Abstract
The stem-loop in the 5′ untranslated region (UTR) of collagen α1(I) and α2(I) mRNAs (5′SL) is the key element regulating their stability and translation. Stabilization of collagen mRNAs is the predominant mechanism for high collagen expression in fibrosis. LARP6 binds the 5′SL of α1(I) and α2(I) mRNAs with high affinity. Here, we report that vimentin filaments associate with collagen mRNAs in a 5′SL- and LARP6-dependent manner and stabilize collagen mRNAs. LARP6 interacts with vimentin filaments through its La domain and colocalizes with the filaments in vivo. Knockdown of LARP6 by small interfering RNA (siRNA) or mutation of the 5′SL abrogates the interaction of collagen mRNAs with vimentin filaments. Vimentin knockout fibroblasts produce reduced amounts of type I collagen due to decreased stability of collagen α1(I) and α2(I) mRNAs. Disruption of vimentin filaments using a drug or by expression of dominant-negative desmin reduces type I collagen expression, primarily due to decreased stability of collagen mRNAs. RNA fluorescence in situ hybridization (FISH) experiments show that collagen α1(I) and α2(I) mRNAs are associated with vimentin filaments in vivo. Thus, vimentin filaments may play a role in the development of tissue fibrosis by stabilizing collagen mRNAs. This finding will serve as a rationale for targeting vimentin in the development of novel antifibrotic therapies.
INTRODUCTION
Fibroproliferative disorders are leading causes of morbidity and mortality globally (1, 5, 11, 20, 28, 38) and pose an enormous threat to human health. There are no effective therapies for fibrotic diseases. Excessive production of type I collagen by activated fibroblasts and myofibroblasts is the hallmark of fibroproliferative disorders. Type I collagen is a heterotrimer composed of two α1(I) chains and one α2(I) chain (29). The increased collagen synthesis can be due to the increased rate of transcription of collagen genes, increased half-life of collagen mRNAs, and their enhanced translation (33, 35, 47, 51). Increased collagen production by activated fibroblasts is primarily due to an increase in stability of collagen mRNAs (16). During activation of hepatic stellate cells, which synthesize collagen in liver fibrosis, a 16-fold prolongation of the half-life of collagen α1(I) mRNAs is primarily responsible for its 50-fold-increased expression (45). The transformation of fibroblasts into myofibroblasts is also associated with increased stability of collagen α1(I) mRNA (37). Transforming growth factor beta (TGF-β), the most potent profibrotic cytokine, induces collagen synthesis by prolonging the half-life of collagen α1(I) mRNA (51). Thus, it is now well established that stability of collagen mRNAs is the predominant mechanism regulating collagen expression (30, 40, 41).
Two cis-acting elements were implicated in regulating the stability of collagen mRNAs. In the 3′ untranslated region (3′ UTR) of collagen α1(I) mRNA, there is a cytosine-rich sequence that interacts with the αCP protein; this interaction stabilizes the mRNA (32, 34). In the 5′ UTR of collagen α1(I) and α2(I) mRNAs, there is a stem-loop sequence (5′SL) (57) that also regulates the stability and translation of collagen mRNAs (9, 10, 46, 48). The 5′SL is located 75 to 85 nucleotides from the cap and includes the translation initiation codon. It is well conserved in evolution, differing by only 2 nucleotides in Xenopus and human collagen mRNAs. LARP6 is the protein that specifically binds the 5′SL with high affinity and is required for high expression levels of type I collagen (9). LARP6 interacts with nonmuscle myosin; this interaction is required for coordinated synthesis of α1(I) and α2(I) polypeptides and their productive folding into heterotrimeric type I collagen (10). Disruption of nonmuscle myosin filaments resulted in secretion of a homotrimer of type I collagen composed of three α1(I) chains in lung fibroblasts or in total lack of secretion of type I collagen in scleroderma fibroblasts (10).
Vimentin is a member of the type III intermediate filaments and is a marker of cells of mesenchymal origin (e.g., fibroblasts and myofibroblasts). Vimentin intermediate filaments are polymers of soluble tetrameric vimentin. Vimentin filaments are involved in motility, maintenance of cell shape, and endurance of mechanical stress of mesenchymal cells (12, 18). Vimentin-deficient mice exhibit delayed wound healing; the phenotype has been attributed to decreased motility of vimentin-null fibroblasts (14). There have been no reports that vimentin can contribute to posttranscriptional regulation of collagen expression.
The present study describes a novel role of vimentin in the stabilization of collagen mRNAs. This stabilization is dependent on the 5′SL and on binding of LARP6 and may contribute to the high level of collagen expression by mesenchymal cells.
MATERIALS AND METHODS
Cells and transfections.
HEK293 cells and human lung fibroblasts immortalized by expression of telomerase reverse transcriptase (56) were grown under standard conditions. HEK293 cells were transfected with 1 μg of plasmid per 35-mm dish using 293TransIT reagent (Mirus). Transduction of lung fibroblasts with adenoviruses was done by adding adenoviruses at a multiplicity of infection (MOI) of 100. With this MOI, between 95% and 100% of the cells were transduced, as judged by expression of the green fluorescent protein (GFP) viral marker. The cells were harvested for analysis 2 to 5 days after virus delivery.
Scleroderma fibroblasts derived from the skin of a scleroderma patient were purchased from the European collection of cell cultures (cell line BM0070). Mouse embryonic fibroblasts (MEFs) were derived from knock-in mice, in which the 5′SL of the collagen α1(I) gene has been mutated, and their wild-type (wt) littermates (36). All cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum for up to 10 passages.
Vim+/+ and Vim−/− MEFs derived from wild-type and vimentin knockout mice were described previously (27) and were kind gifts from Robert Evans, University of Colorado.
Chemicals.
β,β′-Iminodipropionitrile (IDPN) was from Acros Organics (Morris Plains, NJ). Actinomycin D, puromycin, and cycloheximide were purchased from Sigma. For treatment with IDPN, cells were incubated with 1% IDPN for 24 h before analysis. Actinomycin at 10 μg/ml was added, and cells were collected at the indicated time points. Puromycin at 100 μg/ml and cycloheximide at 100 μg/ml were added to cells for 30 min before the cells were fixed for immunostaining or proteins were extracted for immunoprecipitation.
Plasmid constructs and adenovirus preparation.
Hemagglutinin (HA)-tagged wild-type LARP6 and some deletion constructs have been described previously (9). Additional deletion constructs were made by PCR amplification of the regions of LARP6 and by cloning the PCR products into the pCDNA3 vector. Adenovirus expressing dominant-negative desmin was constructed by PCR amplification of the human desmin region from amino acid 1 to amino acid 263 and cloning the PCR product into the pAdCMVTRACK vector, followed by recombination with the pADEasy vector, as described previously (25). Adenovirus was amplified in HEK293 cells and purified with a Virapure kit (Clontech). Expression of the construct was verified by Western blot analysis. This virus expressed both the test protein and GFP, which is encoded by an independent transcription unit (25). Expression of GFP served as a control for viral transduction. Control adenovirus expressed only GFP.
LARP6 siRNA.
A small interfering RNA (siRNA) effective against LARP6, with the sequence 5′-UCCAACUCGUCCACGUCCU-3′, was previously described (9). This siRNA sequence was expressed as short hairpin RNA (shRNA) from adenovirus (9). A control adenovirus contained shRNA with a scrambled sequence (5′-GGAGGGCUUCGAGUUAGGA-3′). The efficacy of LARP6 knockdown was assessed by Western blotting.
RT-PCR analysis.
Total cellular RNA was isolated using an RNA isolation kit (Sigma). Reverse transcription (RT)-PCRs were performed with 100 ng of total RNA or using rTth reverse transcriptase (Boca Scientific, Boca Raton, FL). [32P]dATP was included in the PCR step to label the products, which were resolved on sequencing gels, as described previously (46–50). The number of cycles was adjusted within the linear range of the reaction. The primers used for RT-PCR were as follows: human collagen α1(I), 5′primer (AGAGGCGAAGGCAACAGTCG) and 3′ primer (GCAGGGCCAATGTCTAGTCC); human collagen α2(I), 5′ primer (CTTCGTGCCTAGCAACATGC) and 3′ primer (TCAACACCATCTCTGCCTCG); human fibronectin, 5′ primer (ACCAACCTACGGATGACTCG) and 3′ primer (GCTCATCATCTGGCCATTTT); human GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′ primer (ACCGGTTCCAGTAGGTACTG) and 3′ primer (CTCACCGTCACTACCGTACC); human actin, 5′ primer (GTGCGTGACATTAAGGAGAAG) and 3′ primer (GAAGGTAGTTTCGTGGATGCC); mouse collagen α1(I), 5′ primer (GAGCGGAGAGTACTGGATCG) and 3′ primer (TACTCGAACGGGAATCCATC); mouse collagen α2(I), 5′ primer (CTTCGTGCCTAGCAACATGC) and 3′ primer (TCAACACCATCTCTGCCTCG); mouse actin, 5′ primer (CGTGCGTGACATCAAAGAGAAGC) and 3′ primer (TGGATGCCACAGGATTCCATACC).
Antibodies.
Anti-HA and antivimentin antibodies were obtained from Sigma, anti-collagen α1(I) antibody was obtained from Rockland, anti-collagen α2(I) antibody specific for human polypeptide was from Cell Signaling, the anti-collagen α2(I) antibody to detect mouse polypeptide was obtained from Santa Cruz Biotechnology, antifibronectin and antitubulin antibodies were from BD Biosciences, anti-LARP6 antibody was obtained from Abnova, antiactin antibody was from Novus Biological, and anti-MYH10 antibody was from the hybridoma bank, University of Iowa.
Western blot analysis.
Protein concentrations were estimated with the Bradford assay, with bovine serum albumin (BSA) as the standard. Fifty micrograms of total cellular protein was typically used for Western blot analysis. For Western blot analysis of secreted proteins, equal numbers of cells were seeded, and after 24 to 48 h, serum-free medium was added to the cells and incubation continued for 3 h. The medium was collected, and an aliquot was analyzed directly by Western blot analysis. Use of serum-free medium in collecting secreted proteins was essential, as fetal calf serum contains substantial amounts of collagen and fibronectin.
Immunostaining.
For immunostaining, cells were grown on glass coverslips. After treatment, the cells were fixed with 4% formaldehyde for 30 min at room temperature and permeabilized with 0.5% Triton X-100 in phosphate-buffered saline (PBS) for 10 min. Blocking was done with 10% goat serum-5% bovine serum albumin in PBS for 1 h at room temperature, followed by incubation with primary antibody overnight at 4°C. After washing, proteins were visualized with Alexa Fluor594-, Cy-2-, or Cy5-conjugated secondary antibodies. The cells were mounted using Prolong mounting solution containing 4′,6′-diamidino-2-phenylindole (DAPI) from Invitrogen. Images were taken with the Leica TCS SP2 AOBS laser confocal microscope equipped with a Chameleon Ti-Sapphire multiphoton laser. Optical sections were processed with LCSLite software, and single-plane confocal images are shown.
Immunoprecipitations.
Cell extracts were prepared in lysis buffer (10 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl [pH 7.5], 0.5% NP-40, and 170 μg/ml phenylmethylsulfonyl fluoride). After removal of nuclei by centrifugation, the cleared lysate was incubated with 1 μg of antibody for 1 h at 4°C. Thirty microliters of equilibrated protein A/G-agarose plus beads (Santa Cruz Biotechnology) was added, and incubation continued for an additional 3 h. After the beads were washed three times with PBS, immunoprecipitated complexes were analyzed by SDS-PAGE and Western blotting. For analysis of immunoprecipitated RNA, total RNA was extracted from the immunoprecipitated material.
In reactions in which RNase A digestion of the precipitate was performed, 0.2 μg/μl RNase A was added and incubated with protein A/G-agarose plus beads for 15 min at room temperature, and then the samples were washed two times in PBS before being loaded into the gel.
Determination of RNA stability.
Cells were treated with actinomycin D (10 μg/ml) for 6, 12, or 24 h. After the actinomycin D incubation period, the cells were scraped, and total RNA was extracted and analyzed by RT-PCR. RNA extracted from cells at time point 0 (immediately after the addition of actinomycin D) was used as the initial level of mRNA and arbitrarily set as 100%. The results shown are from 3 independent experiments.
Fractionation of cell extracts into intermediate-filament-rich fractions.
Fractionation of cells into intermediate-filament-rich (detergent-insoluble) and intermediate-filament-poor (detergent-soluble) fractions was done as described previously (54). Briefly, cells were homogenized in a buffer containing 1% Nonidet P-40, 10% glycerol, 20 mM HEPES, pH 7.6, 150 mM NaCl, and protease inhibitors (2 mM benzamidine, 0.5 mM aprotinin, and 1 mM phenylmethylsulfonyl fluoride). The homogenates were incubated in the same buffer at 37°C for 30 min. The soluble fraction (supernatant) and the insoluble fraction (pellet) were collected after centrifugation at 5,200 rpm and 4°C for 30 min. The volumes of both fractions were made the same by adding SDS sample buffer. Equal volumes of the samples were analyzed by Western blotting. For measurement of the distribution of collagen mRNAs into intermediate-filament-rich and intermediate-filament-poor cellular fractions, total RNA was extracted after fractionation and analyzed by RT-PCR.
For immunoprecipitations of collagen mRNAs from the detergent-soluble and detergent-insoluble fractions, the soluble fraction was prepared as described above, and the insoluble fraction was dissolved in an equal volume of buffer containing 0.1% SDS, 10 mM Tris-HCl (pH = 6.8), 10% glycerol, and protease inhibitors. Then, the immunoprecipitation was carried out as described for cell extracts.
RNA fluorescent in situ hybridization (FISH).
For the detection of collagen α1(I) and α2(I)) mRNAs, five fluorescently labeled DNA oligonucleotides were designed for each mRNA. Design of the oligonucleotide probe was done as described previously (23). The probes were 50 nucleotides long and contained four or five amino-modified nucleotides (amino-allyl T) coupled with Cy3 or Cy5. For collagen α1(I) (accession number NM-000088), the sequences of the probes (with modified Ts in boldface) and the nucleotides in the mRNA they are complementary to (in parentheses) are as follows: GTTCTGTACGCAGGTGATTGGTGGGATGTCTTCGTCTTGGCCCTCGACTT (255 to 206), GCAGTTCTTGGTCTCGTCACAGATCACGTCATCGCACAACACCTTGCCG (370 to 321), TGGAGGGAGTTTACAGGAAGCAGACAGGGCCAACGTCGAAGCCGAATTCC (4446 to 4395), GATGATGGGCAGGCGGGAGGTCTTGGTGGTTTTGTATTCAATCACTGTCTT (4531 to 4482), and TGGAATCCATCGGTCATGCTCTCGCCGAACCAGACATGCCTCTTGTCCTT (5030 to 4981). For collagen α2(I) (accession number NM-000089), the sequences of the probes and the nucleotides they are complementary to are as follows: GGACGTGGACACTTTTGAGGCTTTCAAGGGGAAACTCTGACTCGTGTCT (120 to 70), CCGATGTCCAAAGGTGCAATATCAAGGAAGGGCAGGCGTGATGGCTTATT (4520 to 4471), GGGCCAAGTCCAACTCCTTTTCCATCATACTGAGCAGCAAAGTTCCCACC (740 to 691), CATCCAGACCATTGTGTCCCCTAATGCCTTTGAAGCCAGGAAGTCCAGGA (1030 to 981), and AGAAGTCTCCATCGTAACCAAAGTCATAACCACCACCGCTTACACCTGGA (3820 to 3771).
RNA FISH and immunofluorescence (IF)-RNA FISH were done as described previously (23). Briefly, human lung fibroblasts were grown on coverslips for 24 h, followed by fixation with 4% formaldehyde in PBS for 30 min. The cells were rinsed in PBS-gelatin (PBSG) two times for 5 min each time, followed by permeabilization with 0.5% Triton X-100 for 10 min at room temperature. The cells were washed twice in PBSG and equilibrated in prehybridization buffer (50% formamide-2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) for 10 min. For the hybridization step, coverslips were incubated with fluorescently labeled oligonucleotides in hybridization buffer (2× SSC, 10% dextran sulfate, 50% formamide, 1 mg/ml BSA, 1 mg/ml sheared salmon sperm DNA, 1 mg/ml Escherichia coli RNase-free tRNA, and 10 ng/μl of oligonucleotide probe) for 2 h at 37°C. After hybridization, the cells were washed twice in 2× SSC-50% formamide at 37°C for 20 min, in 1× SSC-50% formamide at 37°C for 30 min, then in 1× SSC at room temperature for 15 min, and finally in 0.5× SSC at room temperature for 15 min. The coverslips were mounted with Prolong antifade mounting medium containing DAPI (Invitrogen).
For IF-RNA FISH experiments, human lung fibroblasts were fixed and permeabilized as described above, followed by blocking with FISH-compatible blocking solution (CAS block; Invitrogen). Coverslips were then processed for immunofluorescence using antivimentin antibodies diluted in CAS block containing 10 mM RNase inhibitor (ribonucleoside vanadyl complex [RVC]). After overnight incubation, the cells were washed with PBS containing 0.1% Tween 20 (PBST), and the primary antibody was fixed with 4% paraformaldehyde in PBS for 10 min. After fixation, the cells were washed with PBST for 10 min, followed by FISH hybridization and washing as described above. Following the last wash, the coverslips were incubated with Cy5-labeled anti-rabbit secondary antibody diluted in CAS block with 10 mM RVC. After washing with PBST for 30 min, mounting was done as described above. Fluorescence images were acquired with the DeltaVision image restoration (deconvolution) microscope system (Applied Precision) using a 60× objective (1.42 numerical aperture [NA]; Olympus). Digital images were acquired using a charge-coupled-device camera (Coolsnap HQ; Photometrics), and stacks of 64 images were taken with a Z-step size of 0.2 μm using DeltaVision software and filter sets for DAPI, Cy3, and Cy5. The three-dimensional (3D) stacks were deconvolved to quantitatively improve the signal-to-noise ratio using the SoftWorx (Applied Precision) constrained iterative deconvolution process.
Fractionation for polysomes.
Polysomes were fractionated from 2 × 107 cells on linear 15 to 45% sucrose gradients as described previously (9, 10). In some experiments, puromycin at 100 μM was used to disrupt polysomes. Fractions (0.5 ml) were collected, and RNA was extracted with phenol-chloroform and precipitated with isopropanol. For analysis of the distribution of vimentin and nonmuscle myosin, total protein from the fractions was precipitated with 0.05% deoxycholate and 6.5% trichloroacetic acid, washed with acetone, and analyzed by Western blotting.
Statistical analysis.
Densitometric analysis of scanned images from gels was performed with Image J software. The results were analyzed for statistical significance using Student's t test. P values of <0.05 were considered to be significant. Data are presented as means and standard errors of the mean (SEM).
RESULTS
Vimentin specifically interacts with collagen mRNAs in a 5′ stem-loop-dependent manner.
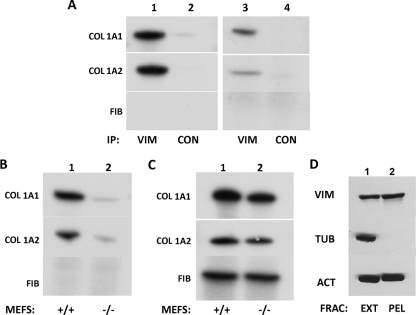
In a previous study, we observed that vimentin copurified with the 5′SL of collagen mRNA in an RNA affinity purification method (10). Therefore, we investigated if collagen mRNAs associate with vimentin filaments in vivo using an RNA immunoprecipitation assay and cellular extracts prepared by hypotonic and detergent lysis. Vimentin is present mostly in the insoluble cellular fraction (see below), but the combination of hypotonic and detergent lysis resulted in the presence of significant amounts of vimentin in the soluble extract (Fig. 1D). This allowed us to do the immunoprecipitation with antivimentin antibody and to analyze collagen mRNAs in the precipitate by RT-PCR (Fig. 1). The immunoprecipitation with an antivimentin antibody in human lung fibroblasts pulled down both collagen mRNAs, α1(I) and α2(I), but not fibronectin mRNA (Fig. 1A, lane 1). Neither collagen nor fibronectin mRNA was pulled down using a control antibody (Fig. 1A, lane 2), indicating the specific association of collagen mRNAs with vimentin. To confirm this result in another collagen-producing cell, we repeated the experiment in primary scleroderma skin fibroblasts. Again, collagen α1(I) and α2(I) mRNAs were pulled down with vimentin, suggesting that this interaction could be a general feature of collagen-producing cells (Fig. 1A, lanes 3 and 4).
Fig. 1.
Collagen α1(I) and collagen α2(I) mRNAs interact with vimentin in a 5′SL-dependent manner. (A) Pulldown of collagen mRNAs with an antivimentin antibody. Immunoprecipitation (IP) with an antivimentin antibody (VIM), followed by RT-PCR analysis of collagen (COL) α1(I), collagen α2(I), and fibronectin (FIB) mRNAs in human lung fibroblasts (lane 1) and scleroderma fibroblasts (lane 3). Lanes 2 and 4, control antibody (CON). Radiolabeled PCR products are indicated. (B) 5′SL-dependent interaction of collagen mRNAs with vimentin. The experiment was as in panel A, except extracts of wild-type (lane 1; +/+) and mutant (lane 2; −/−) mouse embryonic fibroblasts, which carry a mutation of the 5′ stem-loop in the collagen α1(I) gene, were used. (C) Total RNA of wild-type (lane 1; +/+) and mutant (lane 2; −/−) mouse embryonic fibroblasts analyzed by RT-PCR for collagen α1(I), collagen α2(I), and fibronectin mRNAs. (D) Presence of vimentin in the cell extract prepared by hypotonic and detergent lysis. The extract (EXT) and insoluble pellet (PEL) obtained after cell lysis were analyzed for vimentin, actin, and tubulin by Western blotting.
Since vimentin was initially discovered in a complex copurifying with the 5′SL of collagen mRNAs, we determined if the interaction between vimentin and collagen mRNAs depends on the 5′SL. To this end, we utilized MEFs derived from knock-in mice in which the 5′SL of collagen α1(I) mRNA was mutated (denoted here 5′SL−/− MEFs) (36). Immunoprecipitation with antivimentin antibody using extracts from wild-type (5′SL+/+) MEFs pulled down collagen α1(I) and α2(I) mRNAs (Fig. 1B, lane 1). However, only trace amounts of collagen mRNAs were pulled down in 5′SL−/− MEFs (Fig. 1B, lane 2). Fibronectin mRNAs were not precipitated in either cell type. Total steady-state levels of collagen and fibronectin mRNAs from the wild-type and mutant cells are shown in Fig. 1C. The difference in total mRNA levels between 5′SL+/+ and 5′SL−/− MEFs was 2-fold, and this difference cannot account for the dramatic difference in the pulldown of collagen mRNAs (Fig. 1B). In these experiments, collagen α2(I) mRNA contained the wt 5′SL, while only the 5′SL of collagen α1(I) was mutated. Still, collagen α2(I) mRNA failed to interact with vimentin. This suggests that binding of collagen α2(I) mRNAs to vimentin is coupled to the 5′SL-dependent binding of collagen α1(I) mRNA to vimentin.
Collagen mRNAs are enriched in cellular fractions containing vimentin filaments.
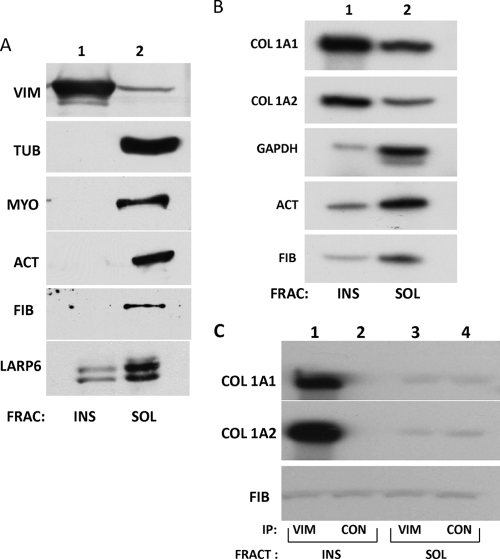
Vimentin exists in the cell in two forms: as soluble tetramers and insoluble filaments (43). Eighty percent of the cellular vimentin is in the form of filaments, and 20% is soluble. In the filamentous form, vimentin is one of the most insoluble proteins, precipitating in buffers of high ionic strength and with high concentrations of nonionic detergents (6, 7). Precipitation in these buffers can separate filaments from the soluble form. To assess which form of vimentin binds collagen mRNAs, we fractionated the cell extract into vimentin filament-rich and vimentin filament-poor fractions using high-salt and nonionic-detergent lysis. This method had been established previously for separating intermediate filaments from other soluble cellular components (53, 54). The fractions were analyzed for the presence of vimentin, tubulin, nonmuscle myosin, actin, fibronectin, and LARP6 by Western blotting (Fig. 2). Most of the vimentin was found in the insoluble fraction (Fig. 2A, lane 1) whereas tubulin, actin, nonmuscle myosin, and fibronectin were all exclusively found in the soluble fraction (Fig. 2A, lane 2). A significant amount of LARP6 was found in the insoluble fraction. The soluble and insoluble fractions were then analyzed for the relative abundance of collagen α1(I) and α2(I) mRNAs by RT-PCR. Actin, GAPDH, and fibronectin mRNAs were analyzed as controls. Figure 2B shows that collagen α1(I) and α2(I) mRNAs were predominantly located in the insoluble fraction (lane 1), whereas actin, GAPDH, and fibronectin mRNAs were enriched in the soluble fraction (lane 2). Preferential localization of collagen mRNAs in the detergent-insoluble fraction containing vimentin filaments suggests that the filamentous vimentin is the main form involved in the interaction with collagen mRNAs.
Fig. 2.
Collagen α1(I) and α2(I) mRNAs cofractionate with vimentin filaments. (A) Insoluble vimentin representing filaments (INS; lane 1) and soluble vimentin (SOL; lane 2) were fractionated by detergent lysis and centrifugation. The fractions (FRAC) were probed for the presence of vimentin (VIM), tubulin (TUB), myosin IIB (MYO), actin (ACT), fibronectin (FIB), and LARP6 proteins by Western blotting. (B) Collagen mRNAs segregate with insoluble vimentin. The fractions in panel A were analyzed by RT-PCR for collagen α1(I), collagen α2(I), GAPDH, actin, and fibronectin mRNAs. Shown is pulldown of collagen mRNAs with an antivimentin antibody. (C) Collagen mRNAs in the insoluble fraction coimmunoprecipitate with vimentin. The insoluble fraction (INS) was redissolved, and immunoprecipitation with vimentin antibody (lane 1) or control antibody (lane 2) was performed. (Lanes 3 and 4) The same experiment using the soluble fraction (SOL). The immunoprecipitated material was analyzed by RT-PCR for the presence of collagen α1(I), collagen α2(I), and fibronectin mRNAs.
In order to further verify the association of the filamentous form of vimentin with collagen mRNAs, we performed experiments with RNA immunoprecipitation from the detergent-soluble and detergent-insoluble fractions. With this goal, we redissolved the insoluble fraction in buffer containing 0.1% SDS. The immunoprecipitation with antivimentin antibody from the detergent-insoluble fraction of human lung fibroblasts pulled down collagen α1(I) and α2(I) mRNAs, but not fibronectin mRNA (Fig. 2C, lane 1). Neither collagen nor fibronectin mRNA was pulled down using a control antibody (Fig. 2C, lane 2). However, immunoprecipitation from the detergent-soluble fraction pulled down only trace amounts of collagen α1(I) and α2(I) mRNAs (Fig. 2C, lane 3). This suggests that collagen α1(I) and α2(I) mRNAs found in the insoluble fraction (Fig. 2B) are associated with vimentin filaments.
Visualization of the interaction between vimentin filaments and collagen α1(I) and α2(I) mRNAs.
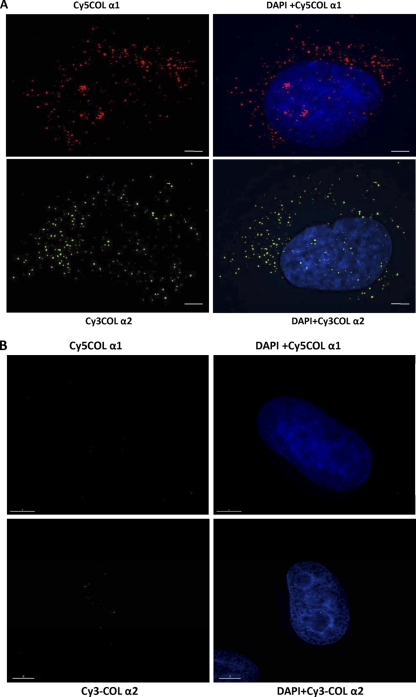
To directly show the association of collagen mRNAs with vimentin filaments in vivo, we utilized RNA FISH. This method can detect single mRNA molecules in the cytoplasm (4, 19, 24, 31, 59). To visualize collagen mRNAs, we designed multiple short 50-mer oligonucleotide probes that specifically hybridize with their target mRNAs. Each probe was labeled with multiple fluorophores to increase the signal-to-noise ratio for detection of individual mRNAs: collagen α1(I) probes were labeled with Cy3 and α2(I) probes with Cy5. RNA FISH experiments using oligonucleotide probes specific for collagen α1(I) mRNA detected multiple spots distributed both in the nucleus and in the cytoplasm of human lung fibroblasts (Fig. 3A, top). The probes spanned the exons of collagen mRNAs, and the signals in the nucleus may be from unspliced mRNA. The cytoplasmic signals appear more concentrated around the perinuclear region, but they can be seen throughout the cytoplasm. At the bottom of Fig. 3A is shown RNA FISH using oligonucleotide probes specific for collagen α2(I) mRNA with a different cell than that shown at the top. The distribution of RNA FISH signals was similar to what was seen for collagen α1(I) mRNA. To verify that our probes specifically detected collagen mRNAs, we stained HeLa cells, which do not express type I collagen. In HeLa cells, neither set of probes showed signal (Fig. 3B), indicating that they specifically detected collagen α1(I) and α2(I) mRNAs.
Fig. 3.
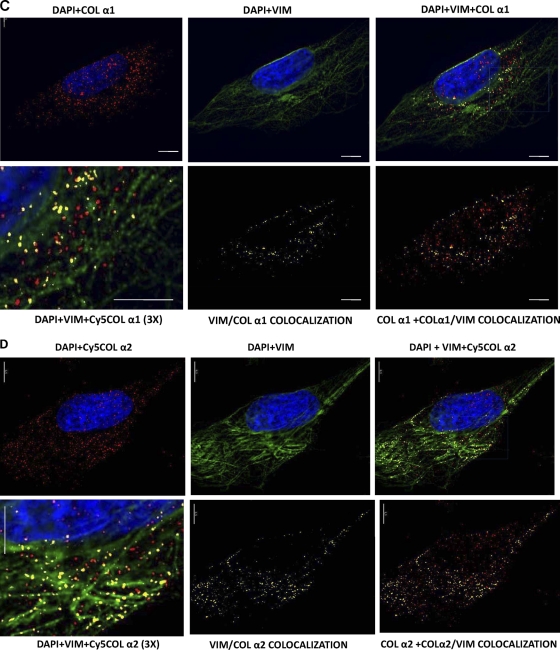
Collagen mRNAs colocalize with vimentin intermediate filaments. (A) Visualization of collagen α1(I) mRNA (top) and collagen α2(I) mRNA (bottom) by RNA FISH. Collagen α1(I)-specific probes (labeled with Cy5; red) and collagen α2(I)-specific probes (labeled with Cy3; green) were hybridized to human lung fibroblasts. (B) The same experiment in HeLa cells, which do not express collagen α1(I) and α2(I) mRNAs. (C) Colocalization of collagen α1(I) mRNA with vimentin. RNA FISH for collagen α1(I) mRNA (upper left), immunostaining of vimentin (upper middle), and the overlaid image (upper right). Magnification (×3) of the selected area of the overlaid image is shown at the bottom left. The yellow dots indicate colocalization. (Bottom middle) Image showing only α1/VIM colocalization. (Bottom right) The image overlaid with total signal for α1(I) mRNA (upper left) to estimate the fraction of colocalized mRNA. Bars, 1 μm. (D) Colocalization of collagen α2(I) mRNA with vimentin. The same experiment as in panel C is shown, except collagen α2(I) mRNA was visualized.
Next, we determined if collagen mRNAs colocalize with vimentin filaments by coupling RNA FISH with immunostaining for vimentin protein (IF-RNA FISH). In these experiments, we labeled collagen α2(I) mRNA with Cy5 oligonucleotide probes to avoid any bleedover of the vimentin immunostaining in the Cy2 channels. Figure 3C, upper right, shows that a significant proportion of collagen α1(I) mRNAs appear to be colocalized with vimentin filaments. The areas of colocalization (yellow) are seen both around the nucleus and throughout the cytoplasm (high-magnification image, lower left). By overlaying the yellow signal of vimentin-associated α1(I) mRNA (Fig. 3C, bottom middle) with the total signal for α1(I) mRNA (red; Fig. 3C, top left), we estimated that about 30% of collagen α1(I) mRNAs are located at or in close proximity to vimentin intermediate filaments (Fig. 3C, bottom right). Similar results were obtained by analyzing multiple cells.
Colocalization of α2(I) mRNA and vimentin is shown in Fig. 3D. Collagen α2(I) mRNA was also colocalized with vimentin filaments; it appears that a larger fraction of this mRNA is found associated with filaments than was seen for α1(I) mRNA (Fig. 3D, top right). The areas of colocalization were distributed throughout the cytoplasm, and some areas clearly show a beads-on-a-string appearance, indicating the association with filaments (Fig. 3D, bottom left, higher-magnification image). The proportion of α2(I) mRNAs colocalized with vimentin filaments was estimated to be about 50% (Fig. 3D, bottom right). Thus, the IF-RNA FISH results provided direct evidence for the in vivo interaction between collagen α1(I) and α2(I) mRNAs and vimentin filaments.
Association of vimentin with collagen mRNAs is LARP6 dependent.
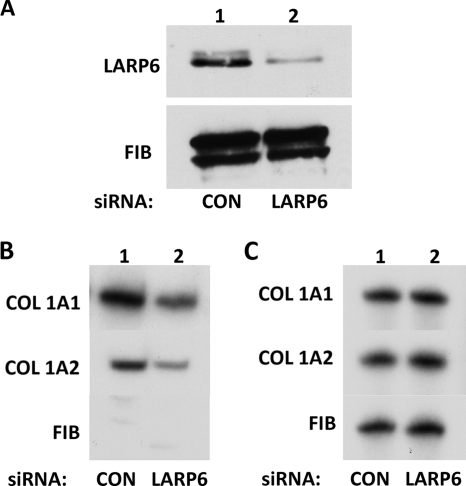
The experiments described above could not distinguish whether the interaction between collagen mRNAs and vimentin is direct or mediated by protein-protein interactions. LARP6 is the protein that directly binds the 5′SL (9), and since collagen mRNAs interact with vimentin in a 5′SL-dependent manner, LARP6 may bridge collagen mRNAs and vimentin filaments. To assess the role of LARP6, we knocked down LARP6 using siRNA (9) and analyzed the association of vimentin with collagen mRNAs. The siRNA was able to deplete LARP6 to 30% of the initial level (Fig. 4A) without drastically affecting the expression of collagen mRNAs, as assessed from RT-PCR analysis of their mRNA input levels (Fig. 4C). When RNA immunoprecipitation was done with anti-vimentin antibody in LARP6-depleted cells, it pulled down less collagen α1(I) and α2(I) mRNAs than was pulled down from cells with control siRNA (Fig. 4B). Considering that not all LARP6 could be depleted (Fig. 4A), this suggested that the interaction of collagen mRNAs with vimentin filaments is LARP6 dependent.
Fig. 4.
LARP6-dependent association of collagen mRNAs with vimentin. (A) Knockdown of LARP6. Control siRNA (lane 1) and LARP6-specific siRNA (lane 2) were expressed in human lung fibroblasts, and the level of LARP6 was determined by Western blotting. Fibronectin was analyzed as a loading control. (B) LARP6 knockdown reduces the association of collagen mRNAs with vimentin. IP with an antivimentin antibody was performed in cells expressing control siRNA (lane 1) or LARP6-specific siRNA (lane 2). Collagen α1(I), collagen α2(I), and fibronectin were analyzed in the immunoprecipitate by RT-PCR. (C) Analysis of the input levels of mRNAs. Total mRNA before the IP was analyzed for expression of collagen α1(I), collagen α2(I), and fibronectin mRNAs by RT-PCR.
LARP6 binds vimentin.
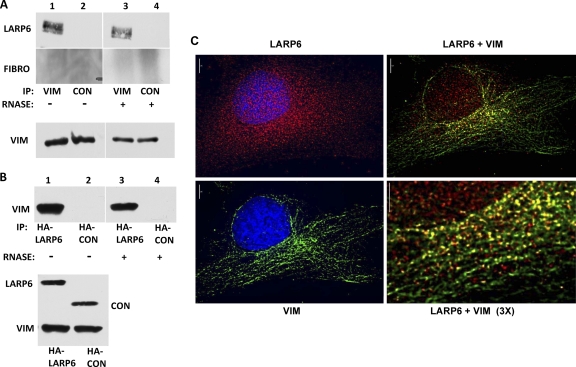
To determine if there is a protein-protein interaction between vimentin and LARP6, we performed immunoprecipitation experiments. The antivimentin antibody pulled down endogenous LARP6 (Fig. 5A, lane 1), while control antibody had no effect (lane 2). The control protein, fibronectin, was not pulled down, suggesting that this interaction is specific for LARP6. We also assessed if the interaction between vimentin and LARP6 is dependent on intact RNA. Treatment with RNase did not release LARP6 from the immunoprecipitate, suggesting that intact RNA is not needed for the interaction between vimentin and LARP6 (Fig. 5A, lane 3).
Fig. 5.
Interaction of LARP6 with vimentin. (A) Pulldown of endogenous LARP6 with anti-vimentin antibody. Shown is immunoprecipitation from lung fibroblasts with antivimentin antibody (lanes 1 and 3) or control antibody (lanes 2 and 4). Immunoprecipitates were left untreated (lanes 1 and 2) or treated with RNase A (lanes 3 and 4) prior to washing and Western blot analysis with anti-LARP6 antibody and antifibronectin (FIBRO) antibody. (Bottom) Input levels of vimentin. (B) Reverse experiment using HA-tagged LARP6. HA-tagged LARP6 (lanes 1 and 3) or control HA-tagged protein (lanes 2 and 4) was expressed in HEK293 cells, and IP was performed with anti-HA antibody without (lanes 1 and 2) or with (lanes 3 and 4) RNase A digestion. The immunoprecipitate was analyzed by Western blotting using antivimentin antibody. (Bottom) Input levels of transfected proteins and endogenous vimentin. (C) Colocalization of LARP6 with vimentin in lung fibroblasts. Immunostaining for vimentin (green; upper left) and endogenous LARP6 (red; upper right) and the merged image (lower left). The areas of colocalization are yellow. (Lower right) Higher magnification of the same image.
To further investigate the interaction between LARP6 and vimentin, we overexpressed HA-tagged LARP6 in HEK293 cells and performed immunoprecipitation with anti-HA antibody. Anti-HA antibody pulled down vimentin in LARP6-transfected cells, but not in the cells transfected with the HA-tagged control protein RBMS3 (21) (Fig. 5B, lanes 1 and 2). Treatment with RNase had no effect on the pulldown (Fig. 5B, lane 3).
To determine if there is cellular colocalization of vimentin and LARP6, we performed double immunostaining using anti-LARP6 and antivimentin antibodies. As previously reported (9), LARP6 exhibited nuclear and cytoplasmic localization (Fig. 5C, upper left), whereas vimentin staining was restricted to the cytoplasm (Fig. 5C, lower left). The merged image of these two channels showed a significant degree of colocalization of LARP6 and vimentin filaments (Fig. 5C, right). This is consistent with the result of cofractionation of LARP6 into the insoluble cellular material containing vimentin filaments (Fig. 2A).
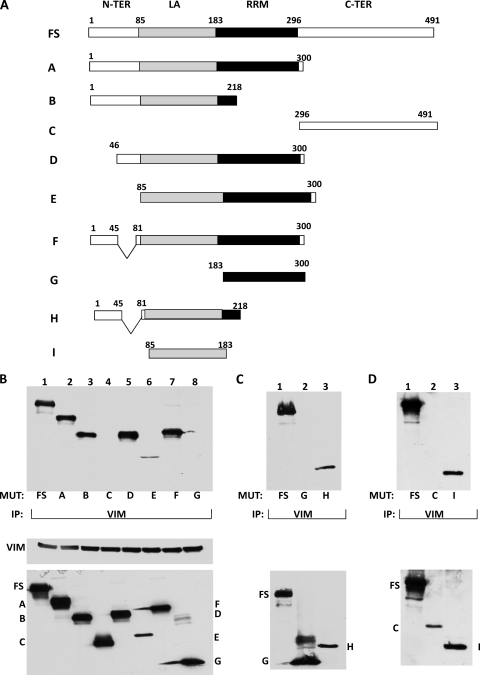
The La domain of LARP6 is required for its interaction with vimentin.
LARP6 has four domains: the N-terminal domain, the La homology domain (55), an RNA recognition motif (RRM), and the C-terminal domain. The La domain and RRM are needed for binding the 5′SL (9), while the C-terminal domain is required for binding to nonmuscle myosin (10). To determine which domain of LARP6 is involved in the interaction with vimentin, we constructed a series of deletion mutants of LARP6 and evaluated their interactions with vimentin in pulldown assays. A schematic representation of full-size LARP6 and LARP6 mutants is shown in Fig. 6A. The constructs were tagged with an HA affinity tag and transfected into HEK293 cells. Western blots showed similar expression levels of all LARP6 mutants, except mutant E, which was expressed at a lower level. The input levels of vimentin were also comparable (Fig. 6B, bottom).
Fig. 6.
LARP6 interacts with vimentin through the La domain. (A) Schematic representation of the constructs used. FS, full-length LARP6. The domains of LARP6 are indicated, with amino acid numbering above. A to I, different deletion mutants of LARP6. All proteins had an HA tag at the N terminus (N-TER). C-TER, C terminus. (B) Interaction of FS LARP6 and deletion mutants with vimentin. (Top) FS LARP6 (lane 1) and the deletion mutants (lanes 2 to 8) were expressed in HEK293 cells. IP was done with antivimentin antibody and Western blotting with anti-HA antibody. (Bottom) Expression of the proteins in the input material. (C) Interaction of the extended La domain with vimentin. A construct containing the intact La domain with additional amino acids at the N terminus (H; lane 3) was analyzed for interaction with vimentin by IP. Lane 1, FS LARP6 as a positive control; lane 2, mutant G as a negative control. (Bottom) Expression of the proteins in the input material. (D) Interaction of the La domain only with vimentin. (Lane 1) Construct I, containing only the La domain, was analyzed for IP with vimentin. (Lane 2) Construct C as a negative control. (Bottom) Expression of the proteins in the input material.
When immunoprecipitation was performed with an anti-vimentin antibody (Fig. 6B, top), the wild-type LARP6 (FS) was pulled down efficiently. Similar coimmunoprecipitation of mutants A and B, which lack the C-terminal domain and the RRM, respectively, suggested that these domains are not needed for interaction with vimentin. The N-terminal deletion mutants (MUT), D and F, also coimmunuprecipitated with vimentin, suggesting that the N terminus is also dispensable for the interaction. Mutant E, which lacks both the N-terminal and the C-terminal domains, showed a similar interaction, although the amount pulled down was less, which is expected given its low expression relative to the other mutants. However, mutant C (containing only the C-terminal domain) and mutant G (containing only the RRM domain) failed to interact with vimentin. Therefore, it appears that all the mutants that contained the La domain of LARP6 (A, B, D, E,F, H, and I) interacted with vimentin, while the mutants lacking the La domain (C and G) failed to do so. This suggests that the La domain of LARP6 is required for interaction with vimentin. Finally, we expressed the La domain only (Fig. 6A, construct I) and performed the immunoprecipitation. The La domain was efficiently pulled down with vimentin (Fig. 6D, lane 3), verifying that the La domain is necessary and sufficient for the interaction.
Vimentin knockout fibroblasts have decreased collagen synthesis.
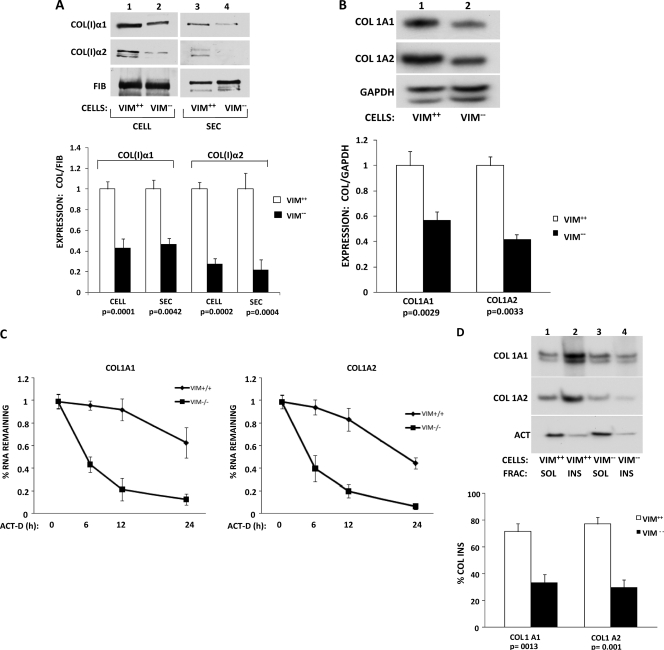
Since LARP6 is needed for high collagen expression (9) and LARP6 and vimentin interact, we determined if collagen expression is altered in cells lacking vimentin. To this end, we measured collagen production from MEFs of vimentin knockout (Vim−/−) mice and wild-type (Vim+/+) mice. (Vim+/+ and Vim−/− MEFs were kind gifts from Robert Evans, University of Colorado Health Sciences Center.) Figure 7A shows that the cellular and the secreted levels of collagen α1(I) polypeptide were decreased 60%, while the levels of α2(I) polypeptide were decreased 70% (Fig. 7A, lanes 2 and 4, bottom). The fibronectin levels in these two cell types were similar, suggesting a specific reduction in collagen expression.
Fig. 7.
Decreased collagen synthesis in vimentin knockout cells. (A) Expression of type I collagen in embryonic fibroblasts from wt (Vim+/+) and vimentin knockout (Vim−/−) mice. (Top) Representative Western blot of cellular (CELL; lanes 1 and 2) and secreted (SEC; lanes 3 and 4) levels of collagen (I) α1 and α2 polypeptides of Vim+/+ cells (lanes 1 and 3) and Vim−/− cells (lanes 2 and 4). Loading control, fibronectin. (Bottom) Quantitation of expression from three independent experiments. The expression of collagen polypeptides was normalized to the expression of fibronectin and arbitrarily set as 1 for Vim+/+ cells. The error bars represent SEM, and statistical significance is indicated. (B) Reduced steady-state levels of collagen α1(I) and α2(I) mRNAs in Vim−/− cells. (Top) Representative RT-PCR analysis of collagen α1(I), collagen α2(I), and GAPDH mRNA levels in Vim+/+ (lane 1) and Vim−/− (lane 2) mouse embryonic fibroblasts. (Bottom) Quantification of mRNA levels in three independent experiments. The expression of collagen mRNAs was normalized to the expression of GAPDH mRNA and arbitrarily set as 1 for Vim+/+ cells. The error bars represent SEM, and statistical significance is indicated. (C) Decay rates of collagen α1(I) and α2(I) mRNAs in Vim+/+ and Vim−/− cells. Transcription was blocked by actinomycin D (ACT-D) in Vim+/+ and Vim−/− cells, and collagen α1(I) and α2(I) mRNA levels were measured at 0 h, 6 h, 12 h, and 24 h after the block by RT-PCR. GAPDH mRNA was measured as a loading control. The expression of collagen mRNAs was normalized to the expression of GAPDH mRNA and set as 1 for Vim+/+ cells. The values for α1(I) mRNA (left) and α2(I) mRNA (right) were plotted for each time point, and error bars representing SEM are shown. (D) Segregation of collagen mRNAs with insoluble vimentin. (Top) Vimentin wt fibroblasts (Vim+/+; lanes 1 and 2) and vimentin knockout fibroblasts (Vim−/−; lanes 3 and 4) were fractionated into soluble (lanes 1 and 3) and insoluble (lanes 2 and 4) fractions. The fractions were analyzed for the presence of collagen α1(I), collagen α2(I), and actin mRNAs by RT-PCR. (Bottom) Percentages of collagen mRNAs found in the insoluble fractions of Vim+/+ and Vim−/− cells, as estimated by three independent experiments. Statistical significance and error bars representing SEM are shown.
The decreased level of collagen protein in vimentin-deficient MEFs may be due to either the decreased mRNA expression, decreased translation, or increased protein turnover. To distinguish between these possibilities, we determined the steady-state levels of collagen mRNAs in Vim−/− MEFs and Vim+/+ MEFs. The steady-state levels of both collagen α1(I) and α2(I) mRNAs were reduced in vimentin-deficient MEFs (Fig. 7B, lane 2). The steady-state level of GAPDH mRNA was unchanged.
Next, we tested if the reduced level was due to increased turnover of collagen mRNA. We determined the stability of collagen mRNAs by assessing their steady-state levels at different time points after transcriptional blocking by actinomycin D. Collagen mRNAs have a half-life of 12 or more hours in various fibroblasts; therefore, there was little decay in wild-type MEFs. However, the decay was much faster in Vim−/− MEFS, where the half-life was measured as 6 h. This result suggests that vimentin is needed for stability of collagen mRNAs and that association of vimentin filaments with collagen mRNAs may serve this purpose.
Vimentin is the only cytoplasmic intermediate-filament system in fibroblasts (17). Since the insoluble fraction of human lung fibroblasts is enriched in collagen mRNAs (Fig. 2), the insoluble fraction from Vim−/− cells should not contain the majority of collagen mRNAs. We prepared detergent-insoluble fractions from Vim+/+ MEFs and Vim−/− MEFs and estimated the abundance of collagen α1(I) and α2(I) mRNAs in these fractions. In vimentin-deficient MEFs, more collagen mRNAs were found in the soluble fraction (Fig. 7D, lanes 3 and 4); in wild-type MEFs, the opposite was found (Fig. 7D, lanes 1 and 2). The total level of collagen mRNAs was lower in Vim−/− MEFS, as shown in Fig. 7B. The distribution of actin mRNA into insoluble and soluble fractions was identical in the Vim+/+ and Vim−/− cells (Fig. 7D, bottom). This finding further corroborates the specific association of vimentin with collagen α1(I) and α2(I) mRNAs.
Disruption of vimentin filaments decreases expression of type I collagen.
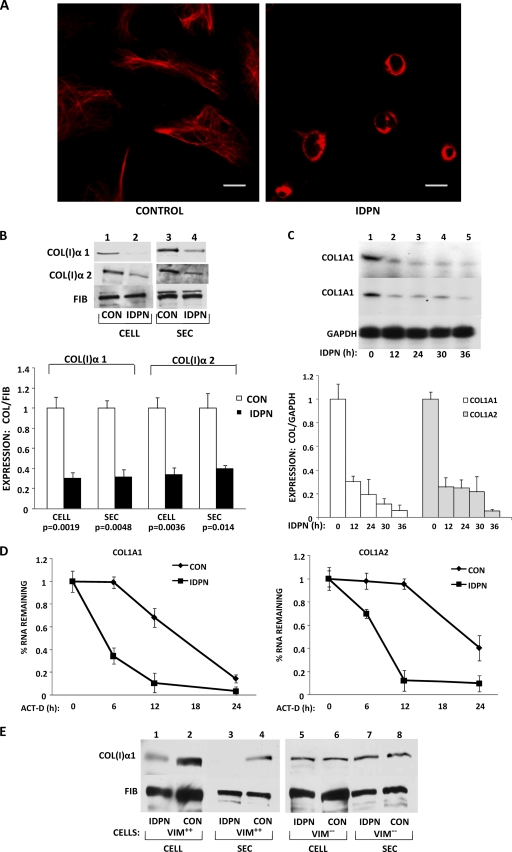
Since the absence of vimentin in fibroblasts resulted in destabilization of collagen mRNAs, we wanted to know if similar effects can be achieved by disrupting vimentin filaments. IDPN is a drug that specifically disrupts vimentin intermediate filaments (22). When human lung fibroblasts were treated with IDPN, complete collapse of the vimentin network was observed 24 h after the treatment (Fig. 8A). Treatment with IDPN also reduced the cellular and secreted levels of collagen α1(I) and α2(I) polypeptides. Here, the effect was more pronounced on α1(I) polypeptides than α2(I) polypeptides, while there was no effect on the control protein, fibronectin (Fig. 8B). Treatment with IDPN significantly decreased the steady-state levels of collagen α1(I) and α2(I) mRNAs, reaching the maximal effect after 12 h (Fig. 8C).
Fig. 8.
Disruption of vimentin filaments by IDPN reduces collagen synthesis. (A) Collapse of vimentin filaments after IDPN treatment. Untreated human lung fibroblasts (HLF) (Control) and HLF treated with 1% IDPN were immunostained with antivimentin antibody. Bars, 1 μm. (B) IDPN reduces collagen protein. (Top) Cellular (lanes 1 and 2) and secreted (lanes 3 and 4) levels of collagen α1(I) and α2(I) polypeptides from control cells (lanes 1 and 3) or IDPN-treated cells (lanes 2 and 4) were analyzed by Western blotting. Loading control, fibronectin. (Bottom) Collagen expression was normalized to fibronectin expression and plotted from three independent experiments. The statistical significance and error bars representing SEM are shown. (C) IDPN decreases the steady-state levels of collagen mRNAs. (Top) Collagen mRNAs from control cells and cells treated with IDPN for the indicated times were analyzed by RT-PCR. Loading control, GAPDH. (Bottom) The expression of collagen mRNAs was normalized to the expression of GAPDH mRNA and plotted from three independent experiments. The error bars represent SEM. (D) Decay of collagen α1(I) mRNA (left) and collagen α2(I) mRNA (right) in control and IDPN-treated HLF. Transcription was blocked with actinomycin D, and the levels of collagen mRNAs were estimated by RT-PCR at the indicated times. The expression at time zero was set as 1. The error bars represent SEM estimated from three independent experiments. (E) IDPN affects collagen expression only in vimentin-expressing cells. Cellular and secreted levels of collagen α1(I) polypeptide were measured in VIM+/+ fibroblasts (lanes 1 to 4) and in VIM−/− fibroblasts (lanes 5 to 8) by Western blotting. The cells were treated with IDPN or left untreated, as indicated. Loading control, fibronectin.
In order to determine if this effect is due to faster decay of collagen mRNAs, we measured the stability of collagen mRNAs after 12 h of IDPN treatment. Figure 8D shows that collagen α1(I) and α2(I) mRNAs decayed with half-lives of 5 h and 8 h in cells treated with IDPN compared to 18 h and 24 h in untreated cells. These half-lives are similar to those obtained in vimentin knockout MEFs (Fig. 7).
The drawback of using drugs to perturb any cellular function is their pleiotropic effect. Therefore, it is possible that IDPN may have had nonspecific effects in our experiments. To verify that the effect of IDPN on collagen production is due to disruption of vimentin filaments, we treated Vim+/+ MEFs and Vim−/− MEFs with IDPN. If the effect of IDPN is vimentin specific, then only Vim+/+ MEFs should show reduced collagen expression, while there should be no effect on Vim−/− MEFs. Treatment of Vim+/+ MEFs with IDPN reduced both cellular and secreted amounts of collagen protein (Fig. 8E, lanes 1 to 4), but the level in Vim−/− MEFs was unchanged (Fig. 8E, lanes 5 to 8). Since IDPN had no effect in the absence of vimentin, we concluded that its effect on collagen expression was specifically due to the disruption of vimentin filaments.
The dominant-negative form of desmin reduces collagen synthesis.
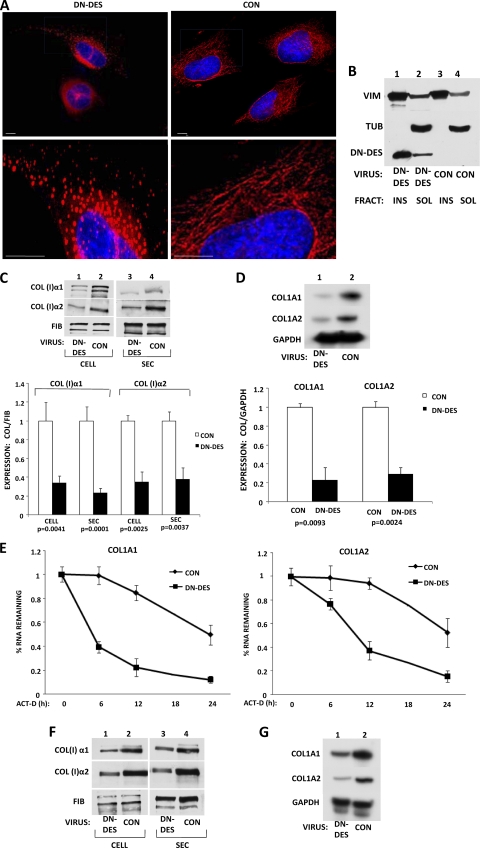
We also used an alternative approach to disrupt vimentin filaments by overexpressing a dominant-negative form of desmin. Desmin is closely related to vimentin and forms filaments, but it is not expressed in fibroblasts (17). Desmin monomers truncated at amino acid 263 act as a dominant-negative protein that interferes with the organization of desmin, as well as vimentin intermediate filaments (39, 42, 58). Therefore, we constructed a recombinant adenovirus expressing this dominant-negative form of desmin. Transduction of human fibroblasts with this adenovirus disrupted vimentin filaments, as assessed by immunostaining (Fig. 9A). We observed granular aggregates of vimentin (Fig. 9A, left) instead of the normal filamentous distribution of the vimentin network (Fig. 9A, right). To verify that the truncated desmin was integrated into the collapsed vimentin filaments, we fractionated the cell lysates into detergent-soluble and -insoluble fractions as described for Fig. 2. Most of the transfected desmin cofractionated into the insoluble fraction, together with vimentin (Fig. 9B). Its presence in the insoluble fraction strongly suggests that it had been incorporated into the endogenous vimentin network.
Fig. 9.
Disruption of vimentin filaments by a dominant-negative mutant of desmin reduces collagen synthesis. (A) A dominant-negative mutant of desmin (DN-DES) disrupts vimentin filaments. HLF were transduced with control adenovirus (right) or adenovirus expressing DN-DES (left), and the cells were immunostained with antivimentin antibody. The lower images show higher magnification. (B) DN-DES cofractionates with insoluble vimentin. The cells in panel A were fractionated into vimentin-soluble and -insoluble fractions and analyzed by Western blotting using antivimentin, antitubulin, and anti-FLAG antibodies, which recognize DN-DES. (C) (Top) Collagen α1(I) and α2(I) polypeptides from HLF transduced with control (lanes 2 and 4) or DN-DES (lanes 1 and 3) adenovirus were analyzed by Western blotting. Loading control, fibronectin. (Bottom) Collagen expression was normalized to fibronectin expression and is shown for three independent experiments. The expression in control cells was set as 1, and statistical significance and error bars representing SEM are shown. (D) DN-DES decreases the steady-state levels of collagen mRNAs. (Top) The cells in panel C were analyzed for expression of collagen α1(I), collagen α2(I), and GAPDH mRNAs by RT-PCR. (Bottom) The expression of collagen mRNAs was normalized to the expression of GAPDH and plotted for three independent experiments. The expression in control cells was set as 1, and the statistical significance and error bars representing SEM are shown. (E) The levels of collagen α1(I) mRNA (left) and collagen α2(I) mRNA (right) were estimated in control and DN-DES-expressing cells after transcriptional blocking with actinomycin D for the indicated times. The expression at time zero was set as 1. The error bars represent SEM estimated from three independent experiments. (F) DN-DES decreases collagen expression in scleroderma fibroblasts. Cellular (lanes 1 and 2) and secreted (lanes 3 and 4) levels of collagen polypeptides of cells expressing DN-DES (lanes 1 and 3) or control protein (lanes 2 and 4) were analyzed by Western blotting. Fibronectin is shown as a loading control. (G) Reduced collagen mRNA levels in scleroderma fibroblasts expressing DN-DES. Total RNA from cells expressing DN-DES (lane 1) or control protein (lane 2) were analyzed by RT-PCR for collagen α1(I), collagen α2(I), and GAPDH mRNAs.
We then assessed the effect of the dominant-negative desmin on collagen expression in two collagen-producing cell types. In human lung fibroblasts, the dominant-negative desmin significantly decreased cellular and secreted levels of collagen α1(I) and α2(I) polypeptides compared to cells infected with control virus (Fig. 9C, compare lanes 1 and 3 to lanes 2 and 4). The level of fibronectin was not affected, suggesting the specificity of the effect on collagen production.
The steady-state levels of both collagen mRNAs, α1(I) and α2(I), were dramatically reduced in lung fibroblasts expressing the dominant-negative desmin (Fig. 9D). In order to determine if this is due to a faster decay of collagen mRNAs, we measured the stability of collagen α1(I) and α2(I) mRNAs in cells expressing dominant-negative desmin. Figure 9E shows that collagen α1(I) and α2(I) mRNAs decayed with half-lives of 5 h and 9 h, respectively, compared to 18 h and 24 h in control cells. This result is similar to the result obtained with vimentin knockout MEFs (Fig. 7) and IDPN-treated cells (Fig. 8).
We also tested the effect of dominant-negative desmin in scleroderma skin fibroblasts. Markedly reduced cellular and secreted levels of collagen α1(I) and α2(I) polypeptides were found (Fig. 9F, compare lanes 2 and 4 with lanes 1 and 3). Cellular and secreted levels of fibronectin were again unaffected. Decreased collagen protein production was correlated with the decreased levels of collagen mRNAs (Fig. 9G).
Taken together, these results strongly indicate that the integrity of vimentin filaments is necessary for the stability of collagen α1(I) and α2(I) mRNAs.
Vimentin filaments stabilize collagen mRNAs that are not actively engaged in translation.
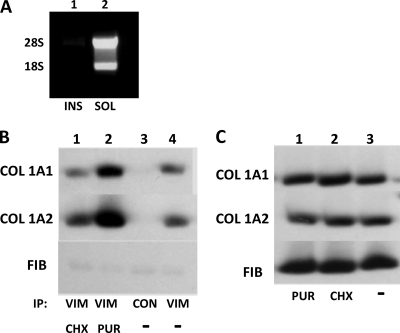
Our results suggest that vimentin filaments regulate the stability of collagen mRNAs but do not indicate if these mRNAs are translated. When we fractionated polysomes on sucrose gradients (9) and analyzed the fractions for vimentin by Western blotting, we could not detect any vimentin in the fractions containing polysomes (not shown). To further corroborate the absence of ribosomes on vimentin filaments, we fractionated cells into soluble and insoluble fractions as described in the legend to Fig. 2 and assessed if any rRNA could be found in the insoluble fraction, which contains the majority of vimentin. No rRNA was found in the insoluble fraction (Fig. 10A), suggesting there was no association of ribosomes with vimentin filaments and that the fraction of collagen mRNAs that associates with these filaments is not translated.
Fig. 10.
Vimentin associates with untranslated collagen mRNAs. (A) Absence of rRNA in the insoluble fraction containing filamentous vimentin. Soluble (lane 2) and insoluble (lane 1) fractions were prepared, and total RNA was extracted and analyzed by agarose gel electrophoresis and ethidium bromide staining. (B) Dissociation of polysomes increases the association of collagen mRNAs with vimentin. Shown is immunoprecipitation with antivimentin antibody (lanes 1, 2, and 4) or no antibody (lane 3) from human lung fibroblasts treated with cycloheximide (CHX) (lane 1) or puromycin (PUR) (lane 2) or from untreated cells (−) (lanes 3 and 4). The immunoprecipitated material was analyzed for collagen α1(I), collagen α2(I), and fibronectin mRNAs by RT-PCR. (C) Cycloheximide and puromycin do not change the total levels of collagen mRNAs. Shown is RT-PCR analysis of collagen α1(I), collagen α2(I), and fibronectin mRNAs in cells treated with puromycin (lane 1) or cycloheximide (lane 2) or untreated cells (lane 3).
If vimentin filaments preferentially bind collagen mRNAs that are not being translated, then dissociation of polysomes should increase their loading onto vimentin filaments. To assess this, we performed immunoprecipitation with vimentin antibody in cells treated with puromycin and cycloheximide and analyzed for pulldown of collagen mRNAs. Twice as much collagen mRNA was pulled down from the puromycin-treated cells than from the cells treated with cycloheximide or from untreated cells (Fig. 10C, compare lane 2 with lanes 1 and 4). The total level of collagen mRNAs was unaffected by cycloheximide and puromycin treatment, suggesting the redistribution of collagen mRNAs onto vimentin filaments upon dissociation of polysomes. Together, the results suggest that vimentin filaments stabilize collagen mRNAs that are not actively engaged in translation.
DISCUSSION
Recent work from our laboratory reported that binding of LARP6 to the conserved 5′SL of collagen mRNAs associates collagen mRNAs with filaments composed of nonmuscle myosin and regulates translation (10). In the present study, we show that (i) collagen mRNAs that are not translating associate with vimentin intermediate filaments, (ii) this association is through the interaction of the La domain of LARP6 with the filaments, (iii) the association with vimentin filaments stabilizes collagen mRNAs, and (iv) one of the roles of vimentin in mesenchymal cells may be to promote collagen synthesis by increasing the level of collagen mRNAs.
There is now substantial evidence that in different cell types mRNAs associate with the cytoskeleton (2). Of the three major filamentous systems, the association of mRNAs with microfilaments and microtubules is well established. There is some evidence that mRNAs may also associate with intermediate filaments. Ultrastructural in situ hybridization study of the distribution of poly(A)+ mRNAs showed about 15% of total poly(A)+ mRNAs are found close to or associated with vimentin filaments (3). In another study, mRNAs for calcitonin gene-related peptide in axons of neurons and interleukin 1β (IL-1β) in monocytes were found associated with vimentin filaments (8, 13). Unlike that of microtubules and microfilaments, however, the functional significance of association of mRNAs with intermediate filaments is not understood. This study gives the first example of a functionally relevant interaction of collagen mRNAs with vimentin intermediate filaments.
Vimentin binding to and stabilizing collagen mRNAs was supported by evidence obtained using different approaches. First, using RNA immunoprecipitation and cellular-fractionation methods, we showed that collagen mRNAs exhibit specific interaction with vimentin filaments (Fig. 1 and 2). Second, using immunofluorescence coupled with RNA FISH, we showed that a significant proportion of collagen mRNAs are colocalized with vimentin filaments (Fig. 3). Third, fibroblasts from vimentin knockout mice have reduced collagen synthesis due to the decreased half-life of collagen mRNAs (Fig. 7). Fourth, disruption of vimentin filaments using either a drug or a dominant-negative mutant of desmin resulted in faster decay of collagen mRNAs (Fig. 8 and 9).
We also provided insight into the mechanism of the interaction between collagen mRNAs and vimentin filaments. The key feature of this interaction is that it is dependent on a cis-acting RNA element of collagen mRNAs, the 5′SL, and its cognate RNA binding protein, LARP6. Possession of cis-acting RNA elements governing interactions with the cytoskeleton is an important characteristic of all mRNAs that localize with the cytoskeleton (26). These cis-acting elements are almost always located in the 3′ UTR. Collagen mRNAs are unique in this respect, because their cis-acting sequence is in the 5′ UTR. That collagen mRNAs require LARP6 for binding vimentin filaments was corroborated by multiple lines of evidence. LARP6 coprecipitates and colocalizes with vimentin, knockdown of LARP6 decreases the association of collagen mRNAs with vimentin, and the interaction of LARP6 with vimentin is 5′SL dependent. Thus, this is a novel function of LARP6 in posttranscriptional regulation of collagen expression.
The lack of association of polysomes with vimentin and the absence of rRNA in the insoluble fraction support the notion that vimentin filaments preferentially bind nontranslating collagen mRNAs. Collagen mRNAs that are released upon dissociation of polysomes seem to be targeted to vimentin filaments (Fig. 10). Thus, vimentin filaments could play a role in storage of untranslated collagen mRNAs. We estimated that 30 to 50% of collagen mRNAs colocalize with vimentin. Based on the polysomal profile of collagen mRNA (9, 10), about 50% of collagen mRNAs are engaged in translation; thus, the rest of the collagen mRNAs appear to be primarily stored on vimentin filaments.
We mapped the domain of LARP6 that binds vimentin to the La domain. The La domain is well conserved in all LARPs (55), so it is possible that other LARPs also bind vimentin filaments. This remains to be verified, but it may provide insight into the functions of other LARP family members. However, no other LARP family member has a high-affinity mRNA target.
The most relevant feature of the interaction of collagen α1(I) and α2(I) mRNAs with vimentin filaments is that this interaction stabilizes collagen mRNAs. Although regulation of the stability of collagen mRNAs has emerged as the predominant means of high-level synthesis of type I collagen, the mechanism of stabilization was only partially understood (16, 46, 47, 52). αCP binds the C-rich region in the 3′ UTR of collagen α1(I) mRNA and contributes to its prolonged half-life (32, 34, 47). This study suggests that an additional mechanism for stabilizing collagen α1(I) and α2(I) mRNAs may utilize the 5′ UTR. The precise mechanism by which vimentin filaments stabilize collagen α1(I) and α2(I) mRNAs is not yet clear. One possibility is that vimentin filaments prevent access of the RNA degradation machinery to collagen mRNAs.
Our finding that vimentin-deficient fibroblasts exhibited markedly reduced collagen synthesis is consistent with the impaired wound healing observed in vimentin knockout mice. In vimentin-deficient mice, wound healing is dramatically impaired due to defective fibroblast invasion into the wound, with the resultant delayed wound contraction (14, 15). Based on our results, we speculate that reduced collagen production by vimentin-deficient fibroblasts may have also contributed to the observed delayed-wound-healing phenotype. Congruent with our hypothesis, the initial collagen deposition in the proliferative phase of wound healing is an important step for the subsequent fibroblast invasion (44). Wound-healing experiments using vimentin-deficient mice, with an emphasis on the amount of collagen deposited, are needed to verify this hypothesis.
From a therapeutic point of view, considering the central role of the increased collagen expression in the development of tissue fibrosis, vimentin filaments may be a target for antifibrotic therapy. The potential utility of targeting vimentin is highlighted by our findings that disruption of vimentin reduced synthesis of type I collagen.
In conclusion, we propose a model for posttranscriptional regulation of collagen expression where LARP6-bound collagen mRNAs are stored and stabilized by vimentin filaments and are available for recruitment by the translational machinery.
ACKNOWLEDGMENTS
This work was supported by NIH grant 5R01DK059466-08 to B.S. and an American Heart Association grant to A.A.C.
Special thanks are due to Amber L. Wells and Robert H. Singer, Albert Einstein College of Medicine, NY, for their help with RNA FISH experiments and for critical reading of the manuscript. We thank our laboratory manager, Lela Stefanovic, for her excellent technical assistance in this project. We also thank Robert Evans, University of Colorado Health Sciences Center, for his kind gift of Vim−/− and Vim+/+ MEFs.
Footnotes
Published ahead of print on 11 July 2011.
REFERENCES
- 1. Anonymous. 2000. The American Association for the Study of Liver Diseases (AASLD) 51st annual meeting and postgraduate courses. October 27-31, 2000, Dallas, Texas, U. S. A. Abstracts. Hepatology 32:163A–655A [PubMed] [Google Scholar]
- 2. Bassell G., Singer R. H. 1997. mRNA and cytoskeletal filaments. Curr. Opin. Cell Biol. 9:109–115 [DOI] [PubMed] [Google Scholar]
- 3. Bassell G. J., Powers C. M., Taneja K. L., Singer R. H. 1994. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J. Cell Biol. 126:863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ben-Ari Y., et al. 2010. The life of an mRNA in space and time. J. Cell Sci. 123:1761–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bitterman P. B., Henke C. A. 1991. Fibroproliferative disorders. Chest 99:81S–84S [DOI] [PubMed] [Google Scholar]
- 6. Blikstad I., Lazarides E. 1983. Vimentin filaments are assembled from a soluble precursor in avian erythroid cells. J. Cell Biol. 96:1803–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bloemendal H., Willemsen M., Groenewoud G., Oomen P. 1985. Isolation of the intermediate filament protein vimentin by chromatofocusing. FEBS Lett. 180:181–184 [DOI] [PubMed] [Google Scholar]
- 8. Böcker U., et al. 2001. Expression and localization of IL-1beta mRNA is interrelated with cytoskeletal rearrangement in monocytes stimulated by adherence: a light microscopy in situ hybridization study. Immunol. Cell Biol. 79:444–453 [DOI] [PubMed] [Google Scholar]
- 9. Cai L., Fritz D., Stefanovic L., Stefanovic B. 2010. Binding of LARP6 to the conserved 5′ stem-loop regulates translation of mRNAs encoding type I collagen. J. Mol. Biol. 395:309–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai L., Fritz D., Stefanovic L., Stefanovic B. 2010. Nonmuscle myosin-dependent synthesis of type I collagen. J. Mol. Biol. 401:564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen A. Y., Zirwas M. J., Heffernan M. P. 2010. Nephrogenic systemic fibrosis: a review. J. Drugs Dermatol. 9:829–834 [PubMed] [Google Scholar]
- 12. Clarke E. J., Allan V. 2002. Intermediate filaments: vimentin moves in. Curr. Biol. 12:R596–R598 [DOI] [PubMed] [Google Scholar]
- 13. Denis-Donini S., Branduardi P., Campiglio S., Carnevali M. D. 1998. Localization of calcitonin gene-related peptide mRNA in developing olfactory axons. Cell Tissue Res. 294:81–91 [DOI] [PubMed] [Google Scholar]
- 14. Eckes B., et al. 2000. Impaired wound healing in embryonic and adult mice lacking vimentin. J. Cell Sci. 113:2455–2462 [DOI] [PubMed] [Google Scholar]
- 15. Eckes B., et al. 1998. Impaired mechanical stability, migration and contractile capacity in vimentin-deficient fibroblasts. J. Cell Sci. 111:1897–1907 [DOI] [PubMed] [Google Scholar]
- 16. Eckes B., Mauch C., Huppe G., Krieg T. 1996. Differential regulation of transcription and transcript stability of pro-alpha 1(I) collagen and fibronectin in activated fibroblasts derived from patients with systemic scleroderma. Biochem. J. 315:549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eriksson J. E., et al. 2009. Introducing intermediate filaments: from discovery to disease. J. Clin. Invest. 119:1763–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Evans R. M. 1998. Vimentin: the conundrum of the intermediate filament gene family. Bioessays 20:79–86 [DOI] [PubMed] [Google Scholar]
- 19. Femino A. M., Fay F. S., Fogarty K., Singer R. H. 1998. Visualization of single RNA transcripts in situ. Science 280:585–590 [DOI] [PubMed] [Google Scholar]
- 20. Friedman S. L. 2010. Evolving challenges in hepatic fibrosis. Nat. Rev. Gastroenterol. Hepatol. 7:425–436 [DOI] [PubMed] [Google Scholar]
- 21. Fritz D., Stefanovic B. 2007. RNA-binding protein RBMS3 is expressed in activated hepatic stellate cells and liver fibrosis and increases expression of transcription factor Prx1. J. Mol. Biol. 371:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galigniana M. D., et al. 1998. Heat shock protein 90-dependent (geldanamycin-inhibited) movement of the glucocorticoid receptor through the cytoplasm to the nucleus requires intact cytoskeleton. Mol. Endocrinol. 12:1903–1913 [DOI] [PubMed] [Google Scholar]
- 23. Grünwald D., Shenoy S. M., Burke S., Singer R. H. 2008. Calibrating excitation light fluxes for quantitative light microscopy in cell biology. Nat. Protoc. 3:1809–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grünwald D., Singer R. H., Czaplinski K. 2008. Cell biology of mRNA decay. Methods Enzymol. 448:553–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He T. C., et al. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 95:2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hesketh J. E. 1996. Sorting of messenger RNAs in the cytoplasm: mRNA localization and the cytoskeleton. Exp. Cell Res. 225:219–236 [DOI] [PubMed] [Google Scholar]
- 27. Holwell T. A., Schweitzer S. C., Evans R. M. 1997. Tetracycline regulated expression of vimentin in fibroblasts derived from vimentin null mice. J. Cell Sci. 110:1947–1956 [DOI] [PubMed] [Google Scholar]
- 28. Kania G., Blyszczuk P., Eriksson U. 2009. Mechanisms of cardiac fibrosis in inflammatory heart disease. Trends Cardiovasc. Med. 19:247–252 [DOI] [PubMed] [Google Scholar]
- 29. Kivirikko K. I. 1998. Collagen biosynthesis: a mini-review cluster. Matrix Biol. 16:355–356 [DOI] [PubMed] [Google Scholar]
- 30. Krupsky M., Kuang P. P., Goldstein R. H. 1997. Regulation of type I collagen mRNA by amino acid deprivation in human lung fibroblasts. J. Biol. Chem. 272:13864–13868 [DOI] [PubMed] [Google Scholar]
- 31. Larson D. R., Singer R. H., Zenklusen D. 2009. A single molecule view of gene expression. Trends Cell Biol. 19:630–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindquist J. N., Kauschke S. G., Stefanovic B., Burchardt E. R., Brenner D. A. 2000. Characterization of the interaction between alphaCP(2) and the 3′-untranslated region of collagen alpha1(I) mRNA. Nucleic Acids Res. 28:4306–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lindquist J. N., Marzluff W. F., Stefanovic B. 2000. Fibrogenesis. III. Posttranscriptional regulation of type I collagen. Am. J. Physiol. Gastrointest. Liver Physiol. 279:G471–G476 [DOI] [PubMed] [Google Scholar]
- 34. Lindquist J. N., Parsons C. J., Stefanovic B., Brenner D. A. 2004. Regulation of alpha1(I) collagen mRNA decay by interactions with alphaCP at the 3′-untranslated region. J. Biol. Chem. 279:23822–23829 [DOI] [PubMed] [Google Scholar]
- 35. Lindquist J. N., Stefanovic B., Brenner D. A. 2000. Regulation of collagen alpha1(I) expression in hepatic stellate cells. J. Gastroenterol. 35:80–83 [PubMed] [Google Scholar]
- 36. Parsons C. J., et al. 2011. Mutation of the 5′ untranslated region stem-loop structure inhibits {alpha}1(i) collagen expression in vivo. J. Biol. Chem. 286:8609–8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parsons C. J., Takashima M., Rippe R. A. 2007. Molecular mechanisms of hepatic fibrogenesis. J. Gastroenterol. Hepatol 22Suppl. 1:S79–S84 [DOI] [PubMed] [Google Scholar]
- 38. Povero D., et al. 2010. Liver fibrosis: a dynamic and potentially reversible process. Histol. Histopathol. 25:1075–1091 [DOI] [PubMed] [Google Scholar]
- 39. Raats J. M., et al. 1992. Biochemical and structural aspects of transiently and stably expressed mutant desmin in vimentin-free and vimentin-containing cells. Eur. J. Cell Biol. 58:108–127 [PubMed] [Google Scholar]
- 40. Ricupero D. A., et al. 2001. Phosphatidylinositol 3-kinase-dependent stabilization of alpha1(I) collagen mRNA in human lung fibroblasts. Am. J. Physiol. Cell Physiol. 281:C99–C105 [DOI] [PubMed] [Google Scholar]
- 41. Rishikof D. C., Kuang P. P., Poliks C., Goldstein R. H. 1998. Regulation of type I collagen mRNA in lung fibroblasts by cystine availability. Biochem. J. 331:417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sjöberg G., et al. 1999. A missense mutation in the desmin rod domain is associated with autosomal dominant distal myopathy, and exerts a dominant negative effect on filament formation. Hum. Mol. Genet. 8:2191–2198 [DOI] [PubMed] [Google Scholar]
- 43. Soellner P., Quinlan R. A., Franke W. W. 1985. Identification of a distinct soluble subunit of an intermediate filament protein: tetrameric vimentin from living cells. Proc. Natl. Acad. Sci. U. S. A. 82:7929–7933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stadelmann W. K., Digenis A. G., Tobin G. R. 1998. Physiology and healing dynamics of chronic cutaneous wounds. Am. J. Surg. 176:26S–38S [DOI] [PubMed] [Google Scholar]
- 45. Stefanovic B., Hellerbrand C., Brenner D. A. 1995. Post-transcriptional regulation of collagen alpha 1(I) mRNA in hepatic stellate cells. Nucleic Acids Symp. Ser. 33:212–214 [PubMed] [Google Scholar]
- 46. Stefanovic B., Hellerbrand C., Brenner D. A. 1999. Regulatory role of the conserved stem-loop structure at the 5′ end of collagen alpha1(I) mRNA. Mol. Cell. Biol. 19:4334–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stefanovic B., et al. 1997. Posttranscriptional regulation of collagen alpha1(I) mRNA in hepatic stellate cells. Mol. Cell. Biol. 17:5201–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stefanovic B., Lindquist J., Brenner D. A. 2000. The 5′ stem-loop regulates expression of collagen alpha1(I) mRNA in mouse fibroblasts cultured in a three-dimensional matrix. Nucleic Acids Res. 28:641–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stefanovic B., Schnabl B., Brenner D. A. 2002. Inhibition of collagen alpha 1(I) expression by the 5′ stem-loop as a molecular decoy. J. Biol. Chem. 277:18229–18237 [DOI] [PubMed] [Google Scholar]
- 50. Stefanovic L., Stefanovic B. 2006. Mechanism of direct hepatotoxic effect of KC chemokine: sequential activation of gene expression and progression from inflammation to necrosis. J. Interferon Cytokine Res. 26:760–770 [DOI] [PubMed] [Google Scholar]
- 51. Tsukada S., Parsons C. J., Rippe R. A. 2006. Mechanisms of liver fibrosis. Clin. Chim. Acta 364:33–60 [DOI] [PubMed] [Google Scholar]
- 52. Tsukada S., Westwick J. K., Ikejima K., Sato N., Rippe R. A. 2005. SMAD and p38 MAPK signaling pathways independently regulate alpha1(I) collagen gene expression in unstimulated and transforming growth factor-beta-stimulated hepatic stellate cells. J. Biol. Chem. 280:10055–10064 [DOI] [PubMed] [Google Scholar]
- 53. Valgeirsdóttir S., et al. 1998. PDGF induces reorganization of vimentin filaments. J. Cell Sci. 111:1973–1980 [DOI] [PubMed] [Google Scholar]
- 54. Wang R., Li Q. F., Anfinogenova Y., Tang D. D. 2007. Dissociation of Crk-associated substrate from the vimentin network is regulated by p21-activated kinase on ACh activation of airway smooth muscle. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L240–L248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wolin S. L., Cedervall T. 2002. The La protein. Annu. Rev. Biochem. 71:375–403 [DOI] [PubMed] [Google Scholar]
- 56. Yamada N. A., Castro A., Farber R. A. 2003. Variation in the extent of microsatellite instability in human cell lines with defects in different mismatch repair genes. Mutagenesis 18:277–282 [DOI] [PubMed] [Google Scholar]
- 57. Yamada Y., Mudryj M., de Crombrugghe B. 1983. A uniquely conserved regulatory signal is found around the translation initiation site in three different collagen genes. J. Biol. Chem. 258:14914–14919 [PubMed] [Google Scholar]
- 58. Yu K. R., et al. 1994. Truncated desmin in PtK2 cells induces desmin-vimentin-cytokeratin coprecipitation, involution of intermediate filament networks, and nuclear fragmentation: a model for many degenerative diseases. Proc. Natl. Acad. Sci. U. S. A. 91:2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zenklusen D., Larson D. R., Singer R. H. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15:1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]