Abstract
Stomata are specialized cellular structures in the epidermis of aerial plant organs that control gas exchange (H2O release and CO2 uptake) between leaves and the atmosphere by modulating the aperture of a pore flanked by two guard cells. Stomata are nonrandomly distributed, and their density is controlled by endogenous and environmental factors. To gain insight into the molecular mechanisms regulating stomatal distribution, Arabidopsis thaliana mutants with altered stomatal characteristics were isolated and examined. The sdd1-1 mutant exhibits a two- to fourfold increase of stomatal density and formation of clustered stomata (i.e., stomata that are not separated by intervening pavement cells), whereas the internal leaf architecture is not altered. The SDD1 gene was identified by map-based cloning. It encodes a subtilisin-like serine protease related to prokaryotic and eukaryotic proteins. We propose that SDD1 acts as a processing protease involved in the mediation of a signal that controls the development of cell lineages that lead to guard cell formation.
Keywords: Arabidopsis mutant, stomata, pattern formation, subtilase, proprotein
During the evolution of terrestrial plants, development of stomata was a key event allowing the balance between the intake of CO2 and the release of water to be maximized. A pair of specialized guard cells wield control over the size of stomatal apertures, responding within minutes to alterations in illumination, water supply, and CO2 levels. Stomatal density is also variable and is set according to the environmental conditions prevailing during leaf development.
For many plant species, stomatal density, or stomatal index (Salisbury 1927), is modulated in response to environmental factors such as humidity (Schürmann 1959), temperature (Srivastava et al. 1995), CO2 partial pressure (Clifford et al. 1995), or light intensity (Gay and Hurd 1975; Schoch et al. 1980; Rahim and Fordham 1991). In addition to these exogenous factors, stomatal density is also subject to genetic (endogenous) control, as illustrated by the differences among varieties of the same species (Reich 1984; Buttery et al. 1992, 1993; Ramos et al. 1992) or among F1 hybrids (Abak and Yanmaz 1985). The multigenic, oligogenic, or monogenic control of stomatal characteristics has also been demonstrated (Jones 1987). The molecular mechanisms controlling stomatal differentiation, however, are poorly understood and no information about the control mechanisms involved was hitherto available.
A level of control beyond density applies to the positioning of stomata relative to each other: Under natural growth conditions stomata are nonrandomly distributed (Willmer and Fricker 1996). The degree of order in stomatal distribution patterns has been quantified by calculation of R values (Clark and Evans 1954) of ∼1.4 (Sachs 1978). By definition, a random distribution of stomata has an R value of 1, whereas a completely ordered hexagonal pattern has a value of 2.15. Stomatal distribution patterns, therefore, bear a limited degree of order. The presence of a stomata-free region surrounding each stoma, and excluding immediate contact with neighboring guard cell pairs, has been identified as the major principle of order (Sachs 1991). Beyond this minimal distance (which in wild-type plants is comprised by at least one epidermal cell) stomata positioning is rather random (Sachs 1991).
In the dicotyledonous Brassicaceae, meristemoids (individual, undifferentiated, mitotically active cells within a tissue of fixed determination; Bünning 1953) undergo three successive unequal divisions, finally resulting in the formation of a centrally located guard cell mother cell surrounded by three neighboring cells. The two guard cells arise from an equal division of the guard cell mother cell (Pant and Kidwai 1967). In accordance with recent cell lineage studies (Larkin et al. 1996), cells belonging to a stomatal complex (guard cells plus neighboring cells) are, in most cases, clonally related derivatives of the meristemoid. Therefore the number and orientation of the cell divisions occurring during stomatal complex formation are critical for correct stomatal distribution establishment. As shown by Pant and Kidwai (1967), in Brassicaceae any one of the subsidiary cells can retain or acquire meristemoidal status leading to the formation of secondary stomatal complexes (satellite stomata). In the same manner tertiary complexes or complexes of an even higher order can be generated. Guard cell pairs in these extended cell lineages are correctly spaced relative to established cells. In addition to cell lineage control, it is likely that cell–cell interactions are involved in the establishment of the final stomatal pattern. Thus, improperly positioned cells that initiated the developmental process toward stomatal formation may be prematurely arrested or even induced to de-differentiate into pavement cells (Sachs et al. 1993; Sachs 1994; Chin et al. 1995).
Because most previous studies on the differentiation of stomatal complexes have been of a descriptive nature, the molecular mechanisms underlying stomatal patterning and regulation of stomatal density are largely unknown. The only hint of the potential involvement of a signaling pathway was obtained when Arabidopsis seedlings were treated with high humidity or with high concentrations of 1-aminocyclopropane-1-carboxylate (ACC), a precursor that is rapidly converted into the phytohormone ethylene (Serna and Fenoll 1997). Such treatments resulted in the occurrence of clustered stomata (guard cell pairs in direct contact with each other with no intervening cells). The only known gene that affects stomatal spacing is Transparent Testa Glabra (TTG; Berger et al. 1998). The influence of TTG on stomatal patterning, however, is confined to hypocotyls and is exerted through the determination of identity of cells belonging to a cell file rather than through the specific control of stomatal complex formation. Several mutants affected in stomatal patterning and differentiation have been isolated in Arabidopsis (Yang and Sack 1995) and other plant species (Zeiger and Stebbins 1972; Maynard et al. 1974). Two of these mutants, designated tmm and flp (Yang and Sack 1995; Geisler et al. 1998), have been studied in detail. Both exhibit the presence of stomatal clusters that, however, differ in size and cell arrangement and in occurrence in different aerial organs. The phenotypes of these mutants suggest that the affected genes control the initiation and repression of precursor cell formation (Larkin et al. 1997). Because neither of the corresponding genes has yet been isolated, the nature of the factors controlling these processes is unknown.
Here, a novel mutant, designated stomatal density and distribution1-1 (sdd1-1) that shows the formation of extra adjacent stomata and a strong increase in stomatal density is described. A comparative analysis of stomatal complex formation in the wt and the sdd1-1 mutant is presented, which demonstrates the involvement of the SDD1 gene product in triggering stomatal initial formation, in controlling cell proliferation during stomatal complex formation (extent of the cell lineages), and in controlling the orientation of cell divisions affecting the positioning of guard cell pairs within stomatal complexes. Using map-based cloning and sequence analysis, the SDD1 gene is shown to encode a protein with homology to subtilisin-like serine proteases. In a manner analogous to that of known eukaryotic subtilisins, SDD1 is proposed to process a proteinacious signal molecule precursor or a precursor of a receptor involved in the transduction of signal(s) controlling meristemoid activity.
Results
To study the molecular processes involved in the control of stomatal spacing (density and distribution), we isolated and analyzed an ethyl methanesulfonate (EMS)-induced mutant called sdd1-1. This mutant, which has a phenotype characterized by increased stomatal density and a disturbed stomatal distribution pattern, was identified in a population of ∼3500 mutagenized transgenic M2 plants, representing ∼1000 M1 plants that were evaluated microscopically for stomatal characteristics. The transgene present in these plants (Müller-Röber et al. 1994) provided a means to stain guard cells specifically for better visibility of stomata. The sdd1-1 mutant was shown to carry a mutation in a gene different from two previously identified loci involved in stomatal complex formation, tmm and flp (Yang and Sack 1995), by crossing sdd1-1 with a homozygous tmm flp double mutant: the resulting F1 plants all displayed a wild-type phenotype (data not shown).
Stomatal density is increased and the stomatal distribution pattern is disturbed in sdd1-1
To examine the expression of the sdd1-1 phenotype, nail polish copies of dental resin imprints taken from various organs were analyzed by light microscopy. As shown in Table 1 and Figure 1, C and D, the density of stomata (number of stomata per square millimeter) in the epidermis of all aerial organs, except cotyledons, is increased in the sdd1-1 mutant two- to fourfold in comparison to the wild type. In the cotyledons, a weak increase of ∼35% was detected in the abaxial surface. In the wild type, the stomatal density is equal on both sides of primary leaves, rosette leaves, and cauline leaves. In contrast, in rosette leaves of sdd1-1 plants, including primary leaves, stomatal density differs. It is doubled in comparison to the wild type in the adaxial surface and is increased three- to fourfold in the abaxial epidermis. The degree of this hypostomatic effect (stronger increase in stomatal density in the abaxial as compared to the adaxial epidermis) is most pronounced in rosette leaves. In wide cauline leaves of sdd1-1, only a weak hypostomatic effect is apparent. In narrow cauline leaves (present in the more apical regions of the shoot), no differences between both epidermis were observed.
Table 1.
Stomatal density in epidermises of different organs of the wild type (SDD1) and the mutant (sdd1-1) at the adaxial and abaxial surfaces
| Organ
|
SDD1
|
sdd1-1
|
||||
|---|---|---|---|---|---|---|
| Adaxial
|
|
Abaxial
|
Adaxial
|
|
Abaxial
|
|
| Carpel | 72 ± 13 | 232 ± 34 | ||||
| Pedicel | 51 ± 8 | 162 ± 12 | ||||
| Sepal | 137 ± 18 | 214 ± 18 | ||||
| Stem | 57 ± 10 | 108 ± 14 | ||||
| Cauline leaf (narrow) | 112 ± 10 | 96 ± 15 | 348 ± 75 | 352 ± 71 | ||
| Cauline leaf (wide) | 98 ± 17 | 89 ± 10 | 157 ± 20 | 230 ± 30 | ||
| Rosette leaf | 160 ± 26 | 153 ± 18 | 326 ± 31 | 582 ± 42 | ||
| Primary leaf | 141 ± 25 | 105 ± 12 | 290 ± 36 | 530 ± 80 | ||
| Cotyledon | 60 ± 21 | 105 ± 13 | 53 ± 12 | 142 ± 18 | ||
Stomatal density, number/mm2; ± s.d. (n = 10).
Figure 1.
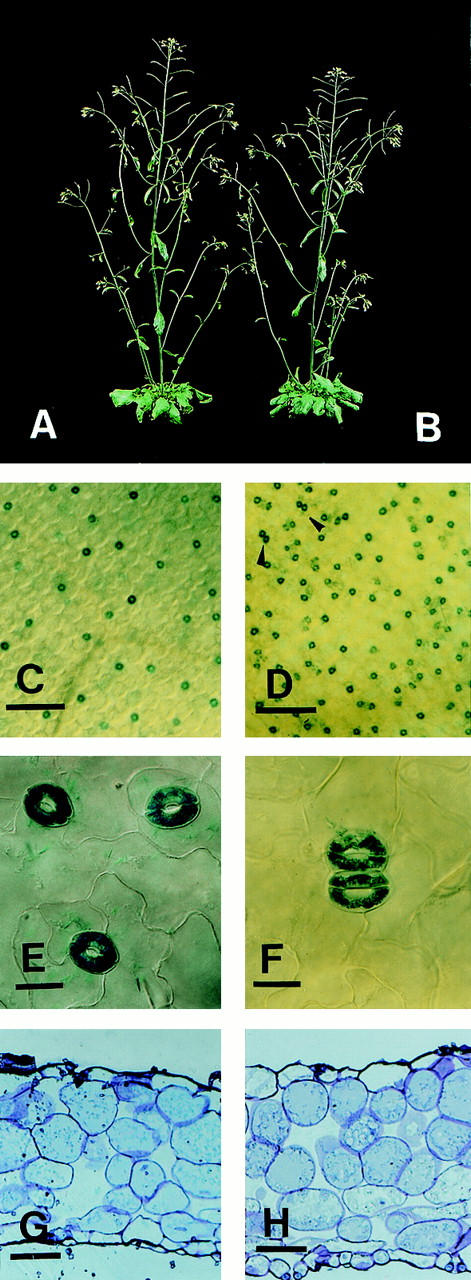
Habitus of wild-type and sdd1-1, increase of stomatal density, formation of paired stomata in leaves of sdd1-1, and cross sections of rosette leaves of wild-type and mutant. Macroscopically, wild type (A) and sdd1-1 (B) show no differences in morphology. Alteration in stomatal density is shown in images of the abaxial surfaces of rosette leaves of the transgenic wild-type control (C) and of the sdd1-1 mutant (D). Bars, 200 μm. Paired stomata in sdd1-1 are indicated in D by arrowheads and shown in higher magnification in F in comparison to single stomata in the wild type (E). Bars, 10 μm. For better visibility, guard cells expressing β-glucuronidase were histochemically stained with X-gluc. Cross sections of rosette leaves of wild type (G) and sdd1-1 (H) demonstrate that the structure or arrangement of internal leaf tissues is not affected by the sdd1-1 mutation. Large substomatal cavities are present at some but not all stomata present in sdd1-1 leaves. Bars, 25 μm.
A further feature characteristic of sdd1-1 is the formation of stomatal clusters (i.e., two or more guard cell pairs placed in direct contact to each other, Fig. 1F). Thus, the major principle of stomatal patterning, the formation of a stomata-free space surrounding each stoma, is aberrant in the mutant. The frequency of stomatal clusters varies between different organs but does not correlate with the degree of stomatal density increase. In sdd1-1 cotyledons, ∼45% of stomata are arranged in clusters, whereas the stomatal density is nearly unchanged. On primary leaves, rosette leaves, cauline leaves, and sepals, 10% of the stomata are arranged in clusters. In carpels and pedicels, where stomatal density is increased approximately threefold, only 5% of the stomata are clustered. Finally, in the epidermis of stems, less than 1% of stomata are placed in direct contact with each other, whereas the density is increased twofold. No difference was detected in the degree of cluster formation in abaxial or adaxial surfaces of leaves (data not shown). In all organs, double clusters of stomata (groups of two adjoining guard cell pairs) are the predominant type (data not shown).
The sdd1-1 mutation affects the stomatal index
Because stomatal density may be affected by alterations in epidermal cell size and/or changes in the ratio of pavement cells per stomata, stomatal indices (si) were determined [si = number of stomata/(number of epidermal cells + number of stomata) × 100; Salisbury 1927]. The stomatal index is shifted from 25% in the abaxial surface of wild-type primary leaves to 37% in sdd1-1. To obtain another measure for this difference, nail polish imprints were taken from the abaxial surface of fully expanded rosette leaves and were evaluated with respect to the fraction of epidermal cells present in stomatal complexes (i.e., being in contact with guard cells). Approximately 40% of the epidermal cells are not in contact with guard cells (pavement cells), whereas ∼60% of the epidermal cells are part of stomatal complexes (neighboring cells). This situation is drastically different in the sdd1-1 mutant. Here, (almost) every epidermal cell is in contact to at least one guard cell. Thus, the cell arrangement is clearly altered in the leaf epidermis of sdd1-1, which according to this criterion is almost entirely composed of stomatal complexes (data not shown).
The morphologic alterations caused by the sdd1-1 mutation are restricted to the epidermis
To determine if the sdd1-1 mutation affects the morphology of the plants in ways other than the cellular structure of the epidermis, sdd1-1 and wild-type plants were further characterized. Comparison of adult plants revealed no differences in the general habitus (Fig. 1A,B). Furthermore, the sizes (leaf area) and the forms (length and width of the leaf blade) of rosette leaves were evaluated and no differences were observed (data not shown). To examine the structure of internal leaf tissues, semithin sections of mature rosette leaves were analyzed by light microscopy. Again, no differences in tissue organization, cell numbers, or cell sizes could be observed between wild-type and sdd1-1 mutant (Fig. 1G,H). In both genotypes, the parenchyma is composed of a single layer of adaxial palisade parenchyma and four to five layers of spongy parenchyma. A rosette leaf thus consists of seven to eight cell layers including the abaxial and the adaxial epidermis.
The alterations in leaf epidermis development caused by the sdd1-1 mutation are not entirely restricted to guard cells. The frequency (number per square millimeter) of pavement cells plus neighboring cells is increased by ∼60% in sdd1-1 in comparison to the wild type. This is probably due to the extension of the stomatal cell lineages (see below). Changes in trichome morphology were not observed. Trichome spacing could not be analyzed in detail, due to a gl1 mutation present in the wild-type C24 (M. Koornneeff, pers. comm.) and in the sdd1-1 mutant, which leads to a strong reduction in the trichome number. Examination of individuals of the mapping population (F2 derived from a cross to the Arabidopsis genotype Columbia-0, GL1), however, did not reveal any gross alterations in trichome distribution.
Initiation of stomatal development is enhanced in sdd1-1 and stomatal cell lineages are extended
To determine the specific changes in the cellular development of the epidermis caused by the sdd1-1 mutation, a set of serial imprints was analyzed with respect to the structure of the cell lineages established in the course of the development of primary leaves. In Arabidopsis thaliana, as in many other species (Sachs and Benouaiche 1978; Sachs et al. 1993), stomata develop through a highly orchestrated set of divisions of a precursor cell (protodermal cell). In the abaxial epidermis of wild type leaf primordia, ∼35% of the protodermal cells enter this developmental pathway (Table 2). Three sequential unequal divisions of the initial cell result in the formation of three neighboring cells that surround the central guard cell mother cell. By a fourth, equal division of the guard cell mother cell, the two guard cells are produced (Fig. 2; Larkin et al. 1997). Each of the neighboring cells surrounding a guard cell pair has the potential to give rise to the formation of satellite (secondary) stomata through further unequal divisions (Pant and Kidwai 1967; Larkin et al. 1997). In the wild type, 77% of satellite stomata (37 cases analyzed) were derived from the youngest neighboring cell (formed through the last unequal division) and 23% were derived from the previously formed neighboring cell. Iteration of this process leads to the development of higher order satellite stomata. Such satellite stomata are produced by one, two, or three sequential unequal cell divisions (see below) followed by the equal division of the newly formed guard cell mother cell. In the wild type, the formation of 64.4% primary, 29% secondary, and 6.6% tertiary stomatal complexes was observed and no quaternary complexes occurred (Table 2).
Table 2.
Characterization of different aspects of stomatal development in wild type plants (SDD1) and in the mutant (sdd1-1)
| Parameters analyzed
|
SDD1
|
sdd1-1
|
|---|---|---|
| Stomata/cell lineage | 2.25 ± 0.75 | 5.09 ± 1.89 (a = 2.26) |
| Primary | 64.44% | 21.16% |
| Secondary | 28.88% | 42.05% |
| Tertiary | 6.66% | 29.9% |
| Quaternary | — | 1.87% |
| Fraction of cell lineages that results in formation of stomatal complexes | 35.3% | 59.45% (b = 1.68) |
| Increase factor | — | 3.79 |
Figure 2.
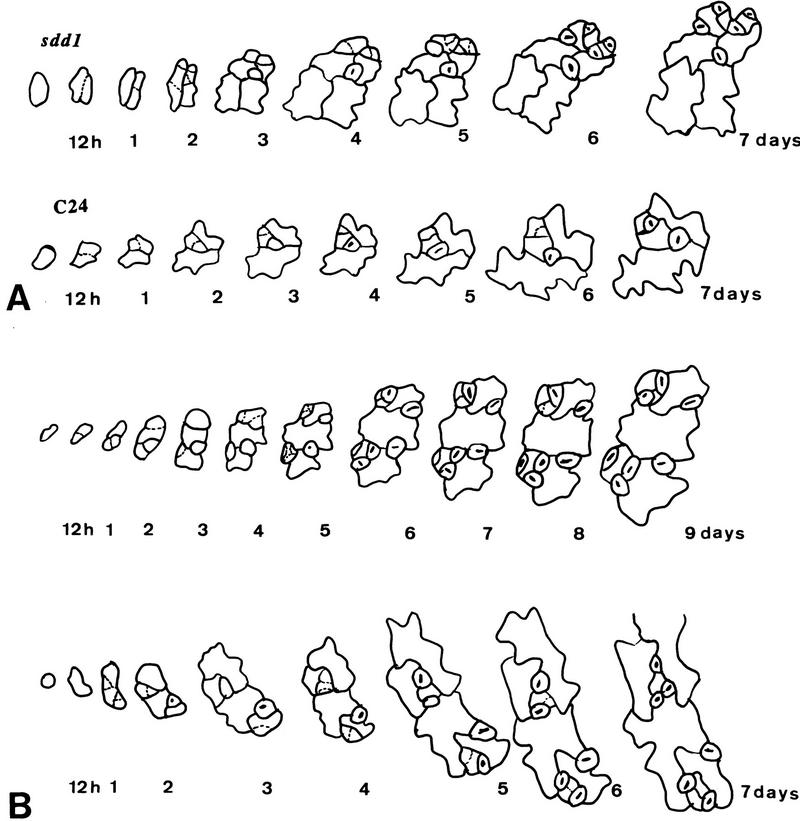
Stomatal complex formation from protodermal cells in the wild-type (C24) and the sdd1-1 mutant (A) and formation of stomatal clusters in sdd1-1 (B). The development of individual stomatal complexes was monitored through serial dental resin imprints repeatedly taken from the abaxial surface of developing (maturing) primary leaves. In sdd1-1, cell lineages are extended leading to the enhanced formation of higher order satellite stomata (A). The first imprint was taken from a leaf ∼0.2 mm in length; the second imprint, 12 hr later. Further imprints were taken at daily intervals. Cluster formation in sdd1-1 occurred exclusively within individual cell lineages (B). The formation of a secondary double cluster with simultaneously maturing guard cells is shown in the top panel, and a nonsynchronous formation of a tertiary double cluster in the bottom panel. Dotted lines indicating the positions of new cell walls formed during the intervals between impressions were introduced for better clarity.
In sdd1-1, the process of stomatal complex formation is modified in several ways. The fraction of protodermal cells that initiate cell lineages leading to the formation of stomatal complexes is elevated to about 60% in the sdd1-1 mutant (Table 2). In addition, more secondary and higher order satellite stomata are formed, 26.1% primary, 42% secondary, 30% tertiary, and even 1.9% quaternary stomatal complexes appeared in the sdd1-1 mutant (Table 2). Accordingly, the extent of the cell lineages is increased (Fig. 2A) and the number of stomata produced by an individual cell lineage is elevated from an average of 2.25 in the wild type to 5.09 in the mutant. Similar to the situation in the wild type, the youngest neighboring cell predominantly forms satellite stomata in sdd1-1. Of 32 secondary stomatal complexes analyzed, 69% were derived from the youngest neighboring cell, 28% from the previously formed neighboring cell, and only 3% from the oldest neighboring cell. Another modification in the development of satellite stomatal complexes caused by the sdd1-1 mutation is a shift toward fewer unequal cell divisions of the neighboring cells: Secondary stomata are formed in the wild type (27 cases analyzed) through three (56%), or two (37%), and rarely through one (7%) cell division(s). In contrast, they are formed in the sdd1-1 mutant (34 cases analyzed) through three (6%), two (76%), and, to a limited extent one (18%) cell division(s). A similar tendency was observed for the formation of tertiary satellite stomata (data not shown).
The two major effects that lead to elevated stomatal density in the sdd1-1 mutant are the 1.68-fold increase in the fraction of cell lineages resulting in the formation of stomatal complexes and the 2.26-fold increase in the number of stomata produced per cell lineage. These two factors multiply to a total increase factor of 3.79, which agrees closely with the observed three- to fourfold increase of the stomatal density in the abaxial leaf epidermis in the mutant.
Stomatal cluster develop through a relaxed control of cell division orientation during higher order satellite stomata formation
Serial imprints were also used to monitor the formation of stomatal clusters in sdd1-1. Among 26 double clusters (the predominant type of cluster found) that were analyzed, all developed within a given cell lineage. All clustered stomata developed through single unequal cell divisions of the youngest neighboring cells of the corresponding satellite stomata. In the wild type, the smaller product of the first (unequal) division of a neighboring cell, which lead to the formation of a higher order stoma, was always positioned at the side opposite to the preestablished (primary or secondary) guard cell pair, thus avoiding the formation of clusters. This control is relaxed in the sdd1-1 mutant. As expected from the cell division patterns described earlier, clustering of stomata belonging to different (adjacent) primary stomatal complexes was not observed. Furthermore, primary guard cell pairs were not included in any of the clusters. Thus, stomatal clusters developed exclusively during the time of satellite stomata formation. Twenty clusters occurred during or after secondary stomatal complex formation and the remaining six during or after tertiary complex formation. In ten clusters, the two guard cell pairs developed simultaneously, four clusters developed successively, (the second guard cell pair was produced after the first pair matured), and in 12 cases one of the two guard cell pairs of the cluster reached maturity earlier than the other (Fig. 2B). We evaluated 37 tertiary stomatal complexes formed through a single unequal division of a neighboring cell. In 13 cases, the smaller daughter cell was positioned adjacent to the preestablished or codeveloping secondary guard cell pair. Therefore, in addition to its role in cell lineage control, the SDD1 gene product also plays a role in determining the proper orientation of cell divisions during the development of higher order satellite stomata.
Map-based cloning of SDD1
Mutant F2 plants that resulted from a cross sdd1-1 (C24) × wild type (Col-0) were used for genetic mapping of the SDD1 locus. Within an F2 population of 2800 individuals, 641 homozygous mutant plants were identified. The segregation of 1:3.4 confirmed the presence of a single nuclear recessive mutation. Using PCR-based, cleaved, amplified, polymorphic sequences (CAPS) markers (Koncienczy and Ausubel 1993), the SDD1 locus was mapped to the upper arm of chromosome 1 into the interval between the markers PVV-4 at 1.9 cM and PAI-1 at position 9 cM (Fig. 3A). The mutant gene was localized in an interval of 1.5 cM bordered by the markers O846A (C. Dean, pers. comm.) and CIC7D5RE (Creusot et al. 1995; data not shown). A contig, consisting of 38 overlapping IGF–BAC clones covering ∼700 kb, was established across this region by the use of the markers and end-fragments of bacterial artificial chromosome (BAC) inserts as hybridization probes against the total IGF–BAC library (Mozo et al. 1998b). By using BAC ends as RFLP and CAPS markers, the SDD1 locus was finally assigned to an estimated interval of 0.59 cM flanked by the ends of the BAC clones F20G19 left end (LE) and F25I3 right end (RE; Fig. 3A). The sequence of two IGF–BAC clones, F20D22 and F21M11, which fully cover this region, was determined by the SPP consortium (http://Sequence-www.stanford.edu/ara/SPP.html) as part of the Arabidopsis Genome Initiative and was made available through public databases (GenBank accession nos. AC003027 and AC002411, respectively). Hence, the 0.59-cM region covers 113 kb of genomic DNA. To identify the SDD1 gene, this 113-kb region was scanned for polymorphisms between sdd1-1 and SDD1 by application of the restriction single-strand conformation polymorphism (SSCP) technique (Iwahana et al. 1992). A set of fifty-seven 2-kb PCR fragments, separately amplified from SDD1 (wtC24) and sdd1-1, was subjected to SSCP analysis. A single SSCP was detected (data not shown) and, by a second round of SSCP the polymorphism was assigned to a 0.5-kb fragment. Sequence analysis of this fragment revealed that the detected SSCP was caused by a single G/C → A/T mutation which created a premature stop codon within a predicted open reading frame (ORF) spanning 2328 bp/775 amino acids (Fig. 3B).
Figure 3.
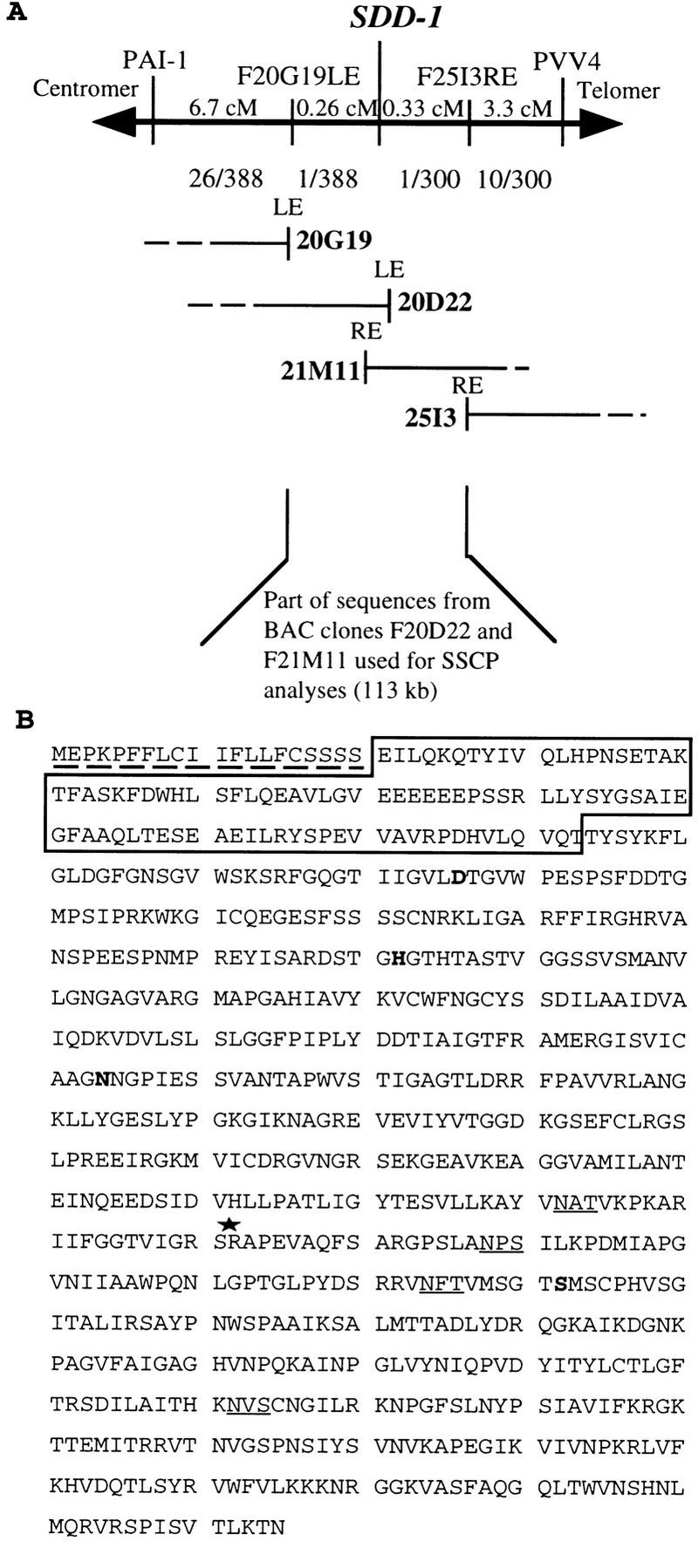
Map-based cloning of the SDD1 gene in the top region of chromosome 1 (A) and the deduced amino acid sequence of SDD1 (B). Two CAPS markers (PAI1 and PVV4) and two RFLP markers (F20G19LE and F25I3RE) used for mapping of SDD1 are shown. The genetic distance in cM between SDD1 and these markers is indicated by numbers above the horizontal line. The numbers below the line represent the fraction of chromosomes that showed recombination between SDD1 and the various markers. Four different, overlapping IGF–BAC clones (20G19, 20D22, 21M11, and 25I3) forming a contig covering the SDD1 locus are shown. The region within the BAC contig, which was subjected to SSCP analysis for the identification of the sdd1-1 mutation, is marked below the BAC contig. At the very bottom, the SDD1 amino acid sequence is shown (B). The putative (amino-terminal) signal sequence for ER uptake is marked by a broken line. The sequence within the box represents the predicted prodomain of the protein, cleavage of which is probably required for activation of the proteolytic activity of the protein. The invariant amino acids in the catalytic triade (D, H, and S domains) and the core region of the substrate binding site (N domain), which are present in all known subtilases, are printed in boldface type. Putative glycosylation sites are underlined. The site of the premature stop caused by the sdd1-1 mutation is marked with an asterisk.
Genetic complementation confirmed the identity of the SDD1 gene
To demonstrate the identity of the 2328-bp ORF as the coding region of the SDD1 gene, two gene constructs were introduced into the sdd1-1 mutant via Agrobacterium tumefaciens in planta transformation (Bechthold et al. 1993). The first construct, G-SDD1, harbored a 7-kb genomic fragment containing the endogenous promotor and the 2.3 kb coding region. The second construct, 35S-SDD1, contained a chimeric gene created by the fusion of the CaMV 35S promoter with the 2.3-kb ORF. Transformed plants selected for Hygromycin resistance were tested for the expression of mutant or wild-type phenotypes by microscopic examination of cotyledons and primary leaves. In cotyledons 7 of 10 and 2 of 10 transformants containing the G-SDD1 or the 35S–SDD1 constructs, respectively, showed a full wild-type phenotype with respect to the presence or absence of clustered stomata (data not shown). Seven 35S–SDD1 transformants showed an intermediate phenotype, probably due to inefficient expression of the transgene in developing cotyledons. No significant reduction of stomatal density below wild-type levels was observed in mutant plants overexpressing SDD1. In primary leaves, complementation (with respect to stomatal density and cluster formation) of the sdd1-1 mutation was observed for each of the 20 transformants carrying either the G-SDD1 or the 35S–SDD1 construct. These data enabled the confirmation of the 2.3-kb DNA fragment as the SDD1 gene.
SDD1 is a subtilisin-like serine protease
The SDD1 gene is predicted to encode a protein 775 amino acids in length with high sequence similarity to the class of subtilisin-like serine proteases (for review, see Barr 1991; Siezen and Leunissen 1997). A number of subtilisin-like serine proteases have been cloned from prokaryotes and eukaryotes and, more recently, a number of plant members of the subtilisin gene family have been isolated (Yamagata et al. 1994; Ribeiro et al. 1995; Jorda et al. 1999; Meichtry et al. 1999).
A common feature of subtilisins is their catalytic triad, consisting of three domains: the D region, the H region, and the S region, the catalytic center. The regions are highly conserved among all known subtilases (Dodson and Wlodawer 1998). A fourth well-conserved domain with a central invariant asparagine residue is the substrate binding site. As shown in Figure 4A, all four domains are conserved in SDD1 and all invariant amino acids are present. Most of the known subtilisins are expressed as pre-pro-protein precursors, which are directed to the endomembrane system or to the extracytoplasmatic phase. The 20 amino-terminal amino acids of SDD1 display the characteristic features of a signal sequence for ER uptake (von Heijne 1986). Sequence comparison to other subtilisins indicates that the putative start of the mature SDD1 protein is at position 113 of the deduced amino acid sequence. No intron was detected in the DNA sequence of SDD1. Southern blot analyses indicated that SDD1 is a single copy, single locus gene (data not shown). The mutation present in the sdd1-1 allele creates a premature stop codon at amino acid 492 leading to the formation of a carboxy-terminally truncated protein lacking the S-domain and, therefore lacking the catalytically active serine residue at position 492 (Fig. 3B).
Figure 4.
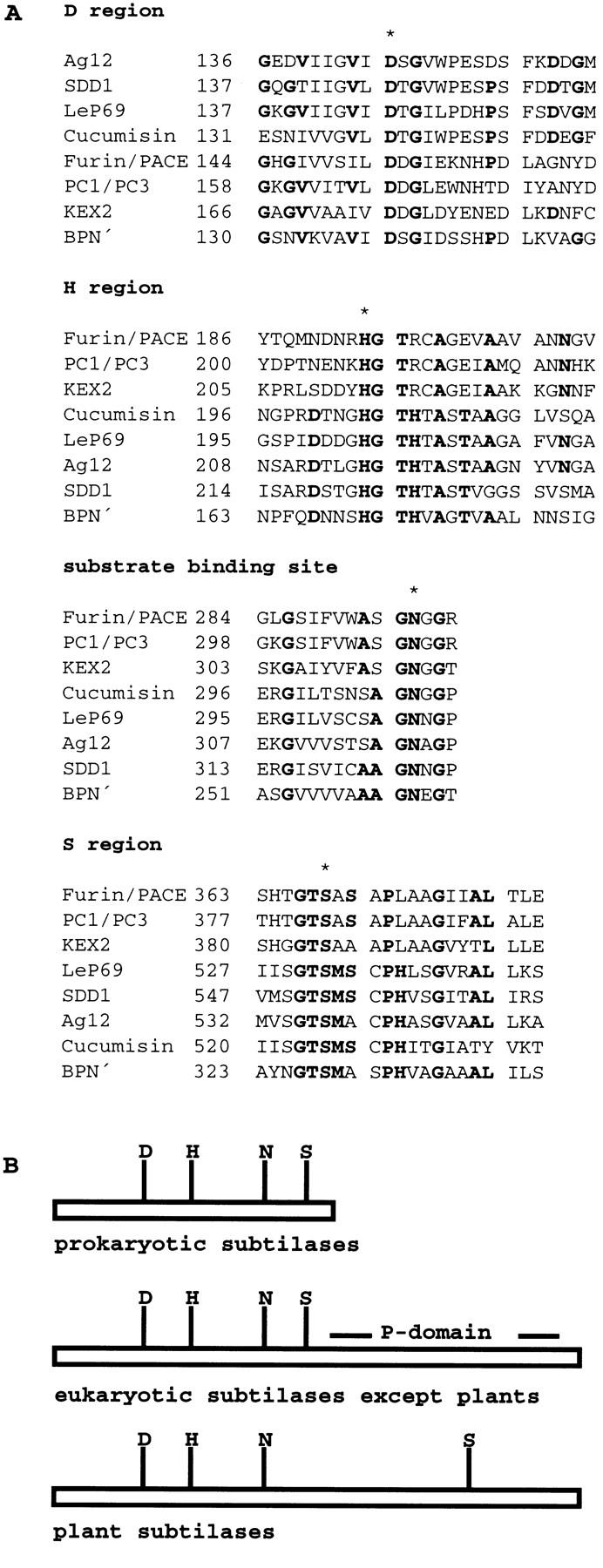
(A) Alignment of the sequences of the characteristic domains of various subtilisin-like serine proteases and SDD1; (B) schematic representation of the overall structure of subtilases. The D, H, and S regions, which together form the catalytic triad, and the substrate-binding site of different subtilisin-like serine proteases—Ag12 from A. glutinosa (Ribeiro et al. 1995), LeP69 from tomato (Jorda et al. 1999), cucumisin from melon (Yamagata et al. 1994), FURIN/PACE (Wise et al. 1990) and PC1/PC3 (Smeekens and Steiner 1990) from human, KEX2 from Sacharomyces cerevisiae (Mizuno et al. 1988), and subtilisin BPN′ from B. amyloliquefaciens (Wells et al. 1983) are shown. Consensus sequences are printed in boldface type, invariant amino acids are marked with an asterisk. (B) The substrate binding sites are marked by an N. The P-domain is exclusively found in nonplant eukaryotic subtilases; the plant representatives are characterized by an extension in the central region of the protein causing a shift of the S-domain toward the amino terminus.
SDD1 is expressed in all aerial organs
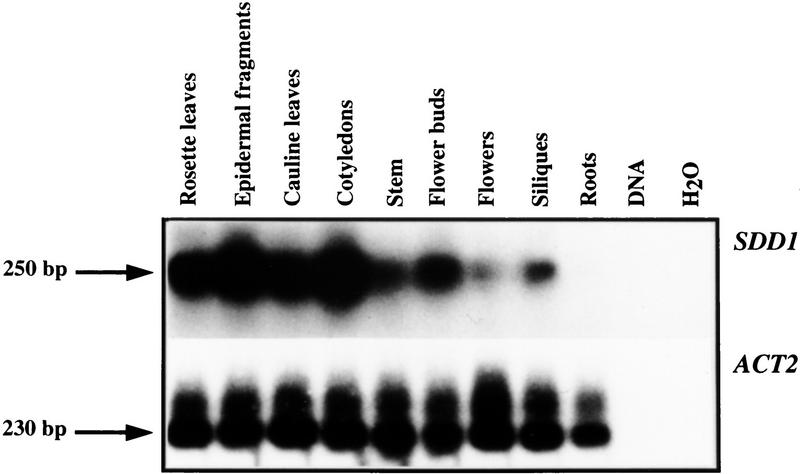
Because RNA-blot analysis indicated the presence of extremely low steady-state SDD1 mRNA levels in wild type plants (data not shown), the expression pattern of SDD1 was investigated by adaptation of the NASBA technique (Leone et al. 1998) for plant tissue analysis. The strongest expression was detected in epidermal fragments of rosette leaves (Fig. 5), an epidermal fraction highly enriched in guard cells (Kopka et al. 1997; K.Raschke, I. Baumann, and R. Hedrich, unpubl.). Relatively strong signals were also detected in the samples extracted from rosette leaves and cotyledons. Lower mRNA levels were detected in floral buds, the stem, and siliques. Expression was the weakest in open flowers and no expression of SDD1 was detected in roots. The Actin2 gene (An et al. 1996), which is constitutively expressed in all of the organs tested, was used to normalize the observed differences in SDD1 expression (Fig. 5).
Figure 5.
Expression analysis of SDD1 (top) by NASBA. SDD1-specific sequences were amplified by NASBA from RNA isolated from various organs, separated by gel electrophoresis, and hybridized with a labeled SDD1 DNA fragment. As control for the amount and the quality of the RNA preparations, mRNA levels of the constitutively expressed ACT2 gene were determined in the same way (bottom). As further control for the specificity of the method for the detection of mRNA, total genomic DNA and a water (mock) control were subjected to the same treatments.
Discussion
Stomata are nonrandomly distributed in the epidermis of green plant organs. In addition to spacing, stomatal density is tightly regulated and is influenced by endogenous as well as exogenous (environmental) factors (for review, see Willmer and Fricker 1996).
This study was devoted to the identification, molecular isolation, and characterization of a gene involved in the regulation of stomatal density and distribution. The SDD1 gene was identified in an Arabidopsis mutant that exhibits increased stomatal density and a disturbed stomatal distribution pattern. The proposed function of the corresponding gene product as a processing protease (a subtilisin-like serine protease) provides the first evidence of the mechanisms involved in the control of these developmental processes.
The sdd1-1 mutation defines a novel factor involved in the control of stomatal distribution
In this study an anisocytic, hemimesogenous, and dolabrate type of stomatal complex formation, as well as the formation of stomatal complexes of higher order (secondary, tertiary, and quaternary complexes, termed satellite stomata by Yang and Sack 1995) was observed. This is in accordance with previous observations of a variety of Brassicaceae (Pant and Kidwai 1967) including Arabidopsis (Yang and Sack 1995; Larkin et al. 1997). In the wild type ∼64% of all stomatal complexes are established through primary cell lineages, whereas ∼29% are secondary, and ∼7% are tertiary stomatal complexes. The sdd1-1 mutant, like the previously identified nonallelic Arabidopsis mutants flp and tmm (Yang and Sack 1995; Geissler et al. 1998), exhibits a number of alterations in stomatal arrangement and positioning of the stomata. Stomatal clusters (groups of guard cell pairs that are not separated by intervening neighboring cells and/or pavement cells) occur in all three mutants. The characteristics of the clusters formed in these mutants differ, however. In flp, guard cell clusters are even or odd numbered (Yang and Sack 1995), whereas sdd1-1 clusters are exclusively even numbered (most frequently composed of four guard cells). In tmm, stomatal clusters occur predominantly in cotyledons and primary leaves and consist of much higher numbers of guard cells (on average, eight guard cells per cluster; Yang and Sack 1995; Geissler et al. 1998). The most striking feature distinguishing sdd1-1 from tmm and flp is sdd1-1's two- to fourfold elevation of stomatal density (number/area) with the concurrent separation of the majority of stomata from each other. Only a minor fraction (except in cotyledons) of the additional stomata occur in clusters in sdd1-1. In flp, the number of guard cells is only moderately increased and the additional stomata are arranged in clusters (Yang and Sack 1995). In tmm, as in sdd1-1, initiation of stomatal complex formation is enhanced and the number of guard cells is strongly elevated. The vast majority of the additional stomata in tmm, however, are arranged in large clusters (Yang and Sack 1995). The proposed functions of TMM and FLP are the organ-dependent initiation and suppression of the formation of stomatal initials, the regulation of the formation and positioning of satellite stomatal complexes, and the control of guard cell/guard mother cell identity (Yang and Sack 1995; Larkin et al. 1997; Geisler et al. 1998).
SDD1 is involved in the regulation of stomatal density and distribution through control of cell fate and orientation of cell divisions
In wild-type Arabidopsis C24 plants, 35.3% of protodermal cells enter a stomatal cell lineage, whereas 59.45% do so in the sdd1-1 mutant (Table 2). Moreover, in the wild type, ∼36% of the stomata are satellite stomata (according to the terminology by Yang and Sack 1995). In contrast, in the sdd1-1 mutant, nearly 74% of the stomata are formed as satellite stomata with a strong shift toward higher order complexes. Consistent with this increase of higher order satellite stomata, the number of stomata produced by individual cell lineages is, on average, more than doubled from 2.25 in wild type to 5.09 in sdd1-1 (Table 2). SDD1, therefore, plays an important role in the initiation of stomatal cell lineages and in the regulation of their lengths. Together, the increase of stomatal initials and the higher number of stomata produced per cell lineage result in a theoretical stomatal density increase of 3.79, which is consistent with the observed three- to fourfold increase.
Clusters in sdd1-1 were produced exclusively by satellite stomatal complexes. Clusters were not formed by two adjacent primary stomata nor did they contain stomata belonging to different, adjacent cell lineages. The guard cell pairs of a given cluster always originated from the same satellite stomata cell lineage and were formed by only one unequal cell division of a neighboring cell. In the mutant, this (first) unequal cell division lacked the tight control of orientation, which always occurs in the wild type and ensures the proper placement of newly formed guard cell mother cells.
It is clear from these results that SDD1 contributes to the control of developmental pathways, including the regulation of the fraction of protodermal cells forming stomatal initials, the cell fate within established cell lineages, and the positioning of stomata. The fraction of protodermal cells forming stomatal initials is elevated 1.7-fold in sdd1-1, yet not all protodermal cells in sdd1-1 develop into stomatal initials. Thus, additional, yet unknown factors together with, but partially independent from SDD1, control protodermal cell fate, specifically the decision between the transition into a meristemoid and the development into a pavement cell. The SDD1 gene product also participates in the control of cell fate within the cell lineages established during stomatal complex formation. Lack of SDD1 activity results in cell lineage extension and the enhanced formation of higher order stomatal complexes. Furthermore, SDD1 affects the positioning of higher order stomata. SDD1 contributes to the control of orientation of unequal neighboring cell divisions in satellite stomatal complexes. These divisions lead to the (direct) formation of higher order guard cell mother cells, a subset of which is improperly positioned in the sdd1-1 mutant.
Given the apparent similarity of the two regulatory pathways determining the fraction of protodermal cells forming stomatal initials and controlling the cell fate within established cell lineages, and given the observation that both pathways involve the activity of SDD1, it is possible that the same mechanism operates in both instances and that protodermal cells and neighboring cells within developing stomatal complexes are developmentally equivalent.
Molecular nature of SDD1 and its possible mode of action
SDD1 is a member of the family of subtilisin-like serine proteases (subtilisins, subtilases), which have been identified in prokaryotes and eukaryotes. Two classes of subtilisins can be distinguished: (1) Degenerative subtilases identified mainly in prokaryotes are responsible for degradation of foreign proteins, and (2) from processing enzymes from eukaryotes; for example from human, mouse, Caenorhabditis, Drosophila, hydra, yeast or, more recently, from plants (for review, see Wells and Estelle 1988; Barr 1991; Steinert et al. 1992; Hook et al. 1994; Siezen and Leunissen 1997). These enzymes are expressed as pre-pro-proteins and are directed to the extracytoplasmic phase via a leader peptide. Here, the prosubtilases are activated and perform their processing function on other pro-proteins (prohormones, proreceptors). Hitherto, little is known about the role of subtilases in plants. A subtilase (cucumisin) has been isolated from the sarcocarp of melon fruits (Yamagata et al. 1994). Four different subtilase genes have been cloned from tomato and analyzed for expression (Jorda et al. 1999). Two of these genes are induced during pathogen attack and two are developmentally regulated. In total, fifteen different subtilisin-like serine protease genes have been cloned from tomato (Meichtry et al. 1999). The subtilase gene, Ag12 is expressed during nodule formation in Alnus glutinosa and a homolog of this gene has been identified in A. thaliana (Ribeiro et al. 1995). The AIR3 gene of Arabidopsis encodes another subtilisin-like serine protease. This gene is expressed in precursor cells that are initiated upon auxin-induced lateral root formation (Neuteboom et al. 1999).
Prokaryotic subtilases are small and terminate immediately behind the S-domain at the carboxyl terminus. These subtilisins are protein-degrading enzymes. In eukaryotes, with the exception of plants, a large domain, the so called P-domain, is a characteristic feature of processing subtilases (Fig. 4B). Deletion of the P-domain leads to the retention of Kex2p in the ER due to a failure in prodomain cleavage (Gluschankof and Fuller 1994). In plants, the distance between the substrate binding site and the S-domain is increased by ∼200 amino acids, whereas the P-domain is lacking. According to Gluschankof and Fuller (1994), processing subtilases have P-domains and degenerative subtilases lack this domain. Plant subtilases would be grouped with the degrading-type subtilases. Recent data, however, indicate that plant subtilases are of the processing type: The substrates for eukaryotic processing subtilases are polypeptides involved in signal transduction. Also in plants, small polypeptides have been identified, for example, systemin (Pearce et al. 1991; Schaller and Ryan 1995) and ENOD40 (Charon et al. 1997; Bladergroen and Spaink 1998), that are involved in the regulation of different processes such as wound signaling or nodulation. Systemin is synthesized as a 200-amino-acid precursor (pro-systemin, McGurl et al. 1992) that is itself processed by SBP50, a pro-systemin-binding protein isolated from membrane fractions of tomato cells, which is immunologically related to subtilisins (Schaller and Ryan 1994). Furthermore, a pathogenesis-induced subtilase isolated from citrus exocortis viroid-infected tomato plants is directed to the extracellular phase and cleaves a pathogen-induced 85-kD leucine-rich repeat protein into its 83-kD active form (Vera et al. 1989; Tornero et al. 1996).
The amino acid sequence near the substrate-binding sites of plant subtilases is well conserved with the consensus sequence VSXSAGNXG. Exceptions are SDD1, with the sequence VICAAGNNG, and AIR3, with the sequence VVCSAGNSG. This could be a hint to different substrates accepted by the various subtilases, which thus might be involved in different signaling pathways.
As outlined above, a major role of SDD1 is that of a negative regulator of the initiation/maintenance of meristemoidal character (the potential to divide and to develop into specialized structures) of cells that lead to guard cell formation. Regulatory factors that participate in similar steps of cell fate control, for example, the switch between the progression into a terminally differentiated state or the maintenance of meristematic activity of cells have been described for other developmental processes in plants. Loci encoding negative regulatory factors are represented by the clavata1, clavata2, and clavata3 mutations (clv1, clv2, clv3; Clark et al. 1993, 1995). All of these mutations cause overproliferation of stem cells leading to enlarged shoot apical meristems and floral meristems and thus to the production of extra organs. CLAVATA 3 encodes a small 78-amino-acid peptide containing a putative dibasic processing site recognized by subtilases (Fletcher et al. 1999), which is probably involved in signal transduction. In a manner similar to that in the clavata mutants, overproliferation of meristemoidal cells occurs in sdd1-1: The fraction of protodermal cells that form stomatal complexes (stomatal initials) is increased and, in addition, the stomatal initial-derived cell lineages are extended.
Acting as a processing protease, SDD1 may activate a proteinaceous signal molecule or a corresponding receptor that mediates communication between developing epidermal cells. The lack of a reduced stomatal density in transgenic Arabidopsis plants overexpressing SDD1 under the control of the CaMV 35S promoter indicates that SDD1 is necessary for proper signal transduction but is not responsible for the tuning of the signal intensity. The perception of the SDD1-dependent signal by protodermal cells or by developing subsidiary cells in young stomatal complexes is likely to contribute to a shift of a developmental program from meristemoidal activity toward epidermal cell differentiation. This perception is likely to be mediated through additional factors.
Materials and methods
Isolation of sdd1
A transgenic Arabidopsis line was generated through an Agrobacterium tumefaciens mediated transfer (according to Schmidt and Willmitzer 1988) of a chimeric uidA gene placed under the control of the guard-cell-selective AGPase S promoter of Solanum tuberosum (Müller-Röber et al. 1994). A line selected for single locus T-DNA insertion, diploidy, and stable transgene expression was mutagenized with EMS (16-hr treatment of seeds with 0.5% EMS). Rosette leaves from the derived M2 plants were stained with x-Gluc (according to Jefferson et al. 1987), cleared with 80% ethanol, and analyzed microscopically. The sdd1-1 mutant was identified among about 3500 plants screened. The mutant was backcrossed four times to the corresponding wild type, A. thaliana cv. C24.
Analysis of stomatal density and stomatal index
Stomatal density and stomatal index were determined for various organs of wild-type (SDD1) and mutant (sdd1-1) plants by light microscopy of nail polish imprints. These imprints were taken from plants germinated and grown in soil [soil types P and T (Einheitserdewerk W. Tantau, Uetersen, Germany) mixed 2:1 and supplemented with 25% sand] under a 16-hr light regimen (140–150 μE fluorescent light, lamp type: TLD36W/840 and TLD36W/830, Phillips, Hamburg, Germany) at 20°C and 60% relative air humidity.
Leaf cross sections
Mature rosette leaves were fixed in 0.2% glutaraldehyde, 4% formaldehyde, 0.5% Triton X-100, 0.1 m Sodium phosphate buffer (pH 7.0) for 12 hr and dehydrated in increasing concentrations of ethanol (75%, 85%, 90%, 95%, and 2 times 100%) for 24 hr. The leaves were kept in Technovit 7100 (Heraeus, Kulzer, Werheim, Germany) overnight and were embedded in Technovit 7100 according to manufacturers instructions. Sections 2- to 5-μm (Leica microtome 2155, Nussloch, Germany) were stained with 0.1% toluidine blue in 0.05% borate for 15–30 sec, excess of dye was rinsed with water, and sections were examined microscopically using Nomarski optics for enhanced contrast (Axiophot, Zeiss, Germany).
Analysis of the formation of individual stomatal complexes using serial imprints
Wild-type and mutant plants were germinated and grown at 80 μE fluorescent light. Dental resin imprints (Kagan et al. 1992) were repeatedly taken from the abaxial surface of primary leaves. The first imprints were taken from leaf primordia (0.5- to 0.8-mm in size). In the first 2 days, imprints were taken every 12 hr; thereafter imprints were taken at daily intervals until the analyzed leaves reached maturity (∼14 days). Nail polish copies prepared from the dental resin imprints were analyzed by light microscopy. In this retrospective analysis, protodermal cells were operationally defined as mitotically active nondifferentiated cells formed through equal cell divisions in the developing epidermis of very young, growing leaves at the developmental stage prior to the initiation of specialized structures (trichomes, stomata). Stomatal initials were defined as those protodermal cells which undergo unequal divisions leading to the formation of stomatal complexes (guard cells plus neighboring cells).
Fine mapping of SDD1
The SDD1 locus was mapped using individuals of an F2 population derived from a cross between sdd1-1 (genetic background C24) and wild-type Col-0. Genetic markers were scored as RFLP through DNA-blot analyses or as CAPS (Konieczny and Ausubel 1993) after PCR amplification. The markers and the restriction enzymes showing polymorphisms used in this study were PVV4 (HaeIII) and PAI-1 (AluI) (Konieczny and Ausubel 1993) and 0846A (BglII) (C. Dean, pers. comm.). Map distances were calculated according to Kosambi (1944). For fine mapping, individuals carrying chromosomes with recombination breakpoints around the SDD1 locus were identified by testing 389 F2 plants for PVV4 and 440 plants for PAI-1. DNA prepared from the identified 36 F2 individuals or from individual pools of corresponding F3 plants was used to test closely linked markers. New genetic markers (F25I3RE and F20G19LE) were developed through recovery of end fragments of genomic DNA cloned in BACs (Mozo et al. 1998b) that were used to identify corresponding RFLPs.
Isolation of YAC ends
Preparation of total yeast DNA and of inverse PCR (iPCR) substrates was carried out according to Gibson and Somerville (1992). Primer sequences for iPCR (Ochmann et al. 1988) were EG1, GGCGATGCTGTCGGAATGGACGATA; EG2, CTTGGAGCCACTATCGACTACGCGATC; EG3, CCGATCTCAAGATTACGGAAT; EG4, TTCCTAATGCAGGAGTCGCATAAG. PCR conditions were as described by Koncieczny and Ausubel (1993), with the following exceptions: annealing temperature for isolation of right ends was 62°C and for isolation of left ends 56°C. PCR was performed in a water bath thermocycler (Biomed, Theres, Germany) with 25 cycles.
Isolation of BAC ends
BAC plasmids were isolated by alkaline lysis (Sambrook et al. 1989). Preparations of iPCR substrates were the same as for isolation of YAC ends with the following exceptions: Relinearization was done by incubation with PvuI (for isolation of left ends) or with BsrBI (for isolation of right ends). PCR was carried out with the following primers: BAC1LE, TTCCCAACAGTTGCGCAGC; BAC2LE, TCTTCGCTATTACGCCAGCT; BAC3RE, TCACACAGGAAACAGCTAT; BAC4RE, ACACAACATACGAGCCGGAA and an annealing temperature of 56°C for the isolation of both ends.
Identification of the sdd1-1 mutation
Bordered by the two closest flanking markers, F20G19LE and F25I3RE, the sdd1-1 mutation was localized within a region of 113 kb, which is covered by the two fully sequenced BAC clones, F21M11 and F20D22 (GenBank accession nos. AC003027 and AC002411). For the identification of the sdd1-1 mutation, restriction SSCP analysis was performed according to Dean and Gerrard (1991) and Iwahana et al. (1992): Fifty-seven 2-kb fragments were amplified by PCR carried out in 100 μl according to Koncieczny and Ausubel (1993). PCR products were purified using the NucleoSpin 2-in-1 kit (Macherey Nagel, Düren, Germany) and digested with the restriction enzymes HaeIII and AluI. After phenol/chloroform/isoamylalcohol extraction, the samples were mixed with one volume denaturation buffer (50% formamide in TE, supplemented with bromphenole blue and xylene xyanole), heated to 95°C for 5 min and chilled on ice. Then, 4 μl were loaded on 12% (29:1 in 1× TBE buffer) nondenaturating PAA gels. After 20–24 hr electrophoresis at 10 V/cm at room temperature, the gels were stained with silver (Silver stain plus kit, Bio-Rad, Hercules, CA).
Subcloning of SDD1 and complementation of sdd1-1
A 7-kb genomic EcoRV–SalI fragment (derived from the BAC clone F20D22, that carries the 2.3-kb SDD1 coding region in addition to 2 kb of upstream DNA, including the SDD1 promoter, and 2.7 kb downstream DNA) was inserted into the SmaI–SalI-digested vector pBIB-Hyg (Becker 1990). This construct was called G-SDD1.
A second construct, called 35S-SDD1, was created by ligation of a fragment (amplified from the BAC F20D22, that covered the 2328-bp SDD1 coding region and was provided with Asp718 linker sequences at the 5′ end and XbaI linker sequences at the 3′ end) into the Asp718- and XbaI-digested vector pBinAR-Hyg (Höfgen and Willmitzer 1990).
G-SDD1 and 35S-SDD1 were transferred into the sdd1-1 mutant through A. tumefaciens mediated in planta transformation (Bechthold et al. 1993). Transgenic seedlings were selected through germination and growth on a sterile medium for 13 days in half-concentrated MS medium (Murashige and Skoog 1962), supplemented with 1% sucrose and 20 mg/liter hygromycin B. Antibiotic-resistant seedlings were transferred to soil (type GS90, Einheitserdewerk W. Tantau, Uetersen, Germany, containing 25% sand) and cultivated for another 6 days under a regimen of 16 hr light (90 μE fluorescent light) with day and night temperatures of 23°C and 20°C, respectively, before fully expanded cotyledons were detached from the seedlings and cleared with 80% ethanol. Nontransformed sdd1-1 mutant plants were grown under the same conditions with the exception of omission of HygromycinB from the germination medium. To analyze primary leaves, nail polish copies of dental resin imprints (Kagan et al. 1992) were taken from the abaxial leaf surface after a further culture of the plants for 2 weeks. Cleared cotyledons and nail polish imprints were examined by standard light microscopy.
NASBA analysis
The NASBA procedure was modified after that of Leone et al. (1998): Each reaction contained 40 mm Tris-HCl (pH 8.8), 12 mm MgCl2, 70 mm KCl, 0.5 mm DTT, 1 mm dNTPs, 2 mm ATP, UTP, and CTP each, 1.5 mm GTP, 0.5 mm ITP in a final volume of 7 μl. After the addition of 100 ng of total RNA isolated according to Logemann et al. (1987), 5 μl of the primer mix containing 15% DMSO and 0.2 μmole of each primer was added. Primers used for amplification of SDD1 were subtilase fwd [5′-CAGCTCTGACATTCTAGCAGCT (nucleotides 807–828)] and subtilase T7 [5′-AATTCTAATACGACTCACTATAGGGGTTGGCTAATCTGACCACAGC (nucleotides 1057–1077), the T7 promoter sequence is underlined]. Primers used for quantification of the constitutively expressed Actin 2 gene (An et al. 1996) were ACT2 fwd [GCCGTTTTGAATCTCCGGCG (nucleotides −589 to −570)] and ACT2 T7 [AATTCTAATACGACTCACTATAGGGGAATATCATCAGCCTCAGCCAT (nucleotides 1–22), the T7 promoter sequence is underlined]. Between nucleotides −453 and −11, an intron is located within the 5′-nontranslated region of Actin 2. The RNA/nucleotide/primer mixture was heated to 65°C for 5 min and chilled on ice for 5 min. After addition of 8 μl of enzyme mix containing 375 mm sorbitol, 2 μg of BSA, 0.1 units of RNase H
(Boehringer Mannheim, Germany), 10 units of avian myeloblastosis virus (AMV) reverse transcriptase (Boehringer Mannheim, Germany) and 30 units T7 RNA polymerase (Boehringer Mannheim, Germany), the samples were incubated at 41°C for 60 min and stored at −20°C for further use.
Post-NASBA analysis
To the NASBA reaction, two volumes of denaturating electrophoresis buffer [1.2× MEN buffer (50% formamide, 6% formaldehyde, 50 μg ethidium bromide, supplemented with bromphenol blue and xylene xyanol)] were added, heated to 75°C for 10 min (denaturation), and kept on ice. Then, 20 μl was loaded onto a 2% denaturing agarose gel (SeaKem agarose, FMC, Rockland, USA) in 1× MEN buffer (200 mm MOPS at pH 7.0, 50 mm sodium acetate, 10 mm EDTA, 6% formaldehyde). After electrophoresis at 120 V for 1–2 hr, nucleic acids were blotted onto a nylon membrane (Hybond N+, Amersham Buchler, Braunschweig, Germany) with 20× SSC [1× SSC (0.15 m NaCl and 15 mm sodium citrate)] by capillary transfer overnight. Nucleic acids were cross-linked to the membrane by baking at 80°C for 2 hr. After washing the membrane with 2× SSC for 5 min, hybridization was performed at 65°C for 16 hr in NSEB buffer (250 mm sodium phosphate at pH 7.2, 7% SDS, 1 mm EDTA, 1% BSA) with a radioactively labeled DNA fragment (ready prime, Amersham Buchler, Braunschweig, Germany) covering the complete sequence of NASBA products. The filters were washed twice with 2× SSC, 0.1% SDS, for 20 min at 65°C. Exposure on X-ray films was carried out at −80°C for 2–16 hr with intensifying screens.
Acknowledgments
We thank L. Willmitzer for his support and his continuous strong interest in this project, B. Marty, B. Hausmann, S. Strähle, F. Huhn, K. Lepa, and B. Altmann for plant care, J. Bergstein for photographic work, and M. McKenzie for critical reading of the manuscript. We are very grateful to O. Leyser for communicating map data and supplying new markers prior to publication, and we thank C. Dean and E. Coen for providing markers. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG), Al 1-1, 1-2, 1-3.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL altmann@mpimp-golm.mpg.de; FAX 49 (331) 567-8250.
References
- Abak K, Yanmaz R. Investigation on the stomatal density in certain pepper lines and their F1 hybrids. Capsicum Newslett. 1985;4:22. [Google Scholar]
- An Y-Q, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant j. 1996;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- Barr PJ. Mammalian subtilisins: The long-sought dibasic processing endoproteases. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Bechthold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci. 1993;316:1194–1199. [Google Scholar]
- Becker D. Binary vectors which allow the exchange of selectable markers and reporter genes. Nucleic Acids Res. 1990;18:203. doi: 10.1093/nar/18.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Linstead P, Dolan L, Haselhof J. Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev Biol. 1998;194:226–234. doi: 10.1006/dbio.1997.8836. [DOI] [PubMed] [Google Scholar]
- Bladergroen MR, Spaink H. Genes and signal molecules involved in the rhizobia-leguminosae symbiosis. Curr Opin Plant Biol. 1998;1:353–359. doi: 10.1016/1369-5266(88)80059-1. [DOI] [PubMed] [Google Scholar]
- Bünning E. Entwicklungs- und Bewegungsphysiologie der Pflanzen. Berlin, Germany: Springer Verlag; 1953. [Google Scholar]
- Buttery BR, Gaynor JD, Buzell RI, Mactavish DC, Armstrong RJ. The effect of shading on kaempherol content and leaf characteristics of five soybean lines. Physiol Plant. 1992;86:279–284. [Google Scholar]
- Buttery BR, Tan CS, Buzell RI, Gaynor JD, Mactavish DC. Stomatal numbers of soybean and response to water stress. Plant Soil. 1993;149:283–288. [Google Scholar]
- Charon C, Johansson C, Kondorosi E, Kondorosi A, Crespi M. enod40 induces dedifferentiation and division of root cortical cells in legumes. Proc Natl Acad Sci. 1997;94:8901–8906. doi: 10.1073/pnas.94.16.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JC, Wan Y, Smith J, Croxdale J. Linear aggregations of stomata and epidermal cells in Tradescantia leaves: Evidence for their group patterning as a function of the cell cycle. Dev Biol. 1995;168:39–46. doi: 10.1006/dbio.1995.1059. [DOI] [PubMed] [Google Scholar]
- Clark PJ, Evans FC. Distance to nearest neighbor as a measure of spatial relationships in populations. Ecology. 1954;35:445–453. [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121:2057–2067. [Google Scholar]
- Clifford SC, Black CR, Roberts JA, Stronach IM, Singleton-Jones PR, Mohamed AD, Azam-Ali SN. The effect of elevated atmospheric CO2 and drought on stomatal frequency in groundnut [Arachis hypogea (L.)] J Exp Bot. 1995;46:847–852. [Google Scholar]
- Creusot F, Fouilloux E, Dron M, Lafleuriel J, Picard G, Billault A, LePaslier D, Cohen D, Chabouté D, Durr E, et al. The CIC library: A large insert YAC library for genome mappimg in Arabidopsis thaliana. Plant J. 1995;8:763–770. doi: 10.1046/j.1365-313x.1995.08050763.x. [DOI] [PubMed] [Google Scholar]
- Dean M, Gerrard B. Helpful hints for the detection of single stranded conformational polymorphisms. BioTechniques. 1991;10:332–333. [PubMed] [Google Scholar]
- Dodson G, Wlodawer A. Catalytic triades and their relatives. Trends Biochem Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- Fletcher LC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decision by CLAVATA3 in Arabidopsis shoot meristem. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Gay AP, Hurd RG. The influence of light on stomatal density in the tomato. New Phytol. 1975;75:37–46. [Google Scholar]
- Geisler MJ, Yang M, Sack FD. Divergent regulation of stomatal initiation and patterning in organ and suborgan regions of the Arabidopsis mutants too many mouths and four lips. Planta. 1998;205:522–533. doi: 10.1007/s004250050351. [DOI] [PubMed] [Google Scholar]
- Gibson SI, Somerville C. Chromosome walking in Arabidopsis thaliana using yeast artificial chromosomes. In: Koncz C, Chua N-H, Schell J, editors. Methods in Arabidopsis research. Singapore: Word Scientific Publishing; 1992. pp. 119–143. [Google Scholar]
- Gluschankof P, Fuller RS. A C-terminal domain conserved in precursor processing protease is required for intramolecular N-terminal maturation of pro-Kex2 protease. EMBO J. 1994;13:2280–2288. doi: 10.1002/j.1460-2075.1994.tb06510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L. Biochemical and genetic analysis of different patatin isoforms expressed in various organs of potato Solanum tuberosum. Plant Sci. 1990;66:221–230. [Google Scholar]
- Hook VYH, Azaryan AV, Hwang S-R, Tezapsidis N. Proteases and the emerging role of protease inhibitors in prohormone processing. FASEB J. 1994;8:1269–1278. doi: 10.1096/fasebj.8.15.8001739. [DOI] [PubMed] [Google Scholar]
- Iwahana H, Yoshimoto K, Itakura M. Detection of point mutations by SSCP of PCR-amplified DNA after endonuclease digestion. BioTechniques. 1992;12:64–66. [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. Gus fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HG. In: Stomatal function. Zeiger E, et al., editors. Stanford, CA: Stanford University Press; 1987. pp. 431–443. [Google Scholar]
- Jorda L, Coego A, Conejero V, Vera P. Genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J Biol Chem. 1999;274:2360–2365. doi: 10.1074/jbc.274.4.2360. [DOI] [PubMed] [Google Scholar]
- Kagan M, Novoplansky N, Sachs T. Variable cell lineages form the functional pea epidermis. Ann Bot. 1992;69:303–312. [Google Scholar]
- Koncieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kopka J, Provart NJ, Müller-Röber B. Potato guard cells respond to drying soil by a complex change in the expression of genes related to carbon metabolism and turgor regulation. Plant J. 1997;11:871–882. doi: 10.1046/j.1365-313x.1997.11040871.x. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombinant values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Larkin JC, Young N, Prigge M, Marks MD. The control of trichome spacing and number in Arabidopsis. Development. 1996;122:997–1005. doi: 10.1242/dev.122.3.997. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Marks MD, Nadeau J, Sack F. Epidermal cell fate and patterning in leaves. Plant Cell. 1997;9:1109–1120. doi: 10.1105/tpc.9.7.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, van Schijndel H, van Gemen B, Kramer FR, Schoen CD. Molecular beacon probes combined with amplification of NASBA enable homogenous, real-time detection of RNA. Nucleic Acids Res. 1998;26:2150–2155. doi: 10.1093/nar/26.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Maynard JC, Mertz SM, Jr, Arntzen CJ, Payne WW. Abnormal guard cell development in an olive necrotic mutant of maize. Am J Bot. 1974;61:580–584. [Google Scholar]
- McGurl B, Pearce G, Orozco-Cardenas M, Ryan CA. Structure, expression and antisense inhibition of the systemin precursor gene. Science. 1992;255:1570–1573. doi: 10.1126/science.1549783. [DOI] [PubMed] [Google Scholar]
- Meichtry J, Amrhein N, Schaller A. Characterization of the subtilase gene family in tomato (Lycopersicon esculentum Mill.) Plant Mol Biol. 1999;39:749–760. doi: 10.1023/a:1006193414434. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Nakamura T, Ohshima T, Tanaka S, Matsuo H. Yeast KEX2 gene encodes an endopeptidase homologous to subtilisin-like serine proteases. Biochem Biophys Res Commun. 1988;156:246–254. doi: 10.1016/s0006-291x(88)80832-5. [DOI] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Meier-Ewert S, Lehrach H, Altmann T. Use of the IGF-BAC library for physical mapping of the Arabidopsis thaliana genome. Plant J. 1998a;16:377–384. doi: 10.1046/j.1365-313x.1998.00299.x. [DOI] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Shizuya H, Altmann T. Construction and characterization of the IGF Arabidopsis library. Mol & Gen Genet. 1998b;258:562–580. doi: 10.1007/s004380050769. [DOI] [PubMed] [Google Scholar]
- Müller-Röber B, La Cognata U, Sonnewald U, Willmitzer L. A truncated version of an ADP-glucose pyrophosphorylase promotor from potato specifies guard cell selective expression in transgenic plants. Plant Cell. 1994;6:601–612. [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Neuteboom LW, Ng JMY, Kuyper M, Clijdesdale OR, Hooykaas PJJ, van der Zaal EJ. Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formation. Plant Mol Biol. 1999;39:273–287. doi: 10.1023/a:1006104205959. [DOI] [PubMed] [Google Scholar]
- Ochmann H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:411–414. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant DD, Kidwai PF. Development of stomata in some Cruciferae. Ann Bot. 1967;31:513–521. [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253:895–898. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- Rahim MA, Fordham R. Effect of shade on leaf and cell size and number of epidermal cells in garlic (Allium sativum) Ann Bot. 1991;67:167–171. [Google Scholar]
- Ramos LJ, Nayaranan KR, McMillan RT., Jr Association of stomatal frequency and morphology in Lycopersicon species with resistance to Xanthomonas campestris pv. vesicatoria. Plant Pathol. 1992;41:157–164. [Google Scholar]
- Reich PB. Leaf stomatal density and diffusive conductance in 3 amphistomatous hybrid poplar cultivars. New Phytol. 1984;98:231–240. [Google Scholar]
- Ribeiro A, Akkermans ADL, van Kammen A, Bisseling T, Pawlowski K. A nodule-specific gene encoding a subtilisin-like protease is expressed in early stages of actinorhizal nodule development. Plant Cell. 1995;7:785–794. doi: 10.1105/tpc.7.6.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T. The development of spacing patterns in the leaf epidermis. In: Subtelny S, Sussex IM, editors. The clonal basis of development. New York, NY: Academic Press; 1978. pp. 161–183. [Google Scholar]
- ————— . Pattern formation in plant tissues. Cambridge, U.K: Cambridge University Press; 1991. [Google Scholar]
- ————— Both cell lineages and cell interactions contribute to stomatal patterning. Int J Plant Sci. 1994;155:245–247. [Google Scholar]
- Sachs T, Benouaiche P. A control of stomata maturation in Aeonium. Isr J Bot. 1978;27:47–53. [Google Scholar]
- Sachs T, Novoplansky N, Kagan M. Variable development and cellular patterning in the epidermis of Ruscus. Ann Bot. 1993;71:237–243. [Google Scholar]
- Salisbury EJ. On the causes and ecological significance on stomatal frequency with special reference to woodland flora. Phil Trans Roy Soc Lond Ser B. 1927;216:1–65. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaller A, Ryan CA. Identification of a 50-kDa systemin-binding protein in tomato plasma membranes having Kex2p-like properties. Proc Natl Acad Sci. 1994;91:11802–11806. doi: 10.1073/pnas.91.25.11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Systemin: A polypeptide defense signal in plants. BioEssays. 1995;18:27–33. doi: 10.1002/bies.950180108. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Willmitzer L. High efficiency Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana leaf and cotyledons explants. Plant Cell Rep. 1988;7:583–586. doi: 10.1007/BF00272763. [DOI] [PubMed] [Google Scholar]
- Schoch P-G, Zinsou C, Sibu M. Dependence of stomatal index on environmental factors during stomata differentiation in leaves of Vigna signensis L. J Exp Bot. 1980;31:1211–1216. [Google Scholar]
- Schürmann B. Über den Einfluss der Hydratur und des Lichtes auf die Ausbildung der Stomata-Initialen. Flora. 1959;147:471–520. [Google Scholar]
- Serna L, Fenoll C. Tracing the ontogeny of stomatal clusters in Arabidopsis with molecular markers. Plant J. 1997;12:747–755. doi: 10.1046/j.1365-313x.1997.12040747.x. [DOI] [PubMed] [Google Scholar]
- Siezen RJ, Leunissen JAM. Subtilases: The superfamily of subtilisin-like serine proteases. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens SP, Steiner DF. Identification of a human insulinoma cDNA encoding a novel mammalian protein structurally related to the yeast dibasic processing protease KEX2. J Biol Chem. 1990;265:2997–3000. [PubMed] [Google Scholar]
- Srivastava A, Lu Z, Zeiger E. Modification of guard cell properties in advanced lines of pima cotton bred for higher yields and heat resistance. Plant Sci. 1995;108:125–131. [Google Scholar]
- Steinert DF, Smeekens SP, Ohagi S, Chan SJ. The new enzymology of precursor processing endoproteases. J Biol Chem. 1992;267:23435–23438. [PubMed] [Google Scholar]
- Tornero P, Mayda E, Gomez MD, Canas L, Conejero V, Vera P. Characterization of LRP, a leucine-rich repeat (LRR) protein from tomato plants that is processed during pathogenesis. Plant J. 1996;10:315–330. doi: 10.1046/j.1365-313x.1996.10020315.x. [DOI] [PubMed] [Google Scholar]
- Vera P, Yago JH, Conejero V. Immunogold localization of the Citrus Exocortis Viroid-induced pathogenesis-related proteinase P69 in tomato leaves. Plant Physiol. 1989;91:119–123. doi: 10.1104/pp.91.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA, Estelle DA. Subtilisins—an enzyme designed to be engineered. Trends Biochem Sci. 1988;13:291–297. doi: 10.1016/0968-0004(88)90121-1. [DOI] [PubMed] [Google Scholar]
- Wells JA, Ferrari E, Henner DJ, Estell DA, Chen EY. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983;11:7911–7919. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmer C, Fricker M. Stomata. London: Chapman and Hall; 1996. pp. 95–125. [Google Scholar]
- Wise RJ, Barr PJ, Wong PA, Kiefer MC, Brake AJ, Kaufmann RJ. Expression of a human proprotein processing enzyme: Correct cleavage of the von Willebrand factor precursor at a paired basic amino acid site. Proc Natl Acad Sci. 1990;87:9378–9382. doi: 10.1073/pnas.87.23.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Masuzawa T, Nagaoka Y, Ohnishi T, Iwasaki T. Cucumisin, a serine protease from melon fruits, shares structural homology with subtilisin and is generated from a large precursor. J Biol Chem. 1994;269:32725–32731. [PubMed] [Google Scholar]
- Yang M, Sack FD. The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell. 1995;7:2227–2239. doi: 10.1105/tpc.7.12.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger E, Stebbins GL. Developmental genetics in barley. A mutant for stomatal development. Am J Bot. 1972;59:143–148. [Google Scholar]