Abstract
High-risk human papillomaviruses (HPVs) infect stratified epithelia to establish persistent infections that maintain low-copy-number episomes in infected basal cells. Amplification of viral genomes occurs upon keratinocyte differentiation, followed by virion synthesis. During persistent HPV infections, viral proteins act to evade surveillance by both innate and adaptive immune responses. One of the primary pathways regulating the innate immune response is the JAK/STAT pathway. Our studies indicate that the expression of STAT-1, but not other members of interferon (IFN)-stimulated gene factor 3 (ISGF-3) complex such as STAT-2 and IFN regulatory factor 9 (IRF9), is selectively suppressed by HPV proteins at the level of transcription. Both E6 and E7 oncoproteins independently suppress the expression of STAT-1, and mutational analyses indicate that the E6 targeting E6-associated protein (E6AP) is responsible for suppression. The levels of STAT-1 proteins increase upon differentiation of both normal and HPV-positive cells but are still significantly reduced in the latter cells. Transient restoration of STAT-1 levels in HPV-positive cells using recombinant retroviruses significantly impaired viral amplification upon differentiation while long-term increases abrogated maintenance of episomes. Similarly, increased levels of STAT-1 induced by gamma interferon treatment inhibited HPV genome amplification upon differentiation. Overall, our findings demonstrate that suppression of STAT-1 expression by HPV proteins is necessary for genome amplification and maintenance of episomes, suggesting an important role for this activity in viral pathogenesis.
INTRODUCTION
Human papillomaviruses (HPVs) are the causative agents of over 98% of cervical cancers, which are the second most common cancers in women worldwide (53). Over 120 HPV types have been identified, and about 30% of HPVs infect the genital epithelia. These genital HPV types are further classified as either high risk (e.g., HPV16, -18, -31, and -35) or low risk (e.g., HPV6 and -11) according to their association with genital cancers (25, 30). The high-risk HPVs are causative agents of cervical cancers and are associated with cancers of the vulva, vagina, anus, and penis as well as the oral cavity. Prior to the development of cancers, HPVs establish persistent infections in the genital tract that successfully evade immune clearance (3, 25, 30).
HPVs infect stratified epithelia and establish their double-stranded DNA genomes as episomes that are replicated in a differentiation-dependent manner (28). During their productive life cycles, these viruses escape host innate immune surveillance as well the adaptive responses through mechanisms that are not fully understood. HPV genomes encode approximately six early genes and two late genes. The E5, E6, and E7 oncoproteins play important roles in regulating the productive life cycle as well as contributing to immune evasion and development of anogenital cancers (28). The high-risk E6 proteins form complexes with the cellular E3 ubiquitin ligase E6-associated protein (E6AP) and p53, resulting in p53 degradation (6, 17, 23, 45). E6 also binds to p300 (36) and blocks p53 acetylation (13), which further inhibits p53 function. E6AP may mediate other cellular events such as activation of expression of the catalytic subunit of telomerase, hTert, and other less-characterized substrates (16, 24, 50). The E7 proteins bind to members of the retinoblastoma protein (Rb) tumor suppressor family (11), resulting in constitutive activation of the E2F family of transcription factors (33), which are critical for host cell cycle progression and differentiation. The binding of E5 to the B cell receptor-associated protein 31 (BAP31) suggests a potential negatively regulatory role of E5 on the interferon (IFN)-inducible trafficking of major histocompatibility complex (MHC) class I proteins (38).
The first line of host defense against viral infections is the innate immune response, which includes the IFN and Janus kinase-signal transducer and activator of transcription (JAK/STAT) pathways (1). The JAK/STAT pathway transmits information from extracellular interferon (5) through kinases to activate expression of over a hundred genes mediated through the translocation of STAT proteins to the nucleus (39, 48). The activation of this pathway involves the phosphorylation and homodimerization of STAT-1 or the heterodimerization of STAT-1 and STAT-2 (44). In cells that stably maintain HPV genomes, the expression of many IFN-inducible genes, such as the myxovirus resistance and protein A gene (MXA, also known as MX1) and 2′-5′-oligoadenylate synthetase 2 (OAS2), was previously shown by microarray analysis to be suppressed (7, 31). STAT-1 was also found to be suppressed by HPV gene products, and it was hypothesized that this may contribute to the repression of downstream interferon-inducible genes (31). The addition of interferon to HPV-positive cells induces expression of a number of interferon-inducible genes (31). Long-term treatment of cells that maintain HPV episomes with IFN-β results in cell death and the appearance of resistant populations that contain integrated HPV genomes (13, 14). The suppression of the interferon-inducible pathway by HPV proteins could be mediated through multiple cellular targets. For instance, E6 has been reported to associate with Tyk2 kinase to interfere with activation of the JAK/STAT pathway (22) as well as bind to the IFN regulatory factor 3 (IRF3), inhibiting its ability to activate interferon expression (41). Furthermore, E7 is able to bind to IRF1 and to inhibit IFN signaling (35). Finally, direct suppression of STAT-1 transcription could result in repression of a number of interferon-inducible genes. Clearly, HPV proteins target the expression and activities of many components of the interferon-inducible pathway, but usually this pathway is targeted at the initial phases of infection. It is less clear is why HPV suppresses the interferon-inducible pathway even after a persistent infection has been established.
In this study we have investigated how HPV proteins suppress STAT-1 levels and whether this downregulation plays an important role in the viral life cycle. We observe that STAT-1 suppression is mediated primarily at the level of transcription and that both HPV oncoproteins E6 and E7 regulate this activity. E6 mediates this suppression through the binding of E6AP, which may be linked to its effects on p53. Significantly, restoration of STAT-1 in HPV-positive cells impaired genome amplification upon differentiation and interfered with stable maintenance of episomes. Furthermore, treatment of differentiating HPV-positive keratinocytes with gamma interferon also inhibited viral genome amplification. Our study provides important insights into suppression of STAT-1 in HPV-positive cells and could provide new targets for treatments to eradicate HPV infections.
MATERIALS AND METHODS
Cell culture.
Human foreskin keratinocytes (HFKs) were isolated from neonatal foreskins using standard methods. HPV genome-expressing cells or E6- or E7-expressing cell lines were selected by G418 (Sigma, St. Louis, MO) using HFKs after transfection with the HPV genome or infection with retroviruses, as previously described (13). All HFKs and HPV-positive cells were cultured in E-medium supplemented with mouse epidermal growth factor (5 ng/ml; Collaborative Biomedical Products, Bedford, MA) along with mitomycin C-treated NIH 3T3 J2 fibroblast feeders, as previously described (13). To induce differentiation in high calcium concentrations, cells were cultured in keratinocyte basal medium (KBM) with growth supplements for at least 24 h and then switched to KBM (without supplements) containing 1.5 mM CaCl2 for 96 h. Alternatively, keratinocytes were grown in organotypic raft cultures to induce differentiation, as previously described (49).
Materials and plasmids.
The antibodies used in this study are as follows: anti-STAT-1, anti-STAT-2, anti-involucrin, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and anti-poly(ADP-ribose) polymerase (PARP) from Santa Cruz (Santa Cruz, CA); anti-p53 from Calbiochem (Gibbstown, NJ); anti-pRb from Cell Signaling (Danvers, MA); and anti-keratin-10 from Abcam (Cambridge, MA). The STAT-1 promoter reporter was a generous gift from J. L. Merchant, University of Michigan (2). The STAT-1 plasmid used for retrovirus infection was generously provided by N. Khodarev and R. Weichselbaum at The University of Chicago (18). The STAT-1 expression plasmid was purchased through Addgene.
Luciferase assay.
HFKs or HPV-positive cells were seeded into six-well plates. The cells were transfected with polyethyleneimine (PEI) and incubated at 37°C for 36 h. Luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI), with Renilla luciferase as an internal control, according to manufacturer's instructions. Significance was determined using a Student's t test.
Western blotting.
Intact HFKs or HPV-positive cells were rinsed with phosphate-buffered saline (PBS) and incubated in PBS containing 0.5 mM EDTA for 2 min to remove J2 feeders at room temperature. The cell lysates were then prepared and transferred to a membrane as previously described (15). The membranes were developed using ECL Plus or ECL reagents (Amersham, Pittsburgh, PA). Chemiluminescence signals were recorded using Eastman Kodak X-ray film.
Reverse transcription-PCR (RT-PCR).
The cells were isolated as described above, and total RNA was extracted using Complete Miniprep (Zymo), purified by RNA concentrator (Zymo, Irvine, CA). Five micrograms of RNA was then transcribed into cDNA using a SuperScript First Round Synthesis System (Invitrogen, Carlsbad, CA). The RT products were mixed with LightCycler 480 SYBR green I Master mix (Roche, Indianapolis, IN), and PCR was performed using a LightCycler 480 instrument. Primer pairs used in the study were designed as follows: for STAT-1, 5′-ATTACTCCAGGCCAAAGGAAGCAC-3′ (forward) and 5′-AGCAAGGCTGGCTTGAGGTTTG-3′ (reverse); for GAPDH, 5′-GAGGACAGAGACCCAGCTGCC-3′ (forward) and 5′-TGGAATTTGCCATGGGTG-3′ (reverse). Results shown are representative of observations from three independent experiments. Normalization to GAPDH is used as a reference. Significance was determined using a Student's t test.
Southern blotting.
The J2 feeders were removed before the processing of HPV-positive cells as described above, and the HPV DNA was isolated as previously described (27). Briefly, following cell lysis, DNA was isolated, and samples were digested with DpnI and XhoI. The DNA samples were electrophoresed in a 0.8% agarose gel at 40 V overnight and then transferred to Gene Screen nylon membranes (Bio-Rad, Hercules, CA) using vacuum transfer according to the manufacturer's protocols. The membrane was hybridized with radioactive probes, washed, and visualized by autoradiography.
RESULTS
HPVs suppress STAT-1 transcription.
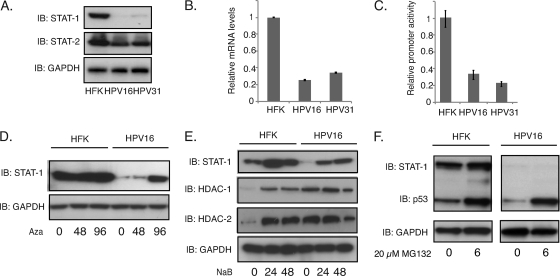
To investigate how and why HPV proteins target STAT-1 proteins during the viral life cycle, we first examined the levels in HPV16- and HPV31-positive cells that stably maintain episomes. The levels of STAT-1 proteins were screened by Western blot analysis of HPV16- and -31-positive human cells grown in monolayer culture and compared them to those seen in normal human keratinocytes (HFKs). Consistent with previous reports (7), we found reduced levels of STAT-1 in HPV-positive cells (Fig. 1A). In contrast, no differences were seen with either STAT-2 (Fig. 1A) or STAT-3 proteins (data not shown), indicating that suppression is specific to STAT-1. Similarly, no differences were seen in the levels of IRF-9 that is present along with STAT-1 and STAT-2 in the IFN-stimulated gene factor 3 (ISGF-3) transcription activation complex (data not shown). To determine if the effects were mediated at the level of transcription or posttranscriptionally, we performed RT-PCR studies on STAT-1 transcripts in HPV-positive and -negative keratinocytes and found comparable reductions to those seen in proteins levels (Fig. 1B). The above analyses do not distinguish between changes at the level of initiation of transcription or posttranscriptional destabilization of messages. To investigate if downregulation of STAT-1 was mediated at the transcriptional level, we transfected STAT-1 reporters in which the promoter of STAT-1 is fused to the luciferase gene (2) into HPV-positive and -negative keratinocytes and assayed for expression levels. As shown in Fig. 1C, luciferase reporter expression was specifically repressed in HPV-positive cells, indicating that the primary mode of suppression is at the level of transcription. In addition, we observed that repression could be significantly reversed by the treatment of HPV-positive cells with DNA modification inhibitors, such as the histone deacetylase inhibitor sodium butyrate (NaB) and the DNA methylation inhibitor 5-aza-2′-deoxycytidine (Aza) (Fig. 1D and E). In contrast, treatment of cells with MG132, an inhibitor of proteasome-mediated degradation, had no effects on STAT-1 protein levels (Fig. 1F). These studies indicate that HPV downregulates STAT-1 expression primarily at the level of transcription.
Fig. 1.
HPV suppresses STAT-1 protein levels and transcription. (A) Western blot analysis of STAT-1, STAT-2, and GAPDH levels in HFK, HPV16, and HPV31 cells. (B) RT-PCR assay of STAT-1 mRNA levels in HFK, HPV16, and HPV31 cells (P < 0.02). (C) STAT-1 promoter activities were characterized using a dual luciferase assay in transfected HFK, HPV16, and HPV31 cells (P < 0.05). (D) Western blot analysis for STAT-1 and GAPDH levels in cells treated with 5-dAza-C (Aza; 2.5 μM) for indicated times (h). (E) Western blot analysis for STAT-1, histone deacetylase 1 (HDAC-1), HDAC-2, and GAPDH levels in cells treated with sodium butyrate (NaB; 6 mM) for indicated times (h). (F) Western blot analysis for STAT-1, p53, and GAPDH levels in cells treated with MG132 (20 μM) for 6 h. All results are representative of observations from three independent experiments. IB, immunoblot.
HPV oncoproteins E6 and E7 individually inhibit the expression of STAT-1.
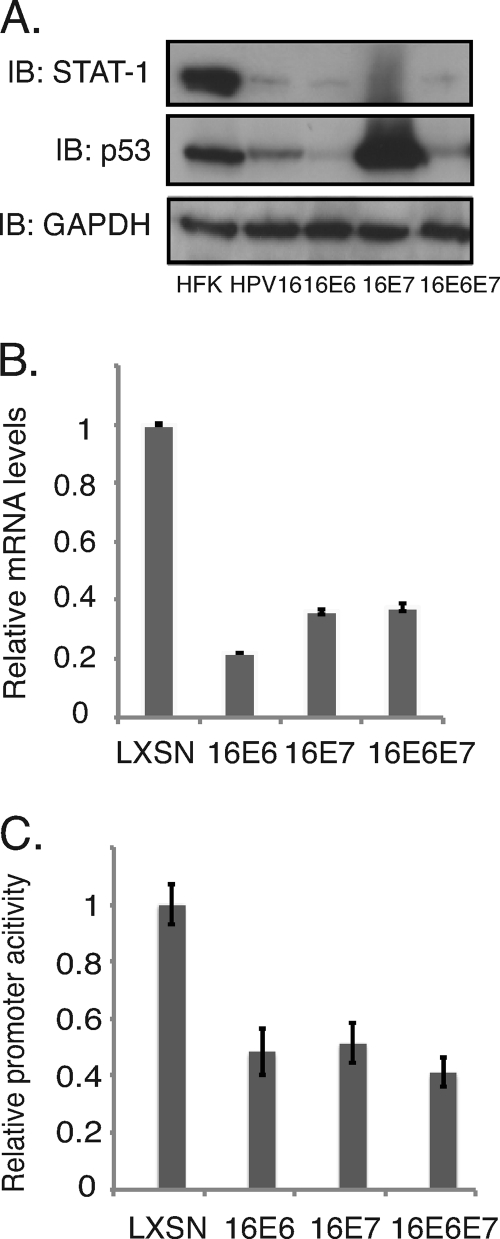
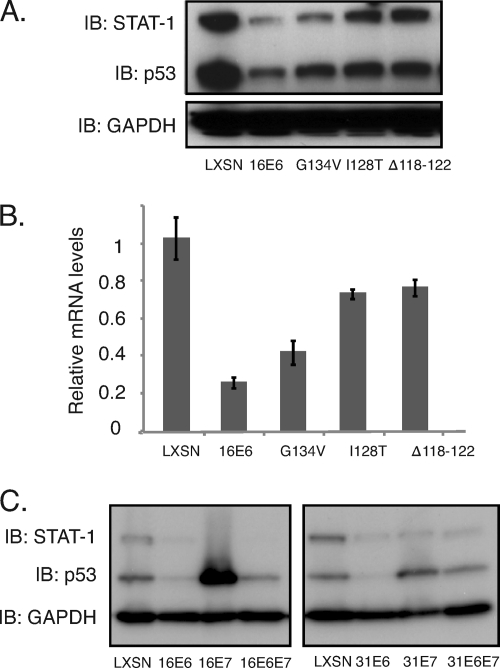
It was next important to determine which HPV proteins were responsible for mediating suppression of STAT-1 expression. For this analysis, we generated retrovirally transduced cell lines expressing either E6 alone or E7 alone and screened for the levels of STAT-1. We observed that the levels of STAT-1 proteins as well as mRNAs were reduced in these oncoprotein-expressing cells compared to those seen in HFKs. Similar effects were seen in cells expressing both E6 and E7 (Fig. 2A and B), and comparable effects were seen in cells expressing either HPV16 or -31 oncoproteins. When STAT-1 luciferase reporters were transfected into cells expressing E6, E7, or E6 and E7, decreased levels of STAT-1 expression were seen, indicating that both factors act at the level of transcription (Fig. 2C). Similar to the effects seen in cells containing HPV episomes, treatment of E6- or E7-expressing cells with NaB or Aza induced the expression of STAT-1 (see Fig. S1 in the supplemental material). It has been reported that p53 can contribute to STAT-1 activation (51), and since E6 targets p53 for degradation as well as blocking its acetylation by p300 (36, 52), we investigated if modulation of p53 by E6 is responsible for STAT-1 suppression. For these studies, several previously characterized E6 mutants (13) were examined for the ability to suppress STAT-1 levels. We infected normal keratinocytes with recombinant retroviruses expressing wild-type or mutant E6 proteins and generated stable cells lines. The G134V mutant fails to bind CBP/p300, resulting in p53 acetylation while maintaining its ability to bind and degrade p53 (13, 47). The deletion of residues 118 to 122 (Δ118–122 mutant) and the I128T mutation inhibit E6 binding to E6AP, therefore preventing p53 degradation (45). We observed that two of the three E6 mutants were able to restore STAT-1 protein levels to those seen in HFKs (Fig. 3). The Δ118–122 and I128T mutants were more effective in restoring STAT-1 protein levels than the G134V mutation, which had only a minimal effect. This suggested that p53 might play a role in regulating STAT-1 levels. However, when we examined the levels of p53 in cells expressing either high-risk E6 or E7, we found significant variability in the levels of p53, yet all were able to reduce STAT-1 to comparable levels (Fig. 3C). This indicates that absolute levels of p53 levels may not be the primary property responsible for STAT-1 repression and suggests that some other activity of p53 or another target of E6AP maybe be important.
Fig. 2.
HPV oncoproteins E6 and E7 individually inhibit the expression of STAT-1. (A) Western blot analysis for STAT-1, p53, and GAPDH levels in HFK cells and cells expressing HPV16, HPV16 E6, and HPV16 E7 as well as HPV16 E6 and E7. (B) RT-PCR assay of STAT-1 mRNA levels in HFK LXSN, HPV16 E6, HPV16 E7, and HPV16 E6 and E7 cells (P < 0.01). (C) STAT-1 promoter activities were characterized using a dual luciferase assay in transfected HFK LXSN, HPV16 E6, HPV16 E7, and HPV16 E6 and E7 cells (P < 0.05). All results are representative of observations from three independent experiments.
Fig. 3.
Mutagenesis studies of E6 indicate a link between STAT-1 suppression and the ability of E6 binding to E6AP. (A) Western blot analysis for STAT-1, p53, and GAPDH levels in HFK cells expressing LXSN vector, HPV16 E6 wild-type and G134V, I128T, and Δ118–122 mutants. (B) RT-PCR assay for STAT-1 mRNA levels in HFK cells expressing LXSN vector, HPV16 E6 wild-type and G134V, I128T, and Δ118–122 mutants (P < 0.02). (C) Western blot analysis for STAT-1, p53, and GAPDH levels in HFK cells expressing LXSN, E6, E7, and E6 and E7 of HPV16 or HPV31. All results are representative of observations from three independent experiments.
STAT-1 levels increase upon differentiation, and this is inhibited in HPV-positive cells.
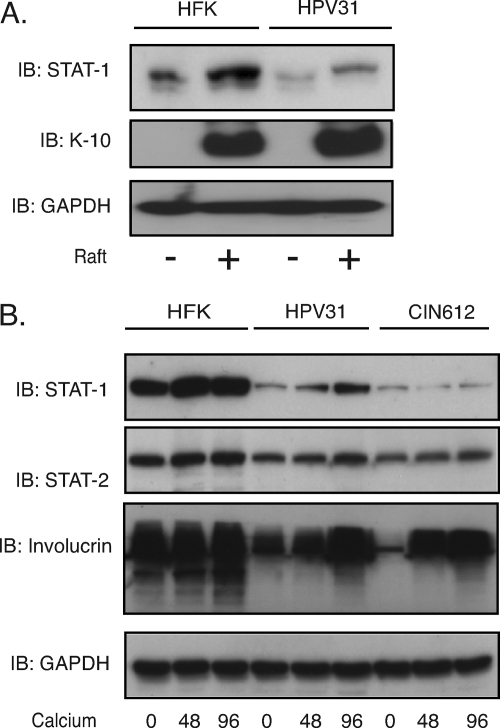
In order to investigate how STAT-1 levels changed during the differentiation-dependent life cycle of human papillomaviruses, we examined the levels of STAT-1 in HPV-positive and -negative keratinocytes grown in organotypic raft cultures or high-calcium medium. As shown in Fig. 4A, the levels of STAT-1 increased upon differentiation of normal foreskin keratinocytes in rafts. In undifferentiated HPV31-positive cells, the levels of STAT-1 are reduced from those seen in HFKs. The levels of STAT-1 also increased upon differentiation of HPV-positive cells but to a lesser extent than seen in HFKs (Fig. 4A). The expression of keratin 10 (K-10), a member of intermediate filaments, was used as a marker of differentiation. We observed similar effects using either HPV16- or HPV31-positive cells as well as CIN612 cells, which are HPV31 positive and were derived from a patient biopsy specimen using another system for differentiation involving growth in high-calcium medium (Fig. 4B). We conclude that while STAT-1 levels increase upon differentiation of HPV-positive cells, they are still significantly reduced from those seen in normal cells.
Fig. 4.
STAT-1 levels increase upon differentiation, and this is reduced in HPV-positive cells. (A) Western blot analysis for STAT-1, K-10, and GAPDH levels in HFK and HPV31 cells grown in organotypic raft culture or in undifferentiated monolayer cultures. (B) Western blot analysis for STAT-1, STAT-2, involucrin, and GAPDH levels in HFK, HPV16, and HPV31 cells differentiated in calcium medium for the indicated times. All results are representative of observations from three independent experiments.
Restoration of STAT-1 levels blocks HPV genome amplification upon keratinocyte differentiation.
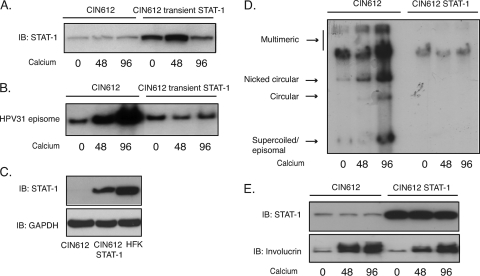
Our studies examining STAT-1 levels in cell lines that stably maintain HPV episomes indicated significant reductions from those seen in HFKs. Activation of the STAT-1 pathway normally occurs during the initial phases of infection so as to inhibit viral spread. After HPV genomes are established and stably maintained as low-copy-number nuclear episomes, it is less clear if STAT-1 levels need to be continuously suppressed. To address this question, we transiently transfected HPV31-positive CIN612 cells with expression vectors for STAT-1 and after 36 h induced these cells to differentiate in high-calcium medium. Amplification of HPV31 genomes begins at approximately 48 h in high-calcium medium and plateaus by 96 h. The total levels of STAT-1 proteins in the transfected cells were increased compared to control CIN612 cells (Fig. 5A) and comparable to amounts seen in HFKs. After 48 and 96 h following addition of high-calcium medium, total DNA was isolated and examined for viral genome amplification by Southern blot analysis. As shown in Fig. 5B, restoring the levels of STAT-1 in HPV-positive cells resulted in significantly reduced amplification of viral genomes upon differentiation.
Fig. 5.
Restoration of STAT-1 levels blocks HPV genome amplification upon keratinocyte differentiation. (A) Western blot analysis of STAT-1 protein levels in CIN612 cells or in cells transiently expressing STAT-1 following differentiation in high-calcium medium for the indicated times. (B) Southern blot analysis for HPV31 episomes in CIN612 cells or the cells transiently expressing STAT-1 following differentiation in high-calcium medium for indicated times (h). (C) Western blot analysis for STAT-1 and involucrin protein levels in CIN612 cells or CIN612 cells stably expressing STAT-1 following differentiation in calcium medium for the indicated times (h). (D) Southern blot analysis for HPV31 genome in CIN612 cells or the cells stably expressing STAT-1 following differentiation in high-calcium medium for the indicated times (h). (E) Western blot assay for STAT-1 expression in CIN612 cells, CIN612 cells stably expressing STAT-1, and HFK cells. All results are representative of observations from two or more independent experiments.
We next wanted to investigate the consequences of long-term restoration of STAT-1 levels on the stable maintenance of HPV episomes as well as differentiation-dependent genome amplification. Monolayer cultures of CIN612 cells, which do not express drug resistance markers, were infected with recombinant retroviruses expressing STAT-1 along with resistance markers, and positive colonies were selected. We examined these selected cells for the levels of STAT-1 by Western analysis and found similar levels to those seen in HFKs (Fig. 5C). These STAT-1-expressing cells maintain the ability to proliferate at a growth rate similar to that of the parental CIN612 cells (data not shown). We next examined the state of viral DNA in these cells and found the genomes to be integrated (Fig. 5D). Consistent with a requirement for episomal genomes for amplification, we detected no change in genome copy number upon differentiation in high calcium concentrations (Fig. 5D). We also observed minimal changes in the expression of the differentiation marker involucrin (Fig. 5E). Similar results were seen in three separate experiments. We conclude that suppression of STAT-1 expression by viral proteins is required for differentiation-dependent genome amplification as well as long-term maintenance of episomal genomes.
IFN-γ treatment blocks HPV amplification upon keratinocyte differentiation.
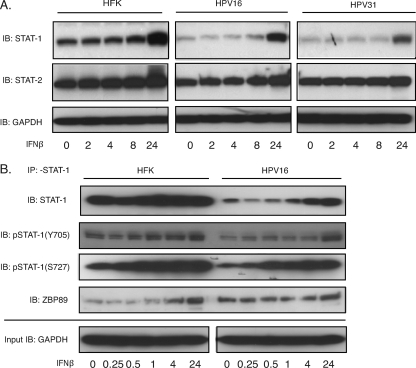
We previously observed that in HPV-positive cells incubated with IFN-β, STAT-1 expression was increased to levels comparable to those seen in treated HFKs (7). We first confirmed that the levels of total STAT-1 proteins increased within 24 h of IFN-β treatment (Fig. 6A). Next, it was important to investigate how the phosphorylated forms of STAT-1, which represent the active forms of the protein, changed with interferon treatment. STAT-1 is phosphorylated at either Y701 or S727, and we found that both forms were reduced in untreated HPV-positive cells compared to HFKs (Fig. 6B). While the total levels of phosphorylated STAT-1 proteins were reduced in HPV-positive cells, the relative phosphorylation ratios in these cells might be high. The significance of this is unclear. Upon exposure to IFN-β, the levels of STAT-1 phosphorylation at both sites increased to those seen in treated HFKs, and these were similar to increases in total STAT-1 protein levels. Surprisingly, the levels of the major positive activator of STAT-1 transcription, ZBP89 (2), were found at higher levels in untreated HPV-positive cells than in HFKs. Upon interferon treatment the levels of ZBP89 increased in HFKs but only minimally in HPV-positive cells. It is not clear why ZBP89 does not increase in HPV-positive cells, but the results suggest that there is some block to activation (Fig. 6B). This suggests that HPV proteins do not act through ZBP89 to repress STAT-1 expression but most likely through modulation of another transcription factor.
Fig. 6.
IFN-β treatment leads to STAT-1 protein induction and activation. (A) Western blot analysis of STAT-1, STAT-2, and GAPDH in HFK, HPV16, and HPV31 cells treated with 500 ng/ml IFN-β for the indicated times (h). (B) Lysates from HFK and HPV16 cells treated with 500 ng/ml IFN-β for the indicated times (h) were immunoprecipitated with anti-STAT-1 and assayed by Western blotting for STAT-1, phospho-STAT-1 (Y701), phospho-STAT-1 (S727), and ZBP-89. Twenty-microliter aliquots from the lysates were assayed for GAPDH as a loading control using Western blotting. All results are representative of observations from three independent experiments.
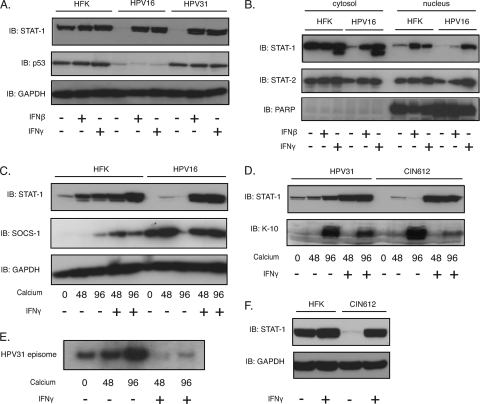
Treatment of cells with gamma interferon results in phosphorylation of STAT-1, homodimerization, and translocation to the nucleus to activate an overlapping but distinct set of responsive genes relative to those activated in response to IFN-α/β (44). We next examined the consequences of treatment of HPV-positive cells with IFN-γ and found increases in total STAT-1 levels similar to those seen following treatment with IFN-β (Fig. 7A). It was important to determine if STAT-1 proteins were present in the cytoplasm of untreated cells and if they translocated to the nucleus upon addition of interferons. For this analysis, we fractionated cells and used poly(ADP-ribose) polymerase (PARP) as a nuclear marker (10). As expected in the absence of added interferons, low levels of STAT-1 were found in the nucleus of HFKs, whereas in HPV-positive cells minimal levels of STAT-1 proteins were observed in either the cytosol or nucleus. Upon IFN-γ treatment, levels of nuclear STAT-1 increased, and similar increases were seen with the cytosolic compartment (Fig. 7B). Comparable effects were seen with IFN-β treatment (Fig. 7B). A slight increase in nuclear STAT-2 was observed upon IFN-γ treatment, but we do not believe that it is significant (Fig. 7B). A previous study reported that HPV16 E6 and E7 proteins reduced levels of nuclear STAT-1 but not those in the cytosol, suggesting that there might be a defect in the process of nuclear translocation (31). Our studies demonstrated equivalent reductions in both nuclear and cytosolic STAT-1, which is consistent with decreases in the levels of gene transcription. The addition of IFN-γ during differentiation resulted in increased levels of STAT-1 in HPV-positive cells to levels comparable to those seen in HFKs (Fig. 7C). Interestingly, the levels of SOCS-1, a direct inhibitor of the JAK2 kinase (19), were found to be elevated in untreated HPV-positive cells compared to levels in HFKs (Fig. 7C). The levels of SOCS-1 increased upon differentiation as well as with IFN-γ treatment to levels significantly greater than those seen in HFKs. This suggests that HPV may act to not only suppress the STAT-1 expression but also downregulate signaling through upstream interferon receptors. The reduced levels at 96 h of interferon treatment are an aberration not seen in other experiments.
Fig. 7.
IFN treatment blocks HPV genome amplification. (A) Western blot analysis of STAT-1, p53, and GAPDH levels in HFK, HPV16, and HPV31 cells treated with 500 ng/ml IFN-β or 50 ng/ml IFN-γ for 24 h. (B) Lysates from the cells processed the same as in panel A were further separated into the cytosol and nucleus fractions. The fractions were assayed by Western blotting for STAT-1, STAT-2, and PARP. (C) Western blot analysis of STAT-1 and SOCS-1 in IFN-γ-treated or untreated HFK and HPV16 cells differentiated in calcium medium for the indicated times (h). (D) Western blot analysis for STAT-1 and K-10 levels in IFN-γ-treated or untreated HFK and CIN612 cells differentiated in calcium medium for the indicated times (h). (E) Southern blot analysis of HPV31 episomes in CIN612 cells or IFN-γ-treated CIN612 cells differentiated in calcium medium for the indicated times (h). (F) Western blot analysis of STAT-1 levels in IFN-γ-treated or untreated HFK and CIN612 cells. All results are representative of observations from two or more independent experiments.
Since treatment of HPV-positive cells with interferon restores STAT-1 levels to those seen in HFKs (Fig. 7A and F), we next investigated the effect of treating differentiated cells with IFN-γ on viral genome amplification. Cells that stably maintain HPV31 genomes were treated with IFN-γ and induced to differentiate in high-calcium medium. After 48 and 96 h, total DNA was isolated and examined by Southern blot analysis. As shown in Fig. 7E, short-term treatment with IFN-γ blocked genome amplification, consistent with the studies described above in which STAT-1 levels were restored through the use of recombinant retroviruses. We conclude that restoration of STAT-1 levels either by interferon treatment or through forced expression from heterologous vectors is deleterious to differentiation-dependent HPV genome amplification.
DISCUSSION
Activation of the JAK/STAT signaling pathway constitutes an important regulatory mechanism by which host cells inhibit viral infections. The central transcriptional activators of this pathway are the STAT proteins. The STAT-1, STAT-2, and STAT-3 proteins are important regulators of the innate immune responses to infections by a variety of RNA and DNA viruses (9, 12, 32, 37). In our study, we observed that HPV suppressed the constitutive expression of STAT-1 but not STAT-2, IRF-9, or STAT-3. STAT-2 acts in a complex with STAT-1, and both are activated in response to IFN-α or IFN-β (20, 44). This suggests that suppression of STAT-1 expression by HPV proteins is not the consequence of a general repression of interferon signaling but, rather, of specific modulation of STAT-1 promoter activity. A model is shown in Fig. S2 in the supplemental material. A number of viruses such as adenoviruses, paramyxovirus, and parainfluenza viruses reduce the levels of STAT-1 and STAT-2 during infection by increasing protein turnover (8, 21, 26, 34, 40), but no effects on protein stability were observed in our studies, and the majority of activity was directed at suppressing transcription.
Both HPV proteins E6 and E7 were found to independently inhibit STAT-1 expression, with the combination being most effective. The mechanism by which E6 suppresses STAT-1 promoter activity was found to be dependent upon the binding of E6AP. One of the consequences of this interaction is the enhanced degradation of p53. p53 has been previously implicated as a regulator of IFN-inducible gene expression (43), but in our studies we could not establish a strict correlation between p53 levels and suppression of STAT-1, suggesting that some other activities may be responsible. E6AP has additional activities beyond the enhancement of p53 degradation, and it is possible that these may play a role in suppressing STAT-1 expression. One potential target of E6 action could be the transcription factor ZBP89 that regulates STAT-1 expression; however, we observed that in HPV-positive cells, the levels of ZBP89 were increased rather than decreased relative to levels in HFKs, as might be expected if this factor were responsible. This suggests that ZBP89 is not responsible for STAT-1 suppression induced by HPV proteins and indicates that other, as yet undefined, transcription factors are targeted by viral proteins for this activity. It is still unclear how E7 contributes to suppression of STAT-1, but we believe it likely to be dependent either on binding of histone deacetylases, as recently shown to be important for HIF-1 activation (4), or through association with the Rb family.
During the HPV life cycle, late functions including genome amplification are induced upon epithelial differentiation. Our studies indicate that while STAT-1 levels increase upon differentiation of HPV-positive cells, they remain significantly reduced from those seen in normal keratinocytes. During initial infection, HPV virions enter basal cells and disassemble, and viral genomes are established in the nucleus as low-copy-number episomes that replicate together with cellular chromosomes. It is understandable why STAT-1 would need to be suppressed during the initial phases of the life cycle but less clear if this downregulation is needed once genomes are stably maintained as nuclear episomes or whether this suppression is a remnant of the initial establishment phase. Our studies indicate that continued suppression of STAT-1 is necessary for differentiation-dependent genome amplification as well as stable maintenance of episomes. In our studies, we transiently restored STAT-1 levels in HPV-positive cells to those seen in normal keratinocytes and found this blocked genome amplification. When STAT-1 proteins were stably increased in HPV-positive cells, the ability even to maintain viral episomes was compromised. This was not the result of a general effect on cell growth or proliferation but was specifically targeted at HPV replication. These studies were, however, performed only in a single HPV-positive cell line, CIN612 cells, that stably maintain HPV31 episomes, and it remains a possibility that the effects of restoring STAT-1 levels on stable HPV replication are restricted to this one cell type.
The question arises why restoring STAT-1 levels to those seen in HFKs is so deleterious to viral replication since increased expression in normal cells has minimal effects on cell viability. STAT-1 along with STAT-2 are major transcriptional activators of the interferon pathway that target over 100 downstream genes, and it is possible that one or more of these factors acts to inhibit HPV genome amplification and plasmid maintenance. One candidate is the p56 protein that has been previously shown to bind to the E1 replication protein and inhibit its helicase activity, as well as its interactions with the second viral replication factor E2 (42, 46). By suppressing STAT-1, HPV proteins could downregulate p56, and understanding if this mechanism is responsible is of high interest. Preliminary studies indicate that p56 RNA levels are efficiently suppressed in HPV-positive cells compared to HFKs but are induced to high levels upon interferon treatment (S. Hong and L. A. Laimins, unpublished data). It is also possible that other STAT-1-regulated factors are responsible for effects on HPV replication or that several proteins act together with p56 to mediate these effects. STAT-1 has also been shown to regulate activation of the DNA damage response, and we have previously shown that this pathway is important for genome amplification upon differentiation (29). However, our initial studies fail to show increased levels of DNA damage response factors in cells with elevated STAT-1 (S. Hong unpublished data). Finally, since STAT-1 is a transcriptional activator, it might have direct or indirect effects on HPV gene transcription that could also impact viral replication.
We also observed that the levels of SOCS-1, a JAK2 kinase inhibitor, were elevated in untreated HPV-positive cells compared to levels in HFKs. The levels of SOCS-1 increased upon differentiation as well as with IFN-γ treatment to levels significantly greater than those seen in HFKs. This indicates that HPV proteins interfere with signaling through the JAK/STAT pathway at multiple points and further underscores the importance of blocking this activity during persistent infections. Our studies identify STAT-1 as a negative regulator of HPV replication and demonstrate the importance of continuous downregulation of the interferon-inducible pathway for persistence of HPV infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Juanita L. Merchant for her generous gift of STAT-1 promoter reporter plasmid. The STAT-1 plasmid using for retrovirus infection was generously provided by. N. Khodarev and. R. Weichselbaum. We also thank Vignesh Gunasekharan for his help on the raft studies and other members of the Laimins lab for helpful discussions.
This work was supported by a grant from the National Cancer Institute to L.A.L.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Aaronson D. S., Horvath C. M. 2002. A road map for those who don't know JAK-STAT. Science 296:1653–1655 [DOI] [PubMed] [Google Scholar]
- 2. Bai L., Merchant J. L. 2003. Transcription factor ZBP-89 is required for STAT1 constitutive expression. Nucleic Acids Res. 31:7264–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bodily J., Laimins L. A. 2011. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol. 19:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bodily J. M., Mehta K. P., Laimins L. A. 2011. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer Res. 71:1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borden E. C., et al. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brimer N., Lyons C., Vande Pol S. B. 2007. Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology 358:303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang Y. E., Laimins L. A. 2000. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J. Virol. 74:4174–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Didcock L., Young D. F., Goodbourn S., Randall R. E. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Durbin J. E., Hackenmiller R., Simon M. C., Levy D. E. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–450 [DOI] [PubMed] [Google Scholar]
- 10. Duriez P. J., Shah G. M. 1997. Cleavage of poly(ADP-ribose) polymerase: a sensitive parameter to study cell death. Biochem. Cell Biol. 75:337–349 [PubMed] [Google Scholar]
- 11. Dyson N., Howley P. M., Munger K., Harlow E. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934–937 [DOI] [PubMed] [Google Scholar]
- 12. Hahm B., Trifilo M. J., Zuniga E. I., Oldstone M. B. 2005. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity 22:247–257 [DOI] [PubMed] [Google Scholar]
- 13. Hebner C., Beglin M., Laimins L. A. 2007. Human papillomavirus E6 proteins mediate resistance to interferon-induced growth arrest through inhibition of p53 acetylation. J. Virol. 81:12740–12747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herdman M. T., et al. 2006. Interferon-beta treatment of cervical keratinocytes naturally infected with human papillomavirus 16 episomes promotes rapid reduction in episome numbers and emergence of latent integrants. Carcinogenesis 27:2341–2353 [DOI] [PubMed] [Google Scholar]
- 15. Hong S., et al. 2007. Lipopolysaccharide, IFN-gamma, and IFN-beta induce expression of the thiol-sensitive ART2.1 Ecto-ADP-ribosyltransferase in murine macrophages. J. Immunol. 179:6215–6227 [DOI] [PubMed] [Google Scholar]
- 16. Howie H. L., Katzenellenbogen R. A., Galloway D. A. 2009. Papillomavirus E6 proteins. Virology 384:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huibregtse J. M., Scheffner M., Howley P. M. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khodarev N. N., et al. 2004. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc. Natl. Acad. Sci. U. S. A. 101:1714–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kubo M., Hanada T., Yoshimura A. 2003. Suppressors of cytokine signaling and immunity. Nat. Immunol. 4:1169–1176 [DOI] [PubMed] [Google Scholar]
- 20. Lehtonen A., Matikainen S., Julkunen I. 1997. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J. Immunol. 159:794–803 [PubMed] [Google Scholar]
- 21. Leonard G. T., Sen G. C. 1996. Effects of adenovirus E1A protein on interferon-signaling. Virology 224:25–33 [DOI] [PubMed] [Google Scholar]
- 22. Li S., et al. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727–5737 [DOI] [PubMed] [Google Scholar]
- 23. Li X., Coffino P. 1996. High-risk human papillomavirus E6 protein has two distinct binding sites within p53, of which only one determines degradation. J. Virol. 70:4509–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu X., et al. 2009. HPV E6 protein interacts physically and functionally with the cellular telomerase complex. Proc. Natl. Acad. Sci. U. S. A. 106:18780–18785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lowy D. R., Solomon D., Hildesheim A., Schiller J. T., Schiffman M. 2008. Human papillomavirus infection and the primary and secondary prevention of cervical cancer. Cancer 113:1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malur A. G., Chattopadhyay S., Maitra R. K., Banerjee A. K. 2005. Inhibition of STAT 1 phosphorylation by human parainfluenza virus type 3 C protein. J. Virol. 79:7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Melar-New M., Laimins L. A. 2010. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J. Virol. 84:5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moody C. A., Laimins L. A. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. 10:550–560 [DOI] [PubMed] [Google Scholar]
- 29. Moody C. A., Laimins L. A. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 5:e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Munger K., Howley P. M. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89:213–228 [DOI] [PubMed] [Google Scholar]
- 31. Nees M., et al. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-κB-responsive genes in cervical keratinocytes. J. Virol. 75:4283–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Gorman W. E., et al. 2010. Alternate mechanisms of initial pattern recognition drive differential immune responses to related poxviruses. Cell Host Microbe 8:174–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pagano M., Durst M., Joswig S., Draetta G., Jansen-Durr P. 1992. Binding of the human E2F transcription factor to the retinoblastoma protein but not to cyclin A is abolished in HPV-16-immortalized cells. Oncogene 7:1681–1686 [PubMed] [Google Scholar]
- 34. Parisien J. P., Lau J. F., Rodriguez J. J., Ulane C. M., Horvath C. M. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park J. S., et al. 2000. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J. Biol. Chem. 275:6764–6769 [DOI] [PubMed] [Google Scholar]
- 36. Patel D., Huang S. M., Baglia L. A., McCance D. J. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 18:5061–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perry S. T., Buck M. D., Lada S. M., Schindler C., Shresta S. 2011. STAT2 mediates innate immunity to dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 7:e1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Regan J. A., Laimins L. A. 2008. Bap31 is a novel target of the human papillomavirus E5 protein. J. Virol. 82:10042–10051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reich N. C., Liu L. 2006. Tracking STAT nuclear traffic. Nat. Rev. Immunol. 6:602–612 [DOI] [PubMed] [Google Scholar]
- 40. Rodriguez J. J., Parisien J. P., Horvath C. M. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476–11483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ronco L. V., Karpova A. Y., Vidal M., Howley P. M. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saikia P., Fensterl V., Sen G. C. 2010. The inhibitory action of P56 on select functions of E1 mediates interferon's effect on human papillomavirus DNA replication. J. Virol. 84:13036–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takaoka A., et al. 2003. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424:516–523 [DOI] [PubMed] [Google Scholar]
- 44. Takaoka A., Taniguchi T. 2003. New aspects of IFN-alpha/beta signalling in immunity, oncogenesis and bone metabolism. Cancer Sci. 94:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Talis A. L., Huibregtse J. M., Howley P. M. 1998. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J. Biol. Chem. 273:6439–6445 [DOI] [PubMed] [Google Scholar]
- 46. Terenzi F., Saikia P., Sen G. C. 2008. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 27:3311–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomas M. C., Chiang C. M. 2005. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol. Cell 17:251–264 [DOI] [PubMed] [Google Scholar]
- 48. van Boxel-Dezaire A. H., Rani M. R., Stark G. R. 2006. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25:361–372 [DOI] [PubMed] [Google Scholar]
- 49. Wilson R., Laimins L. A. 2005. Differentiation of HPV-containing cells using organotypic “raft” culture or methylcellulose. Methods Mol. Med. 119:157–169 [DOI] [PubMed] [Google Scholar]
- 50. Wise-Draper T. M., Wells S. I. 2008. Papillomavirus E6 and E7 proteins and their cellular targets. Front. Biosci. 13:1003–1017 [DOI] [PubMed] [Google Scholar]
- 51. Youlyouz-Marfak I., et al. 2008. Identification of a novel p53-dependent activation pathway of STAT1 by antitumour genotoxic agents. Cell Death Differ. 15:376–385 [DOI] [PubMed] [Google Scholar]
- 52. Zimmermann H., Degenkolbe R., Bernard H. U., O'Connor M. J. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 73:6209–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. 2:342–350 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.