RNA in each of 10 gradient fractions. The arrow indicates the direction of sedimentation. −, fractions without ribosomal components; 40S and 60S, small and large ribosome subunits, respectively; 80S, monosome; LMWP and HMWP, low- and high-molecular-weight polysomes, respectively. In panels A and C, TOP2A levels were quantified by densitometry; results are shown as the means + standard deviations (SD) (n = 5). In panels B and D, mRNA levels are the means + SD (n = 3). In panels F and G, data are representative of three experiments. **, P < 0.01. For all figures, statistical significance was determined using Student's t test.
To test if HuR modulated TOP2A translation, we studied the sizes of polysomes associated with TOP2A mRNA. Cytoplasmic lysates were fractionated through sucrose gradients, wherein the lightest components sedimented at the top (fractions 1 and 2), small (40S) and large (60S) ribosomal subunits and monosomes (80S) appeared in fractions 3 to 5, and progressively larger polysomes, ranging from low to high molecular weight (LMWP and HMWP, respectively), were in fractions 6 to 10 (Fig. 1F). While in Ctrl siRNA cells TOP2A mRNA levels peaked in fraction 9, silencing HuR reduced the average size of polysomes, which peaked in fraction 8 (Fig. 1G). Although this shift in polysome size was seen consistently, it is relatively modest, suggesting that perhaps other steps of translational regulation (such as translational elongation) may be impaired by silencing HuR. Additionally, the TOP2A coding region is long (∼4.5 kb) and could potentially accommodate up to 50 ribosomes; such large TOP2A mRNA-associated polysomes were not detected here, indicating that the TOP2A mRNA was likely not filled to capacity by ribosomes. Instead, the TOP2A mRNA consistently appeared in fractions 8 to 10, suggesting that the TOP2A mRNA may be occupied only sparsely by ribosomes. GAPDH mRNA polysomes largely overlapped between both transfection groups. Together, these findings suggested that HuR promoted the translation of TOP2A, at least in part by enhancing translation initiation.
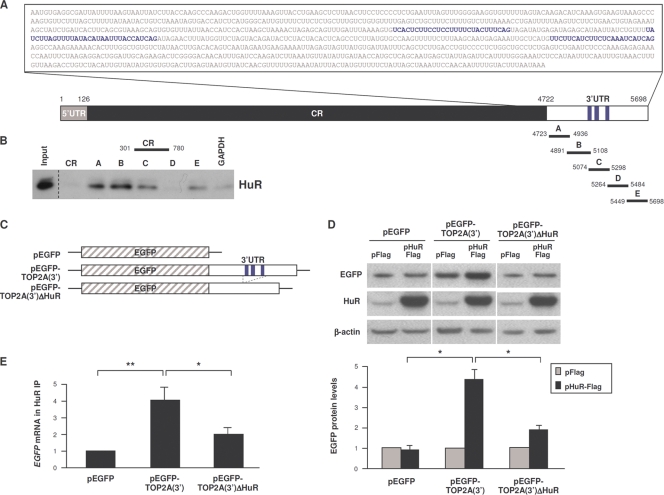
To identify the mechanisms by which HuR enhanced TOP2A translation, we assessed the region of interaction of HuR with TOP2A mRNA. TOP2A mRNA has a short 5′UTR (0.1 kb), a long coding region (CR, ∼4.5 kb) that encodes an ∼170-kDa protein, and an ∼1.0-kb long 3′UTR (Fig. 2A). Photoactivatable-ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) analysis revealed three HuR interaction sites in the TOP2A 3′UTR (J. D. Keene, personal communication); that HuR interacted with these sites was confirmed by biotin pulldown analysis, as biotinylated RNAs B and C, which contained the three CLIP sites, showed affinity for HuR (Fig. 2B). HuR also associated with biotinylated fragments A and E, which lacked HuR sites detectable by CLIP, suggesting that there could be additional sites of association of HuR with the TOP2A 3′UTR (Fig. 2B).
Fig. 2.
HuR binds to the TOP2A 3′UTR. (A) Schematic of the TOP2A 3′UTR depicting HuR interaction sites (blue text and stripes) identified by PAR-CLIP analysis (J. D. Keene, personal communication). Biotinylated transcripts (A to E) were synthesized spanning the TOP2A 3′UTR and a comparable segment of the TOP2A CR; a control biotinylated GAPDH 3′UTR RNA was used to measure background binding. (B) Incubation of equimolar biotinylated RNAs with whole-cell HeLa lysates was followed by pulldown of RNP complexes and HuR detection in the complexes by Western blot analysis. Input, 5 μg of whole-cell lysate. (C) Reporter constructs were engineered by cloning the TOP2A 3′UTR (full length or with deletion of the HuR CLIP sites) into plasmid pEGFP to generate pEGFP-TOP2A(3′) and pEGFP-TOP2A(3′)ΔHuR. (D) Cells were transfected with the plasmids shown in panel C together with either pFlag or pHuR-Flag; 16 h later, the levels of expressed EGFP protein were assessed by Western blot analysis (top), quantified by densitometry, normalized to α-tubulin levels, and plotted (bottom) (n = 5). (E) In cells that were transfected with the EGFP reporter plasmids shown in panel C, the association of HuR with the expressed transcripts [EGFP mRNA and chimeric EGFP-TOP2A(3′) and EGFP-TOP2A(3′)ΔHuR mRNAs] was measured 16 h later by RNP IP analysis. Shown are the means + SD (n = 3). *, P < 0.05; **, P < 0.01.
Further analysis of these interactions was performed using a chimeric reporter construct that expressed the EGFP CR linked to either the full-length TOP2A 3′UTR [pEGFP-TOP2A(3′)] or the TOP2A 3′UTR bearing a deletion of the HuR binding sequences [pEGFP-TOP2A(3′)ΔHuR] (Fig. 2C). Compared with EGFP expressed from the parent plasmid (pEGFP), HuR overexpression increased EGFP reporter protein levels from pEGFP-TOP2A(3′) 4-fold, while EGFP expressed from pEGFP-TOP2A(3′)ΔHuR increased less than 2-fold (Fig. 2D). Accordingly, HuR RNP IP analysis revealed that the association of EGFP-TOP2A(3′) mRNA with HuR was enriched ∼4-fold relative to the control EGFP mRNA, while EGFP-TOP2A(3′)ΔHuR mRNA was enriched only ∼2-fold (Fig. 2E). Together, these findings indicate that HuR enhances TOP2A translation via its interaction with multiple sites of the TOP2A 3′UTR.
Repression of TOP2A translation by silencing HuR is linked to increased colocalization of TOP2A mRNA with RISC and PBs.
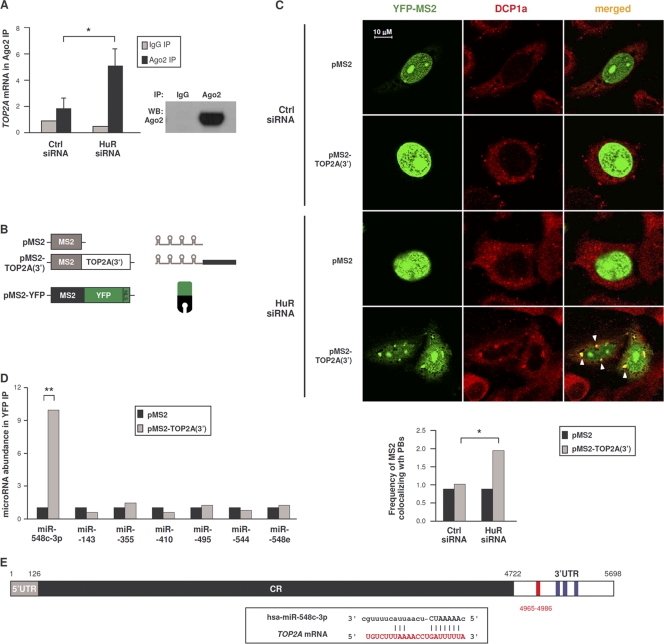
The above-described results suggested that TOP2A mRNA was translationally repressed in the absence of HuR. To test if TOP2A mRNA associated with the RISC machinery, we performed RNP IP analysis of a key RISC component, the protein Argonaute 2 (Ago2) (Fig. 3A). Ago2 RNP IP showed a modest enrichment in TOP2A mRNA in control transfections (Fig. 3A), but this enrichment increased to ∼5-fold after silencing HuR. The finding that silencing HuR increased the association of TOP2A mRNA with Ago2 suggests that HuR silencing may promote translational repression via microRNA-mediated events.
Fig. 3.
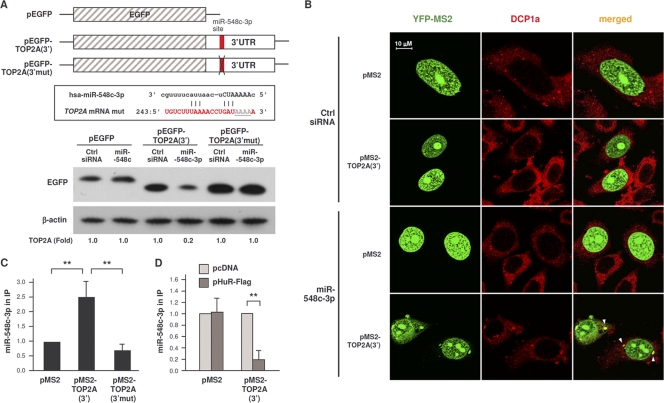
TOP2A mRNA interacts with components of the microRNA decay machinery and with miR-548c-3p. (A) Forty-eight hours after transfection of either Ctrl siRNA or HuR siRNA, the interaction of TOP2A mRNA with Ago2-containing complexes was assessed by RNP IP (see Materials and Methods). (Right) Western blot analysis of Ago2 in the IP samples. (B) Schematic of plasmids expressing MS2-tagged RNA [pMS2, pMS2-TOP2A(3′)] and expressing a fluorescent reporter protein (pMS2-YFP). (C) HeLa cells were transfected with the siRNAs indicated; 48 h later, cells were transfected with the plasmids shown in panel B, and 24 h after that, cells were fixed for analysis. Fluorescent signals from MS2-YFP (green) as well as from DCP1a (red, detected by immunofluorescence) (see Materials and Methods) were analyzed by confocal microscopy. Merged images show the overlap between MS2-YFP and DCP1a; foci of colocalized signals (yellow) are indicated with arrowheads. (Graph) The numbers of cells showing overlapping MS2-YFP and DCP1a signals in each transfection group were counted and plotted. (D) In cells transfected as described for panel C, the interaction of the indicated microRNAs was tested by RNP IP analysis using anti-YFP antibody. The levels of microRNAs in IP samples were assessed by RT-qPCR analysis and plotted as enrichment in the pMS2-TOP2A(3′)-transfected cells relative to pMS2-transfected cells. (E) TOP2A 3′UTR depicting the miR-548c-3p site (red) and the HuR CLIP sites (blue); box, complementarity between TOP2A mRNA and miR-548c-3p. In panels A, C, and D, data are the means + SD from 3 independent experiments. *, P < 0.05; **, P < 0.01.
This possibility was further examined by testing the subcellular localization of a traceable TOP2A mRNA bearing MS2 hairpins [expressed from plasmid pMS2-TOP2A(3′)]. HeLa cells were cotransfected with pMS2-TOP2A(3′) and with plasmid pMS2-YFP, which expressed a chimeric reporter protein (MS2-YFP, containing a nuclear localization signal [NLS]) capable of interacting with MS2-tagged RNA (Fig. 3B). The distribution of MS2-tagged RNA was then studied by fluorescence microscopy. MS2 mRNA expressed from control vector pMS2 and detected by MS2-YFP protein was nuclear (green signals) due to the presence of the NLS (Fig. 3C). MS2-TOP2A(3′) mRNA was also nuclear in control (Ctrl siRNA) cells, but HuR silencing triggered the appearance of punctate green fluorescent foci in the cytoplasm of the HuR siRNA group; these foci colocalized in part with DCP1a (red), another component of RISC which serves as a PB marker, visualized as yellow signals in the merged images (arrowheads). As represented in the graph (Fig. 3C), in HuR-silenced cells MS2 signals colocalized with the Dcp1a PB marker ∼2-fold more frequently than in control cells. These findings indicate that the repression of TOP2A translation that occurs after lowering HuR was associated with the localization of TOP2A mRNA with components of RISC and PBs, sites of translational silencing.
The results further suggested that microRNAs might be involved in TOP2A translational repression that followed HuR silencing. Since several putative microRNAs targeting the TOP2A 3′UTR were predicted computationally, we devised a new method to test the association of the TOP2A 3′UTR with microRNAs in a cell-relevant context. Cells were transfected with the plasmids shown in Fig. 3B, and after RNP IP analysis of MS2-YFP the levels of microRNAs in the IP materials were assessed; among them, only miR-548c-3p was enriched in association with MS2-TOP2A(3′) mRNA relative to MS2 mRNA (Fig. 3D and E). Together, these results support the view that repression of TOP2A expression is linked to the association of TOP2A mRNA with components of the microRNA translational repression machinery.
Repression of TOP2A expression by miR-548c-3p is relieved by HuR.
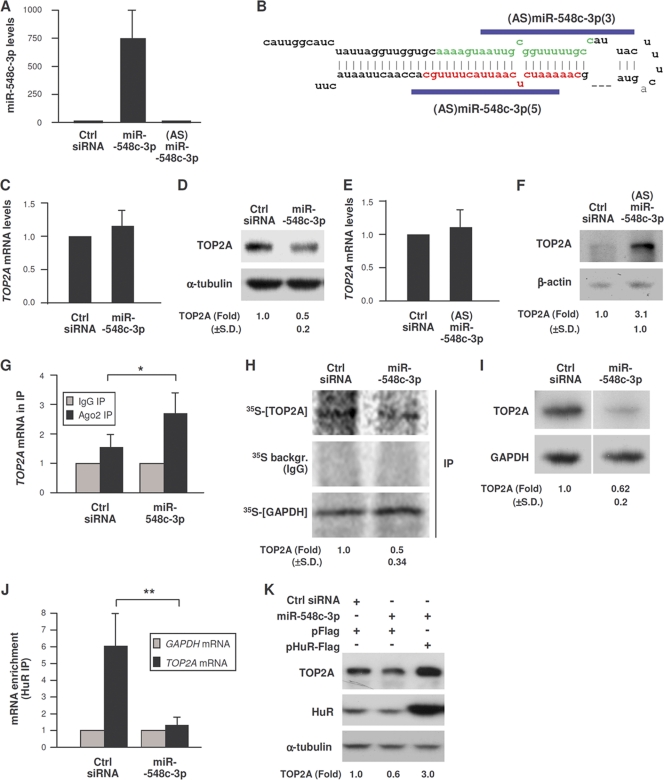
In agreement with the hypothesis that miR-548c-3p can repress TOP2A translation, overexpression of miR-548c-3p by transfection of HeLa cells (Fig. 4A) did not affect the levels of TOP2A mRNA but lowered TOP2A protein levels by 50% (Fig. 4C and D). Conversely, transfection of antisense (AS) RNAs designed to complement Pre-miR-548c-3p (Fig. 4B) increased TOP2A protein levels but did not affect TOP2A mRNA levels (Fig. 4E and F). Commercial antagomirs had no effect (not shown). Evidence that miR-548c-3p recruited the TOP2A mRNA to the RISC was obtained by Ago RNP IP, as overexpression of miR-548c-3p in HeLa cells increased Ago2 association with TOP2A mRNA (Fig. 4G). Consequently, analysis of nascent TOP2A translation by incubation of HeLa cells for a brief time period (15 min) with [35S]Met/Cys followed by detection of newly synthesized TOP2A protein by IP using anti-TOP2A antibody revealed that de novo TOP2A production declined after miR-548c-3p overexpression (Fig. 4H). Similarly, incubation of cells with l-azidohomoalanine followed by labeling of nascent proteins with alkyne-biotin and Western blot analysis to detect newly synthesized proteins (Click-iT method) further showed that miR-548c-3p lowered TOP2A translation (Fig. 4I). In sum, miR-548c-3p interacts with the TOP2A 3′UTR and reduces TOP2A translation.
Fig. 4.
miR-548c-3p represses TOP2A translation. (A and B) miR-548c-3p levels were detected by RT-qPCR analysis in cells transfected with Ctrl siRNA, miR-548c-3p mimic, or (AS)miR-548c-3p (B). (C to F) Seventy-two hours after transfection with Ctrl siRNA, miR-548c-3p mimic (C and D), or siRNAs targeting Pre-miR-548c-3p (E and F), TOP2A mRNA levels were assessed by RT-qPCR analysis and normalized to GAPDH mRNA levels (C and E), and TOP2A abundance was assessed by Western blot analysis and quantified by densitometry (D and F). (G) The association of TOP2A mRNA with Ago2-containing complexes was assessed by RNP IP analysis of cells transfected with miR-548c-3p or control siRNA. (H and I) De novo TOP2A and GAPDH translation in HeLa cells transfected with miR-548c-3p or control siRNA was assessed by 35S-amino acid incorporation (H) or by the Click-iT assay (I). (J) After transfection with either Ctrl siRNA or miR-548c-3p, the enrichment of TOP2A mRNA and GAPDH mRNA in HuR IP samples was tested by RNP IP analysis; samples were normalized to input RNA. (K) Forty-eight hours after transfection of Ctrl siRNA or miR-548c-3p, HeLa cells were transfected with pFlag or pHuR-Flag plasmid, and lysates were prepared 24 h later. The levels of TOP2A, HuR, and control α-tubulin were assessed by Western blot analysis. *, P < 0.05; **, P < 0.01 (3 to 5 independent experiments were performed). For panels C and D, n = 5; for panels E to I, J, and K, n = 3; for panels D, F, H, and I, P < 0.05.
Since miR-548c-3p repressed TOP2A translation while HuR enhanced it, we postulated that these molecules may be functional antagonists. To test this hypothesis, the association of TOP2A mRNA with HuR was measured by RNP IP analysis in cells expressing different levels of miR-548c-3p. miR-548c-3p overexpression markedly reduced the interaction between HuR and TOP2A mRNA (Fig. 4J), while increasing HuR rescued the repression of TOP2A by miR-548c-3p (Fig. 4K) and the inhibition of TOP2A by silencing HuR was rescued if miR-548c-3p levels were simultaneously reduced (data not shown). Together these findings indicate that HuR antagonizes miR-548c-3p, thereby enhancing TOP2A expression.
To test if miR-548c-3p repressed TOP2A expression via its putative TOP2A 3′UTR site, we compared EGFP expression from pEGFP relative to that from pEGFP-TOP2A(3′) (carrying the TOP2A 3′UTR) and to that from a mutant reporter with 4 mismatches in the seed region [pEGFP-TOP2A(3′mut)] to disrupt miR-548c-3p binding (Fig. 5A, top). By Western blot analysis, miR-548c-3p overexpression did not influence EGFP production in the pEGFP transfection group but potently reduced EGFP expressed from pEGFP-TOP2A(3′) (Fig. 5A, bottom). Importantly, mutation of the miR-548c-3p site totally abrogated the repressive influence of miR-548c-3p, indicating that miR-548c-3p acted through the predicted TOP2A 3′UTR site.
Fig. 5.
miR-548c-3p promotes TOP2A localization in PBs. (A) Schematic of plasmids pEGFP, pEGFP-TOP2A(3′), and pEGFP-TOP2A(3′mut); the last plasmid bears four mutations in the seed region of the TOP2A 3′UTR to disrupt its interaction with miR-548c-3p (box). Twenty-four hours after transfection of cells with Ctrl siRNA or miR-548c-3p, cells were transfected with each of the three reporter plasmids; lysates were prepared 24 h after that, and the levels of EGFP were assessed by Western blot analysis. (B) HeLa cells were transfected with Ctrl siRNA or miR-548c-3p; 48 h later, cells were transfected with the plasmids shown in Fig. 3B, and 24 h after that, cells were fixed for analysis. Fluorescent signals from MS2-YFP (green) and DCP1a (red) were detected by confocal microscopy. In the merged images, foci of colonized signals (yellow) are indicated with arrowheads. (C) The levels of miR-548c-3p associated with MS2-TOP2A(3′) and MS2-TOP2A(3′mut) were measured by RNP IP analysis (using IgG or anti-YFP antibodies) in cells that had been transfected with the corresponding plasmids. (D) The abundance of miR-548c-3p associated with MS2-TOP2A(3′) was measured as described for panel C except that the interaction was measured in cells that expressed endogenous HuR (pFlag) or overexpressed HuR (by transfection of pHuR-Flag, as described for Fig. 1C). The data in panel A are representative of 4 experiments, and the data in panels C and D are representative of 3 experiments. **, P < 0.01.
Analysis of MS2-tagged RNA showed that miR-548c-3p overexpression increased the cytoplasmic presence of MS2-TOP2A(3′) mRNA (green), which colocalized with the PB marker DCP1a (red) (Fig. 5B, merged column, arrowheads). Further biochemical analysis by MS2-YFP RNP IP revealed that miR-548c-3p interacted preferentially with MS2-TOP2A(3′) RNA but not with MS2 or MS2-TOP2A(3′mut) RNAs (Fig. 5C). Importantly, the interaction of miR-548c-3p with the TOP2A 3′UTR decreased markedly when HuR was overexpressed, further supporting the notion that HuR might compete with miR-548c-3p for binding to the TOP2A 3′UTR (Fig. 5D). These experiments collectively indicate that miR-548c-3p represses TOP2A expression via a specific TOP2A 3′UTR site, that miR-548c-3p recruits the TOP2A mRNA to cytoplasmic PBs, and that HuR competes with miR-548c-3p for binding to TOP2A mRNA. Based on these results, we propose that the association of TOP2A mRNA with miR-548c-3p represses its translation via Ago/RISC (see model described below), while HuR antagonizes miR-548c-3p, thereby derepressing TOP2A translation and allowing TOP2A to accumulate.
HuR and miR-548c-3p alter TOP2A levels during G2/M and modulate DNA damage by doxorubicin.
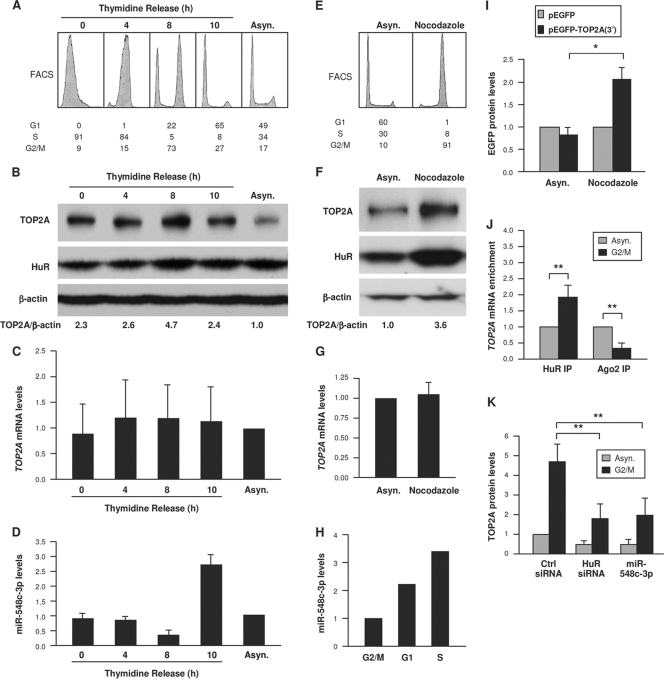
We studied the consequences of these interactions by monitoring TOP2A levels in G2/M, a cell cycle phase in which TOP2A is upregulated (19, 39). In cells released from double thymidine block arrest and progressing through G1, S, and G2/M (Fig. 6A), TOP2A levels were higher when most cells were in G2/M (8 h, Fig. 6B), TOP2A mRNA levels did not appear to change significantly (Fig. 6C), and miR-548c-3p levels correlated inversely with TOP2A (Fig. 6D). In cells arrested in G2/M by 16 h of nocodazole treatment, TOP2A and HuR were elevated, while TOP2A mRNA was unchanged (Fig. 6E to G); miR-548c-3p levels were lowest in G2/M (Fig. 6H). Accordingly, reporter protein expression from pEGFP-TOP2A(3′) was selectively higher (Fig. 6I), indicating that the increased TOP2A expression in G2/M was likely due to enhanced translation through the TOP2A 3′UTR. These results support the hypothesis that low levels of miR-548c-3p and high levels of HuR contribute to elevating TOP2A expression in G2/M. RNP IP analysis further showed that in cells arrested in G2/M by nocodazole treatment, TOP2A mRNA associated more with HuR and less with Ago2 (Fig. 6J). In turn, silencing HuR or overexpressing miR-548c-3p prevented the increase in TOP2A protein seen in G2/M in control (Ctrl siRNA) cells (Fig. 6K). These changes were not due to major alterations in cell cycle distribution (not shown).
Fig. 6.
TOP2A increase in G2/M is influenced by lower miR-548c-3p and higher HuR levels. (A) Fluorescence-activated cell sorter (FACS) analysis of HeLa cells that were synchronized by double thymidine block and released for the times indicated (percentage of cells in each phase is shown). (B to D) Cells treated as described for panel A were used for Western blot analysis to assess the levels of TOP2A, HuR, and control β-actin (B) and for RT-qPCR analysis to quantify the levels of TOP2A mRNA (C) and miR-548c-3p (D). (E) FACS analysis of HeLa cells that were left untreated (asynchronous, Asyn.) or were treated with nocodazole (100 ng/ml, 16 h); the percentage of cells in each cell cycle phase is shown. (F) Lysates were prepared from cells treated as described for panel E, and the levels of TOP2A, HuR, and control β-actin were assessed by Western blot analysis and quantified by densitometry. (G) Total RNA was prepared from cells treated as described for panel E, and the levels of TOP2A mRNA and normalization control GAPDH mRNA were measured by RT-qPCR analysis. (H) The levels of miR-548c-3p were quantified by RT-qPCR (normalized to U6 RNA) in cells that were treated with nocodazole for 16 h (G2/M group) or released from arrest and tested 8 h (G1) and 12 h (S) later. (I) HeLa cells transfected with the plasmids shown in Fig. 2C were treated with nocodazole (100 ng/ml), and lysates were prepared 16 h later. The levels of EGFP were assessed and plotted as described for Fig. 2D. (J) Cells were left untreated (Asyn.) or were treated with nocodazole (100 ng/ml); 16 h later, lysates were prepared and the association of TOP2A mRNA with HuR and Ago was measured by RNP IP analysis. (K) Cells were transfected with Ctrl siRNA, HuR siRNA, or miR-548c-3p; 48 h later, cells were either treated with nocodazole (100 ng/ml, 16 h) or left untreated, and TOP2A levels were assessed by Western blot analysis. For panels B to D, n = 2; for panel I, n = 3. *, P < 0.05; **, P < 0.01.
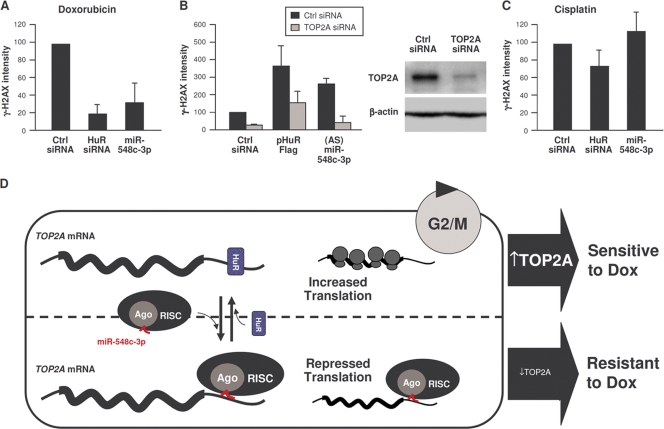
Finally, we studied the impact of modulating TOP2A on the genotoxic damage by doxorubicin (Dox), a DNA intercalator and TOP2 poison that stabilizes the complex of TOP2A bound to the double-strand DNA breaks that TOP2A generates. As Dox treatment is most effective when TOP2A levels are high (29), we postulated that changes in levels of TOP2A elicited by HuR or miR-548c-3p could alter Dox effectiveness. Dox treatment showed extensive DNA damage in control cells, as assessed by monitoring phosphorylated H2AX (γ-H2AX) signals (Fig. 7A). In contrast, lowering TOP2A by overexpressing miR-548c-3p or silencing HuR or TOP2A itself reduced the levels of γ-H2AX, suggesting that decreasing TOP2A ameliorated Dox-induced DNA damage (Fig. 7A and B). This effect was specific for Dox, since chemotherapeutic agents which do not target TOP2A, like cisplatin, did not show differential DNA damage among the transfection groups (Fig. 7C). Moreover, HuR overexpression or reduction of miR-548c-3p increased the Dox-induced γ-H2AX signal, an effect that was rescued by silencing TOP2A (Fig. 7B).
Fig. 7.
TOP2A increase by doxorubicin treatment is influenced by lower miR-548c-3p and higher HuR levels. (A and C) Seventy-two hours after transfection with the small RNAs indicated, cells were treated with doxorubicin (1 μM) (A) or cisplatin (10 μM) (C) or left untreated; 24 h later, γ-H2AX, HuR, and β-actin levels were assessed by Western blot analysis and quantified by densitometry. (B) Cells were transfected with the small RNAs and/or plasmids indicated; 48 h later, cells were treated with Dox (1 μM), and the levels of γ-H2AX were assessed by Western blot analysis 24 h later. (D) Schematic of proposed antagonism between miR-548c-3p, which recruits TOP2A mRNA to microRNA-RISC repression machinery and enables Dox resistance (bottom), and HuR, which competes with miR-548c-3p and allows TOP2A translation to occur (top), as we postulate that TOP2A translation occurs during G2/M and sensitizes cells to doxorubicin. For panels A to C, n = 3.
Based on this evidence, we propose a model whereby the association of TOP2A mRNA with miR-548c-3p leads to its translational repression via Ago/RISC. HuR, likely interacting with multiple sites in the TOP2A 3′UTR, can antagonize miR-548c-3p, thereby derepressing TOP2A synthesis and allowing TOP2A to accumulate (Fig. 7D). In sum, posttranscriptional factors that reduce TOP2A levels (such as miR-548c-3p) in competition with posttranscriptional factors that enhance TOP2A expression (HuR) potently regulate the levels of the major chemotherapeutic target TOP2A and hence the effectiveness of doxorubicin.
DISCUSSION
We report that translation controls TOP2A abundance. The interaction of the TOP2A 3′UTR with HuR promoted TOP2A translation while interaction with miR-548c-3p repressed it. The method used to identify miR-548c-3p as a regulator of TOP2A expression is noteworthy. Although bioinformatic programs can predict microRNA-mRNA interactions, they have substantial limitations. Here, we performed IP of MS2-tagged RNA to validate a number of predicted microRNA-TOP2A mRNA interactions in a meaningful context (Fig. 3B and D). With this approach, we identified miR-548c-3p as a microRNA that associated with a specific site of the TOP2A 3′UTR sequence and selectively repressed its translation (Fig. 4). Analysis of endogenous TOP2A mRNA and heterologous reporters revealed that HuR competed with miR-548c-3p for binding to the TOP2A 3′UTR, since the association of miR-548c-3p prevailed after miR-548c-3p overexpression or after HuR silencing, while binding of miR-548c-3p decreased after HuR overexpression (Fig. 5). We further show that the relative association of the TOP2A mRNA with HuR and with miR-548c-3p potently controls TOP2A translation. It is especially interesting that these TOP2A regulatory factors act upon the TOP2A mRNA and not by affecting TOP2A transcription, as transcription itself could be impaired by genotoxic damage, which may in fact need TOP2A for its resolution. Instead, microRNAs and RBPs can act upon already synthesized TOP2A mRNA to modulate the production of TOP2A protein.
As RBPs and microRNAs coexist on shared target mRNAs, there is increasing interest in elucidating their functional interaction (9, 16, 22). Like most other microRNAs reported to date, miR-548c-3p associated with a specific site of the TOP2A 3′UTR sequence and selectively repressed its translation; mutation of this site abrogated the inhibitory action of miR-548c-3p (Fig. 5A). In contrast, HuR interacted with several regions of the TOP2A 3′UTR (Fig. 2); whether competition by HuR relies on one or several TOP2A 3′UTR interaction regions remains to be determined. The silencing of HuR triggered a reduction in the size of TOP2A mRNA-associated polysomes (Fig. 1G) and caused the mobilization of a traceable TOP2A transcript into PBs (Fig. 3C).
Although a limitation of analyzing tagged RNA is that it must be ectopically expressed, the MS2 tag system has proven to be particularly valuable for investigating the biochemical interactions and subcellular localization of RNAs of interest. For example, it was used to identify the RBPs interacting with a specific mRNA, as well as to investigate the localization and movement of specific transcripts in Saccharomyces cerevisiae and mammalian cells (6, 8, 18, 32). In this report, the cytoplasmic MS2-TOP2A mRNA signals seen after silencing HuR (Fig. 3C) or after overexpressing miR-548c-3p (Fig. 5B) colocalized with the PB marker DCP1a (yellow merged signals). This finding is significant given the link between microRNAs and PBs (10). The fact that some DCP1a signals did not colocalize with MS2-TOP2A(3′) supports the notion that DCP1a-containing foci, such as PBs, are heterogeneous (14, 17).
In closing, our results indicate that the upregulation of TOP2A during G2/M is at least partly due to increased TOP2A translation mediated by its 3′UTR, linked to the increased expression of its translational enhancer HuR and decreased expression of its translational repressor miR-548c-3p. Elucidation of the mechanisms that control expression of TOP2A may also have important implications in cancer therapy. When TOP2 levels are elevated, treatment with the TOP2 poisons doxorubicin and etoposide generates lesions that include DNA breaks and covalently bound protein. High levels of these complexes block DNA replication and transcription, which triggers apoptosis (28). Given this mechanism of action, cancer cells are capable of reducing TOP2 expression levels in order to become resistant to TOP2-targeting drugs (37), although the specific processes needed to achieve this reduction are unknown. In this regard, it will be interesting to test if lowering HuR levels or increasing miR-548c-3p levels contributes to the reduced TOP2A abundance found in cells that become resistant to TOP2 poisons. Conversely, the levels of TOP2 and the efficacy of TOP2 poisons correlate (34), while ectopically lowering TOP2 engenders resistance to doxorubicin (11). In light of this evidence and our results described here, it seems advantageous to selectively increase HuR or decrease miR-548c-3p levels in order to elevate TOP2A abundance in cells exposed to TOP2 poisons. Furthermore, high levels of HuR in cancers might be a predictor of a robust response to doxorubicin, while conversely, TOP2A 3′UTR mutations causing reduced HuR binding could confer resistance to doxorubicin. We expect that these questions will be investigated in the near future. As illustrated with TOP2A, our findings highlight the role of RBPs and microRNAs in the posttranscriptional expression of key factors governing DNA replication and cancer therapy.
ACKNOWLEDGMENTS
We thank J. D. Keene and N. Mukherjee for sharing unpublished results.
This research was supported by the NIA-IRP, NIH. R.K., R.P., and D.C. were supported by the Barr Award and NCI R01CA142698.
Footnotes
Published ahead of print on 18 July 2011.
REFERENCES
- 1. Abdelmohsen K., Gorospe M. 2011. Post-transcriptional regulation of cancer traits by HuR. WIRES RNA 1: 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdelmohsen K., Kuwano Y., Kim H. H., Gorospe M. 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 389: 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abdelmohsen K., et al. 2007. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25: 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abdelmohsen K., Srikantan S., Kuwano Y., Gorospe M. 2008. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. U. S. A. 105: 20297–20302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ambros V. 2004. The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- 6. Bachler M., Schroeder R., von Ahsen U. 1999. StreptoTag: a novel method for the isolation of RNA-binding proteins. RNA 5: 1509–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 8: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bertrand R., et al. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- 9. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- 10. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. 2006. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harbor Symp. Quant. Biol. 71: 513–521 [DOI] [PubMed] [Google Scholar]
- 11. Burgess D. J., et al. 2008. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 105: 9053–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Champoux J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70: 369–413 [DOI] [PubMed] [Google Scholar]
- 13. Chekulaeva M., Filipowicz W. 2009. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 21: 452–460 [DOI] [PubMed] [Google Scholar]
- 14. Decker C. J., Parker R. 2006. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J. Cell Biol. 173: 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dieterich D. C., Link A. J., Graumann J., Tirrell D. A., Schuman E. M. 2006. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U. S. A. 103: 9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eiring A. M., et al. 2010. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140: 652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujimura K., Katahira J., Kano F., Yoneda Y., Murata M. 2009. Selective localization of PCBP2 to cytoplasmic processing bodies. Biochim. Biophys. Acta 1793: 878–887 [DOI] [PubMed] [Google Scholar]
- 18. Fusco D., et al. 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heck M. M., Hittelman W. N., Earnshaw W. C. 1988. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc. Natl. Acad. Sci. U. S. A. 85: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hinman M. N., Lou H. 2008. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65: 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim H. H., et al. 2008. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 22: 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim H. H., et al. 2009. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23: 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kloosterman W. P., Plasterk R. H. 2006. The diverse functions of microRNAs in animal development and disease. Dev. Cell 11: 441–450 [DOI] [PubMed] [Google Scholar]
- 24. Lal A., et al. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23: 3092–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee E. K., et al. 2010. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 17: 732–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. López de Silanes I., et al. 2003. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 22: 7146–7154 [DOI] [PubMed] [Google Scholar]
- 27. López de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. M. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U. S. A. 101: 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nitiss J. L. 2009. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9: 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nitiss J. L. 2009. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9: 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parsons G. G., Spencer C. A. 1997. Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol. Cell. Biol. 17: 5791–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Porter A. C., Farr C. J. 2004. Topoisomerase II: untangling its contribution at the centromere. Chromosome Res. 12: 569–583 [DOI] [PubMed] [Google Scholar]
- 32. Shav-Tal Y., et al. 2004. Dynamics of single mRNPs in nuclei of living cells. Science 304: 1797–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shimogawa M. M., Widlund P. O., Riffle M., Ess M., Davis T. N. 2009. Bir1 is required for the tension checkpoint. Mol. Biol. Cell 20: 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith K., Houlbrook S., Greenall M., Carmichael J., Harris A. L. 1993. Topoisomerase IIα coamplification with erbB2 in human primary breast cancer and breast cancer cell lines—relationship to m-AMSA and mitoxantrone sensitivity. Oncogene 8: 933–938 [PubMed] [Google Scholar]
- 35. Srikantan S., Gorospe M. HuR function in disease. Front. Biosci., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stefani G., Slack F. J. 2008. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9: 219–230 [DOI] [PubMed] [Google Scholar]
- 37. Walker J. V., Nitiss J. L. 2002. DNA topoisomerase II as a target for cancer chemotherapy. Cancer Invest. 20: 570–589 [DOI] [PubMed] [Google Scholar]
- 38. Wang J. C. 1998. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 31: 107–144 [DOI] [PubMed] [Google Scholar]
- 39. Woessner R. D., Mattern M. R., Mirabelli C. K., Johnson R. K., Drake F. H. 1991. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 2: 209–214 [PubMed] [Google Scholar]