Abstract
The interferon (IFN) response is initiated by a variety of triggers, including viruses and foreign RNA, and involves several receptors and intracellular mediators. Although there are common cis-acting consensus sequences in the promoters of many genes stimulated during the IFN response, they exhibit core and context heterogeneity that may lead to differential transcriptional activity. We have developed and validated a live cell-based enhanced green fluorescent protein (EGFP) reporter system employing more than a hundred constructs containing multiple viruses and IFN response elements derived from a variety of promoters involved in immunity to viruses. Common and distinct response patterns were observed due to promoter heterogeneity in response to different stimuli, including IFN-α, TLR3-agonist double-stranded RNA, and several viruses. This information should serve as a resource in selecting specific reporters for sensing nonself ligands.
INTRODUCTION
Interferon (IFN) is produced and secreted by various mammalian cell lines when they are infected by viruses, and it plays a regulatory role in innate immunity against viral infections. It also represents a significant therapeutic molecule in a number of viral diseases and cancers. IFN induces the Jak/STAT pathway leading to the activation and binding of transcriptional activators, e.g., the STAT/IRF9 complex, to the IFN-stimulated response element (ISRE) in the promoters of IFN-stimulated genes (2, 18). The transcription of IFN genes also is mediated via specific virus response elements (VREs) in the promoter; these sequences bind different IFN response factors (IRFs), such as IRF-3 and IRF-7, in the promoters of IFN genes (17). The VRE and ISRE sequences are found in IFN genes and IFN-stimulated genes; they partially overlap with each other, particularly the core AANNGAAA with the following consensus: G(A)AAANNGAAAG/CT/C or A/GNGAAANNGAAACT (also in the complementary strand) (8, 19). Hundreds of virus- and IFN-stimulated genes exist in the human genome (15), and although their promoters harbor specific core sequence consensus elements, these sequences have context heterogeneity, variable reiterations, and distinct transactivation potential. These sequence variations may account for responses to various types of viruses, IFNs, and IRFs. In this study, we have developed and validated a live cell-based enhanced green fluorescent protein (EGFP) reporter system employing 120 constructs containing multiple viruses and IFN response reporters that represent IFN system nucleic acid sequence heterogeneity to monitor and assess promoter sequence-function relationships during innate immunity. Several sequence reporters with highly sensitive and early activation were observed with distinct patterns of responses to different ligands, including IFN, double-stranded RNA, and RNA viruses.
MATERIALS AND METHODS
Cells, IFNs, and viruses.
HuH-7 cells (obtained from Stephen Polyak, University of Washington) were maintained in Dulbecco's modified essential medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. Escherichia coli-derived recombinant human alpha interferon 2a (rIFN-α2a) (Roferon) was from Hoffman-LaRoche, Basel, Switzerland, and had a specific activity of 2 × 108 IU/mg. Recombinant human IFN-γ (1 × 107 U/ml) was from R&D systems. All viruses were obtained from the American Type Culture Collection (ATCC; Manassas, VA) and propagated in the appropriate host cells. Encephalomyocarditis virus (EMCV), vesicular stomatitis virus (VSV; Indiana strain), herpes simplex virus type 1 (HSV-1; strain F), respiratory syncytial virus (RSV), cytomegalovirus (CMV; Towne strain), and human influenza virus (H1N1; A/Puerto Rico/8/34 strain) were obtained from the ATCC. Virus preparations were clarified by low-speed centrifugation, filtered through 0.22-μm membranes for sterility, and titrated on Vero cells (African green monkey kidney cell line; ATCC) or by hemagglutination assay (influenza virus and Newcastle disease virus [NDV]). The titers for the viruses were the following: EMCV, 1.5 × 108 PFU/ml; HSV-1, 3.5 × 109 PFU/ml; VSV, 8 × 108 PFU/ml; influenza virus, 5,120 hemagglutinin units (HA)/ml; and NDV, 2,048 HA/ml. The CMV and RSV stocks were obtained from the laboratory of M. Al-Ahdal (King Faisal Specialist Hospital and Research Center), and titers were monitored for cytopathic effect endpoints (50% tissue culture infective doses [TCID50]). Virus stocks were aliquoted and stored at −70°C until use. Double-stranded RNA as poly(I:C) was obtained from Calbiochem/Merck (Darmstadt, Germany).
Microarray assessment of IFN-stimulated genes in the human transcriptome.
Two whole-genome expression analysis platforms were used, the OpArray whole-transcriptome microarrays (Operon, Inc.) and the whole-transcriptome OneArray (Phalanx, Taiwan). The human liver HuH-7 cell line was treated with rIFN-α2a (100 IU/ml) for a 6-h incubation, which is optimal for the induction of many IFN-stimulated genes (4). Total RNA was extracted using TRI reagent (Molecular Research Center, Cincinnati, OH). Using a Genisphere kit (Genisphere, Inc., Hatfield, PA), the microarrays were cohybridized with labeled cDNA generated from total RNA (20 μg) using Cy3 and Cy5 for control (medium only) and experimental (IFN treatment) procedures; details have been previously described (15). Scanning was performed with a ScanArray scanner (Perkin Elmer, Inc.), and the intensities of green and red fluorescent signals from each spotted cDNA sequence on the microarrays were calculated using an adaptive circle algorithm and the mean intensity of the pixels. Preprocessing, the filtering of erroneous signals, normalization procedures, and the calculation of intensity ratios have been described previously in detail (15).
Bioinformatic analysis.
The IFN-stimulated gene list was utilized to extract the IFN-stimulated gene promoter sequences, in addition to the first intron and exon, using the PromoSer program. PromoSer extracts promoter regions based on transcriptional sites and alignment algorithms (11). Subsequently, a primary list of interferon-stimulated gene (ISG) promoter sequences was used to search for ISREs and VREs (e.g., IRF sites) using the DNA transcription factor binding site prediction (TFSEARCH) program at http://mbs.cbrc.jp/research/db/TFSEARCH.html. Context regions of ∼40 to 60 bases that harbor the ISRE/VRE sequences were extracted, and the information was used for the forward primers' sequences.
Construction of IFN/virus response EGFP reporters.
The mammalian EGFP vector was used for the construction of the reporters. The pUC19-based vector GWIZ-EGFP originally was obtained from Genlantis (San Diego, CA) and comprises the CMV immediate-early (IE) promoter, EGFP, and the stable bovine growth hormone (BGH) 3′-untranslated region (UTR). Destabilized EGFP cloning was previously described (1). Expression-active ISRE/VRE constructs were generated as linear constructs from the EGFP vector using two primers. The forward primer contains 18 bases at the 3′ end, which targets a minimal promoter region of the CMV promoter upstream of the EGFP coding region, and the putative IFN/ISRE sequence context region. The reverse primer contains a sequence complementary to the downstream region of the poly(A) site in the vector. The high-performance liquid chromatography (HPLC)-purified oligonucleotides were custom synthesized by Metabion (Germany). The PCRs were carried out using the following reagents and conditions: 2.5 U HotStart Taq (Qiagen) and 0.2 U Pfx polymerase (Invitrogen, Carlsbad, CA) mix, 2 μl (100 to 200 ng) of the vector template, 1× PCR buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), and 0.2 μM primers. The following cycle conditions were used: 95°C for 12 min, followed by 31 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 3 min, and a final extension at 72°C for 7 min. The PCR products were purified using Qiagen PCR purification columns to eliminate the primers, small PCR products, buffer, and enzymes. The PCR products finally were eluted in sterile water. The PCR products then were run on a 1.2% agarose gel and visualized by ethidium bromide under UV light to verify size and quality. The purified PCR products were used in the transfection experiments.
Transient transfection of ISG promoter-linked EGFP reporter constructs and other constructs.
The promoter-reporter constructs were used in transient transfection at 50 ng per 3 × 104 cell/well in 96-well microplates. The Toll-like receptor 3 (TLR3) expression plasmid was obtained from Invivogen (San Diego) and used to transfect HuH-7 cells to render them responsive to poly(I:C) (added directly to the medium). Transfections of the plasmids or the reporter constructs were performed in serum-free medium using Lipofectamine 2000 (Invitrogen). Transfection efficiency using cells in separate wells was evaluated using red fluorescent protein vector (TurboRFP; Invivogen). After 18 h of incubation, IFNs were added for an additional 6 h or overnight, while viruses were incubated for >20 h. Emissions of green fluorescent levels were visualized by fluorescence microscopy.
Imaging and fluorescence measurement.
The intensity of transfection was measured by monitoring the fluorescence from EGFP constructs (optimum excitation wavelength, 488 nm; emission wavelength, 503 nm). Automated laser-focus image capturing was performed using the high-throughput BD Pathway 435 imager (BD Biosciences, San Jose, CA). In all cases, exposure times and other settings were kept constant to allow equal comparisons between experiments. Automated identification, segmentation, background correction, and quantification were performed using the ProXcell algorithm (13). Further background correction (optional) using basal fluorescence from a mutant ISRE reporter was employed to increase sensitivity. Data are provided as fold increases above the level of the control and are from mean values ± standard errors of the means (SEM) of fluorescence intensity. All transfections were performed in several replicates as indicated in the text. The variance in EGFP fluorescence among replicate microwells was <6%; thus, with this minimum variance, experiments did not warrant transfection normalization. Image processing, segmentation, and fluorescence quantification were as previously described (1). Student's t test was used to compare two data groups, while analysis of variance (ANOVA) was performed for each data set having three or more data groups.
Quantitative real-time PCR.
The expression levels of EGFP mRNA and control (GAPDH) housekeeping mRNA were assessed using a TaqMan expression assay. First, total RNA was extracted from cells by the guanidine isothiocyanate method using TRI reagent (Molecular Research Center). The RNA was subjected to DNase I treatment, followed by chloroform extraction, precipitation, and resuspension in diethyl pyrocarbonate (DEPC)-treated water. Reverse transcription to cDNA was performed using Superscript II and oligo(dT) primer (Invitrogen). A custom-made TaqMan primer and probe set (Applied Biosystems) specific to the EGFP reporter construct was used. The primers span the CMV promoter intron A in the EGFP vector to control DNA contamination. The 6-carboxyfluorescein (6-FAM)-labeled TaqMan probe that targets the CMV exon 1-EGFP (exon 2) junction sequence was used. The probe design allowed further control of DNA contamination. The control GAPDH probe was labeled with a 5′ reporter VIC dye (Applied Biosystems). The specificity for the cDNA of the TaqMan primer was tested on a negative control containing plasmid DNA. The endogenous control was used for normalization. Real-time PCR was performed in multiplex with the Chroma 4 DNA engine cycler (Bio-Rad). The final results are expressed as normalized fold changes compared to levels for the controls.
RESULTS AND DISCUSSION
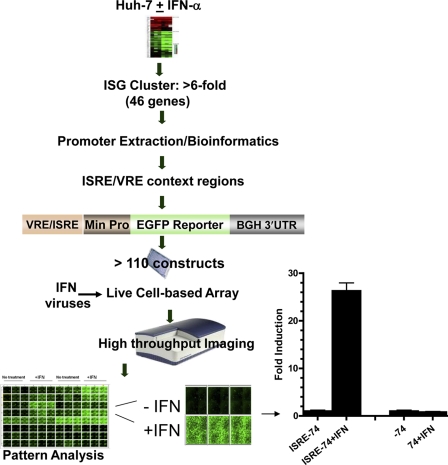
In this work, we have developed an array of optimized reporter constructs that bear natural and artificial VREs and ISREs representing the sequence heterogeneity of IFN-stimulated genes to monitor promoter sequence-function relationships in response to viruses and IFNs (Fig. 1). We utilized the HuH-7 liver cell line, as it is permissive for a number of viruses and is commonly used in viral hepatitis research (5, 9). We first searched for VRE- and ISRE-containing promoters that were activated during the response to human recombinant IFN-α2a by microarray profiling gene expression in the HuH-7 cell line. After a list of 46 strongly induced (>6-fold) genes was generated (see Table S1 in the supplemental material), their promoters were bioinformatically extracted and searched for regions that contain VREs and ISREs (Fig. 1). From each promoter, sequences matching the consensus elements of IRF elements, STATx, and ISREs (>80% match) were extracted, along with their flanking regions of 40 to 60 nucleotides; 110 VRE/ISRE regions were compiled, including introduced repeat variations in ISRE/VRE (see Table S2 in the supplemental material).
Fig. 1.
Schematic representation of the EGFP-based multiple IFN/virus reporter live cell system. HuH-7 cells were treated with recombinant IFN-α2a (100 IU/ml) for 6 h. Total RNA extraction was performed and subjected to whole-genome microarray hybridization and analysis. An IFN-stimulated gene (ISG) cluster was analyzed using bioinformatics by the extraction of the promoters and searching for ISRE/VRE using PromoSer (11) and TFSEARCH (12), respectively. Several variations of ISRE/VRE sequence elements with their context regions were utilized for the construction of EGFP reporters. Cell-based 96-well arrays were assembled for use with various treatments of IFNs and viruses. At the bottom is an image of live cells showing the induction by IFN-α (left) and a mathematical/statistical graph of the quantitative acquisition (right). A reporter with a mutant response element also is shown.
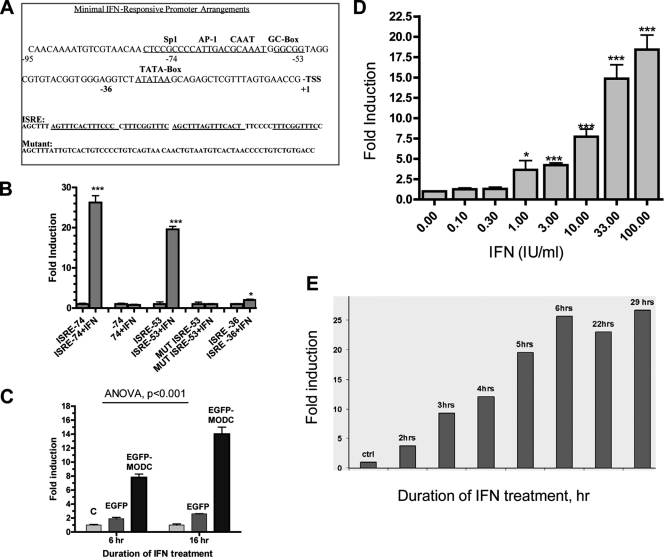
To proceed with constructing the virus/IFN-responsive constructs, we first optimized the VRE/ISRE EGFP reporter backbone by assessing several reporter constructs using a consensus ISRE and a mutant form (Fig. 2A) with different minimal promoters (−36, −53, and −74 from the transcriptional start site of the immediate-early CMV promoter). The optimal −74 CMV reporter response required the following cis-acting elements: the TATA box, GC box, CAAT signal, and AP-1 site. It was necessary to extend the length beyond the TATA box with an additional sequence, since the −36 region that contains only the TATA box failed to induce an IFN response (Fig. 2B, right column). There were higher basal levels (about 2-fold) in the −74 minimal promoter than in the −53 minimal promoter due to the presence of the additional cis factor elements, such as the AP-1 site; however, the response was higher and earlier toward IFN with the −74 ISRE minimal promoter (Fig. 2B). The use of the mouse ornithine decarboxylase (MODC) C-terminal amino acids to destabilize the EGFP protein contributed to better and earlier responses to IFN (Fig. 2C), since MODC contains the protein instability determinant, PEST, known to occur in many proteins with a short half-life (16). For example, as little as 1 U/ml of rIFN-α was able to trigger the reporter activity (Fig. 2D), and as early as 2 h, there was a significant response to rIFN-α at a dose of 100 IU/ml (Fig. 2E). The benefit of earlier response is to allow for flexibility in assay development and alternative drug screening approaches. Thus, the reporter system has an excellent linear dynamic response (1 to 100 IU/ml) sensitivity and rapid kinetics (Fig. 2B to E).
Fig. 2.
Performance of the cell-based IFN-responsive constructs. (A) A graphical scheme showing the minimal promoter of the CMV IE promoter used for the IFN-responsive construct. TSS, transcriptional start site. Numbers are in relation to the TSS. (B) EGFP reporter constructs according to the sequences shown in panel A were used for the transient transfection of HuH-7 cells overnight. rIFN-α2a (100 U/ml) was added for 16 h. Fluorescence was quantified as described in Materials and Methods. Reporter activity as fluorescence was assessed from images captured by a BD automated bioimager and quantitated by ProXcell as described in Materials and Methods. Readings are means ± SEM of the fold increase above the level of the control from fluorescence intensities of quadruplicate wells. (C) Expression constructs containing either wild-type EGFP or unstable EGFP-MODC fusion protein were transfected into HuH-7 cells. Cells were cultured in the presence or absence of 100 U/ml of IFN-α for the indicated period of time. Data are the fold differences in means ± SEM (quadruplicate) from a representative experiment of two. (D) The ISRE-containing destabilized EGFP reporter construct was used for the transient transfection of HuH-7 cells. Increasing doses of IFN were added to the cells for 16 h. (E) HuH-7 cells were transfected with an ISRE-containing EGFP reporter and then treated with IFN-α (100 U/ml) for the indicated periods of time. Fluorescence was quantified as described in Materials and Methods. Reporter activity is given as means ± SEM of the fold increase above the level of the control from fluorescence intensities from quadruplicate wells. P < 0.001 (***) and <0.01 (*) using Student's t test. The designations of sequences, except for those of synthetic variations, were derived from the gene names from which the sequence elements were derived.
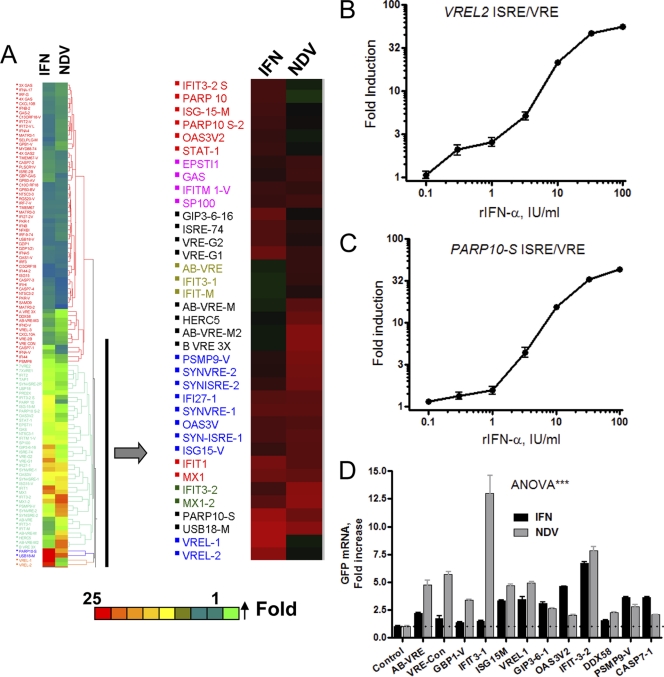
Sequence information was obtained from bioinformatics (see Table S2 in the supplemental material). The ISREs and VREs are heterogeneous in terms of the sequence element reiterations and deviations from their consensuses, numbers of ISRE repeats, and distribution in the entire promoter. Thus, we created 110 IFN- and virus-responsive EGFP constructs representing such sequence heterogeneity (Fig. 1) so that differential fluorescence patterns due to IFN and viruses can be studied and used further as cell-based sensors for IFN triggers. The HuH-7 cell array was used with the VRE/ISRE EGFP constructs and then treated with medium, 100 U/ml of human rIFN-α2a, or 10 hemagglutinin units (HA) (per 3 × 104 cells per well) of Newcastle disease virus (NDV). Two different durations of the IFN-α or NDV treatment were employed: 6 and 16 h, representing early and late responses, respectively. IFN was able to induce a significant subset of both ISRE- and VRE-containing constructs (Fig. 3A). Within the IFN-responsive subset, distinct differential reporter activities ranged from a very weak effect to stronger effects (1.5- to 25-fold). The strongest IFN-selective effect was observed with two synthetic sequences (designated VREL1 and VREL2) that comprise four copies of GGGAAACCGAAACTG or GAAACTAAAGCTG repeats (20- to 25-fold increase). Further evaluation of the VREL2 EGFP promoter was performed in independent experiments; an excellent dose-response curve (correlation coefficient [r], >0.98; P < 0.001) was observed (Fig. 3B). As little as 3 U/ml of IFN-α was able to trigger the VREL2 reporter, and a large dynamic range of 3 logs was observed (Fig. 3B). Using mutant ISRE reporter fluorescence levels as the background, a 50-fold induction can be achieved with the highest IFN-α concentration (100 U/ml) (Fig. 3B). The naturally derived sequences GIP3-6-16, MX1-1, MX1-2, USB18-M, and PARP10 caused strong induction by IFN (>20 fold; P < 0.0001) but variable levels of induction by NDV (Fig. 3A and Table 1). As an example of this reporter group, we selected the PARP10 promoter-derived EGFP construct showing excellent dose-dependent response characteristics (Fig. 3C).
Fig. 3.
Cell-based EGFP array for IFN and virus monitoring. (A) EGFP 96-well microplate arrays contain lyophilized DNA for use in a number of transfection array experiments. HuH-7 cells (4 × 104) were seeded in 96-well microplates and transfected with 50 ng/well of the ISRE/VRE EGFP reporter constructs for 16 h. IFN (100 IU/ml) or NDV (10 HA per well) was added for 16 h; fluorescence was quantitated from captured high-resolution images using a high-throughput BD bioimager and the ProXcell algorithm (13). Fold changes were evaluated by hierarchal clustering. The designations of sequences, except synthetic variations, were derived from the gene names from which the sequence elements were derived. The thick arrow points to an IFN/virus differential response cluster. (B and C) IFN dose-response characteristics of the EGFP reporter, fused with minimal reporters harboring ISRE/VRE derived from the VREL2 sequence (B) and PARP10 gene promoter (C). The treatment of the reporter-transfected HuH-7 cells was with 100 U/ml rIFN-α for 16 h as described above. (D) qPCR evaluation of reporter activity. HuH-7 cells were challenged with the IFN and virus, as described in the legend to Fig. 2. Total RNA was subjected to RT-qPCR using a primer/TaqMan probe specific to the EGFP mRNA and GAPDH control mRNA. Data are given as fold increases compared to the levels for no-IFN treatment.
Table 1.
IFN response cell-based subarray: sequence-function relationships
| Sequence namea | EGFP reporter-linked transcriptional inducible sequence | Typeb | IFNc | NDVc | Pd |
|---|---|---|---|---|---|
| VREL1 | GGGAAACCGAAACTGGGGAAACCGAAACTGGGGAAACCGAAACTGGGAAACCGAAA | Art | 33 | 2.3 | <0.01 |
| VREL2 | GGAAACCGAAAGGGGAAAGTGAAACTAAAGCTGAAACCGAAAGGGGAAAGTGAAACTAAAGC | Art | 27 | 2.8 | <0.01 |
| GP3-6-16 | GGGAAAATGAAACTCGGAGCTGGGAGAGAGGGGAAAATGAAACTGCAGAAATAGAA | Nat | 30 | 3 | <0.01 |
| MX1-1 | GCTAGGTTTCGTTTCTGCGCCCCACAGGGTCTGTGAGTTTCATTTCTTC | Nat | 25 | 7 | <0.01 |
| MX1-2 | TGAGTTTCGTTTCTGAGCTCCTTTCATTTTCACCGGTTTCAATTCTCCTCTGGA | Nat | 22 | 9.5 | <0.01 |
| USB18-M | CTCCCGGCGCGGAGGCCGCTGTAAGTTTCGCTTTCCATTCAGTGGAAAACGAAAG | Mod | 21 | 9 | <0.01 |
| IFIT3-2 | GATTCTGTTTCAGTTTCCCCTCAAGAGGGATCTTGATAGGGTTCCATCAGTTTCACTTTCCTTTCCCCTTTCATCC | Nat | 20 | 15 | <0.01 |
| PARP10 | CCTCCTTCCG TCTTTCAGTT TCACTTTTGTTTTCCTGCTCCTGCTCCCTC | Nat | 20 | 4 | <0.01 |
| IFIT3-1 | GTTTCATTTTCCTCCTCCCAACGATTTTAAATTAGTTTCACTTTCCAGTTTCCTCTTCCTT | Nat | 2 | 7.6 | <0.01 |
| GBP1-V | AAAAAACTGAAACTCAGCCTGAAAGATGAACAGAACAAAACAGAAATCCT | Nat | 0.7 | 2.5 | <0.01 |
| VRE Con | GAAAGTGAAAAGAGAAATGGAAAGTGGAAAAGGAGAAACT | Art | 0.6 | 1.7 | <0.01 |
| AB-VRE | AGAAATGGAAAGTAGAAATGGAAAGTGAGAAGTGAAAGTGAGAAGTGAAAGT | Art | 3.7 | 9 | <0.01 |
| HERC-5 | GTTTCCTTTTCCTTTTCGATTCCGCCCCCTAACATTATGTTTCGTTTTCCACTG | Nat | 4.5 | 8 | <0.01 |
| OAS3-V | AGTGTCTGATTTGCAAAAGGAAAGTGCAAAGACAGCTCCT CCCTTCTGAGG | Nat | 16 | 15 | |
| OAS3-V2 | TTCGGAGAGCCGGGCGGGAAAACGAAACCAGAAATCCGAAGGCCGCGCCAG | Nat | 15 | 16 | |
| ISG15V | TGCCTCGGGAAAGGGAAACCGAAACTGAAGCCAAATTTGGCCAG | Nat | 9 | 6 |
Unique name for the ISRE/VRE sequence.
Nat, wild-type sequence region; Art, artificial sequence.
Fold induction, mean.
Student's t test for comparison between responses to IFN and virus. Standard errors of the means are <10% of mean readings (n = 4).
In many instances, the VREs and ISREs act as a common signature for IFN and virus responses, but there are distinct patterns between the IFN and NDV responses (Fig. 3A). Many NDV-induced reporter activities were due to the same promoter sequences in the IFN-responsive constructs. These activities most likely are due to endogenous IFN, since NDV is a strong inducer of type I IFN (14). Nevertheless, a cluster of sequences caused the EGFP reporter to respond more potently to NDV than to IFN, including IFIT3-1, GPB1-V, and HERC5 (Fig. 3A and Table 1). A number of synthetic sequences also caused a more selective response than IFN-α, such as AB-VRE and VRE-Con, which were derived from the VRE of the IFN-αA1 and IFN-β promoters (Table 1). Using quantitative PCR (qPCR), reporter mRNA levels were evaluated after the transfection and expression of selected constructs (Fig. 3D). Those sequences that were potently responsive toward NDV at the protein fluorescence level also were responsive at the mRNA level (Fig. 3D), although the fold inductions were not similar. The other sequences that were selective by IFN induction at the protein levels were not so at the mRNA levels. This may be due to the general observation that mRNA and protein levels are minimally correlative (10). Effects such as those of coupled transcriptional/posttranscriptional mechanisms can lead to different mRNA and protein level differences. However, the endpoint assay of this approach is at the fluorescent protein level, which is a more differential and simpler approach than the mRNA level determination assay.
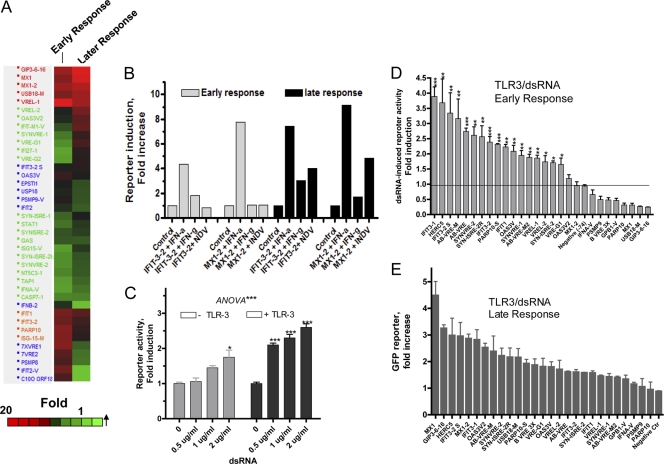
There also are distinct responses among different VRE/ISRE sequences toward IFN during early (6 h) and late (16 to 20 h) responses, as shown in Fig. 4A. Notably, three cluster types were observed. The upper cluster (Fig. 4A) comprised those reporters that are early responders that further increase at later times, such as GP3-6-16, MX1, and USB-18 M reporters. A second cluster group comprised the activities of a group of early responding reporters that decreased at the late time point (Fig. 4A, lower clusters). An example is ISG-15 M, which is modified from ISG-15 VRE/ISRE (see Table S2 in the supplemental material). Other clusters denote mostly reporters that respond only at the 20-h late time rather than the earlier time point. Most of the virus-induced response appears as a later response, as in the case with IFIT3-2 and MX1-2 element-fused reporters (Fig. 4B). Using these same sequence elements, there were strong early responses to IFN-α but late and weak responses to IFN-γ (Fig. 4B).
Fig. 4.
Monitoring of early and late responses to IFN, virus, and dsRNA in the cell-based multiple reporter system. (A) HuH-7 cells (2 × 104 per well) were seeded in 96-well microplates and transfected with 50 ng/well of the ISRE/VRE EGFP reporter constructs for 6 or 16 h of IFN (100 IU/ml) for early times (4 to 8 h) or late times (16 to 20 h); fluorescence was quantitated from captured high-resolution images using a high-throughput BD bioimager and the ProXcell algorithm (13). Fold changes were evaluated by hierarchal clustering centered with Spearman's correlation. (B) Early and late expression of IFN and NDV virus-induced activity. HuH-7 cells (2 × 104) were seeded in 96-well microplates and transfected with 50 ng/well of the indicated ISRE/VRE EGFP reporter constructs for 6 or 16 h with rIFN-α2a (100 IU/ml) or NDV (10 HA per well, 16 h); fluorescence was quantitated from captured high-resolution images using a high-throughput BD bioimager and the ProXcell algorithm. (C) HuH-7 cells were transfected with TLR3 expression vector and ISRE/VRE EGFP reporter construct for 16 h, followed by the addition of increasing amounts of poly(I:C) into the medium for an additional 6 or 16 h. Reporter activity was measured by fluorescence. A EGFP live cell-based subarray consisting of 28 constructs was utilized for early (D) and late (E) responses to dsRNA [poly(I:C); 1 μg/ml] as described above.
Based on the live cell fluorescence pattern in the IFN and NDV response, we further utilized a subarray consisting of 20 constructs representing those of the differential response patterns for use with other IFN triggers, including double-stranded RNA, poly(I:C), and several types of viruses, including DNA and RNA viruses. First, it was necessary to transfect HuH-7 cells with TLR3 expression vector to render the cells responsive to the exogenous addition of double-stranded DNA (dsRNA) to the medium (Fig. 4C). The poly(I:C) was able to trigger, at the early response time (6 h), the EGFP reporter fused with certain transcriptional sequences in cells that were transfected with TLR3 expression plasmid (Fig. 4D). The most inducible transcriptional activity (3- to 5-fold; P < 0.01) to dsRNA is from the EGFP reporters fused with sequences derived from IFIT3-1, HERC5, IFIT3-2, and AB-VRE-M (Fig. 4D). With the later response (24 h), the dsRNA triggers both common (e.g., HERC5, IFIT3-2S, IFIT3-1, and OAS3V2) and different sets of reporters with promoter sequences such as those derived from MX1 and GIP3 (4.4- and 3-fold, respectively) (Fig. 4E). It should be noted that dsRNA can induce genes that are either dependent on or independent from autocrine IFN production (7).
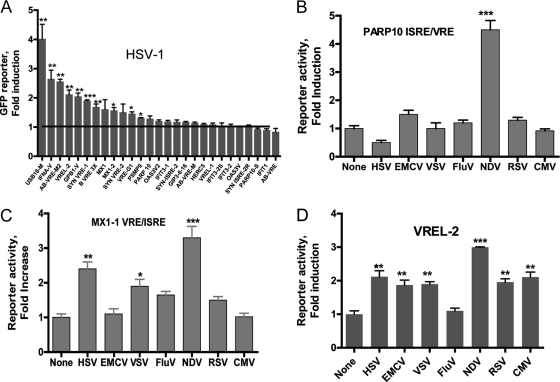
Using a DNA-type virus, herpes simplex virus (HSV-1), and the entire cell-based subarray, we were able to monitor the transcriptional reporter system due to the different ISRE/VRE sequence elements showing maximum fluorescence responses (4-fold; P < 0.001) due to the USB18-M-derived sequence (Fig. 5A). Although HSV-1 is not an RNA virus, the ability of HSV-1 DNA to induce the IFN pathway in HuH-7 in retinoic acid-inducible gene (RIG)-dependent manner has been demonstrated previously (3). The USB18-M sequence is highly inducible by IFN-α (21-fold; P < 0.001) and by NDV (9-fold; P < 0.001); this sequence has two near-tandem antiparallel copies of ISRE/VRE (Table 1). The EGFP reporter responses to different viruses also were evaluated with selected sequences. For example, NDV, but not other viruses, was able to induce reporter fluorescence due to sequence elements derived from the PARP10 gene promoter (Fig. 5B). The MX1-1 sequence was able to trigger EGFP reporter activity due to infection with NDV, HSV-1, and VSV (Fig. 5C). The VREL2 sequence, one of the strongest responsive elements to IFN, was able to modestly trigger the EGFP reporter in cells infected with both DNA and RNA viruses, except for H1N1 human influenza virus (Fig. 5D). This is not surprising, since influenza viruses are known to inhibit IFN production through their nonstructural protein, NS1 (6).
Fig. 5.
EGFP cell-based array for virus response. (A) HSV-1 infection (multiplicity of infection of 10) was performed on the reporter-transfected cells, and fluorescence was measured. HuH-7 cells (3 × 104 per well) transfected with three different EGFP reporters, PARP10 (B), MIX1 (C), and VREL2 (D), were infected with several types of DNA and RNA viruses (NDV, 10 HA; EMVC, MOI of 0.5; VSV, MOI of 25; influenza H1N1 [FluV], 1 HA; HSV-1, MOI of 10; RSV and CMV, 1/10 of TCID50). Cells were infected with the viruses for 20 to 24 h, as cytopathic effects appear largely by 48 h. Reporter activity was quantified by fluorescence quantification as described in Materials and Methods. *, P < 0.01; **, P < 0.001; ***, P < 0.0001.
The data show that the heterogeneity and context sequence of the ISRE and VRE sequences influence the temporal strength and selectivity of the responses of the reporter system toward the activator. The use of EGFP as the reporter, compared to others, such as luciferase, gives further flexibility by allowing monitoring at different time points, i.e., without experimental termination or cell lysis. This is also advantageous in dissecting early and late responses, which can be used to distinguish between IFN response and the later virus response. Among the multiple IFN/virus response reporters described here or from other promoters using the described approach, selecting a reporter for a specific IFN trigger or a group of viruses is possible. To our knowledge, this is the first large-scale evaluation of promoter sequence-function heterogeneity in innate immunity in a reporter live cell-based assay. The resource data here include the functional relationships of more than 110 sequence variations in ISRE/VRE regions in sensing different IFN response triggers, including type I IFN, dsRNA, and DNA and RNA viruses. Further, this study offers a simplified approach to generating resource data for finding biomarker tools for monitoring virus and interferon activity, and their responses, in disease and drug evaluation settings.
Supplementary Material
ACKNOWLEDGMENTS
The study (CEBR2), including open access charges, was funded by the Center for Excellence in Biotechnology Research of King Saud University and the King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 13 July 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Al-Haj L., Al-Ahmadi W., Al-Saif M., Demirkaya O., Khabar K. S. 2009. Cloning-free regulated monitoring of reporter and gene expression. BMC Mol. Biol. 10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borden E. C., et al. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheng G., Zhong J., Chung J., Chisari F. V. 2007. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc. Natl. Acad. Sci. U. S. A. 104:9035–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Der S. D., Zhou A., Williams B. R., Silverman R. H. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Freymuth F., et al. 2005. Replication of respiratory viruses, particularly influenza virus, rhinovirus, and coronavirus in HuH7 hepatocarcinoma cell line. J. Medical Virology. 77:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. García-Sastre A., et al. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330 [DOI] [PubMed] [Google Scholar]
- 7. Geiss G., et al. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178–30182 [DOI] [PubMed] [Google Scholar]
- 8. Génin P., Vacarro A., Civas A. 2009. The role of differential expression of human interferon-A genes in antiviral immunity. Cytokine Growth Factor Rev. 20:283–295 [DOI] [PubMed] [Google Scholar]
- 9. Green J., Khabar K. S., Koo B. C., Williams B. R., Polyak S. J. 2006. Stability of CXCL-8 and related AU-rich mRNAs in the context of hepatitis C virus replication in vitro. J. Infect. Dis. 193:802–811 [DOI] [PubMed] [Google Scholar]
- 10. Greenbaum D., Colangelo C., Williams K., Gerstein M. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biology. 4:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halees A. S., Leyfer D., Weng Z. 2003. PromoSer: a large-scale mammalian promoter and transcription start site identification service. Nucleic Acids Res. 31:3554–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heinemeyer T., et al. 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 26:362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hitti E., et al. 2010. A versatile ribosomal protein promoter-based reporter system for selective assessment of RNA stability and post-transcriptional control. RNA. 16:1245–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ito Y., Nagai Y., Maeno K. 1982. Interferon production in mouse spleen cells and mouse fibroblasts (L cells) stimulated by various strains of Newcastle disease virus. J. Gen. Virol. 62:349–352 [DOI] [PubMed] [Google Scholar]
- 15. Khabar K. S., et al. 2004. Expressed gene clusters associated with cellular sensitivity and resistance towards anti-viral and anti-proliferative actions of interferon. J. Mol. Biol. 342:833–846 [DOI] [PubMed] [Google Scholar]
- 16. Li X., et al. 1998. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 273:34970–34975 [DOI] [PubMed] [Google Scholar]
- 17. Paun A., Pitha P. M. The IRF family, revisited. Biochimie 89:744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sato M., Taniguchi T., Tanaka N. 2001. The interferon system and interferon regulatory factor transcription factors–studies from gene knockout mice. Cytokine Growth Factor Rev. 12:133–142 [DOI] [PubMed] [Google Scholar]
- 19. Savitsky D., Tamura T., Yanai H., Taniguchi T. 2010. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol. Immunother. 59:489–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.