Abstract
Reversible methylation of lysine residues has emerged as a central mechanism for epigenetic regulation and is a component of the “histone code,” which engenders histones with gene regulatory information. KDM4A is a histone demethylase that targets tri- and dimethylation marks on histone H3 lysines 9 and 36. While the abundance of KDM4A oscillates in the cell cycle, little is known how this enzyme is regulated to achieve targeted effects on specific histone residues in chromatin. Here, we report that a previously unstudied SCFFBXO22 ubiquitin ligase complex controls the activity of KDM4A by targeting it for proteasomal turnover. FBXO22 functions as a receptor for KDM4A by recognizing its catalytic JmjN/JmjC domains via its intracellular signal transduction (FIST) domain. Modulation of FBXO22 levels by RNA interference or overexpression leads to increased or decreased levels of KDM4A, respectively. Changes in KDM4A abundance correlate with alterations in histone H3 lysine 9 and 36 methylation levels, and transcription of a KDM4A target gene, ASCL2. Taken together, these results demonstrate that SCFFBXO22 regulates changes in histone H3 marks and cognate transcriptional control pathways by controlling KDM4A levels, and they suggest a potential role for FBXO22 in development, differentiation, and disease through spatial and temporal control of KDM4A activity.
INTRODUCTION
Histones undergo many posttranslational modifications, including acetylation, methylation, phosphorylation, and ubiquitylation (8, 17, 41). These modifications are governed by enzymes that can add or remove these marks, and they are interpreted by proteins containing specific recognition modules whose action on chromatin alters the transcriptional properties or fate of the cell. Histone methylation typically occurs on lysine and arginine residues, and the extent (mono-, di-, or trimethylation) and position of methylation on histones are associated with often opposing biological outcomes (52). Histone methylation is a posttranslational modification regulated by two classes of proteins, histone methyltransferases and histone demethylases, which can add or remove the mark, respectively, often in a spatially and temporally regulated manner. As a result of its role in key cellular processes, misregulation of histone methylation is associated with human diseases, including cancer (33). Despite clear biological roles for histone methyltransferases and demethylases, to date little is known about how these enzymes are regulated, especially at the posttranslational level.
KDM4A (also known as JMJD2A and JHDM3A) belongs to a family of four related proteins—KDM4A, KDM4B, KDM4C, and KDM4D—all of which possess the ability to demethylate histone H3 lysine 9 and 36 tri- and dimethylation (H3K9me3/2 and H3K36me3/2) marks to varied extents and efficiencies (10, 14, 19, 23, 36, 47). They have also been reported to target methylation on H1.4K26, a linker histone isotype associated with transcriptional repression (43). The KDM4 proteins are Fe(II)- and α-ketoglutarate-dependent oxygenases that contain essential catalytic JmjN and JmjC domains, as well as PHD and Tudor domains (except for KDM4D) (22). The tandem Tudor domains located near the C terminus of KDM4A have been shown to bind methylated H3K4 and H4K20 peptides in vitro, and this binding specificity has been speculated to be important for targeting KDM4A to specific chromatin sites in vivo (16, 21, 26). Before its identification as the first trimethylation-targeting histone demethylase, KDM4A was initially identified as a transcriptional repressor that interacts with nuclear receptor corepressor (N-CoR) to selectively repress its target, the achaete scute-like homolog 2 gene (ASCL2) (51). It has also been reported to interact with class I histone deacetylases and Rb to repress E2F promoters (15).
In addition, KDM4A plays a critical role in genome replication and stability (6, 47). Overexpression of KDM4A leads to accelerated passage through S phase, correlating with increased chromatin accessibility, in human cells, while mutation of the Caenorhabditis elegans KDM4A ortholog leads to reduced rates of S phase and DNA damage (6). Interestingly, KDM4A abundance is regulated in a cell cycle-dependent manner in mammalian tissue culture cells; it is abundant in G1- and S-phase cells, but its levels are greatly reduced in G2 and mitotic cells. While alterations in KDM4A levels do not appear to reflect transcriptional control mechanisms, the pathways regulating KDM4A abundance are unknown. To date, mechanisms that control the abundance of demethylases are limited to SMCX orthologs in yeast, which employ the CNOT RING-finger E3 to control their activity (30).
Here, we demonstrate that KDM4A abundance is under the control of the ubiquitin-proteasome system, and its turnover is dependent upon the SCFFBXO22 ubiquitin ligase. SCF (SKP1-CUL1-F-box) complexes are modular ubiquitin ligases wherein the C-terminal cullin homology domain together with the RING protein RBX1 serves to recruit charged E2-conjugating enzymes, while the F-box protein serves as the specificity factor (34). F-box proteins bind to SKP1 via the F-box motif and to substrates with additional protein interaction domains. FBXO22 is a previously uncharacterized F-box protein with no known substrates. The C terminus of FBXO22 contains a partial FIST (F-box and intracellular signal transduction) domain, which was identified bioinformatically as a domain conserved from bacteria to eukarya but for which no function, structure, or target is known (7). Through various in vivo and in vitro assays, we show that FBXO22 is the substrate recognition subunit of the SCFFBXO22 complex that polyubiquitylates KDM4A, targeting it for proteasome-mediated proteolysis. We show that changes in FBXO22 levels by RNA interference (RNAi) or overexpression can affect KDM4A protein levels, and correspondingly, lead to changes in H3K9me3 and H3K36me3 histone marks as well as changes in transcriptional levels of KDM4A's target gene, ASCL2. This study shows that by ubiquitin-mediated targeting of a histone demethylase for proteasomal degradation, the SCFFBXO22 complex regulates changes in histone marks and transcriptional levels of a target gene whose expression is under the control of KDM4A, adding a new dimension to our understanding of how chromatin marks are dynamically regulated.
MATERIALS AND METHODS
Cell culture, drug treatments, cell synchronization, plasmids, and siRNA transfection.
HeLa, HEK293T, and 293T-Rex (Invitrogen) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 incubator. MG132 (Calbiochem), cycloheximide (Sigma), and MLN4924 (Millennium Pharmaceuticals) were resuspended in dimethyl sulfoxide (DMSO; Sigma). Bortezomib (Millennium Pharmaceuticals) and doxycycline (Sigma) were resuspended in sterile phosphate-buffered saline (PBS). Unless otherwise stated, MG132 was used at a final concentration of 10 μM; cycloheximide was used at a final concentration of 100 μg/ml; MLN4924 was used at a concentration of 1 μM; bortezomib was used at a final concentration of 500 nM; doxycycline was used at a final concentration of 4 μg/ml. Cells were synchronized in G1/S by double thymidine treatment and in G2/M by nocodazole treatment. To obtain G1/S synchronized cells, cells were incubated in 2 mM thymidine for 18 h, washed with PBS twice, and supplemented with fresh medium to release the cells for 9 h, followed by an additional treatment of 2 mM thymidine for 18 h before harvesting. To obtain G2/M cells for subcellular fractionation, double thymidine-treated cells were washed twice and released into medium containing 100 ng/ml nocodazole for 10 h before harvesting. To obtain G2/M cells for immunoprecipitation (IP), cells were treated with 100 ng/ml nocodazole for 12 to 16 h before harvesting. Sequence-verified open reading frame clones in pDONR223 or pENTR/D-TOPO were recombined into the following Gateway destination vectors: MSCV-N-Flag-HA-IRES-PURO (long terminal repeat [LTR]-driven expression), CMV-N-Flag-HA, CMV-N-Flag, and CMV-N-MYC, using λ recombinase. Retro- and lentiviruses were packaged in HEK293T cells with their respective helper plasmids and used to infect the indicated cell lines. Selection with 1 μg/ml puromycin was carried out for at least 72 h to obtain stable cell lines. For plasmid transfection, the indicated quantity of DNA was transfected using Lipofectamine 2000 (Invitrogen) for HeLa cells or TransIT-293 (Mirus) for HEK293T cells according to the manufacturers' specifications. Transfected cells were harvested ∼48 h posttransfection for further analysis. For small interfering RNA(siRNA) transfection, 30 nM siRNA was transfected using Lipofectamine RNAiMAX according to the manufacturer's specifications, and cells were harvested ∼72 h posttransfection. Three FBXO22 siRNAs (Dharmacon) targeting different regions of the FBXO22 transcript were used in this study; FBXO22 siRNA_1, 5′-GCACCUUCGUGUUGAGUAA-3′; FBXO22 siRNA_2, 5′-GGUGGGAGCCAGUAAUUAU-3′; FBXO22 siRNA_3, 5′-GUUCGCAUCUUACCACAUA-3′. KDM4A shRNAs were purchased from Open Biosystems in the pLKO.1 vector targeting the following sequences (shKDM4A_1, GCTGCAGTATTGAGATGCTAA; shKDM4A_2, GCACCGAGTTTGTCTTGAAAT; shKDM4A_3, GCCTTGGATCTTTCTGTGAAT). Control shRNA was a pLKO.1 vector containing shRNA sequence targeting green fluorescent protein (GFP; Open Biosystems).
IP, mass spectrometry, and proteomic analysis.
Whole-cell extracts were obtained by lysing cells in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, supplemented with protease inhibitors [Roche]) for 30 min on ice. Lysates were cleared by centrifugation at 4°C for 10 min at 14,000 rpm. Approximately 5 mg of lysate was used for endogenous IP and incubated with 1.5 μg of the indicated antibody or control IgG overnight at 4°C. Protein A or G resin (25 μl) was added, and the IP reaction mixture was incubated for an additional 2 h at 4°C. Beads were washed with lysis buffer, and 2× SDS loading buffer was added to beads prior to boiling for 5 min. Samples were separated on an SDS-PAGE gel prior to immunoblot analysis. For IP of epitope-tagged proteins, agarose beads conjugated with hemagglutinin (HA) antibody (Sigma), or MYC antibody (Sigma) was used.
IP-mass spectrometry was performed as previously described (40). The IP elution was precipitated with trichloroacetic acid (TCA), trypsinized, loaded onto stage tips, and analyzed on an LTQ-Velos linear ion trap mass spectrometer (ThermoFinnigan). Peptides were identified using Sequest and analyzed using CompPASS (4, 40).
Antibodies.
Antibodies used in this work included the following: anti-FBXO22 (WH0026263M1; Sigma), anti-KDM4A (A300-861A; Bethyl Labs), anti-CUL1 (71-8700; Zymed Labs), anti-HA (MMS-101P [Covance] or sc-805 [Santa Cruz Biotechnology]), anti-FLAG (F1804; Sigma), anti-MYC (sc-40; Santa Cruz Biotechnology), antiactin (sc-47778; Santa Cruz Biotechnology), antitubulin (sc-55529; Santa Cruz Biotechnology), anti-H3 (39163; Active Motif), anti-H3K9me3 (ab8898; Abcam), anti-H3K9me2 (ab1220; Abcam), anti-H3K9me1 (ab8896; Abcam), anti-H3K36me3 (ab9050; Abcam), anti-H3K36me2 (ab9049; Abcam), anti-H3K36me1 (ab9048; Abcam), anti-H3K4me3 (07-473; Upstate), anti-H3K27me3 (07-449; Upstate Biotechnology), anti-PCNA (sc-56; Santa Cruz Biotechnology), anti-HSP90A (3670-1; Epitomics), cyclin E (sc-56310; Santa Cruz Biotechnology), cyclin B1 (sc-53236; Santa Cruz Biotechnology).
In vivo ubiquitylation assays.
Forty hours posttransfection, HEK 293T cells expressing HA-Ub and MYC-KDM4A were treated with MG132 for 4 h. After treatment, the cells were washed with PBS supplemented with 200 μM iodoacetamide and 10 mM N-ethylmaleimide (NEM; Sigma). Cells were lysed with lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, supplemented with protease inhibitors [Roche], 200 μM iodoacetamide, and 10 mM NEM) on ice for 30 min. Cleared lysates were quantified, and an equal amount of each lysate was used for immunoprecipitation with HA-agarose beads. Beads were washed with lysis buffer, and 2× SDS loading buffer was added to beads and boiled for 5 min before separation on an SDS-PAGE gel. Subsequent immunoblotting was performed using MYC antibody.
In vitro ubiquitylation assays.
Ubiquitylation assays were performed as described previously (18), with modifications. Briefly, a baculoviral vector expressing HA-KDM4A was transfected into Sf9 cells using a Bac-N-Blue transfection kit (Invitrogen), and Sf9 cells expressing HA-KDM4A were produced according to the manufacturer's protocol. The SCFFBXO22 complex was purified by immunoprecipitation from whole-cell lysates of HEK293T cells stably expressing FLAG-HA-FBXO22. For each reaction, purified SCFFBXO22 complexes and purified HA-KDM4A were incubated at 37°C for 1 h with or without purified E1 (GST-UBA1), E2 (His6-CDC34 and/or His6-UBCH5a), ATP, neddylation apparatus (NAE, NEDD8, UBC12, and ubiquitin. After the incubation, 2× SDS loading buffer was added, and samples were then separated by SDS-PAGE and probed with anti-HA antibody. The same procedure was done for FBXO22ΔF.
RNA extraction, reverse transcription, and qPCR.
Total RNA was obtained by using TRIzol reagent (Invitrogen) lysis, followed by chloroform extraction. RNA was then isolated by isopropanol precipitation, and the pellet was dissolved in RNase-free water. Any genomic DNA contamination was removed by treatment of RNA by using RNase-free DNase I (Ambion), and DNase I was then removed by addition of DNase inactivation reagent as stipulated by the manufacturer. Reverse transcription was performed using SuperScript III (Invitrogen) with 1 to 5 μg total RNA and oligo(dT) to prime the reaction. The cDNA was used for quantitative PCR (qPCR) by employing gene-specific primers and SYBR green for detection on a LightCycler 480 system (Roche). Primers specific to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used for normalization. Primer sequences were as follows: GAPDH_F, ATGCCTCCTGCACCACCAAC; GAPDH_R, GGGGCCATCCACAGTCTTCT; ASCL2_F, TTCCGCCTACTCGTCGGA; ASCL2_R, GCTGAGGCTCATAGGTCGA; FBXO22_F, CTCACTGAAGTAGGTCTTTTAG; FBXO22_R, CCAGCCAAGATGATATTCATATC; KDM4A_F, ATCTAGACTGTCAGTAGCCT; KDM4A_R, GGTATAGTGCAGGCTCAATA.
Cell fractionation.
Cell pellets were incubated with 5 packed cell volumes of hypotonic buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2) at 4°C, 15 min, on a rotator. The swollen cells were passed through a syringe attached to a 26.5-gauge needle 5 times to lyse the cells. The lysate was centrifuged at 3,000 rpm (10 min), and the supernatant was removed as the cytoplasmic fraction. The pellet containing the nucleus was lysed at 4°C for 30 min with nuclear lysis buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 1% NP-40, 2.5% glycerol). After centrifugation, the supernatant was removed as the nuclear fraction. The pellet containing the insoluble chromatin fraction was resuspended in MNase buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 2.5% glycerol, 3 mM CaCl2) and micrococcal nuclease (New England BioLabs) and incubated at 37°C for 10 min with occasional mixing. EDTA (pH 8.0) was added to a final concentration of 5 mM to stop the reaction, and the lysate was centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was removed as the chromatin fraction. All solutions were supplemented with protease inhibitors. Protein content in the fractions was quantified by Bradford assay, and equal amounts of protein were loaded for Western blot analysis.
Histone acid extraction.
Cell pellets were resuspended in PBS with 0.5% (vol/vol) Triton X-100 and protease inhibitors (Roche) at a cell density of approximately 107 cells/ml and rotated at 4°C for 10 min to lyse the cells. The lysate was centrifuged at 4°C for 10 min at 2,000 rpm, and the pellet was rinsed once in the extraction buffer. The histones in the insoluble chromatin pellet were then extracted in 0.2 N HCl overnight at 4°C on a rotator. The lysate was then centrifuged at 4°C for 10 min at 2,000 rpm, and the supernatant containing histones was removed and quantified by Bradford assay. Histone-enriched samples (1 to 5 μg) were subjected to immunoblotting.
ChIP.
Cells were grown on plates and cross-linked by the addition of formaldehyde to 1% (vol/vol) directly to the medium for 10 min. Glycine was then added to a final concentration of 125 mM for 5 min to neutralize the formaldehye. Medium was aspirated, and cells were washed once with cold PBS. Cells were scraped off the plates in PBS and pelleted by centrifugation. Cells were lysed in chromatin immunoprecipitation (ChIP) lysis buffer (50 mM HEPES [pH 7.5], 500 mM NaCl, 1 mM EDTA, 1% [vol/vol] Triton X-100, 0.1% [vol/vol] Na-deoxylcholate, with protease inhibitors [Roche]) for 30 min on ice, and sonicated to obtain approximately 200- to 600-bp DNA fragments (preoptimized in a cell-type-dependent manner). Sonicated lysates were centrifuged at 4°C for 5 min at 14,000 rpm, and supernatants were precleared by incubating with protein A/G-agarose beads for 1 h at 4°C. The protein concentration of the cleared lysates was determined by Bradford assay, and 100 to 500 μg protein was used for each ChIP reaction. ChIP-grade antibodies (1 to 5 μg) specific for H3K4me3 (07-473; Millipore), H3K9me3 (ab8898), H3K36me3 (ab9050), H3 (Active Motif 39163), or Rb IgG (Santa Cruz Biotechnology) were added to each ChIP reaction mixture and incubated with the cleared lysates at 4°C overnight on a rotator. A 25-μl aliquot of protein A/G-agarose-salmon sperm DNA (Millipore) was subsequently added to the reaction mixture for an additional 2 h at 4°C. The beads were washed twice each in (i) cold ChIP lysis buffer, (ii) cold RIPA buffer (50 mM HEPES [pH 7.5], 300 mM LiCl, 1 mM EDTA, 0.5% NP-40, 0.7% Na-deoxylcholate), and (iii) cold Tris-EDTA (TE) buffer with 50 mM NaCl. The ChIP-ed complexes were eluted from the beads by incubation in elution buffer (1% SDS in TE buffer) at 65°C for 15 min twice, and pooled. The supernatant, alongside an input (cleared lysate) sample diluted in elution buffer, was further incubated at 65°C overnight to de-cross-link the protein-DNA complexes. The elution was diluted 1:1 with TE buffer, treated with RNase A, proteinase K, and the DNA remaining was purified by phenol-chloroform-isoamyl alcohol (25:24:1; Sigma-Aldrich) extraction, followed by ethanol precipitation. The ChIP DNA sample was dissolved in 100 μl water and used for qPCR. ChIP samples were normalized against histone H3 levels. ChIP success and efficiency were judged by the percent ChIP-ed DNA relative to input levels (at least 0.1% for H3K4me3 and H3K36me3 ChIPs and at least 2% for H3K9me3 ChIP were obtained), and ChIP background levels were judged by comparison of histone ChIP to Rb IgG ChIP (>20-fold enrichment over Rb IgG). Primers used for ChIP were as follows: ASCL2_promoter_F, CAGGCAGCTGGGTCCACA; ASCL2_promoter_R, AGCCTTTGC CAAGCCGG; Chm4_pericentric_F, CTGTCCATAAAATATCGAAATACCCTA; Chm4_pericentric_R, TACAGTATATAAATACATAATTTGGGC (23).
RESULTS
KDM4A is regulated by the ubiquitin-proteasome pathway in a ubiquitin K48-dependent manner.
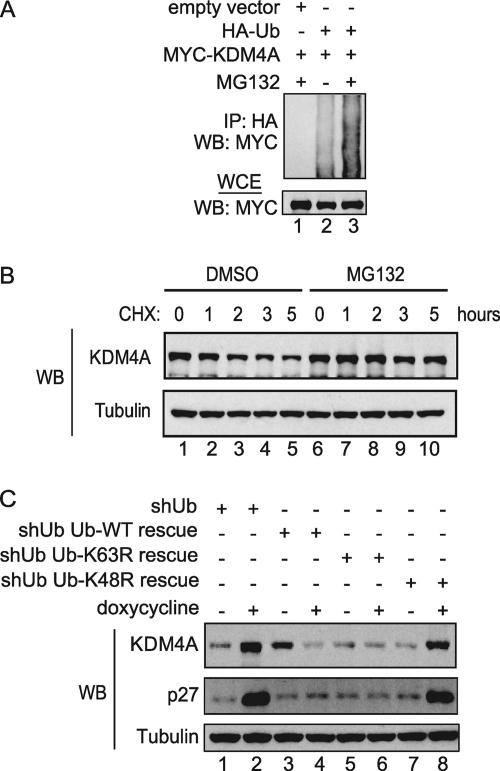
Since histone H3K9 and H3K36 methylation levels are regulated temporally and KDM4A transcript levels do not change significantly throughout the cell cycle, we hypothesized that KDM4A levels may be regulated by the proteasome. To examine whether KDM4A is ubiquitylated in vivo, we ectopically expressed both HA-ubiquitin and MYC-KDM4A in HEK293T cells in the presence or absence of the proteasome inhibitor MG132. Addition of MG132 enhanced the levels of ubiquitin linked to MYC-KDM4A (Fig. 1A). We then examined the effect of proteasome inhibition on the stability of KDM4A. HEK293T cells were treated with either DMSO or MG132 and collected at indicated time intervals after cycloheximide treatment (Fig. 1B). The half-life of KDM4A was ∼2 h in the absence of proteasome inhibition but was extended to >5 h in the presence of MG132, indicating that the proteasome is required for rapid turnover of KDM4A.
Fig. 1.
KDM4A's stability is regulated by the ubiquitin-proteasome system. (A) Immunoprecipitation (IP) and immunoblotting (WB) of lysates of HEK293T transiently transfected with the indicated vectors and treated with or without 10 μM MG132 for 4 h demonstrated that ubiquitylation of MYC-KDM4A is elevated in the presence of MG132. (B) Endogenous KDM4A protein levels were stabilized in a cycloheximide chase experiment performed in HEK293T cells when treated with MG132 and compared to control cells treated with vehicle (DMSO). HEK293T cells were treated with either DMSO or 10 μM MG132 for 4 h before the addition of 100 μg/ml of cycloheximide. Cells were harvested at the indicated times post-cycloheximide treatment, and lysates were immunoblotted using the indicated antibodies. (C) The indicated ubiquitin replacement U2OS cell lines were treated with either PBS or 4 μg/ml of doxycycline for 3 to 5 h, and cell lysates were immunoblotted using the indicated antibodies. Treatment of cells with doxycycline resulted in increased levels of endogenous KDM4A in cells with induced knockdown of endogenous ubiquitin as well as in induced cells with endogenous ubiquitin replaced with ubiquitin K48R mutant compared to their respective uninduced cells.
The canonical route of protein turnover via the proteasome involves the formation of K48-linked ubiquitin chains on target substrates, although it is now recognized that other chain linkages, including K11-linked chains, can also support protein turnover, while K63-linked chains primarily support altered protein function without degradation (3). In order to examine the chain linkage requirements for KDM4A turnover, we employed a previously described ubiquitin replacement cell line (U2OS) (48). The endogenous ubiquitin in these cells can be depleted by tetracycline-induced ubiquitin shRNA while simultaneously replacing it with either wild-type, K48R, or K63R ubiquitin. We treated these cells with either PBS or doxycycline for 3 to 5 days to allow for ubiquitin replacement and then examined the steady-state abundance of KDM4A. As anticipated, the abundance of KDM4A in cells depleted of ubiquitin was significantly elevated (Fig. 1C, lanes 1 and 2). In contrast, KDM4A levels in cells with endogenous ubiquitin depleted and replaced with either wild-type or K63R ubiquitin were essentially unaffected, indicating that K63 in ubiquitin was not required for control of KDM4A turnover (Fig. 1C, lanes 3 to 6). In contrast, the replacement of endogenous ubiquitin with ubiquitin K48R led to accumulation of KDM4A comparable to the levels seen in ubiquitin-depleted control cells (Fig. 1C, lanes 7 and 8). The same trend was observed for p27, a known substrate of the SCFSKP2 complex (9). Taken together, these data indicate that the abundance of KDM4A is controlled by the ubiquitin-proteasome system in a manner that requires K48 in ubiquitin.
KDM4A associates with components of the FBXO22-containing SCF E3 ligase complex.
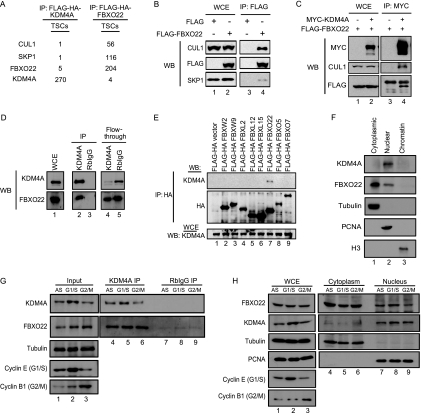
The identification of ubiquitin ligases for particular proteins is challenged by the frequent low affinity between E3s and their substrates. However, recent data (40) suggest that proteomic approaches can be used to recover even weakly associated proteins given the appropriate algorithms for removal of background proteins, which dominate IP-liquid chromatography-tandem mass spectrometry (IP-LC-MS/MS) data sets. To identify E3 ubiquitin ligases responsible for the turnover of KDM4A, we expressed Flag-HA (TAP)-tagged KDM4A in HEK293T cells under the control of an LTR promoter and used these cells for IP-LC-MS/MS, employing our previously described CompPASS platform (40). In this method, the DN score provides an assessment of the uniqueness, abundance, and frequency of candidate interacting proteins for particular bait proteins among a large series of parallel IP-MS/MS experiments, and DN scores of >1 are considered to be high-confidence candidate interacting proteins (HCIPs). Among the proteins with a DN score of >1 was FBXO22 (∼2.5), for which we identified 5 total spectral counts (TSCs) (Fig. 2A). FBXO22 is a previously uncharacterized F-box protein with no known targets. Consistent with KDM4A being associated with an intact SCFFBXO22 complex, we identified 1 peptide each for CUL1 and SKP1, core subunits of the SCF scaffold (Fig. 2A). Recent proteomic studies indicated that FBXO22 associates with CUL1 expressed at endogenous levels (5). Consistent with this, reciprocal tagging and IP-LC-MS/MS analysis of FBXO22 revealed abundant SKP1 and CUL1 peptides, and this was confirmed by immunoblotting (Fig. 2B). Moreover, we observed 4 TSCs for KDM4A (DN score, ∼3.5) (Fig. 2A). Given the absence of other E3s in association with KDM4A in this experiment, SCFFBXO22 emerged as a prime candidate for a KDM4A ubiquitin ligase.
Fig. 2.
KDM4A interacts with components of the SCFFBXO22 complex. (A) Mass spectrometry analyses of FLAG-HA-KDM4A and FLAG-HA-FBXO22 complexes independently showed that KDM4A interacts with FBXO22, SKP1, and CUL1 in HEK293T cells. (B) Ectopically expressed FLAG-FBXO22 immunoprecipitated with endogenous CUL1 and SKP1, affirming the interaction of FBXO22 with components of the SCF complex in HEK293T cells. Extracts of HEK293T cells transfected with either empty FLAG vector or vector expressing FLAG-FBXO22 were immunoprecipitated with anti-FLAG antibody, and immune complexes were blotted (WB) with the indicated antibodies. (C) Coexpression of FLAG-FBXO22 and MYC-KDM4A followed by IP and immunoblotting (WB) with the indicated antibodies verified the interaction of these two proteins together with CUL1 in HEK293T cells. (D) Endogenous IP of KDM4A in MLN4924-treated HEK293T cells confirmed the interaction of KDM4A and FBXO22 in HEK293T cells. HEK293T cells were treated with 1 μM MLN4924 for 4 h, and cell lysates were immunoprecipitated with either anti-KDM4A antibody or rabbit IgG. Immune complexes were blotted (WB) with either anti-KDM4A or anti-FBXO22 antibodies. (E) Eight FLAG-tagged F-box proteins were transiently expressed in HEK293T cells, and cell lysates were immunoprecipitated with anti-HA antibody. Immune complexes were blotted with the indicated antibodies to examine the interaction of each F-box with endogenous KDM4A. (F) Based on cell fractionation and subsequent immunoblotting of the various fractions with the indicated antibodies, endogenous FBXO22 was found in both the nucleus and cytoplasm, and endogenous KDM4A was found predominantly in the nucleus of HeLa cells. (G) HeLa cells were subjected to either double thymidine block to obtain G1/S cells or nocodazole treatment to obtain G2/M cells. The cells were then harvested, and immunoprecipitation was performed using either anti-KDM4A antibody or rabbit IgG. Subsequent immunoblotting with the indicated antibodies showed that the interaction of endogenous FBXO22 and KDM4A is similar in both G1/S and G2/M phases. (H) HeLa cells were arrested either in G1/S or G2/M phase by double thymidine block or by double thymidine block followed by release into nocodazole for 10 h to harvest late G2-to-G2/M cells with intact nuclei. When we used cell fractionation and subsequent immunoblotting of the various fractions with the indicated antibodies, endogenous FBXO22 did not display a significant difference in cellular localization at G1/S and G2/M phases.
In order to verify the interaction between KDM4A and FBXO22, we first performed transient transfections with epitope-tagged versions of FLAG-FBXO22 and MYC-KDM4A. Anti-MYC immune complexes contained FLAG-FBXO22 as well as endogenous CUL1, and these proteins were absent when the MYC-KDM4A vector was omitted from the transfection mixture (Fig. 2C). Next we performed endogenous IP experiments employing anti-KDM4A antibody. Endogenous FBXO22 was detected in anti-KDM4A immune complexes but was absent from control IgG complexes (Fig. 2D). Our mass spectrometry analysis suggested that KDM4A interacts specifically with FBXO22 and not with other F-box proteins. To validate this specificity, we ectopically expressed 8 different FLAG-tagged F-box proteins, including FBXO22, in HEK293T cells and examined their interaction with endogenous KDM4A (Fig. 2E). Only FBXO22 was found to interact with endogenous KDM4A in vivo.
Having validated that KDM4A and FBXO22 interact in vivo, we next examined the localization of FBXO22 and KDM4A in HeLa cells. As antibodies suitable for immunofluorescence of neither KDM4A nor FBXO22 was available, we employed cell fractionation approaches using asynchronous HeLa cells, marking cytoplasmic, nuclear, and chromatin fractions with tubulin, PCNA, and histone H3, respectively (Fig. 2F). While KDM4A was found exclusively in the nucleus, FBXO22 was found primarily in the cytoplasm but also to a lesser extent in the nucleus. Since KDM4A levels fluctuate during the cell cycle (6), we examined whether the FBXO22/KDM4A interaction is cell cycle dependent. Immunoprecipitation of endogenous KDM4A from extracts of HeLa cells arrested at either G1/S or G2/M phase revealed comparable levels of associated endogenous FBXO22, indicating that the interaction does not fluctuate detectably during the cell cycle (Fig. 2G). We also examined the localization of FBXO22 in cells synchronized at G1/S or in cells released into nocodazole for 10 h from the G1/S arrest point, giving primarily a G2/M population with intact nuclei based on maintenance of PCNA in the nuclear compartment (Fig. 2H). We found that the abundance of FBXO22 in the nucleus was comparable at both G1/S and G2/M, indicating the absence of dramatic cell cycle regulation of FBXO22 localization. We also note that depletion of FBXO22 by siRNA failed to result in defects in cell cycle progression (data not shown).
The C-terminal FIST-C domain of FBXO22 interacts with the catalytic domain of KDM4A.
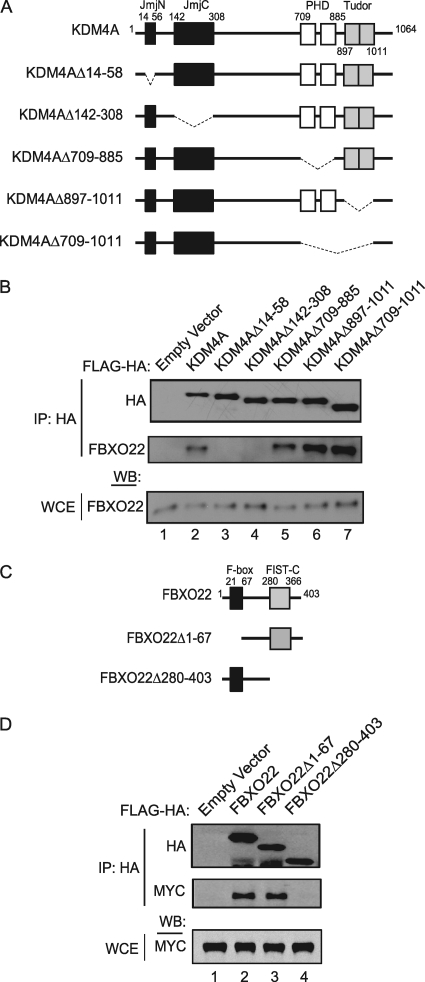
We next sought to identify regions on KDM4A and FBXO22 that are important for their interaction. First, we ectopically expressed KDM4A deletion mutants (Fig. 3A), which had various known domains removed, in HEK293T cells and examined the ability of each mutant to bind endogenous FBXO22 (Fig. 3B). FBXO22 maintained association with all of the mutants tested, with the exception of those lacking the JmjN or JmjC domains (lanes 3 and 4), which together make up the catalytic domain of the KDM4A demethylase.
Fig. 3.
The interaction of FBXO22 and KDM4A is mediated by their FIST-C and Jmj domains, respectively. (A) Schematic diagram of the different FLAG-HA-tagged KDM4A deletion mutants. (B) Immunoprecipitation (IP) of endogenous FBXO22 with either full-length KDM4A or various KDM4A mutants demonstrated a requirement for the JmjN and JmjC domains for interaction. Cell lysates of HEK293T cells transiently expressing either full-length FLAG-HA-KDM4A or various FLAG-HA-KDM4A mutants were immunoprecipitated with anti-HA antibody, and immune complexes were blotted with anti-FBXO22 antibody. WCE, whole-cell extract. (C) Schematic representing the different FLAG-HA-tagged FBXO22 deletion mutants. (D) Immunoprecipitation of MYC-tagged KDM4A with either full-length FBXO22 or various FBXO22 mutants revealed a requirement for the FIST-C domain. Cell lysates of HEK293T cells transiently expressing either full-length FLAG-HA FBXO22 or various recombinant FLAG-HA FBXO22 mutants, together with MYC-KDM4A, were immunoprecipitated with anti-HA antibody, and immune complexes were blotted with anti-MYC antibody.
FBXO22 contains an N-terminal F-box domain responsible for binding SKP1 and a C-terminal domain found in FIST. FIST domains contain recognizable N- and C-terminal components, but FBXO22 contains only a recognizable FIST-C domain (Fig. 3C). While a region of FBXO22 encompassing the N terminus through the F-box did not associate with MYC-KDM4A, the C-terminal region of FBXO22 containing the FIST-C domain associated with MYC-KDM4A to an extent similar to that found with full-length FBXO22 (Fig. 3D). This suggests that the FIST-C domain is a previously unrecognized substrate recruitment module for the SCF pathway, which is conserved in vertebrate FBXO22 orthologs and several other non-F-box-containing proteins (7).
KDM4A levels are regulated by the SCFFBXO22 complex.
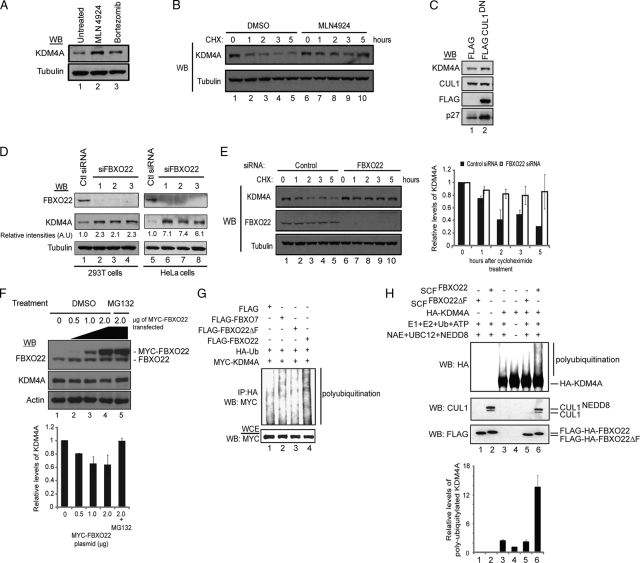
After validating the interaction of KDM4A with components of the SCFFBXO22 complex, we sought to examine whether this interaction was important for KDM4A stability. Neddylation of the cullins is a key event in regulating the ubiquitylation of substrates by cullin-RING ligases (11, 35). MLN4924 is a cell active inhibitor of the NEDD8-activating enzyme and causes accumulation of SCF substrates (39). Thus, if KDM4A levels are regulated by the SCFFBXO22 complex, treatment of cells with MLN4924 should increase the levels of KDM4A in these cells. To test this, we treated cells with either MLN4924 or bortezomib, a potent proteasomal inhibitor (1). Accordingly, both inhibitors resulted in a 2- to 4-fold increase in steady-state KDM4A levels (Fig. 4A). Consistent with a role in turnover, MLN4924 treatment led to stabilization of KDM4A in a cycloheximide chase experiment (Fig. 4B), extending the half-life from ∼2 h to >5 h.
Fig. 4.
SCFFBXO22 regulates KDM4A abundance and ubiquitylation in cells and promotes KDM4A ubiquitylation in vitro. (A) Steady-state levels of KDM4A are increased in HeLa cells treated with MLN4924 (cullin neddylation inhibitor) and bortezomib (proteasomal inhibitor). HeLa cells were treated with either 1 μM MLN4924 or 500 nM bortezomib for 4 h, and cell lysates were immunoblotted (WB) with the indicated antibodies. (B) Stability of KDM4A is increased in HEK293T cells treated with MLN4924 compared to control DMSO-treated cells, as demonstrated in a cycloheximide chase experiment. HEK293T cells were treated with either DMSO or 1 μM MLN4924 for 4 h before the addition of 100 μg/ml of cycloheximide. Cells were harvested at the indicated time points after cycloheximide treatment, and lysates were immunoblotted using the indicated antibodies. (C) Overexpression of CUL1DN increases endogenous levels of KDM4A in HEK293T cells. HEK293T cells were transiently transfected with either empty FLAG vector or vector expressing FLAG-CUL1DN for 48 h, and lysates were immunoblotted with the indicated antibodies. (D) Depletion of FBXO22 using 3 independent siRNAs increased KDM4A levels in both HEK293T and HeLa cells. Cell lysates of HEK293T and HeLa cells transfected with the indicated siRNAs for 72 h were immunoblotted with the indicated antibodies. (E) Depletion of FBXO22 led to stabilization of KDM4A in a cycloheximide chase experiment. HEK293T cells were transiently transfected with either control or FBXO22 siRNAs for 60 h before the addition of 100 μg/ml cycloheximide. Cells were harvested at the indicated time points post-cycloheximide treatment, and lysates were immunoblotted using the indicated antibodies. (Right panel) Graphical representation of data obtained from three independent experiments, using three different FBXO22 siRNAs, as quantified with ImageJ software (NIH). (F) Ectopically expressed MYC-FBXO22 led to a dose-dependent decrease in steady-state KDM4A levels, which could be rescued by the addition of MG132. HEK293T cells were transiently transfected with the indicated amounts of vector expressing MYC-FBXO22, and lysates were immunoblotted with the indicated antibodies to determine levels of endogenous KDM4A. MG132 at 10 μM was added to the last sample, as indicated, for 4 h. (Lower panel) Graphical representation of data obtained from two independent experiments as quantified by using ImageJ software. (G) Expression of FBXO22 but not the F-box deletion mutant led to a significant increase in the polyubiquitylation of MYC-KDM4A. Vectors expressing the indicated proteins were transiently transfected into HEK293T cells. At 48 h after transfection, cells were treated with 10 μM MG132 for 4 h, and cell lysates were immunoprecipitated with anti-HA antibody. Immune complexes were blotted with the indicated antibodies. (H) Ubiquitylation of KDM4A by HEK293T-purified SCFFBXO22 in vitro. (Lower panel) Graphical representation of the average of the relative levels of ubiquitylation of MYC-KDM4A in each sample for two independent in vitro ubiquitylation experiments as quantified with ImageJ software. Error bars represent the standard deviations from averages.
To directly examine SCFFBXO22 in the control of KDM4A stability, we employed both genetic ablation of the pathway as well as overexpression of FBXO22. Overexpression of dominant negative versions of CUL1 (CUL1DN) can lead to stabilization of SCF targets. Upon transient transfection of HEK293T cells with FLAG-CUL1DN, we observed a small (∼2-fold) but reproducible accumulation in the steady-state abundance of KDM4A, compared with vector control transfected cells (Fig. 4C). p27 was used as a positive control and showed about 3-fold enrichment in the presence of FLAG-CUL1DN. To examine the effect of depletion of FBXO22, HEK293T or HeLa cells were transfected with 3 independent siRNAs targeting FBXO22, and the levels of KDM4A were examined by immunoblotting (Fig. 4D). In HeLa cells, a 7-fold increase in KDM4A levels was observed with each of the siRNAs, while in HEK293T cells (lanes 5 to 8), which have higher basal levels of endogenous KDM4A, a 2-fold increase was observed (lanes 1 to 4). Consistent with immunoblotting, siRNAs 1, 2, and 3 depleted FBXO22 transcript levels by more than 80% (data not shown). However, the levels of KDM4A mRNA were unchanged, indicating that the protein level changes observed were not due to transcriptional effects (data not shown).
In order to demonstrate that FBXO22 specifically affects KDM4A turnover, a cycloheximide chase experiment was performed in cells depleted of FBXO22 (Fig. 4E). The half-life of KDM4A increased from ∼2 h to >5 h (compare lanes 1 to 5 with lanes 6 to 10). We repeated this experiment with two other FBXO22 siRNAs that targeted different regions of the FBXO22 transcript. In all three independent experiments, the half-life of KDM4A was at least doubled in cells depleted of FBXO22 compared to that in cells transfected with control siRNA (Fig. 4E, right panel). Finally, we examined the effect of overexpression of MYC-FBXO22, as previous studies had demonstrated that the abundance of F-box proteins is rate limiting for turnover of specific targets (38, 46). We observed a maximal 40% reduction in KDM4A abundance in a dose-dependent manner with increasing levels of MYC-FBXO22 (Fig. 4F, lower panel). Importantly, the addition of MG132 increased the levels of KDM4A even in the presence of high levels of MYC-FBXO22, indicating that the reduction in KDM4A abundance is proteasome dependent (Fig. 4D). Taken together, these results suggest that FBXO22 is required for turnover of KDM4A by the proteasome.
KDM4A is targeted for polyubiquitylation in vivo and in vitro by FBXO22.
The finding that FBXO22 reduces the protein levels of KDM4A in a proteasome- and CUL1-dependent manner and that SCF complexes are able to ubiquitylate their substrates led us to study the ability of FBXO22 to ubiquitylate KDM4A. Cells were transiently cotransfected with control vector or a vector expressing either FLAG-FBXO22, FLAG-FBXO22 lacking the F-box (FBXO22ΔF), and FLAG-FBXO7 as a negative control, together with vectors expressing MYC-KDM4A and HA-ubiquitin. Four hours prior to harvesting cells, MG132 was added to inhibit ongoing KDM4A degradation. The extent of polyubiquitin conjugates seen on MYC-KDM4A was increased, relative to that seen with either FBXO7 or FBXO22ΔF (Fig. 4G).
In order to examine whether SCFFBXO22 was capable of ubiquitylating KDM4A in vitro, SCFFBXO22 complexes were purified from HEK293T cells and incubated together with E1, E2 (CDC34 and UBCH5c), ubiquitin, ATP, and the neddylation apparatus (NEDD8-activating enzyme [NAE], NEDD8, and UBC12), and HA-KDM4A purified from insect cells. Ubiquitin conjugates were increased on KDM4A by 13-fold relative to reaction mixtures lacking SCFFBXO22 in triplicate experiments, while FBXO22 lacking the F-box was inactive in this assay (Fig. 4H). Overall, a 13-fold enhancement of ubiquitination was observed in duplicate experiments (Fig. 4H, lower panel). Taken together, these findings demonstrate that SCFFBXO22 is capable of ubiquitylating KDM4A.
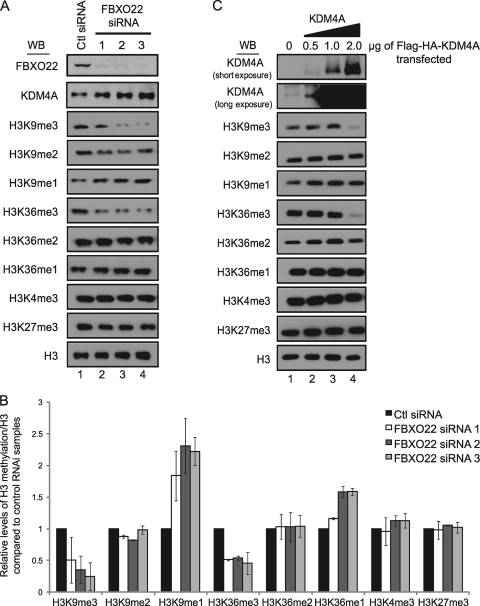
Depletion of FBXO22 leads to global changes in the abundance of histone H3K9 and K36 methylation.
KDM4A is known to promote demethylation of H3K9me3 and H3K36me3 through the dimethyl state to monomethyl state (23, 47). As depletion of FBXO22 leads to an increase in the abundance of KDM4A, the expectation is that this would lead to a reduction in the abundance of H3K9 and H3K36 trimethyl marks. To examine this question, 3 independent FBXO22 siRNAs were transfected into HEK293T cells, and acid-extracted histones were examined by immunoblotting with various antibodies specific to H3 methylation marks and total histone H3 as a loading control (Fig. 5A and B). A 2- to 3-fold increase in KDM4A resulted in a 4-fold reduction in the levels of H3K9me3 with the most potent siRNA (number 3) and a corresponding increase in the abundance of H3K9me1 (Fig. 5B). Likewise, a 2-fold reduction in levels of H3K36me3 resulted in a corresponding increase in the levels of H3K36me1 (Fig. 5B). In contrast, there was no change in the levels of H3K27me3 or H3K4me3 marks (Fig. 5A and B), which are not targeted by KDM4A. Similar extents of global decreases in H3K9me3 and H3K36me3 and global increases in H3K9me1 and H3K36me1 were observed when KDM4A was overexpressed by >100-fold (Fig. 5C). Interestingly, the extent of change in the levels of H3 methylation seen with extensive KDM4A overexpression was significantly muted compared with the effects seen upon stabilization of KDM4A by FBXO22 depletion, suggesting that FBXO22 affects a highly active pool of KDM4A that is not readily represented by overexpression of KDM4A. In contrast with FBXO22 depletion, overexpression of FBXO22 did not result in global changes in H3K9me3 and H3K36me3 levels (data not shown), possibly reflecting the relatively small effect of FBXO22 overexpression on KDM4A abundance (Fig. 4F).
Fig. 5.
Depletion of FBXO22 leads to global changes in KDM4A-targeted histone methylation marks. (A and B) Depletion of FBXO22 with three independent siRNAs led to global decreases in KDM4A-targeted histone H3K9 and H3K36 trimethylation marks and a corresponding increase in global H3K9 and H3K36 monomethylation. HEK293T cells were transfected with the indicated siRNAs for 72 h and harvested. Half the cells for each siRNA transfection were lysed with regular lysis buffer and used to probe for FBXO22 and KDM4A levels, while the remainder was acid extracted to enrich for histones and immunoblotted for the various methylation marks using the indicated antibodies. Quantification of Western blotting (WB) images was performed using ImageJ software (NIH) and is presented in graphical form in panel B. The data in panel B represent averages of two independent experiments, and error bars represent the standard deviations from these averages. (C) Overexpression of KDM4A induces global depletion of H3K9 and H3K36 trimethylation marks. HEK293T cells were transfected with the indicated amounts of FLAG-HA-KDM4A for 48 h and harvested. Cells were processed, and lysates were used for immunoblotting as described for panel A.
Changes in FBXO22 levels affect the histone H3 lysine 9 and 36 methylation status, and transcription levels of the KDM4A target gene ASCL2.
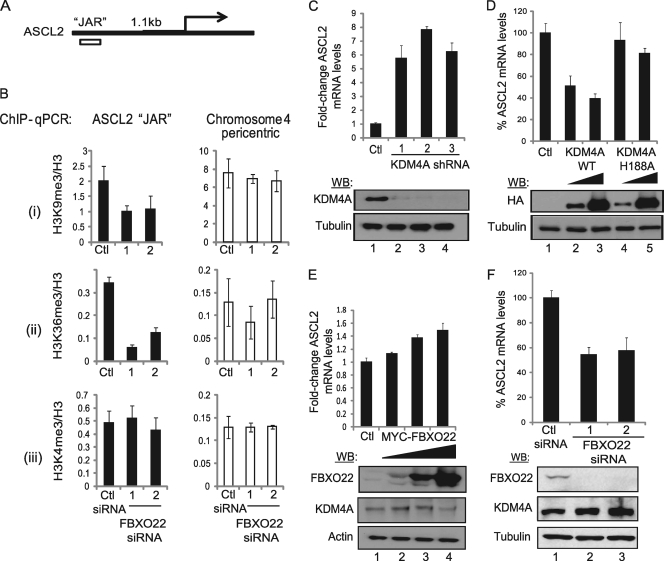
To further interrogate the importance of FBXO22 in the regulation of KDM4A's target histone marks, H3K9me3 and H3K36me3, we performed ChIP experiments to map changes in the methylation status of the ASCL2 promoter region (also known as JAR, or the JHDM3A/KDM4A-associated region) (Fig. 6 A), the only known KDM4A target gene identified to date in humans (23, 51). H3K9me3 and H3K36me3, but not H3K4me3, levels decreased 2- and 5-fold, respectively, on the JAR element upon FBXO22 depletion with 2 independent siRNAs (Fig. 6B). However, a pericentric heterochromatin locus on chromosome 4 did not show significant changes in H3K9me3, H3K36me3, or H3K4me3 levels with FBXO22 RNAi (Fig. 6B), indicating specificity in the target genes affected by FBXO22 loss.
Fig. 6.
Depletion or overexpression of FBXO22 affects mRNA levels and histone methylation levels on a KDM4A target gene, ASCL2. (A) Position of the JAR in the ASCL2 promoter relative to the transcriptional start site. (B) ChIP-qPCR showed that depletion of FBXO22 in HEK293T cells using two independent siRNAs led to depletion of (i) histone H3K9me3 and (ii) H3K36me3, but not (iii) H3K4me3 levels, at the KDM4A target region on ASCL2 but not at a nontargeted region on chromosome 4 pericentric heterochromatin. FBXO22 at 30 nM and control siRNAs were transfected into HEK293T cells and harvested 72 h posttransfection. A small aliquot of cells was used to verify the knockdown efficiency by immunoblotting (>90% knockdown of FBXO22 protein). After sonication and lysate preclearing, the lysate was used for detection of H3, H3K4me3, H3K9me3, H3K36me3, and Rb IgG ChIP. Typically a >20-fold enrichment from Rb IgG ChIP (data not shown) was observed at all regions sampled. Data represent the averages of three independent experiments, and error bars represent the standard deviations from these averages. (C) Depletion of KDM4A using 3 independent shRNAs that produce >95% knockdown of KDM4A protein levels results in ∼6-fold derepression of ASCL2 transcription levels as measured by qPCR. HE293T cells were infected with lentivirus containing the various shRNAs and selected under 1 μg/ml puromycin for >72 h. Protein lysates were used for immunoblotting, and TRIzol extracts were used for qPCR. Error bars for qPCR results represent the standard deviations from the means of two independent experimens. (D) Ectopic expression of KDM4A, but not its catalytic mutant (H188A), leads to repression of ASCL2 transcription in a dose-dependent manner. Two micrograms of FLAG-HA empty vector (Ctl), 0.5 μg or 2 μg of FLAG-HA-KDM4A, or FLAG-HA-KDM4AH188A was transfected into subconfluent HEK293T cells. Cells were harvested 48 h posttransfection and processed as described for panel C. Error bars represent the standard deviations from the means (n = 2). (E) Ectopic expression of MYC-FBXO22 resulted in a dose-dependent activation of ASCL2 transcription levels. Two micrograms of MYC empty vector or 0.5 μg, 1 μg, or 2 μg MYC-FBXO22 was transfected into HEK293T cells and processed as described for panel C. Error bars represent the standard deviations from the means (n = 2). (F) Depletion of FBXO22 by RNAi resulted in repression of ASCL2 transcription levels. HEK293T cells were transfected with 30 nM siRNA, harvested 72 h posttransfection, and processed as described for panel C. Error bars represent the standard deviations from the means (n = 2).
KDM4A functions as a repressor of the ASCL2 promoter via its functional interaction with JAR. As such, the expectation is that alterations in the extent of H3 methylation at JAR would result in altered ASCL2 transcription. Consistent with published results (51), KDM4A depletion with 3 independent siRNAs led to a 6- to 7-fold increase in ASCL2 mRNA levels, and overexpression of the wild type, but not an inactive KDM4A mutant (H188A), resulted in a 2-fold decrease in ASCL2 transcription (Fig. 6C and D). In contrast, modulation of FBXO22 had opposing effects. Overexpression of MYC-FBXO22 led to a 1.5-fold increase in ASCL2 mRNA at the highest level of FBXO22 expression (Fig. 6E), while depletion of FBXO22 with 2 independent siRNAs resulted in a 50% decrease in ASCL2 expression (Fig. 6F). Taken together, these data indicate that FBXO22 controls the transcriptional output of the ASCL2 gene by modulating the abundance of KDM4A.
DISCUSSION
KDM4A is a histone demethylase that plays important roles in a variety of biological processes, including transcription and the response of cells to DNA damage, through the regulation of methylation levels at histone H3 lysines 9 and 36. Posttranslational control of epigenetic regulators has received little attention thus far, despite clear evidence of important functional roles for these proteins and the fact that posttranslational pathways provide a means by which to rapidly change the array of chromatin marks. Here, we report the identification of the first histone demethylase that is posttranslationally regulated by a cullin-RING ligase complex. This is also the first substrate reported and characterized for the F-box protein FBXO22.
KDM4A is regulated by the ubiquitin-proteasome pathway, and this is mediated by the SCFFBXO22 complex.
Our data indicate that KDM4A levels are controlled by the ubiquitin-proteasome system in a manner that depends on K48 in ubiquitin, and we have provided several lines of evidence indicating a role for FBXO22. In particular, FBXO22 interacts with KDM4A endogenously, presumably in the nucleus, where both KDM4A and FBXO22 are localized, and this interaction occurs to similar extents at both G1/S and G2/M, suggesting that association is cell cycle independent. Moreover, depletion of FBXO22 stabilizes KDM4A, while overexpression of FBXO22 reduces the steady-state abundance of KDM4A. Importantly, inhibition of neddylation, like proteasome inhibition, led to the accumulation of KDM4A, consistent with a role for a CRL complex in KDM4A turnover. Finally, SCFFBXO22 promoted the ubiquitylation of KDM4A in vivo and in vitro. The finding that K48 in ubiquitin is required for KDM4A turnover in vivo is consistent with the known specificity of the CDC34 E2 implicated in ubiquitylation of CRL targets (34).
The FIST-C domain of FBXO22 is important for its interaction with KDM4A.
Our studies revealed the importance of a FIST-C domain located at the C terminus of FBXO22 as being required for an interaction with the N-terminal JmjN/JmjC domain of KDM4A. To our knowledge, this is the first demonstrated interaction for a vertebrate FIST domain-containing protein. The FIST domain typically consists of two subdomains: an N-terminal sensory (FIST-N) domain and a C-terminal signaling (FIST-C) domain, with the FIST-C region being more conserved than the FIST-N region (7). Based on bioinformatic analysis, the FIST-C domain is predicted to constitute the effector region responsible for binding to target proteins, which elicits a specific response triggered by the ligand that is detected and presumably transduced by the FIST-N domain. Several questions remain, including whether specific signals exist, such as phosphorylation or other modifications that promote the interaction between KDM4A and FBXO22. Moreover, are their common structural elements in the FIST-C domain that are used to promote the interaction of FBXO22 with other targets, much like the use of WD40 containing F-box proteins for targeting of multiple proteins containing related degrons (45)? The answers to these and related questions will require structural studies coupled to the identification of additional FBXO22 substrates.
FBXO22 regulates cellular H3K9 and H3K36 methylation levels and KDM4A target gene expression.
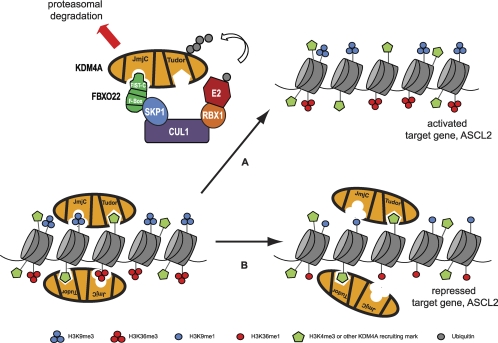
To understand the significance of FBXO22 in control of KDM4A, we examined both the global effects of FBXO22 modulation on histone H3 marks and on expression of the only known KDM4A target gene, ASCL2. Depletion of FBXO22 induced global changes in histone H3 K9 and K36 methylation marks, known targets of KDM4A, under conditions where KDM4A levels increased by only ∼3-fold. This was intriguing, since we observed that gross overexpression of KDM4A (>100-fold above endogenous levels) was required to induce similar changes in the target methylation marks. A likely possibility is that FBXO22 is regulating the stability of not only KDM4A but also other members of the same subfamily, in particular KDM4C, which is the most highly related member. Consistently, our preliminary results suggested that KDM4C steady-state levels are similarly regulated by FBXO22 (data not shown). Whether the more distantly related members, KDM4B and KDM4D, are also regulated by the same mechanism remains to be determined. An alternative possibility is that FBXO22 affects a highly active pool of KDM4A that is not readily represented by overexpression of KDM4A (Fig. 7). In addition, we speculate that FBXO22 targeting could be critical in the regulation of KDM4A's demethylation capacity. This could occur for example if FBXO22 binding to KDM4A was antagonistic to its binding to chromatin, where FBXO22 binding to KDM4A inhibits the localization of KDM4A to chromatin and prevents KDM4A from demethylating its target histone marks. In support of this hypothesis, we noticed that the Tudor domains of KDM4A inhibit the binding of FBXO22 (i.e., KDM4A without its Tudor domains binds about 2- to 3-fold more strongly to FBXO22 relative to full-length KDM4A [Fig. 3B]). The tandem Tudor domains of KDM4A are believed to be important for the recruitment of KDM4A to target histone sites (Fig. 7).
Fig. 7.
Proposed model for how posttranslational turnover of KDM4A by the SCFFBXO22-proteasome pathway leads to histone methylation changes and the target gene transcriptional effects. KDM4A is thought to be recruited to specific gene loci at least partly by its tandem Tudor domains, which bind H3K4me3 marks on chromatin. The catalytic JmjC domain is then able to mediate demethylation of H3K9me3/2 as well as H3K36me3/2 marks, although it is not clear if this can happen on the same histone or nucleosome. Upon an appropriate signal for degradation, KDM4A may be delocalized from its target sites and then be targeted by the SCFFBXO22 ubiquitin ligase complex, as illustrated by arrow A. This interaction is mediated by the JmjN/JmjC domain of KDM4A and the FIST-C domain on FBXO22. Polyubiquitylated KDM4A is then targeted for degradation by the proteasome. In the absence of a degradation signal or when components of the SCFFBXO22 complex are experimentally depleted (arrow B), KDM4A can accumulate to levels sufficient to demethylate its target marks. In the case of FBXO22 RNAi, where there is an overaccumulation of KDM4A, indiscriminate demethylation of its target marks may ensue over many gene loci, leading to global loss of H3K9me3 and H3K36me3 marks while leaving H3K9me1 and H3K36me1 marks in its wake. In the case of ASCL2, loss of H3K9me3 and H3K36me3 at the promoter region leads to transcriptional repression.
To examine the biological impact of FBXO22-dependent alterations in histone methylation, we analyzed the KDM4A target gene, ASCL2 (23, 51). Changes in FBXO22 levels by overexpression or RNAi led to corresponding, dose-dependent changes in transcriptional levels of ASCL2. Transcriptional effects on ASCL2 following FBXO22 depletion were modest despite the multifold increase in KDM4A protein levels, typical of KDM4A's repressor function. By histone ChIP assays, we found that the KDM4A target region of ASCL2 at its promoter reduced H3K9me3 and H3K36me3 levels upon FBXO22 depletion. Interestingly, a region approximately 3 kb from the JAR, in the gene body of ASCL2, also showed a similar reduction in H3K9me3 and H3K36me3 levels by ChIP when FBXO22 was depleted (data not shown). This suggests that when FBXO22 levels are low, KDM4A is able to indiscriminately alter its target methylation marks, even over regions to which it does not usually localize. This result espouses the earlier observation that FBXO22 RNAi leads to global changes in histone H3K9me3 and H3K36me3 levels and suggests that misregulation of KDM4A could result in widespread changes in global chromatin architecture. Changes in methylation were not observed in a pericentric heterochromatin region on chromosome 4, possibly reflecting the ability of KDM4A to target only relatively accessible regions within chromatin.
FBXO22-mediated degradation of KDM4A may play a significant role in development and tumorigenesis.
Although the function(s) of KDM4A in development, differentiation, and disease (such as cancer) is not yet well resolved, there are many indications of its importance in these processes. In chicken embryos, KDM4A regulates neural crest specification (42). In Drosophila melanogaster, inactivation of the KDM4 homolog DmeI reduces the male life span and affects male-specific wing extension/twitching (29). In mammalian cells, KDM4A affects genome replication and stability (6, 47). In addition, initiation and progression of many cancers have been linked to epigenetic misregulation events, and KDM4 proteins are potential oncogenes in certain cancers, such as medullablastomas and esophageal, prostate, and breast cancers, in which they are found to be significantly overexpressed (10, 12, 25, 27, 32, 37, 49, 50). The KDM4A-related protein KDM4C is preferentially expressed in undifferentiated stem cells, where it regulates genes involved in stem cell self-renewal (20, 28).
The importance of H3K9 and H3K36 trimethyl marks in transcription and other processes, which KDM4A regulates, implies that the regulation of KDM4A is necessary and important for normal cellular function. H3K9me3 is a mark characteristic of heterochromatin and gene transcriptional repression, and it is important for chromatin compaction and structure (24, 31). H3K36me3, on the other hand, is a mark that is generally found in the gene body of actively transcribed genes (2). It is not known if these two methylation states are functionally related or coregulated. However, H3K9me2/3 has been reported to localize in the gene body of actively transcribed genes, in a dynamically regulated manner: it increases during gene activation and is rapidly removed upon repression (13, 44). Thus, the abundance and regulatory status of KDM4 proteins are anticipated to be critically linked with multiple biological processes.
As a regulator of KDM4A protein levels, FBXO22 is anticipated to play a key role in KDM4A biology, including development and cancer. Modulation of FBXO22 levels, or alternatively, modulation of yet-to-be-identified signals that promote recognition of KDM4A by SCFFBXO22, is anticipated to alter the balance in repressive and activating marks on KDM4A-regulated promoters (Fig. 7). Posttranslational control of KDM4A, possibly in combination with transcriptional regulation of KDM4A transcript levels, can rapidly regulate KDM4A's levels in the cell or in a tissue-specific manner. Thus, our results suggest that FBXO22 is a key regulator of histone methylation marks, namely, H3K9 and H3K36 methylation, through the regulation of KDM4A protein levels.
ACKNOWLEDGMENTS
We thank Zhijian (James) Chen (University of Texas Southwestern Medical Center) for providing us with the ubiquitin replacement U2OS cell lines, Millennium Pharmaceuticals for providing MLN4924 and bortezomib, and Yang Shi for providing valuable reagents and resources. We also thank members of the Harper lab for their useful advice and suggestions.
This work is supported by NIH grants to J.W.H. M.-K.M.T. and H.-J. L. are supported by predoctoral fellowships from the Agency of Science, Technology and Research (A*STAR), Singapore.
Footnotes
Published ahead of print on 18 July 2011.
REFERENCES
- 1. Adams J., et al. 1999. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 59:2615–2622 [PubMed] [Google Scholar]
- 2. Barski A., et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 3. Behrends C., Harper J. W. 2011. Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 18:520–528 [DOI] [PubMed] [Google Scholar]
- 4. Behrends C., Sowa M. E., Gygi S. P., Harper J. W. 2010. Network organization of the human autophagy system. Nature 466:68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bennett E. J., Rush J., Gygi S. P., Harper J. W. 2010. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143:951–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Black J. C., et al. 2010. Conserved antagonism between JMJD2A/KDM4A and HP1γ during cell cycle progression. Mol. Cell 40:736–748 [DOI] [PubMed] [Google Scholar]
- 7. Borziak K., Zhulin I. B. 2007. FIST: a sensory domain for diverse signal transduction pathways in prokaryotes and ubiquitin signaling in eukaryotes. Bioinformatics 23:2518–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Campos E. I., Reinberg D. 2009. Histones: annotating chromatin. Annu. Rev. Genet. 43:559–599 [DOI] [PubMed] [Google Scholar]
- 9. Carrano A. C., Eytan E., Hershko A., Pagano M. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193–199 [DOI] [PubMed] [Google Scholar]
- 10. Cloos P. A., et al. 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442:307–311 [DOI] [PubMed] [Google Scholar]
- 11. Duda D. M., et al. 2008. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134:995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ehrbrecht A., et al. 2006. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J. Pathol. 208:554–563 [DOI] [PubMed] [Google Scholar]
- 13. Eissenberg J. C., Shilatifard A. 2006. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr. Opin. Genet. Dev. 16:184–190 [DOI] [PubMed] [Google Scholar]
- 14. Fodor B. D., et al. 2006. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20:1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gray S. G., et al. 2005. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J. Biol. Chem. 280:28507–28518 [DOI] [PubMed] [Google Scholar]
- 16. Huang Y., Fang J., Bedford M. T., Zhang Y., Xu R. M. 2006. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312:748–751 [DOI] [PubMed] [Google Scholar]
- 17. Jenuwein T., Allis C. D. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 18. Jin J., Ang X. L., Shirogane T., Wade Harper J. 2005. Identification of substrates for F-box proteins. Methods Enzymol. 399:287–309 [DOI] [PubMed] [Google Scholar]
- 19. Katoh M., Katoh M. 2004. Identification and characterization of JMJD2 family genes in silico. Int. J. Oncol. 24:1623–1628 [PubMed] [Google Scholar]
- 20. Katoh Y., Katoh M. 2007. Comparative integromics on JMJD2A, JMJD2B and JMJD2C: preferential expression of JMJD2C in undifferentiated ES cells. Int. J. Mol. Med. 20:269–273 [PubMed] [Google Scholar]
- 21. Kim J., et al. 2006. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7:397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klose R. J., Kallin E. M., Zhang Y. 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7:715–727 [DOI] [PubMed] [Google Scholar]
- 23. Klose R. J., et al. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442:312–316 [DOI] [PubMed] [Google Scholar]
- 24. Lachner M., Jenuwein T. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286–298 [DOI] [PubMed] [Google Scholar]
- 25. Lapointe J., et al. 2004. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 101:811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee J., Thompson J. R., Botuyan M. V., Mer G. 2008. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 15:109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu G., et al. 2009. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 28:4491–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loh Y. H., Zhang W., Chen X., George J., Ng H. H. 2007. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21:2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lorbeck M. T., et al. 2010. The histone demethylase Dmel/Kdm4A controls genes required for life span and male-specific sex determination in Drosophila. Gene 450:8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mersman D. P., Du H. N., Fingerman I. M., South P. F., Briggs S. D. 2009. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 23:951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113 [DOI] [PubMed] [Google Scholar]
- 32. Northcott P. A., et al. 2009. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat. Genet. 41:465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pedersen M. T., Helin K. 2010. Histone demethylases in development and disease. Trends Cell Biol. 20:662–671 [DOI] [PubMed] [Google Scholar]
- 34. Petroski M. D., Deshaies R. J. 2005. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123:1107–1120 [DOI] [PubMed] [Google Scholar]
- 35. Saha A., Deshaies R. J. 2008. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32:21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shin S., Janknecht R. 2007. Diversity within the JMJD2 histone demethylase family. Biochem. Biophys. Res. Commun. 353:973–977 [DOI] [PubMed] [Google Scholar]
- 37. Shi L., et al. 2011. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 108:7541–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shirogane T., Jin J., Ang X. L., Harper J. W. 2005. SCFβ-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 280:26863–26872 [DOI] [PubMed] [Google Scholar]
- 39. Soucy T. A., et al. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458:732–736 [DOI] [PubMed] [Google Scholar]
- 40. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Strahl B. D., Allis C. D. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 42. Strobl-Mazzulla P. H., Sauka-Spengler T., Bronner-Fraser M. 2010. Histone demethylase JmjD2A regulates neural crest specification. Dev. Cell 19:460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trojer P., et al. 2009. Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9a/KMT1C and the Jumonji domain-containing JMJD2/KDM4 proteins. J. Biol. Chem. 284:8395–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vakoc C. R., Mandat S. A., Olenchock B. A., Blobel G. A. 2005. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19:381–391 [DOI] [PubMed] [Google Scholar]
- 45. Welcker M., Clurman B. E. 2008. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8:83–93 [DOI] [PubMed] [Google Scholar]
- 46. Welcker M., et al. 2003. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell 12:381–392 [DOI] [PubMed] [Google Scholar]
- 47. Whetstine J. R., et al. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467–481 [DOI] [PubMed] [Google Scholar]
- 48. Xu M., Skaug B., Zeng W., Chen Z. J. 2009. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-1β. Mol. Cell 36:302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang Z. Q., et al. 2000. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res. 60:4735–4739 [PubMed] [Google Scholar]
- 50. Yu Y. P., et al. 2004. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 22:2790–2799 [DOI] [PubMed] [Google Scholar]
- 51. Zhang D., Yoon H. G., Wong J. 2005. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol. Cell. Biol. 25:6404–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y., Reinberg D. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343–2360 [DOI] [PubMed] [Google Scholar]