Abstract
The incorporation of histone variants into nucleosomes represents one way of altering the chromatin structure to accommodate diverse functions. Histone variant H2A.Z has specific roles in gene regulation, heterochromatin boundary formation, and genomic integrity. The precise features required for H2A.Z to function and specify an identity different from canonical H2A remain to be fully explored. Analysis of the C-terminal docking domain of H2A.Z in Saccharomyces cerevisiae using epistatic miniarray profile (E-MAP) uncovered nuanced requirements of the H2A.Z C-terminal region for cell growth when additional genes were compromised. Moreover, the H2A.Z(1–114) truncation, lacking the last 20 amino acids of the protein, did not support regular H2A.Z functions, such as resistance to genotoxic stress, restriction of heterochromatin in its native context, GAL1 gene activation, and chromatin anchoring. The corresponding region of H2A could fully rescue the strong defects caused by loss of this functionally essential region in the C terminus of H2A.Z. Despite the dramatic reduction in function, the H2A.Z(1–114) truncation still bound the H2A.Z deposition complex SWR1-C, the histone chaperone Chz1, and histone H2B. These data are consistent with a model in which retaining the variant in chromatin after its deposition by SWR1-C is a crucial determinant of its function.
INTRODUCTION
At its most basic level, the eukaryotic genome is organized as chromatin, which consists of repeating nucleoprotein moieties called nucleosomes. An individual nucleosome is formed from 146 bp of DNA wrapped around an octameric histone core containing two copies each of H2A, H2B, H3, and H4. Several fundamental mechanisms can alter the chromatin structure, including ATP-dependent chromatin remodeling (8), posttranslational modifications of histones (12), and the replacement of canonical histones with nonallelic histone variants that change the protein composition of nucleosomes (17). Whereas canonical histones are deposited into chromatin during DNA replication, histone variants often are deposited in a replication-independent manner by a class of specialized deposition complexes to specific locations in the genome in a nonrandom fashion (21).
One such histone variant, H2A.Z, is conserved from yeast to human and replaces the canonical H2A in 5 to 10% of nucleosomes (54). H2A.Z has roles in regulation of gene expression, maintenance of heterochromatin-euchromatin boundaries, DNA repair, chromosome segregation, and resistance to genotoxic stress (54). In the budding yeast Saccharomyces cerevisiae, H2A.Z is encoded by the nonessential HTZ1 gene, which greatly facilitates the functional analysis of this histone variant. For example, the slow-growth phenotype and drug sensitivity of htz1Δ yeast cells cannot be rescued by overexpression of canonical H2A, suggesting that the variant has specialized and nonredundant functions in the cell (19). The latter may be attributed to the distinct location of H2A.Z in gene promoters and subtelomeric regions, catalyzed by the conserved ATP-dependent chromatin-remodeling complex SWR1-C (2, 15, 25, 27, 30, 38, 42, 53). SWR1-C can replace canonical H2A/H2B dimers with H2A.Z/H2B in a stepwise manner in vitro and is required for H2A.Z chromatin deposition in vivo (34, 38). In addition to SWR1-C, the histone chaperones Chz1 and Nap1 are closely linked to H2A.Z biology. While these two histone chaperones are functionally redundant in aiding the deposition of H2A.Z/H2B into chromatin in vitro (35), they have different binding affinities, with Nap1 capable of binding both H2A/H2B and H2A.Z/H2B dimers (41) and Chz1 having specificity for H2A.Z/H2B (35).
On the amino acid sequence level, H2A.Z shares 60% sequence identity with its canonical cousin, and the three-dimensional structure of an H2A.Z-containing nucleosome is overall similar to that of the H2A nucleosome (49). Similarly to H2A, H2A.Z molecules are engaged in multiple protein-protein and protein-DNA interactions within the nucleosome, with the points of contact being distributed across the length of the protein (33, 49). However, there are subtle differences in specific regions between the structures of the two nucleosomes that might explain their functional differences. One of these is the L1 loop, a region where the two H2A.Z molecules in the nucleosome interact with each other. Another main structural divergence resides in the C-terminal “docking domain,” a region having less than 40% amino acid identity with H2A, which constitutes an interaction surface with H3/H4 and likely provides a binding platform for nucleosome remodeling activities (49). Further supporting the possibility that this region is a major determinant of H2A.Z's identity, amino acids around the αC helix in the docking domain form the M6 region (see Fig. 1A) (49), which is essential for H2A.Z function, as swapping it with its counterpart from H2A results in embryonic lethality of Drosophila melanogaster (5). A similar M6 swap mutant of budding yeast has decreased binding to SWR1-C and causes cellular sensitivity to formamide (52). When it replaces the corresponding region in the canonical histone, the H2A.Z docking domain can confer H2A.Z-like abilities to H2A by supporting induction of the GAL1 gene (1), further suggesting that this region is critical to the function of H2A.Z. The C terminus of H2A.Z is also modified, as K126 and K133 are sites of sumoylation, with this modified form being associated with DNA double-stranded breaks and DNA repair (23). In contrast, canonical H2A in yeast has a SQEL motif at its very C terminus, which, similarly to mammalian H2A.X, is phosphorylated upon DNA damage (7). Lastly, the docking domain also includes acidic surface residues that are part of an extended acidic patch, which may be important for contacting either the N-terminal tail of H4 from a neighboring nucleosome or nonhistone proteins (49). This acidic patch is required for H2A.Z to promote higher-order chromatin folding in higher eukaryotes (9), and specific mutations in the acidic patch result in sensitivity to genotoxic stress (20).
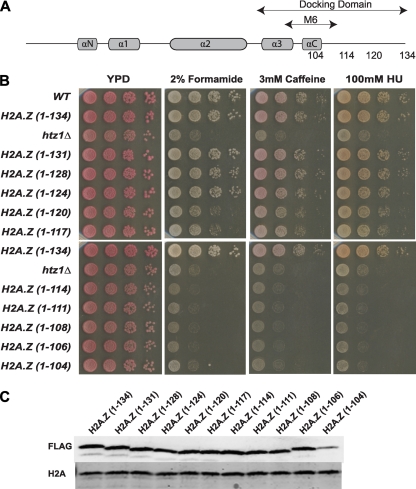
Fig. 1.
The C terminus of H2A.Z is required for H2A.Z function. (A) Schematic representation of the H2A.Z protein, indicating the positions of the alpha helices, the M6 domain (5), and the docking domain (49). The important C-terminal positions from which H2A.Z was truncated in this study are also shown. (B) The H2A.Z C terminus is required for resistance to genotoxic stress. Ten-fold serial dilutions of strains containing the indicated H2A.Z truncations with C-terminal 3×FLAG tags were plated and incubated on YP-dextrose medium containing the indicated concentrations of formamide, caffeine, and HU. WT, wild type. (C) FLAG-tagged H2A.Z truncations were expressed to wild-type levels, except for a slight decrease in H2A.Z(1–104). Protein expression levels were analyzed by immunoblotting of whole-cell extracts of the indicated strains with an anti-FLAG antibody. Antibody against histone H2A was used as a loading control.
Here, we further explore two critical questions concerning H2A.Z biology, specifically, the identification of regions that are required for H2A.Z function and the determination of regions that distinguish H2A.Z from its canonical counterpart H2A. Although different functions have been attributed to the C terminus of H2A.Z, the necessity of the docking domain has not been studied extensively with regard to H2A.Z binding to chromatin, cellular functions, and genetic interactions. A recent comprehensive alanine scan of yeast H2A.Z identified relatively few residues that resulted in sensitivity to genotoxic stressors (24). Pointing toward an important contribution of the C terminus for H2A histone family function, a recently published deletion analysis of human H2A identified its C terminus as being required for nucleosome stability, chromatin remodeling, and binding to histone H1 (50). Thus, to better understand the docking domain of H2A.Z in yeast, we created C-terminus truncation mutants. Using epistatic miniarray profile (E-MAP) (45) analyses of H2A.Z truncation alleles, we revealed nuanced requirements for the H2A.Z C terminus and demonstrated the effectiveness of this method in identifying differential sensitivities of genetic interaction profiles. Furthermore, we determined that the last 20 amino acids in the C terminus distal to amino acid 114 were critical for H2A.Z functions in resistance to genotoxic stress, restriction of heterochromatin at several loci in their native context, and GAL1 gene activation but not for retaining the ability to bind SWR1-C, H2B, and Chz1. However, this region was not unique to H2A.Z, as it could be replaced by the corresponding region in H2A. Consistent with this region being required for chromatin anchoring, it contained three important amino acids that facilitated H2A.Z residence in nucleosomes and associated functions, as well as H2A function in vivo.
MATERIALS AND METHODS
Yeast strains, plasmids, and yeast techniques.
All strains used in this study are listed in Table 1 and were created using standard yeast genetic techniques (3). Complete and partial deletion of genes and integration of a vesicular stomatitis virus (VSV) tag (11) or a 3×FLAG tag (13) in frame at the 3′ end of genes were achieved using one-step gene integration of PCR-amplified modules (32).
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype |
|---|---|
| Strain | Relevant genotype |
| MKY5 | W303, MATα ade2Δ1 can1Δ100 his3Δ11 leu2Δ3,112 trp1Δ1 ura3Δ1 LYS2 |
| MKY357 | MKY5, htz1Δ::NATMX |
| MKY583 | BY4741, MATα his3Δ1 leu2Δ0 LYS2 ura3Δ0 can1Δ::MATαPr-HIS3 lyp1Δ::MATαPr-LEU2 |
| MKY1133 | MKY5, H2A.Z(1-134)-3×FLAG::NATMX |
| MKY1134 | MKY5, H2A.Z(1-131)-3×FLAG::NATMX |
| MKY1135 | MKY5, H2A.Z(1-128)-3×FLAG::NATMX |
| MKY1136 | MKY5, H2A.Z(1-124)-3×FLAG::NATMX |
| MKY1137 | MKY5, H2A.Z(1–120)-3×FLAG::NATMX |
| MKY1138 | MKY5, H2A.Z(1-117)-3×FLAG::NATMX |
| MKY1139 | MKY5, H2A.Z(1–114)-3×FLAG::NATMX |
| MKY1140 | MKY5, H2A.Z(1-111)-3×FLAG::NATMX |
| MKY1141 | MKY5, H2A.Z(1–108)-3×FLAG::NATMX |
| MKY1142 | MKY5, H2A.Z(1–106)-3×FLAG::NATMX |
| MKY1143 | MKY5, H2A.Z(1–104)-3×FLAG::NATMX |
| MKY1144 | MKY5, htz1Δ::HYGMX |
| MKY1145 | MKY5, SWC2-VSV::KANMX |
| MKY1146 | MKY5, H2A.Z(1–134)-3×FLAG::NATMX SWC2-VSV::KANMX |
| MKY1147 | MKY5, H2A.Z(1–124)-3×FLAG::NATMX SWC2-VSV::KANMX |
| MKY1148 | MKY5, H2A.Z(1–117)-3×FLAG::NATMX SWC2-VSV::KANMX |
| MKY1149 | MKY5, H2A.Z(1–114)-3×FLAG::NATMX SWC2-VSV::KANMX |
| MKY1150 | MKY5, H2A.Z(1–108)-3×FLAG::NATMX SWC2-VSV::KANMX |
| MKY1151 | MKY5, H2A.Z(1–106)-3×FLAG::NATMX SWC2-VSV::KANMX |
| MKY1152 | MKY5, H2A.Z(1–104)-3×FLAG::NATMX SWC2-VSV::KANMX |
| MKY1153 | MKY5, SWC3-VSV::KANMX |
| MKY1154 | MKY5, H2A.Z(1–134)-3×FLAG::NATMX SWC3-VSV::KANMX |
| MKY1155 | MKY5, H2A.Z(1–124)-3×FLAG::NATMX SWC3-VSV::KANMX |
| MKY1156 | MKY5, H2A.Z(1–117)-3×FLAG::NATMX SWC3-VSV::KANMX |
| MKY1157 | MKY5, H2A.Z(1–114)-3×FLAG::NATMX SWC3-VSV::KANMX |
| MKY1158 | MKY5, H2A.Z(1–108)-3×FLAG::NATMX SWC3-VSV::KANMX |
| MKY1159 | MKY5, H2A.Z(1–106)-3×FLAG::NATMX SWC3-VSV::KANMX |
| MKY1160 | MKY5, H2A.Z(1–104)-3×FLAG::NATMX SWC3-VSV::KANMX |
| MKY1161 | MKY5, SWC4-VSV::KANMX |
| MKY1162 | MKY5, H2A.Z(1–134)-3×FLAG::NATMX SWC4-VSV::KANMX |
| MKY1163 | MKY5, H2A.Z(1–124)-3×FLAG::NATMX SWC4-VSV::KANMX |
| MKY1164 | MKY5, H2A.Z(1–117)-3×FLAG::NATMX SWC4-VSV::KANMX |
| MKY1165 | MKY5, H2A.Z(1–114)-3×FLAG::NATMX SWC4-VSV::KANMX |
| MKY1166 | MKY5, H2A.Z(1–108)-3×FLAG::NATMX SWC4-VSV::KANMX |
| MKY1167 | MKY5, H2A.Z(1–106)-3×FLAG::NATMX SWC4-VSV::KANMX |
| MKY1168 | MKY5, H2A.Z(1–104)-3×FLAG::NATMX SWC4-VSV::KANMX |
| MKY1169 | MKY5, ARP4-VSV::KANMX |
| MKY1170 | MKY5, H2A.Z(1–134)-3×FLAG::NATMX ARP4-VSV::KANMX |
| MKY1171 | MKY5, H2A.Z(1–124)-3×FLAG::NATMX ARP4-VSV::KANMX |
| MKY1172 | MKY5, H2A.Z(1–117)-3×FLAG::NATMX ARP4-VSV::KANMX |
| MKY1173 | MKY5, H2A.Z(1–114)-3×FLAG::NATMX ARP4-VSV::KANMX |
| MKY1174 | MKY5, H2A.Z(1–108)-3×FLAG::NATMX ARP4-VSV::KAN |
| MKY1175 | MKY5, H2A.Z(1–106)-3×FLAG::NATMX ARP4-VSV::KANMX |
| MKY1176 | MKY5, H2A.Z(1–104)-3×FLAG::NATMX ARP4-VSV::KANMX |
| MKY1177 | MKY5, CHZ1-VSV::KANMX |
| MKY1178 | MKY5, H2A.Z(1–134)-3×FLAG::NATMX CHZ1-VSV::KANMX |
| MKY1179 | MKY5, H2A.Z(1–124)-3×FLAG::NATMX CHZ1-VSV::KANMX |
| MKY1180 | MKY5, H2A.Z(1–117)-3×FLAG::NATMX CHZ1-VSV::KANMX |
| MKY1181 | MKY5, H2A.Z(1–114)-3×FLAG::NATMX CHZ1-VSV::KANMX |
| MKY1182 | MKY5, H2A.Z(1–108)-3×FLAG::NATMX CHZ1-VSV::KANMX |
| MKY1183 | MKY5, H2A.Z(1–106)-3×FLAG::NATMX CHZ1-VSV::KANMX |
| MKY1184 | MKY5, H2A.Z(1–104)-3×FLAG::NATMX CHZ1-VSV::KANMX |
| MKY1185 | MKY1144, [pRS314] |
| MKY1186 | MKY1144, [pRS314 HTZ1] |
| MKY1187 | MKY1144, [pRS314 HTZ1-3×FLAG::KANMX] |
| MKY1308 | MKY583, htz1Δ::NATMX |
| MKY1309 | MKY583, H2A.Z(1–134)-3×FLAG::NATMX |
| MKY1310 | MKY583, H2A.Z(1–120)-3×FLAG::NATMX |
| MKY1311 | MKY583, H2A.Z(1–117)-3×FLAG::NATMX |
| MKY1312 | MKY583, H2A.Z(1–114)-3×FLAG::NATMX |
| MKY1313 | MKY583, H2A.Z(1–111)-3×FLAG::NATMX |
| MKY1314 | MKY583, H2A.Z(1–108)-3×FLAG::NATMX |
| MKY1315 | MKY583, H2A.Z(1–106)-3×FLAG::NATMX |
| MKY1316 | MKY583, H2A.Z(1–104)-3×FLAG::NATMX |
All plasmids used in this study are listed in Table 2 and were created using standard molecular biology techniques. The parental HTZ1 pRS314 plasmid (4) was used for subsequent manipulations, including site-directed mutagenesis and construction of the HTA1 and ZA hybrid (an H2A.Z derivative containing the corresponding C terminus of H2A distal to amino acid 114). The HTA1-FLAG vector was created by amplification of the entire HTA1 gene using primers containing overhangs that anneal to the promoter region of HTZ1 and the beginning of the 3×FLAG tag. This double-stranded product was gel purified and used as primers in a site-directed mutagenesis reaction with the HTZ1-FLAG vector as a template, using the QuikChange method (Stratagene) following the protocol of the manufacturer (primers used are available upon request). All mutations were confirmed by DNA sequencing. The hybrid vector ZA was created in a similar manner, except that the amplified region of HTA1 corresponded to the last 20 amino acids in H2A.Z.
Table 2.
Plasmids used in this study
| Plasmid | Relevant genotype | Reference/source |
|---|---|---|
| pMK148 | pRS314, HTZ1 | 4 |
| pMK149 | pRS314, HTZ1-3×FLAG::KANMX | This study |
| pMK418 | pRS314, HTA1-3×FLAG::KANMX | This study |
| pMK420 | pRS314, ZA-3×FLAG::KANMX | This study |
| pMK509 | pRS314, htz1-H118A,I119A,N120A- 3×FLAG::KANMX | This study |
| pMK400 | pRS316, HTA1-HTB1 | 39 |
| pMK429 | pRS413, HTA1-HTB1 | 39 |
| pMK510 | pRS413, HTA1-3×FLAG-HTB1 | This study |
| pMK511 | pRS413, hta1-N112A-3×FLAG-HTB1 | This study |
| pMK513 | pRS413, hta1-H114A-3×FLAG-HTB1 | This study |
Mutations in specific amino acids of HTZ1 and HTA1 were also generated by site-directed mutagenesis (primers available upon request).
MKY985 has chromosomal deletions of the HTA-HTB genes (hta1htb1Δ::NAT hta2htb2Δ::HYG) that are complemented by the plasmid pMK400 (39), containing genomic HTA1-HTB1 on the URA CEN/ARS plasmid pRS316. The plasmid shuffle experiments were performed by transformation of MKY985 with HTA1-HTB1, HTA1-FLAG-HTB1, or various hta1 mutant genes on plasmids containing a HIS3-selective marker (pRS413) (39) and subsequent counterselection of the URA3 marker on synthetic complete (SC) medium plates containing 5-fluoroorotic acid (5-FOA) (48).
To determine the expression level of the truncated H2A.Z proteins, yeast whole-cell extracts (WCE) were prepared using the NaOH extraction protocol as previously described (28). Immunoblotting was performed using anti-FLAG M2 (Sigma) and anti-H2A (Upstate) antibodies.
Analytical-scale affinity purifications.
Coimmunoprecipitation assays were performed as described previously (25). Briefly, yeast cells were harvested, lysed in immunoprecipitation (IP) buffer {50 mM Tris (pH 7.8), 150 mM NaCl, 1.5 mM magnesium acetate [Mg(OAc)2], 0.15% NP-40, 1 mM dithiothreitol (DTT), 10 mM NaPPi, 5 mM EGTA, 5 mM EDTA, 0.1 mM Na3VO4, 5 mM NaF, Complete protease inhibitor cocktail} using acid-washed glass beads, and mechanically disrupted using a bead beater (BioSpec Products, Bartlesville, OK). FLAG-tagged fusion proteins were captured using FLAG M2 agarose beads (Sigma) and subsequently washed in IP buffer. Captured material was analyzed by immunoblotting, and copurifying proteins were detected with anti-VSV (Applied Biological Materials), anti-Nap1 (Santa Cruz Biotechnology), and anti-H2B (Upstate) antibodies. Bands were visualized using an Odyssey infrared imaging system (Licor).
Chromatin association assays.
Chromatin association assays were performed as previously described (51). Briefly, yeast cells were incubated in prespheroplast buffer {100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)]–KOH (pH 9.4), 10 mM DTT, 0.1% sodium azide} for 10 min at room temperature and spheroplasted with 20 mg/ml Zymolyase-100T (Seikagaku Corporation) in spheroplast buffer (50 mM KPO4 [pH 7.5], 0.6 M sorbitol, 10 mM DTT) at 37°C for 15 min. Spheroplasts were washed with wash buffer (50 mM HEPES-KOH [pH 7.5], 100 mM KCl, 2.5 mM MgCl2, 0.4 M sorbitol), resuspended in equal volumes of elution buffer (EB; 50 mM HEPES-KOH [pH 7.5], 100 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitor cocktail), and lysed with 1% Triton X-100). WCE was saved, and the remaining lysate was separated into chromatin pellet and supernatant fractions by centrifugation through EBSX (EB plus 30% sucrose and 0.25% Triton X-100. WCE and pellet and supernatant fractions were subjected to SDS-PAGE and immunoblotted with anti-FLAG M2 (Sigma), anti-H2A (Upstate), and anti-Pgk1 (Sigma) antibodies. Immunoblots were scanned with an Odyssey infrared imaging system (Licor).
Yeast cultures.
Yeast cultures for GAL1 induction were performed as previously described (16). Briefly, seed cultures were grown in yeast extract-peptone (YP)-dextrose (d-glucose, 2%) overnight with shaking at 30°C. Forty optical density at 600 nm (OD600) units of cells was harvested by centrifugation at 3,000 rpm, washed with sterile water, and resuspended in 200 ml of YP-galactose (2% galactose). The volume of culture removed at each time point was replaced with the same volume of YP-galactose.
RT-PCR.
Cells were grown in YP-dextrose to an OD600 of 0.5. Ten OD600 units of cells was harvested for RNA extraction and purification using a Qiagen RNeasy minikit as per the manufacturer protocol. RNA was digested with RNase-free DNase I (Qiagen). cDNA was synthesized using a SuperScript III first-strand synthesis system for reverse transcriptase PCR (RT-PCR) and oligo(dT) (Invitrogen). cDNA was analyzed using a Rotor-Gene 6000 (Corbett Research) and PerfeCTa SYBR green FastMix (Quanta Biosciences). mRNA levels were normalized to that of ACT1 mRNA. Samples were analyzed in triplicate for three independent RNA preparations. Primer sequences are listed in Table 3.
Table 3.
ChIP-qPCR and RT-qPCR primers
| Primer name | Forward sequence | Reverse sequence |
|---|---|---|
| PRP8-ChIP | GGATGTATCCAGAGGCCAAT | AACCCGCGTATTAAGCCATA |
| GIT1-ChIP | TTCATGAATTTCCTTACTGGAC | GTTGACTAGTCACAAGAAACAG |
| YCR095c-ChIP | TACCGTATGCGGTATAATGA | GTCTCCACTTTAGAACATCT |
| YCR100c-ChIP | CTCGTTATGCCCGTCATCTT | CAGGTCATGTTCGTGCTTCT |
| RDS1-ChIP | TGTGCTATCTAAGAGGATGGTTCA | CAGCAGCCAATTTCATGTTC |
| YIR042c-ChIP | TACGCCACTCGCTGAATTTG | TTGTAAGCCCAGTAAACAGCTTC |
| GAL1-ChIP | GGGTAATTAATCAGCGAAGCGATG | TGCGCTAGAATTGAACTCAGGTAC |
| ACT1-RT | TGTCCTTGTACTCTTCCGGT | CCGGCCAAATCGATTCTCAA |
| GIT1-RT | ATCGGTTCTGTAGTAGGCG | TTACCAGTCCAGCCATTGG |
| YCR095c-RT | CTTTGCAGAGAGCCAGAAGTG | TCAAATCGTCTCCTAGAACTCCAC |
| YCR100c-RT | CCAGATGGATCAGGCTCAAA | TCGATCGCATACAGGACACT |
| RDS1-RT | AAGCCGTGAGATTGAAATGG | CTCCATCTGGCACAACAGAA |
| YIR042c-RT | TGCGGGACCATTCACTAAC | AGTGAACCGTTGGCTTCATC |
| GAL1-RT | GGTGGTTGTACTGTTCACTTGGTTCC | TCATATAGACAGCTGCCCAATGCTG |
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (46). In brief, yeast cells (500 ml) were grown in a rich medium to an OD600 of 0.5 to 0.6 and were cross-linked with 1% formaldehyde for 20 min before chromatin was extracted. The chromatin was sonicated (10 cycles, 30 s on/off, high setting [Bioruptor; Diagenode, Sparta, NJ]) to yield an average DNA fragment of 500 bp. Anti-FLAG antibody (4 μl; Sigma) was coupled to 60 μl of protein A magnetic beads (Invitrogen). After reversal of the cross-linking and DNA purification, the immunoprecipitated and input DNA was analyzed by quantitative PCR (qPCR). Samples were analyzed in triplicate for at least three independent ChIP experiments. Primer sequences are listed in Table 3.
E-MAP.
E-MAP screens were performed as described previously (45). Briefly, HTZ1 truncation alleles were crossed, using a Singer robot, to a library of 1,536 mutants covering a number of processes, including RNA processing and chromatin biology. All strains were screened three to four times, and scores were calculated as previously described (6, 45).
RESULTS
The C terminus of H2A.Z is required for H2A.Z function.
To more closely dissect the requirements of the docking domain for H2A.Z function in yeast, we constructed a set of HTZ1 truncation alleles encoding successively shortened versions of H2A.Z distal to the C-terminal α-helix containing the M6 region (Fig. 1A). To facilitate functional and biochemical analyses, the alleles were generated through insertion of a 3×FLAG epitope tag followed by a stop codon. For simplicity, we omit writing out the 3×FLAG tag in all subsequent descriptions of H2A.Z truncations and mutants. We first tested the ability of the truncation alleles to confer resistance to genotoxic stress, as cells lacking H2A.Z are sensitive to genotoxic stressors, such as formamide, caffeine, and hydroxyurea (HU) (19, 25). Yeast cells containing the full-length H2A.Z [H2A.Z(1–134)] and the longer truncations had normal growth comparable to that of wild-type cells (Fig. 1B). The short truncations up to and including H2A.Z(1–114) (lacking the last 20 amino acids of the protein) exhibited retarded growth on all three drugs, similarly to the htz1Δ mutant (Fig. 1B). Cells containing H2A.Z(1–120) and H2A.Z(1–117) had noticeable growth defects on all three drugs, suggesting that the effects that these two truncations had on H2A.Z function were intermediate in nature. The growth defects observed for the H2A.Z truncations were not due to lower expression levels, as all FLAG-tagged proteins were expressed similarly to the wild type, except for a slight decrease in H2A.Z(1–104) (Fig. 1C). Furthermore, the same phenotypic pattern of drug sensitivity was observed in the S288c strain background, suggesting that it was a general feature rather than one specific to the W303 strain background (data not shown).
Genetic dissection of HTZ1 truncation alleles revealed distinct requirements for the C terminus in H2A.Z function.
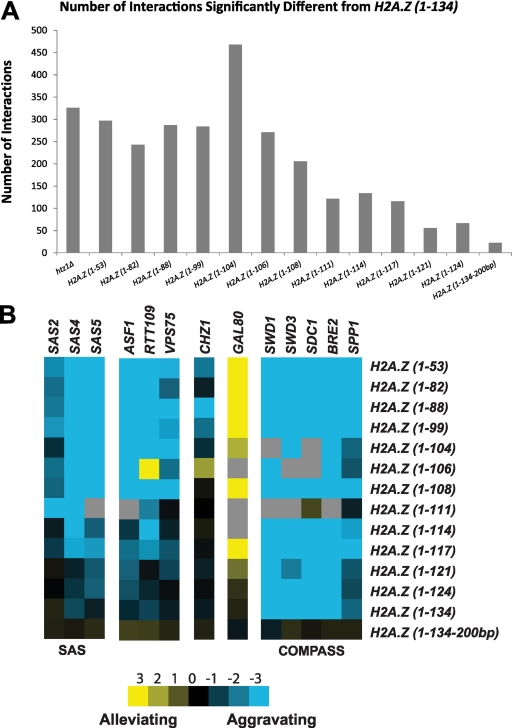
To extend our analysis of the H2A.Z C terminus, we adapted the epistatic miniarray profile (E-MAP) approach (6, 45) to determine requirements of the H2A.Z C-terminal region for cell growth when additional genes were compromised. To quantitatively analyze global genetic interaction patterns, query strains containing sequentially shorter HTZ1 truncation alleles generated by placing a TAG stop codon followed by a NAT resistance marker at the appropriate open reading frame (ORF) location were analyzed using a library of 1,536 mutants representing various processes, including chromatin biology and RNA processing (6, 45). To control for effects of the truncation strategy, we included two additional allelic variants, H2A.Z(1–134) and H2A.Z(1-134-200bp) mutants, which, respectively, contain the NAT marker immediately adjacent to the natural stop codon or 200 bp downstream. Comparing the genetic profiles of the H2A.Z(1-134) and H2A.Z(1-134-200bp) mutants, we determined that integration of the NAT marker immediately following the ORF generated a “decreased abundance by mRNA perturbation” (DAmP)-like allele (44), suggesting that the interactions observed in the E-MAP were a combination of loss of function due to the truncation and a decrease in mRNA stability. To evaluate the extent of the DAmP effect in our data, we counted the number of interactions that have a magnitude of S ≥ 2 or S ≤ −2.5 compared to those for the H2A.Z(1-134) mutant (10) (Fig. 2A). This analysis revealed that for the H2A.Z(1-134-200bp) mutant only 23 interactions meet these criteria, suggesting that the DAmP effect in our data is minimal (Fig. 2A). Conversely, the shorter truncations, starting with the H2A.Z(1-117) mutant, have upwards of 100 interactions, demonstrating that the influence of the truncations was much greater than that of the DAmP.
Fig. 2.
Genetic dissection of H2A.Z truncation alleles revealed distinct requirements for the C terminus in H2A.Z function. (A) Bar graph illustrating the number of genetic interactions that differ significantly from those for the H2A.Z(1-134) mutant, with thresholds of S ≥ 2 and S ≤ −2.5 (10). (B) Individual interaction profiles of H2A.Z truncation alleles with various complexes. The intensities of blue and yellow represent the strength of aggravating and alleviating interactions, respectively. Gray represents missing data. Note that the nomenclature indicates the H2A.Z protein truncated at the corresponding amino acids.
The E-MAP approach allowed us not only to identify specific interactions that were dependent on the H2A.Z C terminus but also to dissect the strength of a number of negative and positive genetic interactions. Several distinct interaction patterns were revealed. For example, loss of genes encoding the histone chaperone/H3K56 acetylase complex ASF1/RTT109/VPS75, the SAS silencing complex, and the H2A.Z-specific histone chaperone Chz1 resulted in increasingly aggravating interactions as H2A.Z was shortened (Fig. 2B). In contrast, loss of GAL80, the gene encoding the repressor of the Gal4 transcriptional activation domain (22), resulted in progressively alleviating interactions with the truncated alleles (Fig. 2B). Representing a different pattern, loss of genes encoding members of the COMPASS complex, which is involved in transcription regulation, resulted in similar aggravating interactions regardless of H2A.Z length, suggesting that this interaction was not dependent on the C terminus of H2A.Z and instead was highly sensitive to reduction in H2A.Z protein caused by the destabilization of its mRNA (Fig. 2B).
The H2A.Z C terminus is required for the role of H2A.Z in heterochromatic boundary function.
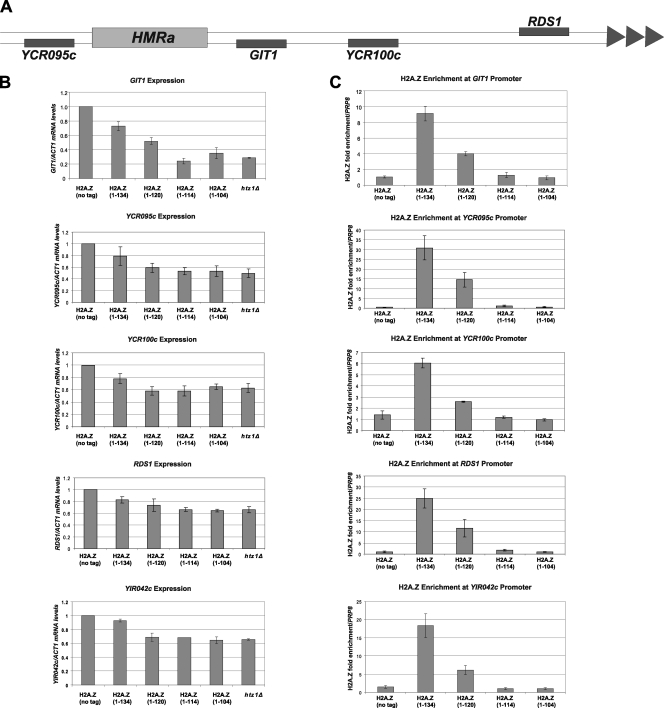
H2A.Z is required to restrict the spread of heterochromatin at boundaries with euchromatin (37). To test whether its C terminus is important for this boundary function in a native context, we measured the expression of two ORFs near the left and right boundaries of the silent HMR locus on chromosome III (YCR095c and GIT1, respectively), as well as three ORFs in the subtelomeric regions of chromosomes III (YCR100c and RDS1) (Fig. 3A) and IX (YIR042c). In the absence of H2A.Z, the Sir protein complex spreads from either HMR or telomere into the respective ORFs, causing decreased mRNA levels to be transcribed from these genes (4, 37). At the boundaries of HMR and in subtelomeric regions, htz1Δ, H2A.Z(1–114), and H2A.Z(1–104) mutant cells had similar defects in boundary function, as reflected by the reduction in mRNA expression of all genes tested compared with the results for the wild type (Fig. 3B). The H2A.Z(1–120) mutant was more variable in its effect on boundary function, with GIT1, YCR095c, and RDS1 mRNA expression levels showing intermediate reduction, whereas YCR100c and YIR042c mRNA expression was reduced to levels similar to that of the htz1Δ mutant.
Fig. 3.
The H2A.Z C terminus is required for the role of H2A.Z in heterochromatic boundary function. (A) Schematic representation of the tested ORFs located at the boundaries of the HMR locus and the telomere of chromosome III. Note that YIR042c is located at the telomere on the right arm of chromosome IX. (B) qRT-PCR of the mRNA levels of the indicated genes, normalized to levels of ACT1. (C) ChIP analysis of H2A.Z enrichment at the indicated promoters, normalized to the level for the PRP8 ORF. Error bars represent standard deviations of values from three replicates.
We next used chromatin immunoprecipitation (ChIP) followed by qPCR to determine if the decreased expression of these genes at the heterochromatin boundaries was due to loss of binding of H2A.Z truncations at the respective promoter regions. H2A.Z(1–114) and H2A.Z(1–104) were completely lost from all five promoters, while H2A.Z(1–120) enrichment was intermediate, decreasing by more than half compared to the wild-type level (Fig. 3C).
The H2A.Z C terminus is required for the role of H2A.Z in transcriptional activation of the GAL1 gene.
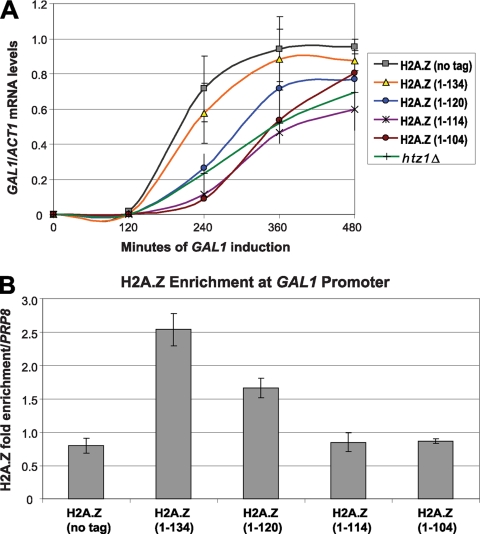
In addition to its role in restricting heterochromatin spread, H2A.Z has more direct roles in regulating gene expression. Among the best documented is the requirement for H2A.Z in GAL1 induction after cells grown under long-term repressing conditions (glucose) are shifted to galactose medium. This requirement is mediated by H2A.Z-containing nucleosomes in the divergent GAL1/GAL10 promoter (14, 16, 29, 43). To determine whether the C terminus of H2A.Z facilitated GAL1 expression, mRNA levels were monitored at 2-h intervals by qRT-PCR after cells were shifted from long-term growth in glucose to galactose-containing medium. Similarly to previous reports (16), the htz1Δ mutant induced GAL1 expression at a significantly lower rate than wild-type or H2A.Z(1–134) cells (Fig. 4A). The GAL1 induction patterns of cells containing H2A.Z(1–114) and H2A.Z(1–104) were similar to that of the htz1Δ mutant, whereas cells containing H2A.Z(1–120) had an intermediate phenotype, with induction being faster than that in htz1Δ mutants but slower than that in wild-type and H2A.Z(1–134) strains (Fig. 4A). Consistent with the expression results, H2A.Z(1–114) and H2A.Z(1–104) were completely lost from the GAL1/10 promoter, while the level of H2A.Z(1–120) was reduced by about one-third compared to that of H2A.Z(1–134) (Fig. 4B).
Fig. 4.
The H2A.Z C terminus is required for the role of H2A.Z in activation of the GAL1 gene. (A) qRT-PCR of GAL1 mRNA performed on the indicated cultures, which were grown long-term in YP-glucose (2%) prior to being transferred to YP-galactose (2%) and collected at 120-min intervals. GAL1 mRNA levels were normalized to levels of ACT1. Solid lines represent the smoothed curves through the averages of three replicates. (B) ChIP analysis of H2A.Z enrichment at the GAL1/10 promoter, normalized to the level for the PRP8 ORF. Error bars represent standard deviations of values from three replicates.
Loss of H2A.Z truncations from chromatin is SWR1-C and H2B independent.
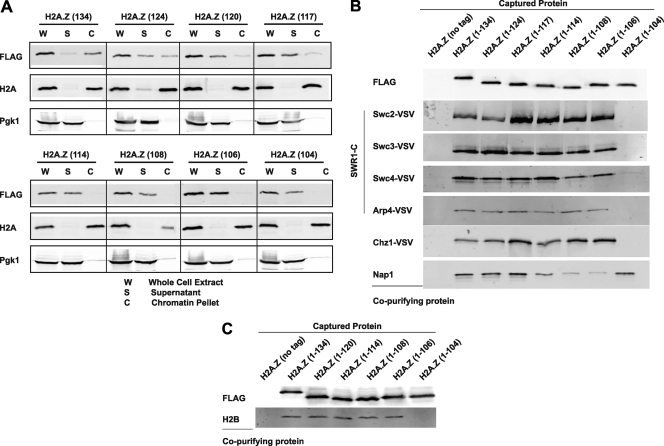
Having established that the H2A.Z C terminus was required for binding to specific promoter regions, we tested whether this reflected a general inability of the truncated proteins to incorporate into chromatin. Cellular fractionation assays (31) revealed that H2A.Z(1–134) was present predominantly in the chromatin fraction, with low levels found in the nonchromatin fraction (Fig. 5A). In contrast, the short truncations, including H2A.Z(1–114), were completely lost from chromatin, while H2A.Z(1–120) and H2A.Z(1–117) were partially associated with chromatin (Fig. 5A). The bulk chromatin association assay thus recapitulated the findings from the gene-specific location analysis. The chromatin association of H2A and the nonchromatin association of Pgk1 were unchanged for all experiments (Fig. 5A).
Fig. 5.
Loss of H2A.Z truncations from chromatin is SWR1 and H2B independent. (A) Bulk fractionations were performed, and the amount of H2A.Z-FLAG in each fraction was determined by immunoblotting in the different strains. Antibodies against histone H2A and Pgk1 were used as loading controls for the chromatin pellet and supernatant, respectively. (B) Analytical-scale affinity purifications of FLAG-tagged H2A.Z truncations from cells containing affinity-tagged versions of SWR1-C subunits, Chz1, and Nap1 were performed, followed by immunoblotting for the indicated copurifying proteins. (C) Analytical-scale affinity purifications of the indicated FLAG-tagged H2A.Z truncations were performed, followed by immunoblotting for histone H2B.
We next set out to determine if the loss from chromatin of the short H2A.Z truncations was due to a loss of binding to known factors involved in H2A.Z biology, including the ATP-dependent remodeler SWR1-C and the histone chaperones Nap1 and Chz1. We performed analytical-scale affinity purifications of truncated versions of H2A.Z from cells containing VSV epitope-tagged SWR1-C subunits Swc2, Swc3, Swc4, and Arp4. All truncations copurified with SWR1-C components, with the exception of H2A.Z(1–104) (Fig. 5B), consistent with the overlapping M6 region being important for engagement of SWR1-C (52). This suggested that the loss of H2A.Z truncations from chromatin was not due to an inability to associate with SWR1-C. Similarly to SWR1-C, Chz1 bound all truncations except H2A.Z(1–104), while Nap1 copurified with all H2A.Z truncations but had reduced affinity to H2A.Z(1–106) and H2A.Z(1–108) (Fig. 5B).
Given that H2A.Z is deposited into chromatin as a dimer with H2B (25, 27, 38), we tested how this interaction was affected by the H2A.Z truncations. Interestingly, all truncations bound to H2B, except for H2A.Z(1–104), mimicking the binding pattern for SWR1-C and Chz1 (Fig. 5C).
The C terminus of histone H2A replaced its variant counterpart in H2A.Z function.
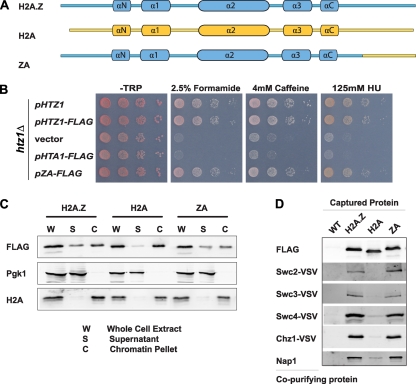
Our results revealed that the last 20 amino acids of the H2A.Z C terminus were required for function of the histone variant. To test whether this region also contributed to the differences between the variant and its canonical cousin, we replaced it with the corresponding region from canonical H2A. To this end, a plasmid-borne allele encoding an H2A.Z derivative containing the corresponding C terminus of H2A distal to amino acid 114 (here termed “ZA”) was compared to wild-type H2A.Z and H2A (Fig. 6A). All plasmids were driven by the endogenous HTZ1 promoter and furnished with a C-terminal 3×FLAG tag. In contrast to H2A or the empty control vector, ZA complemented the sensitivity of the htz1Δ mutant to formamide, caffeine, and HU similarly to H2A.Z (Fig. 6B).
Fig. 6.
The C terminus of histone H2A replaced its variant counterpart in H2A.Z function. (A) Schematic representation of the ZA hybrid, constructed with regions originating from H2A.Z (blue) and H2A (yellow). (B) ZA was able to substitute for H2A.Z function in sensitivity to drugs. Tenfold serial dilutions of htz1Δ strains carrying the indicated plasmids were plated and incubated on plates lacking tryptophan and containing the indicated concentrations of formamide, caffeine, and HU. (C) ZA was incorporated into chromatin. Bulk fractionations were performed, and the amounts of FLAG-tagged proteins in each fraction were determined by immunoblotting. Antibodies against histone H2A and Pgk1 were used as loading controls for the chromatin pellet and supernatant, respectively. (D) H2A.Z and ZA copurified with SWR1-C subunits, while H2A did not. Analytical-scale affinity purifications of FLAG-tagged proteins from cells containing VSV-tagged SWR1-C subunits, Chz1-VSV, and Nap1 were performed, followed by immunoblotting for the indicated copurifying proteins.
These results suggested that the last 20 amino acids of H2A.Z could functionally be replaced by the corresponding region from H2A, likely through restoring the ability to bind chromatin. To more formally test this possibility, we performed chromatin fractionation assays of htz1Δ cells containing plasmid-borne versions of H2A.Z, H2A, and ZA. The ratio of ZA between the chromatin-bound and unbound fractions was similar to that of wild-type H2A.Z (Fig. 6C). In contrast, more H2A was found in the chromatin-bound fraction than in the nonchromatin fraction, a pattern seen also for the endogenous H2A control (Fig. 6D). Further supporting the conclusion that ZA behaved like H2A.Z, the hybrid protein copurified SWR1-C, Chz1, and Nap1 in analytical-scale affinity purification assays similarly to H2A.Z, whereas these proteins had much lower binding to H2A (Fig. 6D).
Specific amino acids of the H2A.Z C terminus are important for H2A.Z chromatin association and function in vivo.
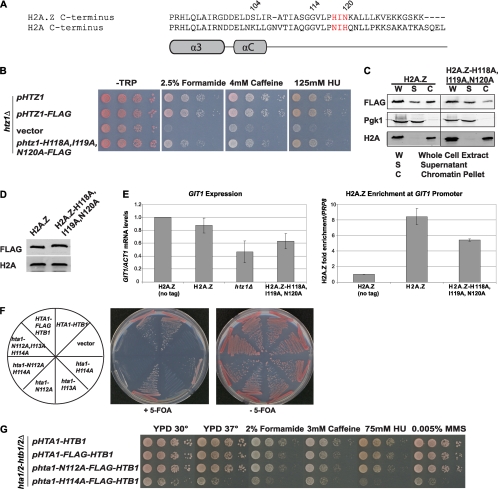
Since our data thus far suggested that amino acids between 114 and 120 were critical for H2A.Z function and chromatin binding, we inspected this region more closely in the mouse H2A.Z nucleosome (49). The crystal structure shows that the C terminus is anchored in the nucleosome, at least in part through hydrophobic interactions of side chains and backbone residues with histone H3. Specifically, we identified three corresponding amino acids (H118, I119, and N120) in this region of yeast H2A.Z that likely perform an analogous anchoring function (Fig. 7A). Functional analysis of a mutant strain having these residues changed to alanine (htz1-H118A,I119A,N120A mutant) revealed an intermediate phenotype compared to that of the htz1Δ mutant when exposed to genotoxic stress (Fig. 7B), most likely due to the decreased incorporation into chromatin of the mutant protein (Fig. 7C), as the protein level of the htz1-H118A,I119A,N120A mutant was normal (Fig. 7D). In addition, the htz1-H118A,I119A,N120A mutant caused an intermediate defect in heterochromatic boundary function as measured by mRNA levels of the GIT1 gene, which coincided with reduced binding to the GIT1 promoter (Fig. 7E). Thus, these three residues contributed to the association of H2A.Z with chromatin, thereby likely regulating its function.
Fig. 7.
Specific amino acids of the H2A.Z C terminus are important for H2A.Z chromatin association and function in vivo. (A) Schematic representation and sequence alignment of the H2A.Z and H2A C termini, with the H2A.Z amino acids H118, I119, and N120 highlighted in red. (B) The htz1-H118A,I119A,N120A mutant had an intermediate phenotype on plates containing formamide, caffeine, and HU. (C) The H2A.Z-H118A,I119A,N120A mutant had reduced binding to chromatin in the bulk fractionation assay. (D) Protein expression of the H2A.Z-H118A,I119A,N120A mutant was the same as wild-type levels, as analyzed by immunoblotting of whole-cell extracts of the indicated strains with an anti-FLAG antibody. Antibody against histone H2A was used as a loading control. (E) The same htz1 mutant had an intermediate defect in heterochromatic boundary function, as measured by mRNA levels of GIT1 normalized to ACT1, and reduced binding to the GIT1 promoter, as measured by ChIP followed by qPCR. (F) Mutation of the same amino acids in H2A to alanine resulted in lethality of yeast cells. The abilities of the hta1 mutants to complement the HTA1/2-HTB1/2 chromosomal deletion were tested by plasmid shuffling on medium containing 5-FOA to counterselect the URA marker. (G) hta1-H114A mutant cells were sensitive to higher temperature and genotoxic stress.
Given that the region distal to amino acid 114 was required for H2A.Z function but not unique to the variant, we tested whether the importance of the H2A.Z H118, I119, and N120 residues in chromatin binding was conserved in the corresponding amino acids of H2A (N112, I113, and H114) (Fig. 7A). We tested the abilities of various hta1-HTB1 mutants to complement the chromosomal deletion of the two copies of genes encoding H2A and H2B in yeast. Strains containing both the wild-type HTA1-HTB1 on a plasmid with a UR3A-selective marker and various hta1-FLAG-HTB1 mutants on plasmids with a HIS3 marker were counterselected on medium containing 5-fluoroorotic acid (5-FOA) to test for viability upon loss of the URA3 plasmid, an indication of complementation (26). The triple mutant, the hta1-N112A,H114A double mutant, and the hta1-I113A single mutant were unable to confer viability on the requirement for HTA1/2, while the hta1-H114A mutant was viable but had slow growth (Fig. 7F). In contrast, the hta1-N112A mutant fully complemented the chromosomal deletion of HTA1/2. To investigate the viable hta1-N112A and hta1-H114A mutants further, we tested their growth at high temperature and in the presence of the drugs formamide, caffeine, HU, and methyl methanesulfonate (MMS). As expected from the initial complementation assays, hta1-H114A mutants were very sick under all conditions tested, whereas hta1-N112A mutants grew robustly (Fig. 7G).
DISCUSSION
Here we have identified key functional regions located in the C-terminal docking domain in histone variant H2A.Z. Importantly, the last 20 amino acids of the C terminus were essential for H2A.Z function, as they were required for resistance to genotoxic stress, heterochromatin boundary function at HMR and subtelomeres, activation of the GAL1 gene, and incorporation of H2A.Z into chromatin. Thus, our data strongly suggested that these important aspects of H2A.Z function were closely related. In spite of its profound loss of functions, H2A.Z(1–114) was fully capable of binding to the key H2A.Z interaction partners SWR1-C, Chz1, and histone H2B. Interestingly, the corresponding region from canonical H2A could functionally replace this essential 20-amino-acid region in H2A.Z. Supporting the functional relevance of this region, we identified 3 specific amino acids in this region that facilitated chromatin anchoring and function of H2A.Z and were important for H2A to function in vivo. Further illuminating the importance of the C terminus for H2A.Z biology, we have uncovered a more nuanced requirement of this region for cell growth by using E-MAP, establishing the utility of this method for dissecting the sensitivity of genetic interaction profiles.
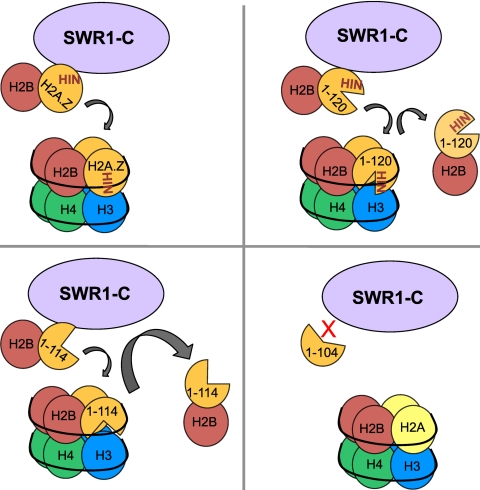
Given the extensive contacts made by H2A.Z with other histones and DNA in the nucleosome, it was surprising that removal of the last 20 amino acids caused a dramatic reduction in the amount of H2A.Z associated with chromatin. Specifically, these molecular interactions not involving residues located in the C-terminal region include the acidic patch that interacts with histone H4, the α3 helix that forms a β-sheet with the H4 C-terminal tail, and the α1 helix/L1/L2 that interacts with DNA (33, 49). Contrasting the complete loss of function associated with H2A.Z(1–114), H2A.Z(1–120) generally had intermediate defects in conferring resistance to genotoxic stress, heterochromatin boundary formation, transcriptional activation, and chromatin binding. Multiple studies show that SWR1-C is required for chromatin deposition of H2A.Z/H2B dimers, with this process likely being stimulated by H2A (25, 27, 34, 38). The fact that both H2A.Z(1–114) and H2A.Z(1–120) retained binding to SWR1-C and H2B suggests that anchoring of H2A.Z to chromatin through its C terminus is at least equally important for function in vivo. Based on our data, we propose a model for the sequential nature underlying the stable formation of H2A.Z nucleosomes. Upon deposition by SWR1-C, the H2A.Z C terminus is required to retain the histone in chromatin, with failure to do so leading to grave consequences for the ability of H2A.Z to perform its function. Accordingly, we postulate that based on their ability to interact with SWR1-C and H2B, both H2A.Z(1–114) and H2A.Z(1–120) are efficiently deposited as dimers with H2B into chromatin by SWR1-C but dissociate thereafter, resulting in decreased residency in nucleosomes (Fig. 8). This is likely in part due to weakened interactions with other nucleosomal histones, specifically H3, as indicated by the effect of the htz1-H118A,I119A,N120A mutant on chromatin binding.
Fig. 8.
Schematic diagram depicting models of wild-type H2A.Z, H2A.Z(1–120), H2A.Z(1–114), and H2A.Z(1–104) nucleosomes. SWR1-C binds to H2A.Z, H2A.Z(1–120), and H2A.Z(1–114) and deposits them into chromatin. However, H2A.Z(1–120) and H2A.Z(1–114) are not stably anchored into the nucleosome. The gray arrows represent the relative amounts of H2A.Z that dissociate from the nucleosome. H2A.Z(1–104) cannot bind to SWR1-C and thus cannot be deposited into chromatin in the first place. HIN represents the H2A.Z residues H118, I119, and N120. The diagram does not necessarily represent real intranucleosomal structure and interactions.
Support for our model is provided by the ability of the corresponding region of H2A to fully rescue the strong defects caused by loss of the last 20 amino acids in H2A.Z. These data suggested that this region of the docking domain might play a similar role in chromatin anchoring of both histone H2A forms. Consistent with this, the Xenopus laevis H2A docking domain is critical to stabilize its interactions within the nucleosome in vitro (47), and the human H2A C terminus is required for proper nucleosome stability in vivo (50). The general functional requirement of this region is further supported by our own data showing that the H2A residues corresponding to H2A.Z H118, I119, and N120 were important in vivo. Specifically, the H2A hta1-I113A mutant was lethal, while the hta1-H114A mutant had strong sensitivity to genotoxic stress. While this result was in slight contrast to published point mutant analyses of H2A reporting that both hta1-I113A and hta1-H114A mutants were viable but had strong phenotypes on HU and MMS, the difference may be due to strain background, as all of the previous studies were performed with S288c strains (18, 36, 40).
The intermediate effects of the htz1-H118A,I119A,N120A mutant and of the H2A.Z(1–120) derivative indicate that additional residues distal to amino acid 120 were required for proper H2A.Z function in chromatin. While the nature of these effects is unclear at present, the crystal structure of the mouse H2A.Z suggests that L124 makes a polar contact with H3 within the nucleosome (49). In contrast to H118, I119, and N120 residues, this interaction involves the backbone and not the side chain of the amino acid. In addition, K126 and K133 of H2A.Z may be functionally important, as these residues can be sumoylated during DNA double-strand breaks and DNA repair (23), although H2A.Z derivatives lacking just this region did not have appreciable defects in our assays. Despite our clear evidence for the last 20 amino acids being important for function, the C-terminal truncation approach employed here does not exclude the possibility of residues located before amino acid 114 also contributing to H2A.Z function. As such, an alanine scan of H2A.Z identified mutants with mutations in two residues, I109 and G113, in the C terminus that have mild sensitivity to genotoxic stressors (24), as do some mutants with mutations in the H2A.Z acidic patch region (20).
In contrast to the clear boundary at H2A.Z amino acid 114 derived from the growth assay and biochemical measures, E-MAP analysis uncovered a more intricate and nuanced nature of this region. In addition to the robust binding of derivatives longer than H2A.Z(1–104) to SWR1-C, Chz1, and H2B, the gradual loss of function caused by progressively longer C-terminal truncations suggests that removal of these residues did not result in structurally defective or unfolded proteins. We dissected the sensitivity of biologically interesting negative and positive interactions to distinct HTZ1 truncation alleles and highlighted the differences in strength of genetic interaction that a few amino acids can make. For example, the HTZ1 truncation alleles exhibited a very good correlation between their progressively negative genetic interactions with the silencing factors Asf1 and SAS complex and their ability to restrict the spread of heterochromatin at GIT1, suggesting that perhaps H2A.Z acts together with other silencing factors to maintain the boundaries of heterochromatin. Similarly, the progressive positive-interaction profile with GAL80 was consistent with the requirement of the last 20 amino acids for GAL1 activation, which is regulated by Gal4 in concert with Gal80 (22). Lastly, CHZ1 had a transition from no interaction to negative interaction at H2A.Z(1-106) and H2A.Z(1–104) alleles, correlating well with H2A.Z(1–104) losing binding to Chz1. The interaction profiles described here suggested that the drug phenotypes observed for the HTZ1 alleles were a result of specific and not global functional changes.
ACKNOWLEDGMENTS
We thank J. E. Krebs, J. E. Babiarz, and J. Rine for generously providing plasmids, M. Shales for critical reading of the manuscript, and L. P. McIntosh for helpful discussion.
Work in N.J.K.'s laboratory is supported by NIH grants GM084448, GM084279, and GM081879. N.J.K. is a Searle Scholar and Keck Young Investigator Fellow. C.R. is supported by the IRCSET-funded Ph.D. program in Bioinformatics and Systems Biology. Work in M.S.K.'s laboratory is supported by Canadian Institutes of Health Research (CIHR) grant MOP-79442. A.Y.W. and M.J.A. are supported by Frederick Banting and Charles Best Canada graduate scholarships from the CIHR. M.S.K. is a Scholar of the Canadian Institute for Advanced Research and of the Mowafaghian Foundation.
Footnotes
Published ahead of print on 26 July 2011.
REFERENCES
- 1. Adam M., Robert F., Larochelle M., Gaudreau L. 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21:6270–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albert I., et al. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446:572–576 [DOI] [PubMed] [Google Scholar]
- 3. Ausubel F. M. 1987. Current protocols in molecular biology. J. Wiley, New York, NY. [Google Scholar]
- 4. Babiarz J. E., Halley J. E., Rine J. 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20:700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clarkson M. J., Wells J. R., Gibson F., Saint R., Tremethick D. J. 1999. Regions of variant histone His2AvD required for Drosophila development. Nature 399:694–697 [DOI] [PubMed] [Google Scholar]
- 6. Collins S. R., Roguev A., Krogan N. J. 2010. Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 470:205–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Downs J. A., Lowndes N. F., Jackson S. P. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001–1004 [DOI] [PubMed] [Google Scholar]
- 8. Eberharter A., Becker P. B. 2004. ATP-dependent nucleosome remodelling: factors and functions. J. Cell Sci. 117:3707–3711 [DOI] [PubMed] [Google Scholar]
- 9. Fan J. Y., Rangasamy D., Luger K., Tremethick D. J. 2004. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol. Cell 16:655–661 [DOI] [PubMed] [Google Scholar]
- 10. Fiedler D., et al. 2009. Functional organization of the S. cerevisiae phosphorylation network. Cell 136:952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Funakoshi M., Hochstrasser M. 2009. Small epitope-linker modules for PCR-based C-terminal tagging in Saccharomyces cerevisiae. Yeast 26:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gelato K. A., Fischle W. 2008. Role of histone modifications in defining chromatin structure and function. Biol. Chem. 389:353–363 [DOI] [PubMed] [Google Scholar]
- 13. Gelbart M. E., Rechsteiner T., Richmond T. J., Tsukiyama T. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gligoris T., Thireos G., Tzamarias D. 2007. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol. Cell. Biol. 27:4198–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guillemette B., et al. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Halley J. E., Kaplan T., Wang A. Y., Kobor M. S., Rine J. 2010. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 8:e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henikoff S., Ahmad K. 2005. Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 21:133–153 [DOI] [PubMed] [Google Scholar]
- 18. Huang H., et al. 2009. HistoneHits: a database for histone mutations and their phenotypes. Genome Res. 19:674–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackson J. D., Gorovsky M. A. 2000. Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 28:3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jensen K., Santisteban M. S., Urekar C., Smith M. M. 2011. Histone H2A.Z acid patch residues required for deposition and function. Mol. Genet. Genomics 285:287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jin J., et al. 2005. In and out: histone variant exchange in chromatin. Trends Biochem. Sci. 30:680–687 [DOI] [PubMed] [Google Scholar]
- 22. Johnston M. 1987. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 51:458–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kalocsay M., Hiller N. J., Jentsch S. 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 33:335–343 [DOI] [PubMed] [Google Scholar]
- 24. Kawano A., et al. 2011. Global analysis for functional residues of histone variant Htz1 using the comprehensive point mutant library. Genes Cells 16:590–607 [DOI] [PubMed] [Google Scholar]
- 25. Kobor M. S., et al. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kolodrubetz D., Rykowski M. C., Grunstein M. 1982. Histone H2A subtypes associate interchangeably in vivo with histone H2B subtypes. Proc. Natl. Acad. Sci. U. S. A. 79:7814–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krogan N. J., et al. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565–1576 [DOI] [PubMed] [Google Scholar]
- 28. Kushnirov V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857–860 [DOI] [PubMed] [Google Scholar]
- 29. Lemieux K., Larochelle M., Gaudreau L. 2008. Variant histone H2A.Z, but not the HMG proteins Nhp6a/b, is essential for the recruitment of Swi/Snf, Mediator, and SAGA to the yeast GAL1 UAS(G). Biochem. Biophys. Res. Commun. 369:1103–1107 [DOI] [PubMed] [Google Scholar]
- 30. Li B., et al. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. U. S. A. 102:18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang C., Stillman B. 1997. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11:3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Longtine M. S., et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 33. Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- 34. Luk E., et al. 2010. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143:725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luk E., et al. 2007. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell 25:357–368 [DOI] [PubMed] [Google Scholar]
- 36. Matsubara K., Sano N., Umehara T., Horikoshi M. 2007. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 12:13–33 [DOI] [PubMed] [Google Scholar]
- 37. Meneghini M. D., Wu M., Madhani H. D. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725–736 [DOI] [PubMed] [Google Scholar]
- 38. Mizuguchi G., et al. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343–348 [DOI] [PubMed] [Google Scholar]
- 39. Moore J. D., Yazgan O., Ataian Y., Krebs J. E. 2007. Diverse roles for histone H2A modifications in DNA damage response pathways in yeast. Genetics 176:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakanishi S., et al. 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15:881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park Y. J., Chodaparambil J. V., Bao Y., McBryant S. J., Luger K. 2005. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 280:1817–1825 [DOI] [PubMed] [Google Scholar]
- 42. Raisner R. M., et al. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Santisteban M. S., Kalashnikova T., Smith M. M. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411–422 [DOI] [PubMed] [Google Scholar]
- 44. Schuldiner M., et al. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123:507–519 [DOI] [PubMed] [Google Scholar]
- 45. Schuldiner M., Collins S. R., Weissman J. S., Krogan N. J. 2006. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods 40:344–352 [DOI] [PubMed] [Google Scholar]
- 46. Schulze J. M., et al. 2009. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell 35:626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shukla M. S., et al. 2010. The docking domain of histone H2A is required for H1 binding and RSC-mediated nucleosome remodeling. Nucleic Acids Res. 39:2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sikorski R. S., Boeke J. D. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302–318 [DOI] [PubMed] [Google Scholar]
- 49. Suto R. K., Clarkson M. J., Tremethick D. J., Luger K. 2000. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 7:1121–1124 [DOI] [PubMed] [Google Scholar]
- 50. Vogler C., et al. 2010. Histone H2A C-terminus regulates chromatin dynamics, remodeling, and histone H1 binding. PLoS Genet. 6:e1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang A. Y., et al. 2009. Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation. Proc. Natl. Acad. Sci. U. S. A. 106:21573–21578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu W. H., et al. 2005. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 12:1064–1071 [DOI] [PubMed] [Google Scholar]
- 53. Zhang H., Roberts D. N., Cairns B. R. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zlatanova J., Thakar A. 2008. H2A.Z: view from the top. Structure 16:166–179 [DOI] [PubMed] [Google Scholar]