Abstract
The gene M94 of murine cytomegalovirus (MCMV) as well as its homologues UL16 in alphaherpesviruses is involved in viral morphogenesis. For a better understanding of its role in the viral life cycle, a library of random M94 mutants was generated by modified transposon-based linker scanning mutagenesis. A comprehensive set of M94 mutants was reinserted into the MCMV genome and tested for their capacity to complement the M94 null mutant. Thereby, 34 loss-of-function mutants of M94 were identified, which were tested in a second screen for their capacity to inhibit virus replication. This analysis identified two N-terminal insertion mutants of M94 with a dominant negative effect. We compared phenotypes induced by the conditional expression of these dominant negative M94 alleles with the null phenotype of the M94 deletion. The viral gene expression cascade and the nuclear morphogenesis steps were not affected in either setting. In both cases, however, secondary envelopment did not proceed in the absence of functional M94, and capsids subsequently accumulated in the center of the cytoplasmic assembly complex. In addition, deletion of M94 resulted in a block of cell-to-cell spread. Moreover, the dominant negative mutant of M94 demonstrated a defect in interacting with M99, the UL11 homologue of MCMV.
INTRODUCTION
Based on their different biological properties, herpesviruses are divided into three subfamilies: Alpha-, Beta-, and Gammaherpesvirinae (35). Cytomegaloviruses (CMVs) belong to the betaherpesvirus subfamily. The human cytomegalovirus (HCMV) is a leading cause of congenital infection and of complications in immunocompromised patients. The murine cytomegalovirus (MCMV) serves as a model of its human counterpart both in vivo and in vitro. As in other herpesvirus subfamilies, infectious particles of cytomegaloviruses are assembled along a complex morphogenesis pathway which includes capsid assembly, genome packaging in the nucleus (5), primary envelopment at the nuclear membrane (1, 30), and secondary envelopment in the cytoplasm (5, 12), resulting in either release of infectious particles or virus transmission by cell-to-cell spread (10, 40, 41). More than 50 viral proteins as well as numerous cellular proteins are involved in this process. The maturation of virions is spatially and temporally tightly regulated, and the basic mechanisms by which these specific morphogenesis steps are carried out seem to be conserved among herpesviruses (17, 26); still, they are not completely understood. According to the established model, there is a set of about 40 essential core genes conserved among all herpesviruses which govern basic functions of the lytic cycle (13).
Recently, we developed a genetic approach to characterize essential genes and to study functions of core genes in MCMV (7, 37). By using this approach, we wanted to characterize the gene M94 and its gene product pM94. It belongs to the UL16 family of conserved tegument proteins. This core gene is encoded in gene block 6 within the intron of the conserved herpesvirus spliced gene (M89 in MCMV) and is shared by all herpesviruses studied so far (32). Its homologues in HCMV and herpes simplex virus type 1 (HSV-1) are expressed with true late kinetics (31, 46). However, unlike the HSV-1 gene UL16 (2), UL94 of HCMV was found to be essential for virus replication in tissue culture by a genome-wide screen based on a molecular clone of the Towne strain (14). Interestingly, another genetic screen based on the strain AD169 found UL94 dispensable for HCMV replication in vitro (48). pUL94 interacts with pUL99, an interaction which can also be found between their alphaherpesvirus homologues pUL16 and pUL11 (18, 20, 23). Both pUL99 and pUL11 play a crucial role in secondary envelopment (3, 6, 43). The most extensively studied homologue of pM94, the HSV UL16 gene product, has been associated with virus entry and nuclear and cytoplasmic egress (23, 31, 32, 47). It has been shown to localize in the nucleus and the cytoplasm of infected cells (31, 47) and is tightly associated with capsids (23). In addition to pUL11, it also interacts with the core gene product pUL21 (15). Confirmatory work on other homologues of pUL16 was reviewed recently in reference (17).
M94 therefore represented an interesting candidate as no structural information was available, and none of its functional sites were characterized. We hypothesized that one of its functions is essential in MCMV, allowing us to dissect this function from nonessential ones by our genetic approach. To this end, we performed a random, transposon-based mutagenesis of M94 in combination with two successive genetic screens which were previously able to identify functionally important sites or domains of essential genes (reviewed in reference 38). The first screen, which we termed a cis-complementation assay, tests for the ability of insertion mutants to complement the null mutant of a wild-type (wt) gene. The second assay, which we termed an inhibitory screen, identifies loss-of-function mutants, which can inhibit virus reconstitution from wt viral bacterial artificial chromosomes (BACs). Most of the mutants which appear to be inhibitory in the second screen could be verified as dominant negative (DN) mutants by further analysis of their phenotypes in the virus context (37).
DN mutants have been used for gene inactivation for decades (16). A DN mutant is defined as an inhibitory variant of the wt gene product that inhibits the wt activity when it is overexpressed. To achieve this, a DN protein must fulfill at least two functions. One function, which is usually based on biomolecular interactions, must remain intact, whereas another essential function must be disrupted by either deletion of a protein domain or its mutation, resulting in loss of function (29).
Here, we show that M94 is essential for MCMV replication, and we report on the analysis of a mutant library of the M94 gene which was constructed by random transposon mutagenesis. We tested a set of 74 insertion and frameshift mutants of M94 for their ability to complement a M94 deletion mutant and subsequently for their inhibitory capacity in the presence of the wt M94 allele. We identified an essential activity, located in the C-terminal domain of M94, which is conserved among its homologues (24, 46). Additionally, we could assign another crucial site to its N-terminal end. Two insertion mutants within this N-terminal region were identified as DN alleles. By inserting these mutants into an inducible overexpression cassette and comparing the overexpression phenotype with the deletion mutant of M94 (27), we could elucidate the essential role of M94 in MCMV envelopment. Moreover, M94 deletion as well as overexpression of the DN mutants blocked secondary envelopment, and this was accompanied by a lack of pM99 interaction.
MATERIALS AND METHODS
Cells and viruses.
Mouse embryonic fibroblasts (MEFs), NIH 3T3 murine fibroblasts (ATCC CRL-1658), M2-10B4 bone marrow stromal cells (ATCC CRL-1972), and 293 human epithelial kidney cells (ATCC CRL-1573) were propagated as described before (36). The M94 complementing cell line NT/M94-7 was propagated as described previously (27). All viruses used in this study were derived from BACs. MCMV-FRT (where FRT is Flp recombinase target), which is derived from the pSM3fr-FRT BAC (8), served as the wild-type genome. MCMVΔ1-16-FRT was derived from pSM3fr-Δ1-16-FRT featuring a genome with deletion of genes m01 to m16 and an FRT site between the left terminal region and gene m17. It replicated like the wt virus in tissue culture and offered space for large insertions (28) (see Fig. S1 in the supplemental material). The virus genome with the M94 deletion, MCMVΔM94tTA, was derived from pSM3fr-flox-ova-ΔM94, in which the m157 gene was replaced by a floxed ova cassette and the M94 gene was replaced by a tetracycline-controlled transactivator (tTA) expression cassette (27).
To serve as an acceptor for cis-complementation, the pSM3fr-FRT BAC was deleted of M94 by insertion of a kanamycin cassette. This M94 deletion BAC was also used to generate the rescue viruses MCMVΔM94-EPM94 and MCMVΔM94-EHAM94. The MCMVΔM94-EPM94 BAC was generated by inserting M94 ectopically under the control of its own promoter introducing pOriR6K-PM94 (see “Plasmid construction,” part i, below) at the FRT site by Flp recombination (7). Accordingly, the MCMVΔM94-EHAM94 BAC was generated by inserting a hemagglutinin (HA)-tagged M94 open reading frame ([ORF] HA-M94) under the control of the HCMV immediate-early promoter/enhancer (CMV promoter) using pOriR6K-HAM94 (see “Plasmid construction,” part ii, below). The inhibitory mutants M94i7 and M94i13 were subcloned into the conditional expression cassette (see “Plasmid construction,” part iii, below) and ectopically inserted into pSM3fr-FRT and pSM3fr-Δ1-16-FRT, resulting in MCMV-RM94i7, MCMV-RM94i13, MCMVΔ1-16-RM94i7, and MCMVΔ1-16-RM94i13, respectively. To control for the expression of M94, two recombinants were constructed: (i) MCMVΔ1-16-RHAM94 was generated by introducing HA-M94 into the BAC in a manner similar to that used for the constructs for conditional expression of the inhibitory mutants; (ii) MCMVΔ1-16-EHAM94 was constructed by inserting pOriR6K-HAM94 into pSM3fr-Δ1-16-FRT. BACs were reconstituted by transfection of mouse embryonic fibroblasts (MEFs) using SuperFect Transfection Reagent (Qiagen) according to the manufacturer's instructions. Reconstituted viruses were propagated on M2-10B4 cells as described previously (25) and counted by standard plaque assay (34). MCMVΔM94tTA was propagated and assessed on NT/M94-7 as described previously (27).
Growth analysis.
The in vitro growth characteristics of MCMV mutants were analyzed by multistep growth curves. MEFs were infected at a multiplicity of infection (MOI) of 0.1. Supernatants of infected cells were then harvested from day 0 (input virus) until day 5 postinfection (p.i.) every 24 h. Supernatants of each sample were taken, and the release of infectious particles was analyzed by a standard plaque assay in duplicates on MEFs (37) or by 50% tissue culture infective dose (TCID50) assay on NT/M94-7 (27). In the case of viruses with inducible expression cassettes, doxycycline (Dox) was added at 1 h postinfection (hpi) at a final concentration of 1 μg/ml. Three days later the Dox-containing medium was replenished.
Plasmid construction.
The plasmids used in this study were constructed as follows. (i) To construct pSM3fr-ΔM94-EPM94, the M94 ORF together with its 300-bp upstream region was amplified from pSM3fr (44) using primers M94Pfor (GGTGGATCCGGTTCGCCGTGATCTGGT) and M94rev (ACTCTAGAGTCGACTTCACATGTGCTCGAGAAC). The PCR product was digested with BamHI and XbaI and inserted into the BamHI/SpeI linearized rescue plasmid pOriR6K-pA (GenBank accession number HQ232343), resulting in pOriR6K-PM94. (ii) The second ΔM94 rescue plasmid encoding an HA-tagged M94 was constructed by PCR amplification of the M94 ORF from pSM3fr using primers HAM94 (GTGGGATCCACCATGTACCCCTACGACGTGCCCGACTACGCCACGTCCAGACTATCC) and M94rev and was used for the construction of pSM3fr-ΔM94-EHAM94. The forward primer carried the coding sequences for an N-terminal HA tag. The PCR product was cloned into pCR3 by BamHI/XbaI, resulting in pCR3-HAM94. HA-M94 was then subcloned into pOriR6K-zeo-ie (7) by NdeI/ApaI, resulting in pOriR6K-HAM94. (iii) The mutant HA-M94 ORFs (M94i7 and M94i13), which were derived from the library of transposon insertion mutants (see “Linker scanning mutagenesis” below), were subcloned for conditional expression under the control of the SVT promoter (37). For this, the mutant M94 ORFs were amplified by PCR using the primers M94-MluI-for (GTGTACGCGTACCATGTACCCCTACGACGTGCCC) and M94-Pvu2-rev (ACACCAGCTGTCGACTTCACATGTGCTCGAGAAC), followed by restriction digestion with MluI and PvuII. The vector pO6-TR (37) was cut with AscI and HpaI, and ligation resulted in pO6TR-M94i7 and pO6TR-M94i13. (iv) For generation of the control BAC pSM3fr-Δ1-16-RHAM94, which encodes HA-tagged M94 in a conditional expression cassette, the HA-M94 ORF was amplified from the plasmid pOriR6K-HAM94 using the primers HAM94-Asc-for (GGTGGCGCGCCTCACTATAGGGAGACCCAAGCT) and HAM94-Sma-rev (GTGGCCCGGGAGAGTCGACTTCACATGTGCTC). The PCR product was cut with AscI and SmaI and ligated to pO6-TR linearized by AscI and HpaI. (v) For studies on the pM99 protein, an expression vector was designed encoding M99 with a C-terminal FLAG affinity tag connected by a 3-amino-acid (aa) flexible linker region. The M99 ORF was amplified from pSM3fr-Δ1-16-FRT using primers M99for (GAGAGGTACCGCTCTTCATGGGTGCAGAGTGCTGTAAAC) and M99Frev (AGAGCTCGAGTTATTTGTCGTCGTCGTCTTTGTAGTCGCCGGAGCCCAAGGCCCTGACTTTTTTCTTCAC). The reverse primer carried the FLAG tag coding sequence. The PCR product was inserted into pOriR6K-zeo-ie by KpnI/XhoI, resulting in pOriR6K-M99FLAG.
Linker scanning mutagenesis.
HA-M94 was subcloned from pOriR6K-HAM94 by KpnI and SalI digestion into Litmus28 (NEB), opened with KpnI and XhoI. The resulting plasmid, Litmus28-HAM94, was subjected to in vitro GPS-LS Linker Scanning mutagenesis (NEB) according to the manufacturer's manual, using pGPS4 (NEB) as a transposon donor. Escherichia coli XL1-Blue bacteria (Stratagene) were transformed with the transposition reaction, and minitransposon insertions were selected by ampicillin and chloramphenicol. Plasmid DNA was isolated from the mutant pool, and the 2,488-bp fragment containing HA-M94 including the transprimer element was subcloned by KpnI and NsiI into pOriR6K-zeo-ie. Recombinants were selected by zeocin and chloramphenicol. DNA of this secondary mutant pool was isolated, and the transprimer elements were removed by PmeI digestion and religation and subsequently reintroduced into E. coli PIR1 cells (Invitrogen), resulting in the final M94 library. Individual clones from this library were picked and analyzed by sequencing.
Construction of recombinant BACs.
E. coli strain DH10B (Invitrogen) containing either pSM3fr-FRT, pSM3fr-Δ1-16-FRT, or pSM3fr-ΔM94 and the Flp-expressing plasmid pCP20 (11) were transformed with pOriR6K-zeo-ie-based constructs, incubated at 30°C for 1 h in solution, plated out, and incubated at 43°C overnight (7). The resulting constructs were tested by restriction analysis to exclude multiple insertions of pOriR6K-based plasmids. Mutants carrying single insertions were selected and further analyzed in the cis-complementation assay (7) as well as in the inhibitory screen (36).
cis-Complementation assay and screening for inhibitory mutants.
The cis-complementation assay and inhibitory screen, both of which are based on virus reconstitution, were performed as described earlier (33). The success of virus reconstitution after transfection of MEFs with purified BAC DNA was assessed by checking cultures regularly for cytopathic effect over a period of 6 weeks after transfection. As a positive control for successful reconstitution, indicated by total cell lysis, we always transfected wt BAC DNA. Lack of detectable cell lysis after 6 weeks of culture indicated failure of virus reconstitution. For each mutant, two independent clones were analyzed in virus reconstitution, and conflicting results were resolved by isolation and reconstitution of two additional clones. Constructs which induced only subtotal cell lysis or confined plaques would have been recorded as attenuated mutants. As a control for attenuated reconstitution, pΔM53MCMV-EK128A DNA, a BAC carrying an attenuated allele of M53 (21), was used as described previously (33). A virus reconstitution experiment was considered valid (i) if plaques of wt MCMV-BAC (pSM3fr-FRT BAC)-derived virus occurred after 1 week and grew exponentially, leading to total cell lysis after 6 weeks (positive control), (ii) if transfection with pΔM53MCMV-EK128A BAC resulted in plaque formation in between the second and fourth week (attenuated control), and (iii) if no plaques were observed in pSM3fr-ΔM94-transfected cultures after a total of 6 weeks (negative control).
Western blot analysis.
NIH 3T3 cells were infected at an MOI of 0.5 for 24 h in the absence or presence of 1 μg/ml Dox and/or 300 μg/ml phosphonoacetic acid (PAA). Cells were lysed in total lysis buffer (62.5 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 6 M urea, 5% β-methanol [MetOH], 0.01% bromphenol blue, 0.01% phenol red) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred onto Hybond-P membranes (GE Healthcare) using blotting buffer (25 mM Tris, 192 mM glycine, 20% methanol). Membranes were blocked in TBS-T (150 mM NaCl, 10 mM Tris, pH 8.0, 0.05% Tween 20) containing 5% nonfat dry milk at 4°C overnight, followed by incubation with the primary antibody in TBS-T for 2 h at 4°C. For detection of the immediate-early protein pp89, a specific mouse monoclonal antibody (CROMA101; provided by S. Jonijic, University of Rijeka, Rijeka, Croatia) was used. pM50 was detected by using a specific polyclonal rabbit serum (30). A pM94-specific immune serum was generated by immunization of rabbits with synthetic peptides representing M94 aa 25 to 39 (CMLVNKSARYREFRAV) and aa 225 to 238 (CRDSGRHVDTGRFV) (Metabion, Munich, Germany). For detection of the HA tag, a peroxidase-conjugated rat anti-HA monoclonal antibody (Roche) was used. The FLAG tag was detected using a rabbit monoclonal antibody (Sigma). The binding of the primary antibodies was detected by horseradish peroxidase-conjugated secondary antibodies of corresponding affinity (Dianova). Signals were visualized using an ECL Plus Western blotting detection system (GE Healthcare).
HA/FLAG tag pulldown assays.
Subconfluent M2-10B4 cells in 10-cm dishes were transfected with 8 μg of total DNA of pOriR6K-zeo-ie, pOriR6K-M50HA, pOriR6K-HAM94, or pO6TR-M94i13 alone or in combination with pOriR6K-M99FLAG using Lipofectamine LTX (Invitrogen) according to the manufacturer's instruction. Cultures were treated with 1 μg/ml Dox if necessary. At 48 h after transfection, cells were washed with Dulbecco's phosphate-buffered saline (D-PBS), scratched from the plates, resuspended in 1.5 ml of lysis buffer (1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 20 mM Tris, pH 7.5), and supplemented with 650 units of Benzonase (Novagen). After 2 h of digestion in a rolling incubator, the lysates were cleared by centrifugation (30 min at 22,000 × g at 4°C) to remove insoluble matter. Ten percent of the clarified protein solution served as the input control, and 90% was subjected to pulldown analysis using an EZview Red Anti-HA Affinity Gel (Sigma-Aldrich). The gel matrix was washed three times using buffer 1 (150 mM NaCl, 5 mM EDTA, 20 mM Tris, pH 7.5) and once using buffer 2 (20 mM Tris pH 7.5). The bound proteins were eluted under denaturing conditions using total lysis buffer and were separated by SDS-PAGE, followed by Western blotting as described above.
Cleavage-packaging assay.
The packaging of MCMV genomes was analyzed by a specifically designed Southern blot assay which utilized the specific properties of the unit length genome. The DNA probe was generated by PCR using the primers Apa2-for (ATCGGGTCACAGTCCTCACGCC) and Apa2-rev (CCAACATCCGTGGGTCGACACC) utilizing a PCR digoxigenin (DIG) probe synthesis kit (Roche). M2-10B4 cells were infected at an MOI of 0.5, if necessary in the presence of 1 μg/ml of Dox, and total cellular DNA was isolated at 48 hpi. One microgram of DNA was digested by ApaLI overnight and subjected to agarose gel electrophoresis. The DNA fragments from the gel were transferred onto Hybond-N membrane (GE Healthcare) by capillary blotting according to a standard protocol (39). Subsequently, the DNA was cross-linked by UV light (0.125 J) and prehybridized for 4 to 5 h at 60°C in hybridization buffer (DIG Easy Hyb Granules; Roche). Fifty microliters of probe volume (25 ng/μl) was added, and hybridization was performed overnight. DIG-labeled probes were detected using a DIG Luminescent Detection Kit (Roche) according to manufacturer's instructions and quantified on a chemiluminescence scanner (Typhoon 9400; Amersham Bioscience) with Image Quant software.
Transmission electron microscopy (EM).
NIH 3T3 cells were grown on fibronectin-carbon-coated sapphire discs and infected with the respective viruses at an MOI of 1 using centrifugal enhancement (30 min at 1,200 × g at room temperature). Afterwards, cells were incubated for another hour before the virus suspension was replenished with fresh medium. Starting with the addition of virus, cells were incubated with or without 1 μg/ml Dox. At 48 hpi cells were fixed by high-pressure freezing with an HPF 01 instrument (Engineering Office, M. Wohlwend GmbH), freeze-substituted, and plastic embedded as described previously (45).
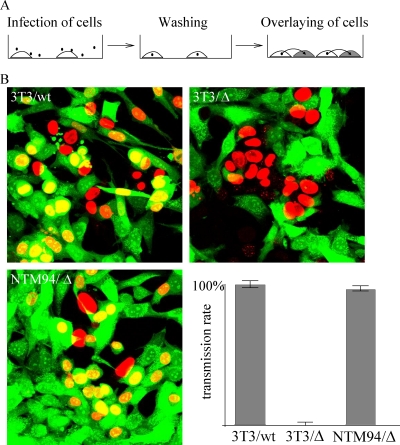
Spread assay.
NIH 3T3 and NT/M94-7 cells were plated and infected at an MOI of 0.25 for 1 h and then washed twice with D-PBS. Cells were incubated for 6 h and afterwards washed four times with D-PBS. Equal numbers of noninfected cells were stained with 5 μM carboxyfluorescein succinimidyl ester (CFSE) for 8 min and blocked by 2% fetal calf serum (FCS)–D-PBS, then washed twice with 2% FCS–D-PBS, and subsequently seeded on top of the unstained but infected cells. Cells were fixed at 48 hpi with 4% paraformaldehyde (PFA) in D-PBS for 10 min at 37°C and washed and permeabilized with 0.1% Triton X-100 for 10 min. After a triple washing, cells were blocked with 3% bovine serum albumin (BSA)–D-PBS for 1 h. Staining of immediate-early gene products was performed by incubating fixed cells with a monoclonal antibody to MCMV pp89 in 3% BSA–D-PBS. After three D-PBS washes, cells were incubated with an Alexa Fluor 555-coupled anti-mouse secondary antibody (Invitrogen) in 3% BSA–D-PBS. Finally, cells were washed three times and imaged by confocal microscopy using an LSM 510 Meta (Zeiss). Virus transmission was determined by counting pp89- and CFSE-positive cells using the ImageJ cell counter plugin (W. S. Rasband, U.S. National Institutes of Health, Bethesda, MD [http://rsb.info.nih.gov/ij/]).
RESULTS
M94 is essential for virus growth in tissue culture.
We recently published genetic screens to identify loss-of-function and DN mutants which allowed us to assign novel functional sites within the MCMV proteins M50 and M53 (7, 21). The M94 gene of MCMV was an interesting candidate, as no structural information on the gene product was available, and the functional sites of the protein were not characterized. In order to use M94 in our genetic screens, we first had to determine if M94 was indispensable for virus replication in tissue culture as our screens are applicable only for essential genes. This is due to the fact that both screening assays are based on a scoring that is dependent on the capability of the tested recombinants to reconstitute from BAC-DNA and to further replicate in cell culture.
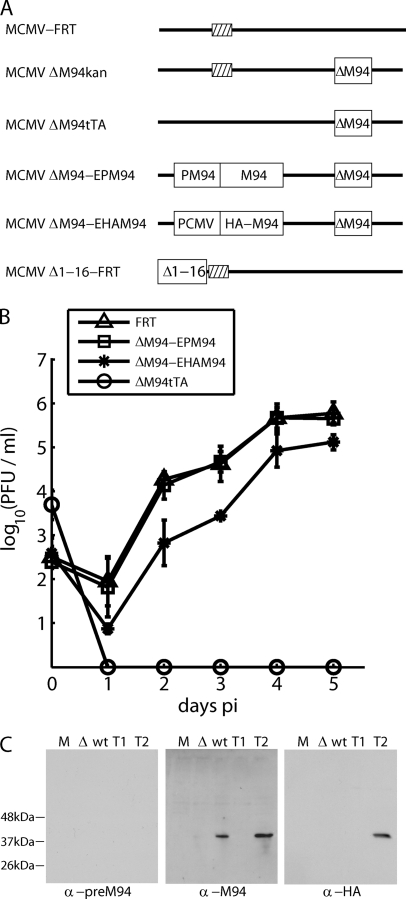
Deletion of the M94 gene by replacing the M94 ORF by a kanamycin cassette resulted in pSM3fr-ΔM94kan, which could not be reconstituted in MEF cells. To ensure that the null phenotype of the M94 deletion mutant was due to the lack of the M94 protein (pM94), the deletion was rescued by an ectopic insertion of either the wt M94 under the control of its own promoter (MCMVΔM94-EPM94) or an HA-tagged M94 under the control of the HCMV immediate-early promoter (MCMVΔM94-EHAM94). Both constructs led to virus progeny after reconstitution, and their growth kinetics were analyzed (Fig. 1B). They replicated similarly to wt MCMV-FRT, demonstrating that the deficiency in M94, and not an overlapping cis element, rendered the pSM3fr-FRT-ΔM94 nonviable. These data also showed that neither the fusion of an HA tag to M94 nor the expression of this fusion under the control of the HCMV immediate-early promoter inhibited the rescue. We also confirmed the null phenotype of the M94 deletion by analyzing the growth characteristics of MCMVΔM94tTA, an M94 deletion virus, which could be propagated on the complementing cell line NT/M94-7 (27). After infection of noncomplementing MEF cells with this virus, we did not observe any evidence of viral growth (Fig. 1B).
Fig. 1.
Ectopic rescue of the M94 deletion mutant. (A) Schematic representation of the analyzed mutants. The MCMV genome is presented as a bold line, and deletions of the M94 gene, either done by replacement with a kanamycin cassette (kan) or a tTA cassette (tTA) are indicated by a white box (ΔM94). The tTA cassette is a constitutive transcription unit expressing a tetracycline-regulated transcriptional activator. It is required for the transcriptional activation of the complementing gene in NT/M94-7 cells. For ectopic rescues, an FRT site (striped box) was utilized between the genes m16 and m17. Both MCMVΔM94-EPM94 and MCMVΔM94-EHAM94 carry a functional copy of the M94 gene under the control of its own or the HCMV immediate-early promoter, respectively. The CMV promoter-driven M94 ORF was tagged with an HA epitope. The MCMVΔ1-16-FRT genome carries a deletion of a nonessential, subterminal region coding for the genes m1 to m16. (B) Analysis of the growth kinetics of MCMV-FRT (FRT), MCMVΔM94tTA (ΔM94tTA), MCMVΔM94-EPM94 (ΔM94-EPM94), and MCMVΔM94-EHAM94 (ΔM94-EHAM94) determined using multistep growth conditions. (C) Western blots demonstrating the specificity of the anti-pM94 immune serum recognizing pM94. Samples after mock treatment (M), MCMVΔM94tTA (Δ), or MCMVΔ1-16-FRT (wt) infection as well as after transfection of pOriR6K-zeo-ie (T1) or pOriR6K-HAM94 (T2) were analyzed. Anti-M94 and anti-HA staining resulted in the expected bands. α-, anti; PCMV, porcine CMV.
To confirm that the genetic changes at the M94 locus indeed resulted in the loss of the gene product, a pM94-specific immune serum was generated. To detect pM94, NIH 3T3 cells were infected with either MCMVΔM94tTA or wt virus. To exclude cross-reacting viral proteins, 293 cells were transfected with either pOriR6K-zeo-ie or pOriR6K-HAM94 as a control. Cell lysates were prepared and analyzed by SDS-PAGE. The blotted proteins were stained with preimmune serum, pM94-specific immune serum, or an antibody detecting the HA tag (Fig. 1C). The predicted size of pM94 is 37.7 kDa. The staining against the HA tag resulted in one band at the predicted size of about 37 kDa, which could be detected solely in cell lysates from cells transfected with the HA-M94 construct. The staining of pM94 with the specific immune serum showed a single band in the wt infection and pOriR6K-HAM94 transfection. The pM94-specific signal was missing in cells infected with MCMVΔM94tTA, confirming the absence of the M94 gene product. Taking these results together, we concluded that M94 is essential for MCMV growth.
Random mutagenesis of the M94 ORF.
To map sites and domains which are essential for the functionality of pM94 and to identify DN mutants of M94, we generated a library of M94 mutants. To this end, we subcloned the HA-tagged M94 ORF into the Litmus28 vector and obtained a transposon acceptor construct (Litmus28-HAM94), and that was subjected to Tn7-mediated random mutagenesis. Then, the entire pool of the mutated M94 ORFs was directly recloned into the pOriR6K-zeo-ie rescue plasmid (see Materials and Methods and Fig. S2 in the supplemental material for details). After recovery of the Tn7-containing M94 mutant library in the rescue vector, the operational minitransposon sequences were removed. This resulted in 15-nucleotide (nt) insertions coding for either in-frame 5-aa insertions or a stop codon within the M94 coding sequence (4). The final mutant library of M94 contained 32,000 primary clones, from which 613 randomly picked clones were sequenced. A total of 494 (80%) of the sequenced mutants contained a single 15-bp insertion in the M94 ORF. The remaining 20% could be attributed to incorrect transpositions (10%), to mutations in sequences flanking the M94 ORF or within the HA tag (5%), or to mixed clones (5%). With this optimized protocol, we achieved an increase in the number of correct mutants from the previously reported 28% to 80% (21). It resulted also in a better coverage of the M94 coding region and allowed us to select a mutant set for further analysis in which the average distance between two insertions was 5.75 aa (compared to 7.2 aa in the previously published M53 library).
Identification of inhibitory mutants of pM94.
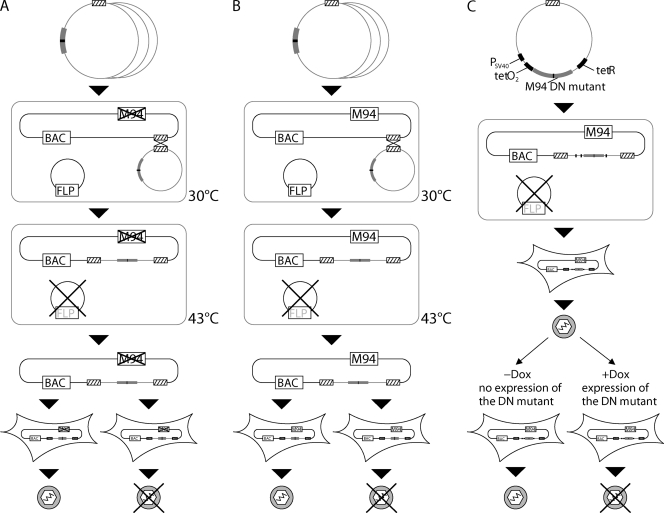
The analysis of phenotypes induced by DN mutants proved to be extremely useful in genetics, cell biology, signaling, and biochemistry. Therefore, we recently established a screening protocol which allows the selection of DN mutants of viral genes from a comprehensive set of random insertion mutants (36). First, M94 mutants were analyzed for their capacity to rescue the M94 deletion mutant in the viral context. This was achieved by ectopic insertion (cis-complementation) of the M94 mutant rescue plasmids into pSM3fr-ΔM94 (Fig. 2A). All M94 mutants that could not recover virus growth were termed loss-of-function mutants. Next, those mutants were reinserted into the wt pSM3fr-FRT BAC as a second M94 allele, and the viability of these constructs was assessed (Fig. 2B). In a subsequent inhibitory screen, lack of virus reconstitution was observed only if the introduced mutant interfered with virus reconstitution in a dominant negative fashion. Dominant negative M94 mutants were confirmed and further characterized using our viral conditional expression system (37), which allowed the analysis of their inhibitory effect during the virus life cycle (Fig. 2C) upon induction of their expression.
Fig. 2.
Strategy for the analysis of M94 mutants in the viral context. The cis-complementation assay (A), the inhibitory screen (B), and the conditional expression (C) are illustrated. pOriR6K-zeo-ie-based vectors containing individual M94 mutants (gray box with black line) were inserted into the MCMV BAC via a Flp-mediated recombination of their FRT sites (striped boxes). The Flp-recombinase was expressed from a helper plasmid containing a temperature-sensitive origin of replication. When the temperature is shifted to 43°C, this helper plasmid is lost. The MCMV BAC lacked the M94 gene in the cis-complementation assay but coded for wt M94 in the inhibitory screen. Also the BAC used to conditionally express inhibitory mutants of M94 contained a wt copy of M94. The reconstitution on MEFs resulted in either viral progeny or failure in viral reconstitution, leading to the identification of essential regions in the cis-complementation assay or inhibitory mutants in the inhibitory screen. Reconstitution of viruses coding for mutant genes that could be regulated conditionally was carried out in the absence of Dox (off state). Therefore, initial growth was not affected by the inhibitory alleles. The inhibitory phenotype of the DN alleles was analyzed by comparing these mutants in the absence (off state) and in the presence (on state) of Dox when the DN mutants inhibited virus growth.
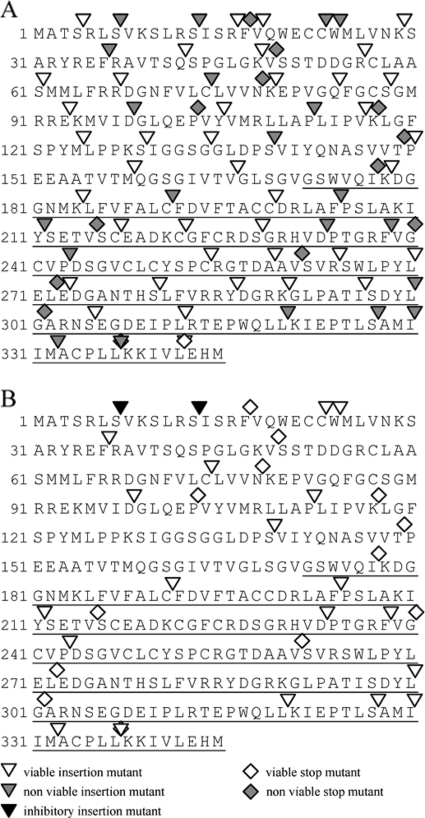
In order to cover the M94 sequence thoroughly with insertion sites, we selected 60 insertion and 14 frameshift mutants of M94 spanning the whole protein evenly. We tested these 74 mutants in the cis-complementation assay (33) as described above (for more details, see Materials and Methods). All frameshift mutants, except the one deleting the last 3 aa of the protein, failed to complement for the M94 deletion in the cis-complementation assay (Fig. 3A). This confirmed the importance of the conserved C terminus as already proposed in previous studies (46). Also, transfection of several mutants which had an insertion in the C terminus and elsewhere in the sequence did not give rise to viral progeny, thereby identifying essential regions within amino acid positions 7 to 37, 191 to 212, 233 to 244, and 320 to 338 of pM94. Subsequently, all nonfunctional insertion and frameshift mutants were used in the inhibitory screen. As illustrated in Fig. 3B, the analysis of these 34 loss-of-function mutants resulted in identification of two inhibitory insertion mutants, named M94i7 and M94i13. Interestingly, we did not observe attenuated mutants of M94 in any of the above described assays. Figure 3 illustrates the result of both screens.
Fig. 3.
Analysis of M94 mutants in the virus context. Sixty insertion mutants and 14 stop mutants spanning the entire M94 ORF evenly were analyzed in two subsequent assays. (A) Results of the cis-complementation assay probing the ability of a given mutant to rescue an M94 deletion genome. Twenty-one noncomplementing insertion mutants were identified. (B) Together with 13 stop mutants, these were tested in the inhibitory screen for their potential to inhibit the reconstitution of a wt genome. Displayed is the amino acid sequence of the M94 protein. Arrowheads indicate approximate insertion sites, and diamonds represent truncations due to stop mutants. Open symbols indicate growth of viral progeny, gray symbols represent failure to reconstitute, and black characters represent inhibitory mutants. The C-terminal domain, as defined by Wing et al. (46), is underlined.
Conditional expression of M94 inhibitory mutants.
In order to study the phenotype of DN mutants in the context of virus replication, they need to be delivered by conditional expression. To this end, we used a system previously described by us (37). In this system, a constitutively expressed Tet repressor (tetR) binds a tet operator (tetO2) in the promoter region of the target gene. The expression of the target gene can be induced by addition of Dox, upon which the binding of tetR to the tetO2 is disrupted. The regulated expression of the inhibitory mutants M94i7 and M94i13 as well as of controls was achieved by subcloning them into the conditional expression cassette and inserting these constructs into MCMVΔ1-16-FRT, which lacks the nonessential genes m01 to m16 but grows like wt MCMV in tissue culture (see Fig. S1 in the supplemental material) (28). This virus provided enough coding space to circumvent growth inhibition due to an oversized genome resulting from insertion of the large regulation cassette (37).
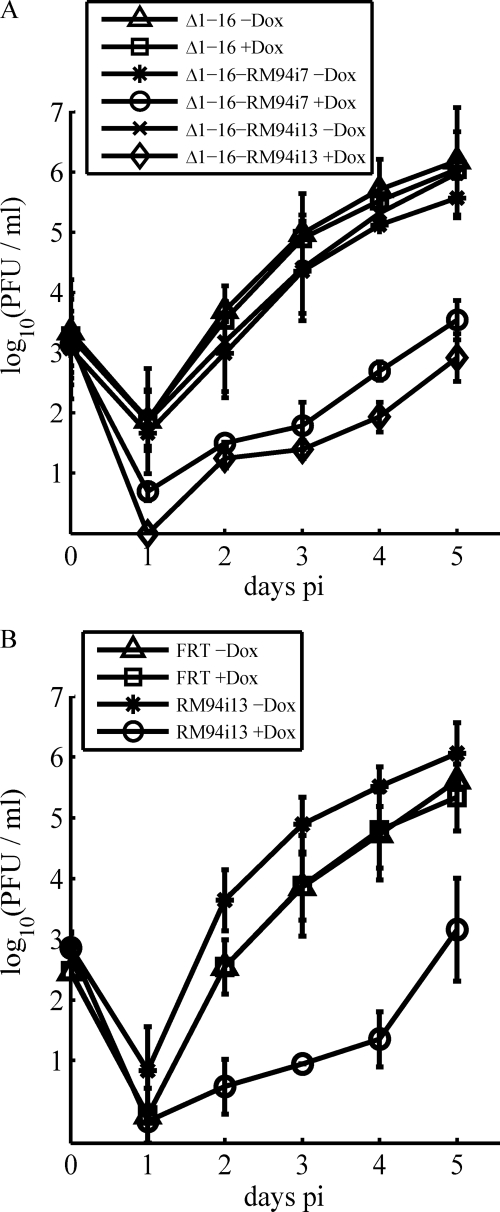
To study the inhibitory potential of these DNs mutants, we used multistep growth curves. The viral titers on day 5 p.i. (Fig. 4A) for MCMVΔ1-16-FRT in the absence and presence of Dox as well as for MCMVΔ1-16-RM94i7 and MCMVΔ1-16-RM94i13 in the absence of Dox were nearly the same. In contrast, in the presence of Dox, the titer of MCMVΔ1-16-RM94i7 decreased by 2.5 orders of magnitude, and the titer of MCMVΔ1-16-RM94i13 decreased by 3 orders of magnitude. This phenotype could also be confirmed by independent clones of both mutants (data not shown) and by growth analysis in the MCMV-FRT background (Fig. 4B). These results demonstrated that the mutants M94i7 and M94i13 indeed interfered with viral growth and that they did not induce a failure of virus reconstitution by toxicity. As the insertions in these two mutants lay closely together, we hypothesized that they interfere with the same protein function, and therefore we chose the slightly stronger inhibitory mutant M94i13 for all assays described in the following.
Fig. 4.
Conditional growth of recombinant MCMVs in primary fibroblasts. (A) Conditional growth of mutants based on the MCMVΔ1–16 genome. The release of infectious viral progeny at the indicated days was quantified under multistep growth conditions after infection of MEFs at an MOI of 0.1 with MCMVΔ1–16 (Δ1–16), MCMVΔ1-16-RM94i7 (Δ1-16-RM94i7), or MCMVΔ1-16-RM94i13 (Δ1-16-RM94i13) in the absence or presence of Dox. (B) Conditional growth of mutants based on the MCMV FRT genome. Infectious viral progeny released after infection with MCMV-FRT (FRT) and MCMV-RM94i13 (RM94i13) in the absence or presence of Dox are depicted.
The true late kinetics of M94 expression are not affected by M94 DN expression.
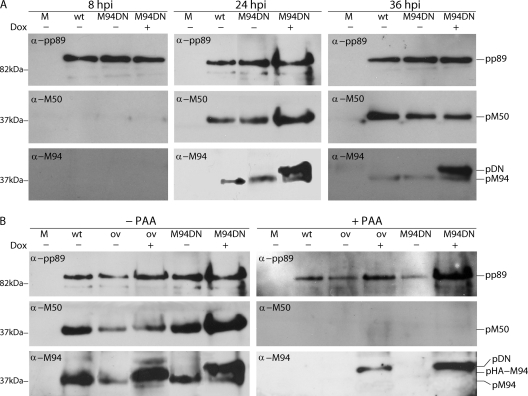
To study whether the inhibition of viral growth was a consequence of the expression of the inhibitory mutants upon Dox induction, the expression levels of the DN proteins were analyzed. Simultaneously, the expression kinetics of M94 were studied to analyze possible effects of the DN mutation on wt protein expression. To this end, NIH 3T3 cells were mock treated or infected with MCMVΔ1-16-FRT and MCMVΔ1-16-RM94i13 at an MOI of 0.5 in the absence and presence of Dox (Fig. 5A). Cell lysates were prepared the time points indicated on Fig. 5, and proteins were separated by SDS-PAGE. pM94 was visualized by Western blotting using a specific immune serum. To probe the viral gene expression cascade in general and to control for protein load, the signals of pp89 and pM50 were analyzed as well. Starting at 8 hpi we could detect a pp89 signal, as expected, for an immediate-early gene product of MCMV, whereas the late protein pM50 was first detected at 24 hpi. The 37-kDa band representing the wt pM94 also became detectable at 24 hpi. The dominant negative mutant of M94 pM94i13 (pDN) was slightly bigger than its wt counterpart because it carried an additional 5-aa insertion as well as an HA tag. Correspondingly, we could discern both forms as the pDN form ran marginally higher than wt pM94 on a blot. The pDN was clearly detectable at 24 hpi and 36 hpi upon addition of Dox at an enhanced rate compared to pM94.
Fig. 5.
Expression kinetics of pM94 and pHA-M94. (A) NIH 3T3 fibroblasts were mock treated (M) or infected with MCMVΔ1-16-FRT (wt) and MCMVΔ1-16-RM94i13 (DN) in the absence or presence of Dox. Cell lysates were prepared at the indicated time points. Proteins were separated by SDS-PAGE, and pp89, pM50, and pM94 were detected by Western blotting using specific immune sera. (B) NIH 3T3 fibroblasts were infected under the same conditions and with the same viruses as described for panel A and additionally with MCMVΔ1-16-RHAM94 (ov). Infections were performed in the absence and presence of PAA. Cell lysates were prepared at 24 hpi and separated by SDS-PAGE, and signals for pp89, pM50, and pM94 were visualized by Western blotting using specific immune sera. The different M94 protein versions are labeled as follows: pM94 for the wt M94 protein; pHA-M94 for the not mutated, HA-tagged M94; and pDN for the HA-tagged M94i13.
To confirm the true late expression profile of pM94, the viral genome replication was blocked by the addition of PAA to infected cells (Fig. 5B). As in the previous experiment, NIH 3T3 cells were mock treated or infected with MCMVΔ1-16-FRT and MCMVΔ1-16-RM94i13 in the absence and presence of Dox. As a further control, MCMVΔ1-16-RHAM94 was used, which carries an HA-tagged wt M94 allele at an ectopic position. The blots were analyzed as above. Here, the pp89 staining served as a loading control, and pM50 expression was used to monitor the efficiency of PAA treatment. As anticipated, the endogenous pM94 was detectable in the absence of PAA at 24 hpi but not in the presence of PAA. In contrast, the expression of the HA-tagged wt M94 (pHA-M94) and the HA-tagged M94i13 were not influenced by PAA treatment. The analysis of the protein expression profiles confirmed that expression of the DN M94 mutant was Dox dependent and that overexpression of M94i13 did not influence the expression of other genes like pp89 or pM50. In addition, the induction of the DN M94 did not affect the kinetics of wt pM94 expression from the native locus. Altogether, these data suggested that M94i13 had no negative effects on the expression of other genes.
M94 plays an essential role in secondary envelopment.
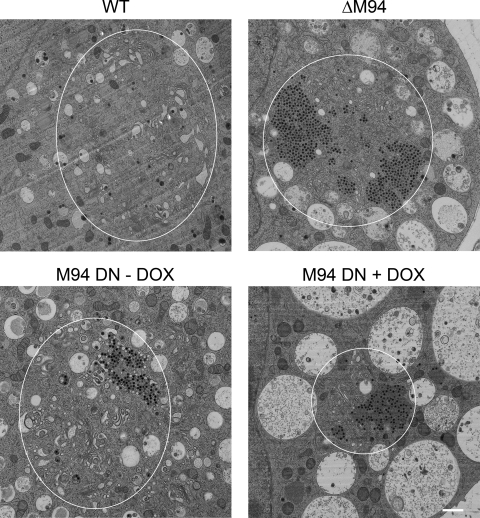
To characterize the inhibitory phenotype of the DN mutant during morphogenesis, transmission electron microscopy (EM) was used. NIH 3T3 cells were infected with wt MCMV, M94 deletion virus, and MCMVΔ1-16-RM94i13 in the absence or presence of Dox (Fig. 6). Mutant viruses showed a wt-like nuclear phenotype as well as comparable primary envelopment mainly at nuclear infoldings, as described earlier for MCMV (9) (see Fig. S3 in the supplemental material). However, no clear evidence of secondary envelopment could be found in cells infected with the M94 deletion virus or DN M94-expressing virus in the presence of Dox. Instead, only capsid accumulations in the center of the cytoplasmic structure, a recently described assembly complex (12), were found. Therefore, we quantified the relative abundance of cytoplasmic capsids and categorized them in three classes. These classes represented the main stages of cytoplasmic morphogenesis, i.e., nonenveloped capsids, capsids in the envelopment process, and enveloped capsids. Filled cytoplasmic capsids without any membrane wrapping were classified as nonenveloped. Capsids next to cellular membranes inducing a membrane bending were classified as in the envelopment process while those inside a completely closed membrane wrap were categorized as enveloped. Examples of these three different classes are depicted in Fig. 7A.
Fig. 6.
Ultrastructural analysis of M94 mutants. NIH 3T3 cells were infected with MCMVΔ1-16-FRT (WT), MCMVΔM94tTA (ΔM94), and MCMVΔ1-16-RM94i13 in the absence (M94 DN −DOX) or presence (M94 DN +DOX) of doxycycline at an MOI of 1 followed by centrifugal enhancement. Cells were fixed 48 h after infection and prepared for electron microscopy by high-pressure freezing. The samples were freeze-substituted, plastic embedded, thin sectioned, and analyzed by transmission electron microscopy. Representative examples show the characteristic cytoplasmic phenotypes of MCMV with Golgi membranes next to cytoplasmic capsids and surrounded by multivesicular membranous compartments. White ellipses indicate the approximate outer boundary of assembly compartments as defined by Golgi membranes and surrounding multivesicular membranous compartments. Scale bar, 1 μm.
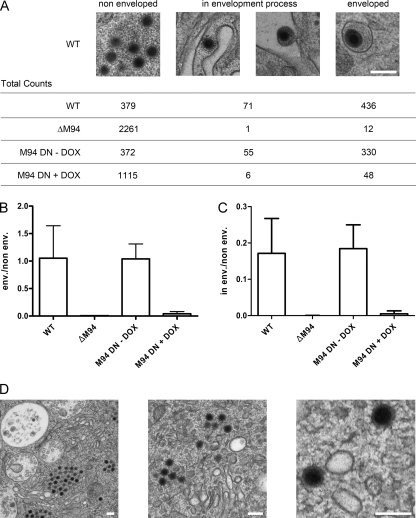
Fig. 7.
Quantification of phenotypes in electron micrographs. Cells were treated as described in the legend of Fig. 6, and morphogenesis stages of individual capsids were classified and subsequently counted. Three morphogenesis stages were defined. Filled cytoplasmic capsids without any membrane wrapping were classified as nonenveloped. Capsids next to cellular membranes inducing a membrane bending were classified as in envelopment process while those inside a completely closed membrane wrap were categorized as enveloped. (A) Example pictures from wt-infected cells, depicting the three categories that were counted. Summarizing total counts from two to three independent experiments for each virus and category are given. The ratio between counts for enveloped to nonenveloped is depicted in panel B while the ratio between counts for capsids in the envelopment process to nonenveloped capsids is given in panel C. Bars indicate the standard deviation from independent experiments. (D) Representative magnifications of capsids near cellular membranes that show no sign of interaction or membrane bending from cells infected with MCMVΔM94tTA (ΔM94) in comparison to wt shown in panel A. Scale bar, 200 nm.
We counted all cytoplasmic capsids within views derived from 13 randomly chosen cells infected with wt MCMV, 9 cells after infection with the M94 deletion mutant, and 18 cells infected with MCMVΔ1-16-RM94i13 in the absence of Dox and 14 cells in the presence of Dox. For all analyzed conditions, cells were selected from at least two independent experiments. Then, we counted all cytoplasmic capsids and clustered them into the categories described above. Next, we determined the ratios between counts of enveloped and nonenveloped capsids (Fig. 7B) and between counts of capsids in the envelopment process and nonenveloped capsids (Fig. 7C). The deletion of the M94 gene or the overexpression of DN M94 resulted in a drastic drop of both ratios (Fig. 7A, B, and C). Additionally, no substantial evidence for particles being in the process of secondary envelopment could be detected in cells which were infected with the deletion mutant, even if capsids were located in close proximity to membranes (Fig. 7D). Taken together, these findings indicated a central role for M94 in secondary envelopment.
M94 is not involved in viral cleavage-packaging.
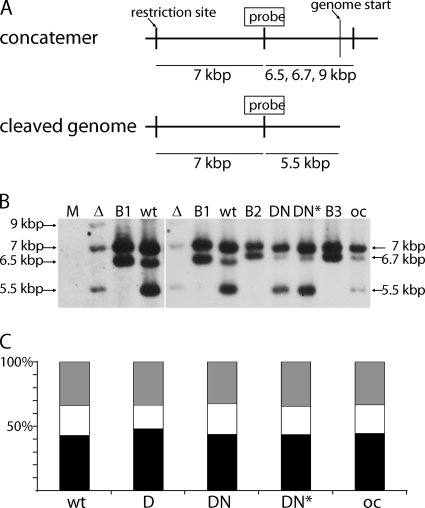
The EM analysis of the phenotypes induced by both the deletion mutant and the expression of the DN mutant had no apparent effect on the nuclear morphogenesis of MCMV. Nevertheless, studies on Alphaherpesvirinae homologues of pM94 suggested that UL16 family proteins may play a role in genome cleavage-packaging (31, 32). Therefore, a second function of pM94 in MCMV DNA packaging was specifically addressed. To monitor whether absence of the M94 gene product or expression of the DN allele interfered with DNA packaging, we analyzed the cleavage of genomes from their postreplication concatemeric form to the unit length form found in mature capsids. In herpesviruses, this reaction is coupled with DNA packaging (22). To this end, M2-10B4 cells were mock treated or infected at an MOI of 0.5 with either MCMVΔM94tTA, MCMVΔ1-16-FRT, MCMVΔ1-16-EHAM94, or MCMVΔ1-16-RM94i13 in the absence or presence of Dox. At 48 hpi total cellular DNA was extracted from infected cells, and equal amounts of DNA were digested with ApaLI and subjected to Southern blot analysis (Fig. 8). The cleavages induced by both the restriction enzyme and the viral terminase resulted in fragments of different sizes detected by a corresponding probe (Fig. 8A). The 7-kbp fragment (nucleotides 217741 to 217940) was utilized as a loading control, whereas the 5.5-kbp fragment (nucleotides 224735 to 230278) represents the ApaLI-digested, cleaved genomes. The fragments defining the concatemeric form differed in length due to the insertions from 6.5 kbp over 6.7 kbp to 9 kbp. All three bands representing the different genome forms could be visualized by our specific probe (Fig. 8B).
Fig. 8.
Analysis of MCMV DNA cleavage-packaging in pM94 mutant viruses. (A) Schematic representation of the concatemeric and cleaved genome forms. The probe binding site used for Southern blotting as well as the utilized restriction sites, genome starts, and detected fragments are indicated. (B) The detection of different genome fragments in DNA isolated from mock-treated M2-10B4 cells (M) or from cells infected with MCMVΔM94tTA (Δ), MCMVΔ1-16-FRT (wt), or M2-10B4 cells infected with MCMVΔ1-16-RM94i13 in the absence (DN) or presence (DN*) of Dox, as well as MCMVΔ1-16-EHAM94 for overexpression control (oc) is shown. The BACs MCMVΔ1-16-FRT (B1), MCMVΔ1-16-RM94i13 (B2), and MCMVΔ1-16-EHAM94 (B3) served as negative controls. Due to their circular form, digested BAC DNA results in all fragments except the one indicating the cleaved terminal fragments. The left part of the image depicts the same first three lanes also shown in the right image but acquired with an increased exposure time. The sizes for the following fragments are indicated: 7 kbp, loading control fragment; 6.5/6.7/9 kbp, concatemeric fragment; 5.5 kbp, terminal fragment characteristic for the unique length genomes. (C) Quantification of signal intensities. The intensities of individual bands were quantified luminometrically. The ratios of control fragment (black), concatemeric fragment (white), and cleaved genomes (gray) are given as relative intensities in percentages.
The obtained chemiluminescence signal was quantified in order to compare the amount of the different genome forms in different samples. All samples were normalized and compared to each other based on the intensity of the corresponding 7-kbp loading control fragment. The results were plotted as relative intensities of the three above mentioned fragments (Fig. 8C). The ratios between the cleaved and concatemeric forms were almost identical in every sample, indicating no influence of the induction of the M94 DN mutant or M94 deletion on the cleavage-packaging process. Therefore, we concluded that M94 of MCMV played no major role in genome cleavage and packaging.
MCMV lacking M94 is spread deficient.
Lack of secondary envelopment, as detected by EM analysis, surprisingly does not necessarily result in loss of cell-to-cell spread, as reported for a UL99-negative HCMV mutant (42). Therefore, we analyzed the phenotype of the M94 deletion in cell-to-cell spread. This was investigated by infection of NIH 3T3 and NT/M94-7 (NTM94) cells with wt MCMV or MCMVΔM94tTA, followed by extensive washing to remove excess virus. Next, CFSE-stained NIH 3T3 cells were added, and after an additional 48 h, the culture was fixed and stained for pp89. This resulted in cells which were either pp89 positive, CFSE positive, or positive for both stains (Fig. 9A). Stained cells were counted, and cell-to-cell spread was determined by calculating the ratio between pp89-positive/CFSE-stained cells and pp89-positive/CFSE-negative cells (Fig. 9B). The spread rate of the wt MCMV was set at 100%.
Fig. 9.
Quantification of virus transmission. (A) NIH 3T3 (3T3) and NT/M94-7 (NTM94) cells were infected with wt MCMV (wt) or MCMVΔM94tTA (Δ) at an MOI of 0.25. CSFE-stained NIH 3T3 cells were added at 6 h postinfection. After incubation for a further 42 h, the virus-infected cells were visualized by pp89-staining. (B) CFSE-stained cells, pp89-positive cells, and cells showing both signals were counted. Virus transmission was determined by calculating the ratios between pp89-positive/CFSE-stained cells and pp89-positive/CFSE-negative cells (lower right quadrant). The spread rate of the wt MCMV was set to 100%.
wt MCMV infection spread rapidly throughout the cell culture, as indicated by the large number of double-stained nuclei (Fig. 9). In contrast, the M94 deletion mutant was unable to infect the newly added cells. Only one double-stained nucleus was seen after counting 416 pp89-positive/CFSE-negative cells. Its ability to infect fresh cells was, however, restored to a transmission rate of 97% when the mutant was grown on complementing NT/M94-7 cells. We concluded that the effect of the M94 deletion on secondary envelopment of MCMV also resulted in a deficiency to spread from cell to cell.
M94 DN mutant fails to interact with M99.
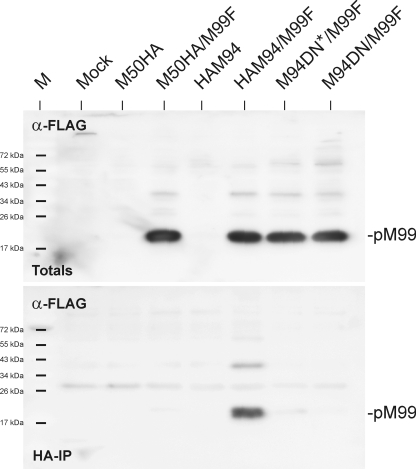
A specific interaction between pUL16 (the HSV-1 homologue of pM94) and pUL11 (the HSV-1 homologue of pM99) could be established by Loomis et al. (20). The same could be shown for the HCMV homologues later (18). This indicated conserved features among the UL16 family proteins of herpesviruses. The HSV-1 and HCMV homologues of pM99 are believed to be involved in secondary envelopment (43). Because our EM data showed a lack of secondary envelopment, we decided to check whether the DN M94 affected the pM94-pM99 interaction. To probe the ability of the pM94 variants to interact with pM99, a FLAG-tagged version of pM99 was used. M2-10B4 cells were transfected with constructs coding for pHAM94, pHAM94-DN, or pM50HA as a negative control alone or in combination with pM99FLAG and subsequently used for HA pulldown assays (Fig. 10). In each reaction mixture, 10% of the input material was used to determine the total pM99 expression (Fig. 10A). While pHAM94 and pM99FLAG did copurify upon HA pulldown, no unspecific interaction of pM99FLAG with the HA matrix was detected upon expression of the pM50HA control protein. Remarkably, the pM94 DN mutant was unable to pull down the M99 protein. This implied that the DN mutant of pM94 lost the ability to form a detectable pM99/pM94 complex.
Fig. 10.
Binding of pM94 and the pM94 DN mutant to pM99. Lysates were prepared from mock-treated M2-10B4 cells and cells transfected with pM50HA (M50HA), pHAM94 (HAM94), and the pHAM94 DN mutant (M94DN) alone or in combination with pM99FLAG (M99F). A 10% fraction of the total lysates served as an input control (Totals) while 90% of the lysates were subjected to pulldowns with an HA matrix (HA immunoprecipitation [HA-IP]). The samples were separated by SDS-PAGE and were immunoblotted using FLAG-specific antibodies as indicated.
DISCUSSION
In this study we showed (i) that the M94 gene product was essential for MCMV replication in cell culture, (ii) that pM94 played a key role in secondary envelopment of MCMV, (iii) that at least two domains of pM94, located at the N and C termini, were crucial for the function of this protein, (iv) that two insertion mutants at the N-terminal domain were dominant negative to MCMV replication, and (v) that the DN mutations impaired the interactions with the MCMV tegument protein pM99. In addition, our data indicated that at least one other site of pM94, located within its conserved C-terminal domain, was required for an essential activity of pM94.
In a deletion-based strategy the HCMV homologue of pM94, pUL94, was shown to be essential (14). However, a functional screen utilizing transposon mutagenesis reported a small-plaque phenotype of UL94 null mutants, arguing against essentiality of pUL94 (48). Thus, it was therefore not predictable if pM94 was essential in MCMV. However, as shown by data presented here, pM94 was indeed essential for growth of MCMV in tissue culture.
As M94 was essential for growth in cell culture, we could subject it to a three-step mutagenesis and screening protocol that is designed to identify dominant negative mutants. First, we used a transposon-based random mutagenesis approach to generate a library of M94 insertion and stop mutants. Then, we chose 74 mutants from the library which covered the whole M94 open reading frame and used them in two successive assays. With the first assay, termed cis-complementation assay (7), we were able to map two essential regions located in the N-terminal variable part and the conserved C-terminal domain of pM94. All mutants which could not rescue the M94 deletion virus in the first screen were then used in the second assay, termed the inhibitory screen. This assay is tailored to identify inhibitory and dominant negative mutants (36). DN mutants are those inhibitory mutants that act within the pathway in which the wt protein is involved (16, 29). With this assay we were able to find two N-terminal insertion mutants of pM94 which showed dominant negative activity. Interestingly, we did not observe attenuated M94 mutants in this study in either the cis-complementation assay or in the inhibitory screen, as was the case for comparable studies on the MCMV proteins M53 and M50 (21, 36). This was surprising as both assays are suitable to identify such mutants as we reported previously (7, 33).
We then cloned these DN mutants into a regulation cassette to conditionally induce their expression in the virus context. Both DN mutants inhibited MCMV growth upon Dox-induced overexpression in the presence of the wt protein. Upon expression of the M94i13 mutant, no detectable influence on viral gene expression was observed. Even the expression of the wt M94 protein was not affected.
To cause DN effects, a mutant must meet at least two requirements. First, it must be able to interact with its protein partners and at the same time lack an essential function, which leads to interference with or full blockage of wild-type protein function. M94 is a homologue of the pUL16 family of proteins which interact with the pUL11 family of viral proteins (18, 20). As shown here, this was also true for MCMV as we could establish an interaction between pM94 and pM99. Both pUL11 and its homologue pUL99 contain a membrane anchor and are involved in secondary envelopment (19, 43) and interact with envelope glycoproteins (17). Therefore, the UL16 family proteins might act as bridging factors, connecting capsids to the site of secondary envelopment by means of the membrane-anchored pUL11 family members (17, 47).
Notably, the pM94 DN mutant failed to interact with pM99. We therefore concluded that one of the essential functions missing in the M94 DN mutant is located in the N terminus of pM94. This finding is remarkable as sequence comparisons indicate no N-terminal conservation of the UL16 protein family. Studies on pUL16 homologues utilized N- and C-terminal deletion mutants. Interestingly, both truncations destroyed the interaction with pUL11 homologues (15, 18, 47). It is therefore possible that, besides the N-terminal feature, there is a conserved motif located at the conserved C-terminal region, which is also involved in the interaction between the pUL16 and pUL11 homologues.
The second requirement a DN mutant has to fulfill, besides lacking an essential function, is its capacity to interact with its other protein partners in a wt manner. This second, intact functional feature of the DN M94 still remains to be identified. Binding to viral capsids could be one possibility as transient binding of pUL16 to the viral nucleocapsid has been established (24). Moreover, C-terminal cysteine residues are conserved throughout the pUL16 homologues and are involved in capsid binding (23). Therefore, binding to the nucleocapsid, as the second functional feature of pM94, might be connected with its C terminus.
Taken together, conditional expression of the DN mutant leads to essentially the same phenotype as observed for the null mutant of M94. Thus, the N-terminal insertion mutant M94i13 was deficient in at least one functional site, which was required specifically for the secondary envelopment. This allowed us to characterize the M99 interaction as a functional feature of M94 which seemed to be crucial for secondary envelopment. On the other hand, our data pointed to the fact that DN M94i13, acting specifically on secondary envelopment, was still able to enter the same pathway where the wt protein acts. This indicates that M94i13 was capable to interact with another, yet unidentified, activity such as a viral or cellular protein or with wt M94 itself.
In the light of the findings discussed above, one can speculate that at least in MCMV, and possibly in other betaherpesviruses, the pM99/pM94 (pUL99/pUL94) complex is a central entity in MCMV secondary envelopment (17–19). Interestingly, the pUL99 deletion mutant of the HCMV AD169 strain (which should also lack this complex) shows no secondary envelopment but can, nevertheless, spread from cell to cell (42). In our experiments, the M94 deletion mutant did not spread to noninfected cells. This could imply that in some strains of HCMV, alternative protein complexes exist in addition to the pUL94/pUL99 complex, which can take over its function at least in some egress pathways. In this case, alternative viral protein complexes consisting of strain-specific viral glycoproteins might be able to direct HCMV morphogenesis toward cell-to-cell spread rather than release of virions (41). These alternative protein complexes probably do not exist in MCMV (at least not in the BAC derived from the Smith strain).
pUL16 is not essential in the Alphaherpesvirinae (2). Here, the link between the viral capsid and its secondary envelopment apparently involves functionally redundant protein complexes (for a review, see reference 17). Consequently, the study and mechanistic dissection of secondary envelopment in the alphaherpesvirus model are technically demanding and require complex genetic engineering. Therefore, it is remarkable that a deletion of only a single gene or overexpression of a single DN allele already resulted in a complete block of secondary envelopment in a betaherpesvirus model. Thus, we believe that further characterization of the pM94/pM99 complex in MCMV and definition of its connections either to the capsid and/or to the envelope are needed and will provide highly valuable insights into the principles of herpesvirus secondary envelopment.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Thomas Mertens for his generous help in EM studies. We thank Sigrid Seelmeir, Simone Boos, Natalie Röder, and Eberhard Schmid for their excellent technical assistance.
This work was supported by grants from DFG priority research program SPP1175.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 29 June 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Baines J. D. 2007. Envelopment of herpes simplex virus nucleocapsids at the inner nuclear membrane, p. 144–150 In Arvin A., et al. (ed.), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 2. Baines J. D., Roizman B. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baines J. D., Roizman B. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biery M. C., Stewart F. J., Stellwagen A. E., Raleigh E. A., Craig N. L. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28:1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Britt B. 2007. Maturation and egress, p. 311–323 In Arvin A., et al. (ed.), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 6. Britt W. J., Jarvis M., Seo J. Y., Drummond D., Nelson J. 2004. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bubeck A., et al. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J. Virol. 78:8026–8035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bubic I., et al. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buser C., Walther P., Mertens T., Michel D. 2007. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J. Virol. 81:3042–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campadelli-Fiume G., Menotti L. 2007. Entry of alphaherpesviruses into the cell, p. 93–111 In Arvin A., et al. (ed.), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 11. Cherepanov P. P., Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 12. Das S., Vasanji A., Pellett P. E. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81:11861–11869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davison A. J. 2010. Herpesvirus systematics. Vet. Microbiol. 143:52–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunn W., et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harper A. L., et al. 2010. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J. Virol. 84:2963–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Herskowitz I. 1987. Functional inactivation of genes by dominant negative mutations. Nature 329:219–222 [DOI] [PubMed] [Google Scholar]
- 17. Johnson D. C., Baines J. D. 2011. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9:382–394 [DOI] [PubMed] [Google Scholar]
- 18. Liu Y., et al. 2009. The tegument protein UL94 of human cytomegalovirus as a binding partner for tegument protein pp28 identified by intracellular imaging. Virology 388:68–77 [DOI] [PubMed] [Google Scholar]
- 19. Loomis J. S., Bowzard J. B., Courtney R. J., Wills J. W. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209–12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loomis J. S., Courtney R. J., Wills J. W. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417–11424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lotzerich M., Ruzsics Z., Koszinowski U. H. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 80:73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McVoy M. A., Adler S. P. 1994. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 68:1040–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meckes D. G., Jr., Wills J. W. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 81:13028–13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meckes D. G., Jr., Wills J. W. 2008. Structural rearrangement within an enveloped virus upon binding to the host cell. J. Virol. 82:10429–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menard C., et al. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mocarski E. D. J. 2007. Comparative analysis of herpesvirus-common proteins, p. 177–203. In Arvin A., et al. (ed.), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 27. Mohr C. A., et al. 2010. A spread-deficient cytomegalovirus for assessment of first-target cells in vaccination. J. Virol. 84:7730–7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mohr C. A., et al. 2008. Engineering of cytomegalovirus genomes for recombinant live herpesvirus vaccines. Int. J. Med. Microbiol. 298:115–125 [DOI] [PubMed] [Google Scholar]
- 29. Mühlbach H., Mohr C., Ruzsics Z., Koszinowski U. 2009. Dominant-negative proteins in herpesviruses—from assigning gene function to intracellular immunization. Viruses 1:420–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857 [DOI] [PubMed] [Google Scholar]
- 31. Nalwanga D., Rempel S., Roizman B., Baines J. D. 1996. The UL 16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226:236–242 [DOI] [PubMed] [Google Scholar]
- 32. Oshima S., et al. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch. Virol. 143:863–880 [DOI] [PubMed] [Google Scholar]
- 33. Popa M., et al. 2010. Dominant negative mutants of the murine cytomegalovirus M53 gene block nuclear egress and inhibit capsid maturation. J. Virol. 84:9035–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reddehase M. J., et al. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roizman B., et al. 1981. Herpesviridae. Definition, provisional nomenclature, and taxonomy. The Herpesvirus Study Group, the International Committee on Taxonomy of Viruses. Intervirology 16:201–217 [DOI] [PubMed] [Google Scholar]
- 36. Rupp B., et al. 2007. Random screening for dominant-negative mutants of the cytomegalovirus nuclear egress protein M50. J. Virol. 81:5508–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rupp B., Ruzsics Z., Sacher T., Koszinowski U. H. 2005. Conditional cytomegalovirus replication in vitro and in vivo. J. Virol. 79:486–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruzsics Z., Koszinowski U. H. 2008. Mutagenesis of the cytomegalovirus genome. Curr. Top. Microbiol. Immunol. 325:41–61 [DOI] [PubMed] [Google Scholar]
- 39. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Scrivano L., et al. 2010. The m74 gene product of murine cytomegalovirus (MCMV) is a functional homolog of human CMV gO and determines the entry pathway of MCMV. J. Virol. 84:4469–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scrivano L., Sinzger C., Nitschko H., Koszinowski U. H., Adler B. 2011. HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog. 7:e1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Silva M. C., Schroer J., Shenk T. 2005. Human cytomegalovirus cell-to-cell spread in the absence of an essential assembly protein. Proc. Natl. Acad. Sci. U. S. A. 102:2081–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Silva M. C., Yu Q. C., Enquist L., Shenk T. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594–10605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wagner M., Jonjic S., Koszinowski U. H., Messerle M. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Walther P., Ziegler A. 2002. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J. Microsc. 208:3–10 [DOI] [PubMed] [Google Scholar]
- 46. Wing B. A., Lee G. C., Huang E. S. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 70:3339–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yeh P. C., Meckes D. G., Jr., Wills J. W. 2008. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J. Virol. 82:10693–10700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu D., Silva M. C., Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.