Abstract
Monkeypox virus (MPXV) is an orthopoxvirus closely related to variola virus, the causative agent of smallpox. Human MPXV infection results in a disease that is similar to smallpox and can also be fatal. Two clades of MPXV have been identified, with viruses of the central African clade displaying more pathogenic properties than those within the west African clade. The monkeypox inhibitor of complement enzymes (MOPICE), which is not expressed by viruses of the west African clade, has been hypothesized to be a main virulence factor responsible for increased pathogenic properties of central African strains of MPXV. To gain a better understanding of the role of MOPICE during MPXV-mediated disease, we compared the host adaptive immune response and disease severity following intrabronchial infection with MPXV-Zaire (n = 4), or a recombinant MPXV-Zaire (n = 4) lacking expression of MOPICE in rhesus macaques (RM). Data presented here demonstrate that infection of RM with MPXV leads to significant viral replication in the peripheral blood and lungs and results in the induction of a robust and sustained adaptive immune response against the virus. More importantly, we show that the loss of MOPICE expression results in enhanced viral replication in vivo, as well as a dampened adaptive immune response against MPXV. Taken together, these findings suggest that MOPICE modulates the anti-MPXV immune response and that this protein is not the sole virulence factor of the central African clade of MPXV.
INTRODUCTION
Monkeypox virus (MPXV) is a member of the Orthopoxviridae family of poxviruses and is closely related to variola virus, the causative agent of smallpox (32). MPXV is endemic in the rainforests of central and western Africa and is believed to be primarily a zoonotic disease with sporadic infection of humans due to exposure to infected animals or animal tissues (17). The clinical signs of human MPXV disease closely mimic those seen with smallpox infection and include the development of fever, rash, disseminated pox lesions, and respiratory symptoms (9). MPXV infection can be fatal, with mortality rates as high as 10 to 17% (3, 9, 12). Although MPXV can infect a wide array of animal species, including nonhuman primates (NHP) and rodents, it appears that wild squirrels may be the natural reservoir of the virus, with NHP and humans being incidental hosts (13, 18).
Although genetically very similar, all strains of MPXV examined thus far can be separated into two distinct clades based on comparison of genomic sequences (21). The more deadly outbreaks of monkeypox in central Africa have been determined to be caused by strains belonging to a distinct central African clade, while viruses belonging to the west African clade have been responsible for less severe outbreaks. The virus strain responsible for a U.S. outbreak of MPXV in 2003 was determined to belong to the west African clade based on genetic analysis (28), and in fact, no cases of human-to-human transmission or deaths were reported in this outbreak, further suggesting a less severe disease associated with west African strains of MPXV in humans. In vivo studies conducted in cynomolgus monkeys, prairie dogs, and ground squirrels appear to support this observation, with central African strains displaying higher virulence than west African strains of MPXV in these animal models (7, 14, 29, 30).
These observations led to the conclusion that the genetic differences between the two clades of MPXV likely account for their variable pathogenesis and transmission rates. Based on genomic comparisons of several MPXV strains of both west African and central African origin, five genes—D10L (host range protein), D14L (complement inhibitor), B10R (apoptotic regulator), B14R (interleukin [IL]-1β binding protein), and B19R (serine protease inhibitor-like protein)—have been speculated to be most likely responsible for the increased virulence of central African strains of MPXV (7, 21). Since D14L is completely absent from west African strains of MPXV (7, 36), it has been suggested to be a leading candidate to explain the difference in virulence between MPXV clades. D14L encodes the monkeypox inhibitor of complement enzyme (MOPICE) and is an ortholog of other poxvirus inhibitors of complement enzymes (PICEs), including the vaccinia complement control protein (VCP), the cowpox inflammation modulatory protein (IMP), and the smallpox inhibitor of complement enzyme (SPICE) (31, 36). The PICEs are highly related proteins that also share significant homology to human regulators of complement activation (RCA) and contain tandem short consensus repeat (SCR) domains that are found in RCA proteins (8, 36). Despite a truncation in one of the SCR domains (7), MOPICE was shown to be a functional inhibitor of complement activation in vitro, displaying cofactor activity for C3 and C5 convertases intermediate to that of VCP and SPICE, although it does appear to lack decay-accelerating activity that is associated with other PICEs (7, 22). Due to its complement regulatory activity, MOPICE may play a major role in limiting the host immune response during MPXV infection, which could ultimately lead to an increase in pathogenesis compared to that of MPXV lacking this protein. Although MOPICE has been suggested to be one of the main virulence factors of central African strains of MPXV (7), this hypothesis has not been directly examined in vivo.
In order to gain a better understanding of the primary immune response to MPXV infection and to assess the role of MOPICE in MPXV disease, we further developed the rhesus macaque (RM) (Macaca mulatta) model of MPXV infection utilizing the pathogenic Zaire strain of MPXV (MPXV-Z) and a recombinant D14L “knockout” MPXV-Z strain (D14L KO MPXV-Z). Previous in vivo studies utilizing NHP have indicated that aerosolized or intravenous MPXV-Z infection yields a clinical outcome that parallels those of human infections with MPXV (7, 33, 38). For our studies we utilized intrabronchial (i.b.) infection of RM with purified virions, which allows for the delivery of a defined dose of virus directly into the lungs, thus mimicking an aerosol exposure while also providing more consistent dosing. This method of inoculation has recently been shown to recapitulate many aspects of human monkeypox disease in a cynomolgus macaque model of MPXV infection (16). Similarly, findings presented in this study indicate that i.b. inoculation with MPXV-Z in RM recapitulates the hallmarks of human clinical infection, with the development of fever, rash, and respiratory symptoms. Furthermore, we present the first detailed characterization of the primary adaptive immune response to MPXV infection, and we demonstrate that RM generate a robust T- and B-cell response to MPXV-Z after infection, which coincides with the resolution of clinical symptoms. More importantly, using the RM i.b. MPXV infection model system, we were able to directly test the unanswered hypothesis that MOPICE is a major determinant of central African MPXV virulence and to demonstrate that animals infected with a recombinant form of MPXV-Z lacking MOPICE expression (D14L KO MPXV-Z) exhibit higher viral loads, as well as a delayed and reduced immune response, compared to animals infected with wild-type MPXV-Z. These data suggest that MOPICE is likely not the leading determinant of increased virulence of central African strains of MPXV and that MOPICE might actually play a role in the generation of a successful adaptive immune response to infection.
MATERIALS AND METHODS
Viruses and purification.
MPXV-Zaire (V79-I-005) was kindly provided by Inger Damon (Centers for Disease Control and Prevention, Atlanta, GA), and all work with this virus and recombinant derivatives was conducted in accordance with institutional guidelines for biosafety at the Oregon Health & Science University. All viruses were grown in BSC40 monkey kidney cells, and the intracellular mature form of virus (IMV) was purified by sucrose gradient ultracentrifugation as described elsewhere (23). Titers of viral stocks were determined by standard plaque assay on BSC40 cells.
Generation of recombinant D14L KO MPXV-Z.
To generate a form of MPXV-Zaire (V79-I-005) devoid of D14L, we constructed a knockout virus using standard recombination techniques. Briefly, 600-bp fragments of MPXV genomic sequence directly flanking either side of MPXV Zaire D14L (nucleotides 19834 to 19184) were amplified by PCR using primers engineered to contain restriction sites for cloning. The following upstream D14L primers amplified nucleotide (nt) 20433 to nt 19835 and contain a 5′ EcoRI site and a 3′ XbaI site, respectively: primer 1, 5′-CGAATTCAGACTTCCAAACTTAATCAC-3′; primer 2, 5′-GCTCTAGAATTTATTTATCCGTAAAATG-3′. The following downstream primers amplified nt 18585 to nt 19183 and contain a 5′ HindIII site and a 3′ XbaI site, respectively: primer 1, 5′-GACAAGCTTTAACCAATTTATATTCTCGC-3′; primer 2, 5′-GCTCTAGAAAGATCATACTCTCATACAA-3′. Restriction sites are underlined, and the genomic sequence is in italics. The resulting products were digested with HindIII, EcoRI, and XbaI and cloned into plasmid pSP73 (Promega, Madison, WI), generating a plasmid containing the D14L flanking sequences linked by a central XbaI site. The resulting plasmid was sequenced to confirm the lack of mutations to the MPXV-Z genomic sequence. Next, the XbaI site between the two flanking PCR fragments was digested to allow the insertion of the enhanced green fluorescent protein (EGFP)-guanosine phosphoribosyltransferase (GPT) expression cassette from the plasmid pT7 E/L EGFP-GPT (a generous gift from G. McFadden) between the two products. pT7 E/L EGFP-GPT contains the EGFP gene under the control of a poxvirus synthetic early/late promoter and the Escherichia coli guanosine phosphoribosyltransferase (GPT) gene driven by the vaccinia virus (VV) 7,500-molecular-weight (7.5K) promoter (pT7 E/L EGFP-GPT), allowing for selection (GPT expression) and/or visual identification (EGFP expression) of recombinant virus in culture (5). BSC40 cells were transfected with the recombination plasmid, infected at a multiplicity of infection (MOI) of 0.1 with MPXV-Zaire (V79-I-005), and grown in the absence of drug selection, and cultures were collected after development of full cytopathic effect (CPE). Next, plaque assays were performed with the resulting virus culture, and an EGFP-positive plaque was identified, isolated, and passed to fresh BSC40 cells. This virus was then collected and plaque purified 4 more times until a single EGFP-positive viral isolate was obtained, and a stock of this virus was grown in BSC40 cells. The resulting recombinant virus was named D14L KO MPXV-Z, while the parental wild-type MPXV-Zaire (V79-I-005) virus is referred to throughout as WT MPXV-Z.
Viral DNA was obtained by infecting BSC40 cells with WT MPXV-Z or D14L KO MPXV-Z and purifying DNA using a standard alkaline lysis method. For PCR analysis, 100 ng of each viral DNA sample was used per reaction with primers that bind 10 nt outside the region of recombination: primer 1, 5′-ATGGCTTGTATATCAAAGAT-3′; primer 2, 5′-TATTTGAAACACGGCACTTC-3′. The resulting PCR products were sequenced to identify the correct insertion of the EGFP/GPT cassette in place of D14L in the MPXV Zaire genome. To obtain the complete genomic sequence of each virus, short read sequencing analysis was performed by the OHSU Massively Parallel Sequencing Shared Resource using purified viral DNA, and the resulting reads were assembled into a consensus sequence for each viral genome. An estimated 300-fold depth of coverage was achieved for each viral genome. Genomic sequences were deposited in GenBank. Whole-genome sequence alignments were performed with ClustalW using MacVector software, and detailed viral genome analyses were performed using the Viral Genome Organizer (VGO), Genome Annotation Transfer Utility (GATU), and Base-By-Base (BBB) tools available online from the Viral Bioinformatics Resource Center (http://athena.bioc.uvic.ca/) (4, 34, 35). Outside the region containing D14L sequence (196,169 bp total), only 183 nucleotides were identified as differing between the parental WT MPXV-Z and D14L KO MPXV-Z genomes, with 178 of these differences being attributed to the presence of ambiguous base calls that are equally dispersed throughout the consensus genomic sequences of both viruses. Although their identities are unconfirmed, for the purposes of our analyses, we considered all ambiguous bases locations of actual nucleotide variation between sequences.
For Western blot analysis, 1.5 × 105 BSC40 cells in a well of a 12-well plate were infected at an MOI of 5 with WT MPXV-Z, D14L KO MPXV-Z, or VVWR, and total cell lysates and supernatants were collected 24 h postinfection. Supernatants (0.5 ml total volume per well) were collected and cleared by centrifugation at 28,000 × g for 1 h, and the cells in each well were rinsed with phosphate-buffered saline (PBS) and lysed in 50 μl of RIPA buffer (1× PBS, 1% NP-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate). Then, 10 μl of total cell lysate or 30 μl of supernatant was run on a 10% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with rabbit antiserum which cross-reacts with both VCP and MOPICE (a generous gift from J. Atkinson, Washington University, St. Louis, MO). Membranes were then stripped and reprobed with a polyclonal anti-vaccinia virus A33R antibody (NR-628, BEI Resources), which also cross-reacts with monkeypox A35R, as a loading control for the presence of viral proteins.
For reverse transcriptase PCR (RT-PCR) analysis, BSC40 cells were infected with either WT MPXV-Z or D14L KO MPXV-Z at an MOI of 2.5, and RNA was collected at 6 and 24 h postinfection using a High Pure RNA isolation kit (Roche). RT-PCR was performed with a Superscript III RT-PCR kit (Invitrogen) using primers specific for D13L or D15L transcripts (D13L primer pair, 5′-ATGGATACTATTAAAATATT-3′ and 5′-CTATTCATAATTGATATTGA-3′; D15L primer pair, 5′-CGTTCGTGTGTTCTTGTGGT-3′ and 5′-TGCTCAAAAACAATGGCAGT-3′).
Single-step growth curve analysis was performed by infecting BSC40 cells with WT MPXV-Z or D14L KO MPXV-Z at an MOI of 2.5, collecting samples at the indicated times postinfection, and performing plaque assays to measure the total amount of PFU in each sample.
Animal inoculations and sample collection.
All aspects of the animal studies were conducted in accordance with institutional guidelines for animal care and use at the Oregon National Primate Research Center. Adult male and female rhesus macaques (5 to 9 years old) were utilized for infection studies. Cohort 1 (WT MPXV-Z) included animals 23358 (4-year-old male), 23218 (4-year-old male), 24739 (5-year-old female), and 25510 (4-year-old female); cohort 2 (D14L KO MPXV-Z) included animals 20405 (9-year-old female), 21965 (7-year-old female), 26315 (8-year-old female), and 22665 (8-year-old female). Animals were infected with 2 × 105 PFU of MPXV-Z or D14L KO MPXV-Z intrabronchially, which was delivered in a 1-ml bolus of phosphate-buffered saline (PBS). Back titers were determined from samples of all viral inocula to confirm the delivery of the correct dosage during infection. Blood and bronchoalveolar lavage (BAL) fluid samples were obtained 4 and 2 weeks prior to infection, on the day of infection (day 0), and then on a biweekly basis thereafter. Skin punch biopsy specimens of the pock lesions were collected on day 7 postinfection. Disease severity was determined based on several physiological parameters, including body temperature and activity collected via telemetry implants (Mini Mitter, Bend, OR), body weight, and number of lesions. Complete blood counts were obtained at every time point to monitor changes in immune cell numbers/μl of blood. Peripheral blood mononuclear cells (PBMC) were isolated from whole blood (WB) by centrifugation over Histopaque gradient (Histopaque, Sigma-Aldrich, St. Louis, MO). Total cells were isolated from BAL fluid samples by centrifugation followed by resuspension in RPMI supplemented with 10% fetal bovine serum (FBS), glutamine, and Pen/Strep.
Real-time PCR analysis of viral load.
MPXV viral loads were measured by real-time PCR using primers and probes specific for MPXV-Zaire genomic sequence corresponding to open reading frame (ORF) F3L (interferon [IFN] resistance gene). DNA was purified using a Puregene DNA purification kit (Gentra Systems, Inc., Minneapolis, MN), and 100 ng of DNA was analyzed by real-time PCR on an ABI Prism 7700 DNA sequence detector (Applied Biosystems, Foster City, CA).
Measuring MPXV-specific T-cell responses.
PBMC and BAL fluid cells were stained with surface antibodies CD4, CD8β, CD28, and CD95 to delineate naïve, central memory (CM), and effector memory (EM) T-cell subsets. All antibodies, with the exception of CD8b (Beckman Coulter), were purchased from Biolegend (San Diego, CA). The cells were then fixed and the nuclear membrane permeabilized as per the manufacturer's recommendation (BD Pharmingen, San Diego, CA) before staining with anti-Ki67 (BD Pharmingen, San Diego, CA). Samples were acquired using the LSRII instrument (BD Bioscience, San Jose, CA), and data were analyzed by FlowJo (TreeStar, Ashland, OR). Intracellular cytokine staining (ICCS) was carried out as previously described (11). Briefly, PBMC and BAL fluid cells were stimulated with simian varicella virus (negative control), WT MPXV-Z (MOI, 1), or anti-CD3 (positive control) overnight. Brefeldin A (Bfa) was then added for an additional 6 h. PBMC were first stained with surface antibodies CD4, CD8β, CD28, and CD95 (Biolegend, San Diego, CA) and then permeabilized and stained with antibodies directed against IFN-γ and tumor necrosis factor alpha (TNF-α) (Biolegend, San Diego, CA).
Measuring MPXV-specific B-cell response.
PBMC and BAL fluid cells were stained with surface antibodies directed against CD20 (Beckman Coulter), IgD (Southern Biotech), and CD27 (Biolegend, San Diego, CA) to delineate naive, marginal zone-like, and memory B cells. The cells were then fixed and the nuclear membrane permeabilized as per the manufacturer's recommendation before the addition of Ki67 (BD Pharmingen, San Diego, CA) specific antibodies. Samples were acquired using the LSRII instrument (BD Bioscience, San Jose, CA), and data were analyzed by FlowJo (TreeStar, Ashland, OR).
Antiviral IgG levels were measured in circulating plasma using a standard enzyme-linked immunosorbent assay (ELISA) using plates coated with varicella virus VV-WR viral lysate, which reacts with MPXV antibodies (Abs). In these experiments, serial 3-fold dilutions of plasma were incubated in triplicate VV-WR virus lysate-coated ELISA plates for 1 h prior to washing, staining with detection reagents (horseradish peroxidase [HRP]-anti-IgG), and addition of chromogen substrate to allow for detection and quantitation of bound antibody molecules. Log-log transformation of the linear portion of the curve was then performed, and 0.1 optical density (OD) units was used as the cutoff point to calculate endpoint titers. Each plate always contained a positive-control sample that was used to normalize the ELISA titers between assays and a negative-control sample to ensure the specificity of the assay conditions.
Nucleotide sequence accession numbers.
Genomic sequences determined in this study were deposited in GenBank and are available under accession numbers HQ857562 (WT MPXV-Z) and HQ857563 (D14L KO MPXV-Z).
RESULTS
Generation of a MOPICE KO MPXV-Z.
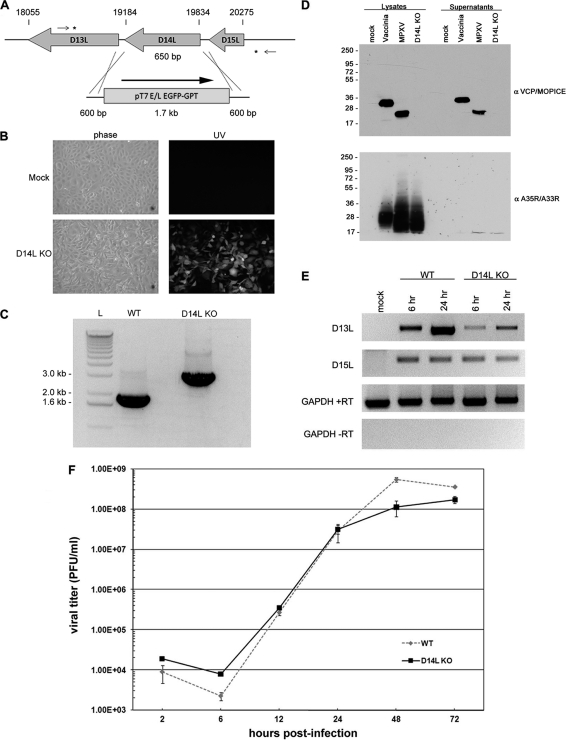
The MPXV-Z D14L ORF encodes a 216-amino-acid (aa) secreted protein termed MOPICE, which possesses significant homology to other poxviral inhibitors of complement enzymes (PICEs) and has been shown to possess complement regulatory activity in vitro (22). To investigate the role of MOPICE in MPXV pathogenesis and its impact on the development of the immune response to MPXV infection, we generated a recombinant MPXV-Z knockout in which the D14L gene is completely absent from the viral genome. To achieve this, D14L was removed from the MPXV-Z genome and replaced with an EGFP-GPT cassette via homologous recombination (Fig. 1 A), and an EGFP-expressing plaque isolate was identified and purified by standard plaque assay to obtain a purified isolate lacking D14L (D14L KO MPXV-Z) (Fig. 1B). The correct deletion of D14L in this virus, with no other rearrangements to the surrounding genomic sequence, was confirmed by PCR of viral genomic DNA and sequence analysis of the resulting product (Fig. 1C and data not shown).
Fig. 1.
Construction of D14L KO MPXV-Z. (A) Overview of recombination approach utilized to remove D14L from the MPXV-Z genome. Asterisks indicate the location of recombination within the genomic DNA, small arrows indicate location of primers used in PCR analysis of recombinant virus DNA, and nucleotide numbers of D14L and neighboring ORFs are noted by hash marks. The orientation of the EGFP-GPT cassette is noted by an arrow. (B) EGFP expression in BSC40 cells infected with D14L KO MPXV-Z isolate. (C) PCR analysis of genomic DNA from WT MPXV-Z and D14L KO MPXV-Z demonstrates the complete removal of D14L sequence (650 bp) and the insertion of an EGFP-GPT cassette (∼1.7 kb). (D) Anti-VCP/MOPICE Western blot analysis of lysates and supernatants from vaccinia virus (Western Reserve)-, WT MPXV-Z-, and D14L KO MPXV-Z-infected BSC40 cells, demonstrating production of MOPICE in WT MPXV-Z-infected cells and lack of MOPICE expression in D14L KO MPXV-Z-infected cells. Membranes were probed with anti-VCP antibody and then stripped and reprobed with antibody cross-reactive with vaccinia virus A35R and MPXV-Z A33R as a control for virus infection. (E) RT-PCR analysis of D13L and D15L transcript levels in WT or D14L KO MPXV-Z-infected BSC40 cells, indicating that deletion of D14L and insertion of the EGFP/GPT-expressing cassette in the MPXV-Z genome do not prevent the expression of neighboring genes. Cells were infected at an MOI of 2.5, and total RNA was collected at 6 and 24 h postinfection for analysis using primers specific for D13L or D15L transcripts. RT-PCR for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) serves as a control for equal RNA levels (+RT) and lack of DNA contamination (−RT) in each sample. (F) Single-step growth curve analysis (MOI, 2.5) of WT and D14L KO MPXV-Z in BSC40 cells.
Due to the large size of poxvirus genomes, the potential for the introduction of undesired random genetic changes and point mutations during the selection, growth, and passage of a virus is essentially unavoidable and thus will always be a factor that should be considered when generating a recombinant poxvirus. To address this issue directly, and to confirm that no major genetic changes or genomic alterations occurred during the process of generating D14L KO MPXV-Z, whole-genome sequence analysis was performed using viral DNA purified from both parental WT MPXV-Z and D14L KO MPXV-Z, allowing for the direct comparison of the nucleotide sequences of both viral genomes. Alignment of the consensus genomic sequences generated from this analysis indicates that the viruses are ≥99.91% identical at the nucleotide level across their entire genomic length, when excluding the region containing D14L sequence (data not shown; sequences deposited in GenBank under accession numbers HQ857562 [WT MPXV-Z] and HQ857563 [D14L KO MPXV-Z]). In addition, analysis of the resulting sequences using viral genome analysis tools (4, 34, 35) further indicates that outside D14L, the genomic organization of both viruses is identical and that no genetic alterations predicted to significantly alter genome structure or viral gene expression are present in the recombinant D14L KO MPXV-Z. A description of the potential single-nucleotide polymorphisms identified in D14L KO MPXV-Z relative to predicted coding regions was generated from this analysis (see Table S1 in the supplemental material). As was anticipated, there is a low level of nucleotide variation between WT and D14L KO MPXV-Z, although overall, the results obtained from genomic sequence analyses indicate that the D14L KO virus is highly similar at the genetic level to the parental WT MPXV-Z, aside from the desired deletion of the D14L gene and insertion of the EGFP/GPT cassette.
To confirm the lack of MOPICE expression by the D14L KO virus, lysates and supernatants from WT MPXV-Z- or D14L KO MPXV-Z-infected BSC40 cells were analyzed by Western blotting using an antibody specific for VCP, which also cross-reacts with MOPICE (22). An approximately 23-kDa band corresponding to MOPICE (216 aa) was detected in MPXV-Z-infected cell lysates and supernatants but not in cells infected with D14L KO MPXV-Z (Fig. 1D), demonstrating lack of MOPICE production by the D14L KO virus.
Next, to determine whether the deletion of D14L and insertion of an EGFP/GPT expressing cassette in the MPXV-Z genome had any effects on transcription of the neighboring genes D13L (putative IL-1 receptor antagonist) and D15L (Kelch-like protein), RT-PCR analysis was performed using primers specific for transcripts for each gene and RNA isolated from WT or D14L KO MPXV-Z-infected BSC40 cells at 6 and 24 h postinfection (Fig. 1E). Compared to levels in WT MPXV-Z infection, transcript levels of D15L are unchanged during the course of D14L KO MPXV-Z infection, while the levels of D13L transcript appear to be somewhat lower in cells infected with D14L KO MPXV-Z. Although decreased levels of D13L transcript do not necessarily indicate lower levels of protein expression from this gene during infection with D14L KO MPXV-Z in vivo, it is possible that some variation could exist compared to levels in WT MPXV-Z. However, since the actual function of MPXV-Z D13L has not been examined, the effects that decreased transcript production from this gene might have during in vivo infection are unclear.
Finally, the effects of D14L deletion and EGFP/GPT cassette insertion on the in vitro growth characteristics of MPXV-Z were also analyzed by performing a single-step growth curve in BSC40 cells (Fig. 1F). These results indicate that D14L KO MPXV-Z and WT MPXV-Z replicate with identical kinetics and that deletion of D14L from the MPXV-Z genome does not alter the growth properties of MPXV-Z in vitro.
Intrabronchial infection of rhesus macaques with MPXV-Z and D14L KO MPXV-Z.
We utilized an intrabronchial (i.b.) model of MPXV infection of RM to represent an aerosol route of infection while also delivering a defined dose of virus for our comparative infection studies. Thus, two cohorts consisting of four animals each were inoculated with 2 × 105 PFU of WT MPXV-Z (cohort 1: animals 23358, 23218, 24739, and 25510) or D14L KO MPXV-Z (cohort 2: animals 20405, 21965, 26315, and 22665). This dose was determined based on a prior trial study in which animals received 107 or 106 PFU of virus, both of which were determined to be 100% lethal in all animals at 10 days postinfection (dpi) (data not shown). Animals were observed twice daily for signs of disease. Blood and BAL fluid samples were collected at defined dpi to determine the kinetics of virus replication and the kinetics and magnitude of the adaptive immune response. MPXV-Z inoculation resulted in the appearance of numerous widespread pox lesions, which appeared more concentrated on the flanks (Fig. 2 A). These lesions first developed as 2- to 4-cm-wide pustules on skin and oral mucosa at 7 to 12 dpi and then progressed from pustular to crusting stages at 12 to 14 dpi, finally becoming scabbed at 18 to 21 dpi. Lesions were healed and completely resolved by 28 dpi in all animals (Fig. 2A). Animals also developed coughs and symptoms of labored breathing between days 7 and 14. The temperature of each animal was recorded throughout the study using telemetry implants, and the results indicated that all infected animals developed a fever after infection, with the highest temperature being detected from 7 to 14 dpi and remaining elevated until week 3 or 4 pi (Table 1). Animals were euthanized at 35 or 49 dpi and, at time of necropsy, did not harbor any overt evidence of MPXV-associated disease.
Fig. 2.
Development of pox lesions in WT and D14L KO MPXV-Z-infected rhesus macaques. Images of skin lesions demonstrating the development of pox lesions on skin (flank and trunk) of infected RM over the course of infection. (A) WT MPXV-Z-infected animal 24739, and (B) D14L KO MPXV-Z-infected animal 22665. (C) Pox lesions from D14L KO MPXV-infected animals 21965 and 20405, demonstrating EGFP expression within pox lesions due to the presence of virus. Right panels depict the same region shown in left panels exposed to UV light (+UV).
Table 1.
Temperatures of study animals
| Cohort | Animal no. | Sex/age/wta | Temp (°C)b |
||
|---|---|---|---|---|---|
| Baseline (day 0) | Peakc | Resolvedd | |||
| WT MPXV | 23218 | M/4/5.8 | 35.5 | 39.8 (day 11) | 36.2 (day 27) |
| 23358 | M/4/6.4 | 34.7 | 38.8 (day 9) | 34.4 (day 24) | |
| 25510 | F/4/3.5 | 37.1 | 40.4 (day 7) | 35.8 (day 22) | |
| 24739 | F/5/4.7 | 36.0 | 40.8 (day 14) | 36.0 (day 28) | |
| D14L KO | 20405 | F/9/7.7 | 35.6 | 39.0 (day 10) | ND |
| 21965 | F/7/6.8 | 35.6 | 39.6 (day 17) | ND | |
| 22665 | F/8/6.4 | 36.6 | 39.8 (day 8) | 36.5 (day 30) | |
| 26315 | F/8/4.3 | 35.7 | 39.9 (day 7) | ND | |
M, male; F, female; age in years; weight in kilograms.
Recorded telemetry temperature at 1 a.m. on indicated day postinfection.
Highest temperature reading recorded over course of infection.
Recorded temperature closest to or below baseline value that occurred after the date of peak temperature. ND, not determined. No temperature readings were available after day 17 for animals 20405 or 21965; animal 26315 died on day 17 postinfection.
In general, the disease manifestations associated with D14L KO MPXV-Z infection were similar to those observed with WT MPXV. However, unlike infection with WT MPXV-Z, in which all animals survived the infection, one D14L KO MPXV-Z-infected animal (26315) succumbed to MPXV-related disease complications at 17 dpi, while animals 20405 and 21965 experienced such low oxygen saturation levels at 14 to 28 dpi that we could not collect BAL fluid samples. Moreover, although the lesions in the remaining three D14L KO MPXV-Z-infected animals exhibited kinetics similar to those of WT MPXV-Z-infected animals (7 to 12 dpi) (Fig. 2B), two animals (20405 and 21965) experienced a delay in lesion scabbing, which did not occur until 21 to 25 dpi. The lesions in these two animals developed a more flattened and moist appearance than lesions seen in WT MPXV-Z-infected animals (Fig. 2C). Due to insertion of the EGFP-expressing cassette in place of the D14L ORF in the KO virus, all lesions in D14L KO MPXV-Z-infected animals were EGFP positive when exposed to UV light, demonstrating the presence of the D14L KO virus in the lesions. With the exception of animal 26315, no animals harbored any evidence of MPXV-associated disease at the time of necropsy, except for fibrosis in the lungs, similar to findings in the WT MPXV-Z-infected animals.
Viral loads in whole blood (WB) or peripheral blood mononuclear cells (PBMC) and in cells isolated from bronchial alveolar lavage (BAL) fluid were determined by real-time PCR using primers and probes specific for the MPXV-Z F3L gene. In general, elevated viral loads paralleled the appearance of pox lesions, as well as the development of fever and other clinical symptoms, demonstrating a direct correlation between increased viral load in BAL fluid and blood and severity of disease symptoms. In WB (animals 24739 and 25510) or PBMC (animals 23358 and 23218) of WT MPXV-Z-infected animals, viral DNA was first detected at 4 (23218), 7 (25510), 10 (23358), and 14 (24739) dpi (Fig. 3 A). Despite the variation in onset, viremia lasted approximately 10 days and declined to undetectable levels by 21 dpi in all animals (Fig. 3A). Viral loads in BAL fluid were examined only in animals 24739 and 25510 due to the paucity of cells recovered from animals 23218 and 23358. MPXV viral DNA was first detected in BAL fluid cells at 7 to 10 dpi, remained elevated with some fluctuations, and eventually returned to undetectable levels by 42 dpi, with no viral DNA being detected by time of necropsy at day 49 (Fig. 3C). Peak viral loads in BAL fluid samples of animals 24739 and 25510 were higher than those in WB, which may partly reflect the route of infection directly into the lungs and viral replication at the site of inoculation.
Fig. 3.
Viral loads in WT and D14L KO MPXV-Z-infected rhesus macaques. Quantitative real-time PCR analysis was performed using samples collected at the indicated days postinfection (dpi) to determine viral genome copy numbers in whole blood (WB) or peripheral blood mononuclear cells (PBMC) (A and B) and in bronchial alveolar lavage (BAL) fluid cells (C and D). Filled symbols represent WT MPXV-Z-infected animals (A and C), and open symbols depict D14L KO MPXV-Z-infected animals (B and D). DNA from WB was analyzed for all animals except 23358 and 23218, in which cases DNA was obtained from PBMC purified from WB samples. BAL fluid DNA samples for real-time PCR analysis were available only for WT MPXV-Z-infected animals 24739 and 25510. D14L KO MPXV-Z-infected animal 26315 succumbed to disease 17 dpi. BAL fluid samples from D14L KO MPXV-Z-infected animals 21965 and 20405 were not collected 17 and 21 dpi due to low oxygen saturation levels. The highest viral load was detected 7 dpi in BAL fluid of animal 26315 (1.2 × 108), which succumbed to disease on day 17 pi.
Examination of viral loads in animals infected with D14L KO MPXV-Z revealed a pattern similar to that seen during WT MPXV-Z infection, with higher viral loads being detected in BAL fluid than in WB. However, the levels of peak viral loads in both WB and BAL fluid samples of D14L KO MPXV-Z-infected animals were higher than those detected in animals infected with WT MPXV-Z (Fig. 3B and D). Interestingly, animal 26315, which succumbed to disease on day 17 after D14L KO MPXV-Z infection, displayed the highest viral load overall, with 1.21 × 108 genome copies/100 ng DNA being detected in BAL fluid at 7 dpi, a level 100-fold higher than any peak viral load detected in WT MPXV-Z-infected animals (Fig. 3D). BAL fluid samples were not obtained between 14 and 28 dpi from animal 21965 due to very low oxygen saturation levels. Taken together, the differences in viral loads between WT and D14L KO MPXV-Z in both WB/PBMC and BAL fluid suggest that the lack of MOPICE expression allows MPXV-Z to replicate to higher levels in vivo.
B-cell and antibody responses to MPXV infection.
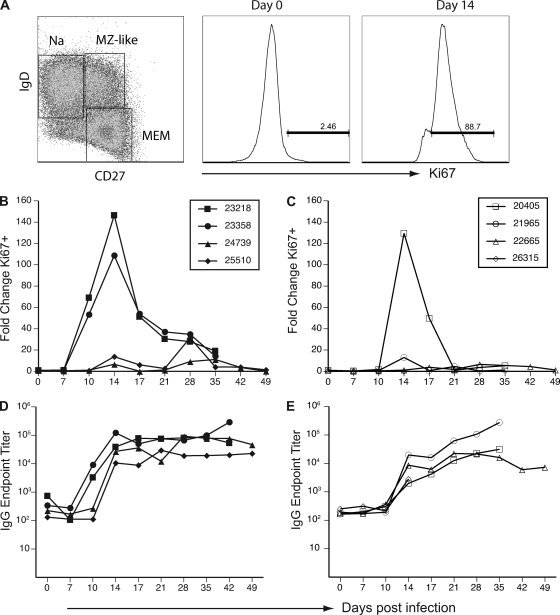
To analyze the contributions of MOPICE to the host immune response, we compared the kinetics and magnitude of the B-cell proliferative burst in peripheral blood of animals infected with WT or D14L KO MPXV-Z. B-cell responses in the BAL fluid were not analyzed due to the extremely low numbers of B cells detected (>2% of total lymphocytes; data not shown). B cells can be broadly divided into three subsets based on the expression of IgD and CD27: naïve (IgD+ CD27−), marginal-zone like (IgD+ CD27+), and isotype-switched memory (IgD− CD27+) B cells, as shown in Fig. 4 A (left panel). The IgD− CD27+ subset harbors the majority of the isotype-switched, antibody-secreting memory B cells. When naïve B cells encounter antigen, they undergo a proliferative burst while simultaneously acquiring memory markers. Therefore, to measure the kinetics and magnitude of the B-cell proliferative burst in response to MPXV-Z antigen encounter, PBMC were stained with antibodies directed against surface markers CD20, CD27, and IgD to identify B-cell subsets and were simultaneously stained for expression of Ki67, a nuclear protein that indicates recent entry into cell cycle (27). Figure 4A illustrates an example of subset identification (left panel), and Ki67 staining on day 0 (middle panel) and 14 dpi (right panel), using PBMC isolated from animal 23218.
Fig. 4.
D14L KO MPXV-Z infection results in diminished B-cell response in rhesus macaques. (A) PBMC from infected animals were analyzed by flow cytometry (FCM) to identify three B-cell subsets: naïve (CD27− IgD+), marginal zone-like (CD27+ IgD+), and memory (CD27+ IgD−). A representative example from animal 23218 obtained on day 0 is shown in the left panel. B-cell proliferation within the memory cell population was measured by staining for the nuclear protein Ki67, which is upregulated at 14 dpi (right panel) relative to 0 dpi (middle panel). Percentage of Ki67+ memory B cells was converted at every time point to absolute number of Ki67+ memory B cells based on complete blood counts (CBC), and fold increase over baseline (average of three preinfection time points) was calculated. (B) B-cell proliferation in response to WT MPXV-Z infection peaked at 14 dpi. (C) A similar pattern of B-cell proliferation was observed in D14L KO MPXV-Z-infected animals, although only two animals displayed a measurable proliferative burst, which also peaked at 14 dpi. Orthopoxvirus-specific IgG responses in WT (D) and D14L KO MPXV-Z-infected (E) animals were analyzed by endpoint ELISA and were first detectable by 14 dpi in all animals. IgG endpoint titers in D14L KO MPXV-Z-infected animals were on average a log lower than those achieved with WT MPXV-Z infection at 17 dpi.
WT MPXV-Z infection resulted in an increase in the frequency of proliferating memory B cells in PBMC that peaked at 14 dpi in all animals (Fig. 4B) and eventually returned to baseline levels at 35 dpi in all animals (Fig. 4B). Interestingly, animals 23218 and 23358 experienced an earlier and greater level of B-cell proliferation than animals 24739 and 25510 (approximately 100- to 150-fold increases versus 10-fold). The earlier proliferation appears to directly correlate with earlier detection of viral DNA in PBMC of these animals (Fig. 3A). In contrast, animals infected with D14L KO MPXV-Z exhibited an overall reduced level of B-cell proliferation compared to that of WT MPXV-Z-infected animals. Specifically, two of the four animals infected with D14L KO MPXV-Z (22665 and 26315) exhibited essentially no B-cell response, whereas animals 21965 and 20405 displayed transient 13-fold and 130-fold increases, respectively, in proliferating B cells 14 dpi (Fig. 4C). Although the magnitude of the proliferative burst correlated with viral loads within each group, the overall proliferation observed in each group as a whole negatively correlated with peak viral loads. Specifically, D14L KO MPXV-Z-infected animals exhibited higher peak viral loads and lower overall proliferative burst than WT MPXV-Z-infected animals. These observations suggest that lower levels of B-cell proliferation are associated with increased viral loads in D14L KO MPXV-Z-infected animals.
We also measured the IgG antibody response against MPXV-Z using an endpoint titer ELISA. All WT MPXV-Z-infected animals developed an orthopox-specific IgG response that peaked by 14 dpi and remained stable for the duration of the study (Fig. 4D). Interestingly, MPXV antibodies were detected earlier in animals 23218 and 23358 (10 dpi) than in animals 24739 and 25510 (14 dpi), which correlates closely with the observed differences in kinetics of memory B-cell proliferation between these animals. Although all D14L KO MPXV-Z-infected animals developed an IgG response with kinetics similar to those for WT MPXV-Z-infected animals, the IgG titers were approximately one log lower than those observed in WT MPXV-Z-infected animals at 14 dpi. The IgG titers continued to increase in these animals, ultimately reaching levels that were similar to those seen in WT MPXV-Z-infected animals at 35 dpi. Taken together, these data suggest that in the absence of MOPICE, B-cell and antibody responses directed against the virus are dampened. Further, the observed delay in IgG response may contribute to increased viral loads seen in D14L KO MPXV-Z-infected animals. It is unclear at this point whether MOPICE directly modulates B-cell activation or whether it is a target of the B-cell response during MPXV-Z infection.
T-cell responses to MPXV infection.
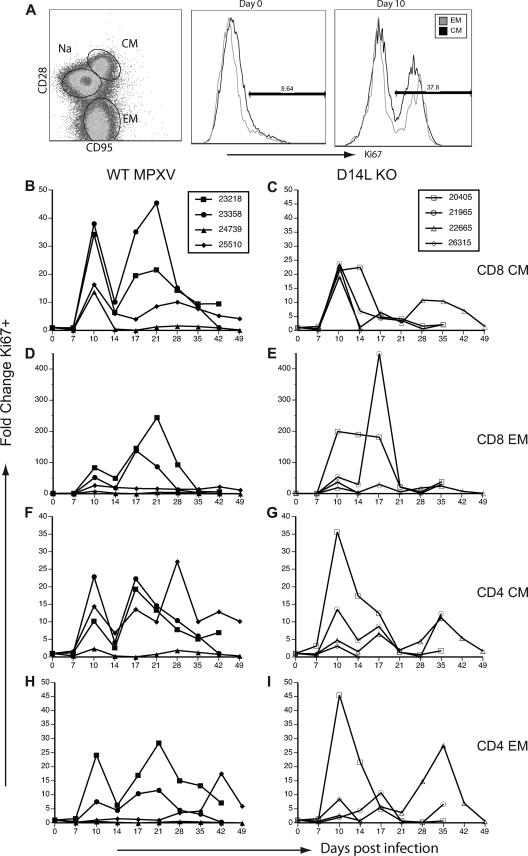
T cells play a critical role in controlling and eliminating viral infections; thus, we analyzed the T-cell response in RM infected with WT and D14L KO MPXV-Z. T cells can also be subdivided into three major subsets based on the expression of CD28 and CD95: naïve (CD28+ CD95−), central memory (CM) (CD28+ CD95+), and effector memory (EM) (CD28− CD95+) (Fig. 5 A). Similarly to B cells, T cells undergo a robust proliferative burst and acquire memory markers following pathogen encounter that can be measured by determining changes in Ki67 expression within EM and CM T-cell subsets (25). To measure the kinetics and magnitude of the proliferative T-cell response after MPXV-Z infection, we measured changes in the frequency of Ki67+ CM and EM CD4 and CD8 T cells in both PBMC and BAL fluid samples. In PBMC from WT MPXV-Z-infected animals, the frequency of Ki67+ CM and EM T cells rapidly increased at 10 dpi before dramatically declining at 14 dpi (Fig. 5B, D, F, and H). Following this initial decline, a second burst of proliferation was detected at 17 dpi in most subsets, which was more sustained and remained elevated until 35 dpi. As described for the B-cell response, animals 23218 and 23358 displayed a more robust T-cell proliferative response than animals 24739 and 25510.
Fig. 5.
D14L KO MPXV-Z-infected rhesus macaques show decreased T-cell proliferation in peripheral blood compared to WT MPXV-Z-infected animals. PBMC were analyzed by FCM in to delineate CD4 and CD8 T-cell subsets: naïve (CD28+ CD95−), central memory (CM) (CD28+ CD95+), and effector memory (EM) (CD28− CD95+). T-cell proliferation was measured by staining for nuclear protein Ki67. (A) A representative example of CD8 T-cell subsets in PBMC from animal 23218 at 0 dpi is shown in the left panel. An example of Ki67 expression within CM and EM population at 0 (middle panel) and 10 dpi (right panel) is shown. Percentage of Ki67+ T cells was converted to numbers of proliferating cells based on CBC values, and fold increase over baseline (average of three preinfection time points) was then calculated. (B, D, F, and H) An initial proliferative burst was detected at 10 dpi in CM and EM CD4 and CD8 subsets in WT MPXV-Z-infected animals, followed by a dramatic decrease in frequency of proliferating cells detected at 14 dpi and a secondary proliferative burst at 17 dpi in most animals. Animals 23218 and 23358 displayed a greater proliferative burst than animals 24739 and 25510. (C, E, G, and I) An increase in Ki67+ T cells was detected at 10 dpi in D14L KO MPXV-Z-infected animals, followed by a gradual return to baseline proliferation levels at 21 dpi, with no consistent secondary proliferative burst occurring. Overall, D14L KO MPXV-Z-infected animals experienced a proliferative burst of lower magnitude than that of WT MPXV-Z-infected animals.
A different pattern was observed for animals infected with D14L KO MPXV-Z. In this case, infection resulted in a T-cell proliferative burst that was first detected at 10 dpi; however, the biphasic proliferative pattern observed in WT MPXV-Z-infected animals was not as evident (Fig. 5C, E, G, and I). All T-cell subsets from animals 20450 and 21965 displayed a single proliferative burst that was sustained from 10 to 21 dpi. T cells from animal 22665 underwent two small proliferative bursts at 10 and 17 dpi followed by a third larger burst that peaked at 35 dpi. Overall, there was a trend toward a more sustained proliferative burst in the WT-infected animals than in the D14LKO-infected animals.
We also characterized the T-cell responses in the BAL fluid of animals, as the BAL fluid is populated only by memory T cells (Fig. 6A). In WT MPXV-Z-infected animals, all T-cell subsets, with the exception of CD4 EM, displayed a biphasic response similar to that seen in PBMC, with a rapid increase in Ki67+ cells occurring 10 dpi in all animals, followed by a rapid decline at 14 dpi, a subsequent increase at day 17 pi, and a gradual return to baseline levels by 35 dpi (Fig. 6B, D, and F). Interestingly, although CD4 EM T cells in most animals displayed a rapid increase in Ki67+ cells at 10 dpi (17 dpi in animal 24739), this cell type exhibited a series of fluctuations in the frequency of proliferating cells followed by a gradual decline to baseline levels by 42 dpi (Fig. 6H). The analysis of T-cell proliferation in the BAL fluid of D14L KO MPXV-Z-infected animals was more complicated, since BAL fluid samples could not be collected from animals 20405 and 21965 at 17, 21, and 35 dpi, due to low oxygen saturation levels (indicative of more severe infection in the lungs). Further, animal 26315 was euthanized at 17 dpi due to complications associated with MPXV infection. Thus, a complete time course of T-cell responses in BAL fluid is available only for animal 22665. T-cell proliferation was first detected at 10 dpi in all subsets, with the exception of the CD4 EM subset, where an increased frequency of Ki67+ cells was detected at 7 dpi. An increased frequency of proliferating CD8 CM T cells was detected only in samples from animal 22665, whereas robust proliferation of CD8 EM T cells that was sustained until 28 dpi was detected in all four D14L KO MPXV-Z-infected animals. T-cell proliferation within the CD4 CM subset was also detected to various degrees in all four animals at 10 dpi. Proliferation of CD4 CM T cells returned to baseline levels in all animals at 28 dpi, with the exception of T cells from animal 22665, which continued to proliferate until 42 dpi. CD4 EM T cells from 20405 and 21965 showed an early proliferative burst that peaked at 10 dpi followed by a return to the baseline level at 28 dpi. On the other hand, CD4 EM T cells from animal 22665 displayed proliferation at 17 dpi returning to baseline levels 48 dpi. Overall, there was a trend toward lower levels of T-cell proliferation in BAL fluid in D14L KO MPXV-Z-infected animals compared to those of WT MPXV-Z-infected animals.
Fig. 6.
T-cell proliferation is lower in BAL fluid of D14L KO MPXV-Z-infected rhesus macaques compared to WT MPXV-Z-infected animals. In contrast to PBMC, BAL fluid contains only CM and EM T cells. (A) A representative example of CD8 T-cell subsets and frequency of Ki67 cells at 0 (middle panel) and 10 (right panel) dpi from animals 23218 is shown. (B, D, F, and H) In BAL fluid of WT MPXV-Z-infected animals, proliferation in all subsets of T cells followed a pattern similar to that described for PBMC, with an initial proliferative burst first being detected at 10 dpi, a dramatic decrease in frequency of proliferating T cells occurring at 14 dpi, followed by the onset of a second proliferative burst at 17 dpi. The frequency of proliferating T cells returned to baseline at approximately 35 dpi in all animals. (C, E, G, and I) The patterns of proliferation in T-cell subsets from BAL fluid of D14L KO MPXV-Z-infected animals displayed a more sporadic profile, but proliferation was first detected 10 dpi in all subsets and most animals. Data from later time points were not available for animal 26315, which succumbed to disease 17 dpi, or for animals 20405 and 21965 at 17 and 21 dpi.
To better characterize the host immune response to MPXV-Z infection and to define how MOPICE impacts the adaptive T-cell response, we enumerated the frequency of MPXV-specific CD4 and CD8 T cells in PBMC and BAL fluid of all MPXV-Z-infected animals using intracellular cytokine staining (ICS), as has been described previously (11). PBMC and BAL fluid cells isolated at different dpi were stimulated with MPXV-Z, simian varicella virus (SVV) (a negative control), or anti-CD3 (positive control), and the frequency of TNF-α- and IFN-γ-producing T cells was determined using flow cytometry (FCM) as described in Materials and Methods. As depicted in a representative example of CD8 T-cell responses from animal 24739 at 21 dpi (Fig. 7A), CD8 and CD4 T cells from WT and D14L KO MPXV-Z-infected animals secrete TNF-α and IFN-γ only in response to MPXV, and not SVV, indicative of the development of an orthopoxvirus-specific T-cell response in infected animals. The panels in Fig. 7 show the averages of the responses generated by PBMC CD8 CM and EM T cells in each cohort.
Fig. 7.
Frequency of MPXV-specific T cells in peripheral blood is higher in WT MPXV-Z-infected animals. MPXV-specific CD4 and CD8 T cells were detected by intracellular cytokine staining (ICCS). PBMC from WT and D14L KO MPXV-Z-infected animals were stimulated with purified MPXV-Z virus (MOI, 1) or simian varicella virus (SVV) (negative control) or anti-CD3 (positive control) as described in Materials and Methods. MPXV-specific T cells were identified based on the production of TNF-α and IFN-γ. (A) Representative example of CD8 CM and EM responses from animal 24739 at 17 dpi, indicating that specific T-cell responses are detected only in response to MPXV-Z stimulation. The average percentage of responding (TNF-α+ IFN-γ+ double-positive or IFN-γ+ only) cells in each subset for all four animals is shown for WT MPXV-Z (B, D, F, and H) and D14L KO MPXV-Z (C, E, G, and I). The T-cell response in all subsets of WT MPXV-Z-infected animals peaked by 21 dpi, while the responses in D14L KO MPXV-Z-infected animals were much lower in most subsets.
Orthopox-specific CD8 CM T cells were detected with low frequency at 7 dpi in WT MPXV-Z-infected animals, before reaching a peak value at 21 dpi, after which the frequency of responding T cells declined but remained detectable. Interestingly, 21 dpi correlates with the onset of the second proliferative peak in T cells that occurred following WT MPXV-Z infection. The frequency of MPXV-specific CD8 EM T cells also peaked 21 dpi and reached higher levels than CD8 CM T cells. Similar to previous observations made in PBMC from humans vaccinated with vaccinia virus (1), the frequency of MPXV-specific CD4 T cells was significantly lower than that of MPXV-specific CD8 T cells in PBMC WT MPXV-Z-infected animals (Fig. 7F and H). MPXV-specific CD4 EM T cells were detected at 7 dpi (Fig. 7H), whereas CD4 CM T cells were detected at 14 dpi (Fig. 7F). However, the peak of the CD4 response in PBMC still occurred at 21 dpi as described for CD8 T cells. Interestingly, compared to those in WT MPXV-Z-infected animals, the frequency of MPXV-specific CD8 and CD4 T cells was lower in the D14L KO-infected animals (Fig. 7C, E, G, and I). This was especially evident for the CD8 and CD4 CM (Fig. 7C and I) subsets, in which very few MPXV-specific T cells were detected in D14L KO animals. This decreased frequency of responding T cells could contribute to the increased viral loads and lesion severity observed in D14L KO MPXV-Z-infected animals.
We also measured the frequency of MPXV-specific T cells in BAL fluid (Fig. 8). However, we were able to study samples from only two WT MPXV-Z-infected animals (24739 and 25510) and one D14L KO MPXV-Z-infected animal (22665), due to a lack of cells recovered (animals 22318 and 22358) or an inability to obtain BAL fluid samples due to low oxygen saturation levels (animals 21965 and 20405). In general, however, the frequency of MPXV-specific T cells in BAL fluid was similar to that observed in PBMC, with the exception of an apparent delayed but robust increase in the frequency of responding T cells at approximately 49 dpi.
Fig. 8.
MPXV-Z-specific T cells in BAL fluid of WT and D14L KO MPXV-Z-infected animals. ICCS was performed as described for Fig. 7 to measure frequency of MPXV-specific T cells in BAL fluid of WT and D14L KO MPXV-Z-infected animals. (A, C, E, and G) Average percentage of responding (TNF-α+ IFN-γ+ double-positive or IFN-γ+ only) T-cell subsets in BAL fluid of WT MPXV-Z-infected animals 24739 and 22510. (B, D, F, and H) Frequency of responding T cells from D14L KO MPXV-Z-infected animal 22665.
DISCUSSION
Orthopoxviruses encode numerous proteins capable of modulating the host immune response. Among these proteins are the poxviral inhibitors of complement enzymes (PICEs). These secreted viral proteins share structural and functional homology with mammalian regulators of complement activation, proteins which protect host cells from damage due to uncontrolled complement activation (31). Activation of the complement cascade in an infected host is generally thought to contribute to resolution of viral infection due to mechanisms such as increased viral opsonization, lysis of infected cells, and production of the proinflammatory anaphylatoxins C5a and C3a (20). Moreover, complement activation has been found to enhance both B- and T-cell responses to some pathogens (6). The vaccinia complement control protein (VCP) has been shown to protect both virus-infected cells and virions from host complement attack, and it appears to be important for the virulence of vaccinia virus (15). Further, VCP has recently been shown to limit the adaptive immune response against vaccinia virus in an in vivo model of intradermal infection (10). In the case of cowpox, in vivo studies indicate that the cowpox inflammation modulatory protein is important for preventing tissue damage and swelling associated with virus infection (26). Therefore, PICE proteins may limit the extent of host complement activation, not only as a mechanism to promote increased viral persistence and limiting clearance of the virus but also to prevent extensive injury to host tissue and cells, thus maintaining an optimal environment for viral replication in an infected host. Based on this information, it was hypothesized that the monkeypox complement regulator, MOPICE, is one of the main virulence factors of central African strains of MPXV, since it is completely absent in west African strains of MPXV (7, 21).
To determine how MOPICE modulates MPXV pathogenesis and the adaptive immune response, we first generated a D14L knockout (D14L KO) MPXV in which the D14L open reading frame encoding MOPICE was deleted from the MPXV-Zaire genome and replaced with an EGFP-expressing cassette. Importantly, complete genomic sequence analysis of the resulting virus indicates that outside the region containing D14L sequence, D14L KO MPXV-Z is highly similar to the parental WT MPXV-Z, with no major genetic changes likely to affect gene expression being identified. Further, deletion of D14L from the MPXV-Z genome does not abrogate transcription of the neighboring genes D13L (putative IL-1 receptor antagonist) and D15L (Kelch-like protein), although lower levels of D13L transcript production by D14L KO MPXV-Z were observed. It is currently unknown if a decrease in D13L transcript production actually affects protein expression levels of this gene during D14L KO MPXV-Z infection or whether this might have any effects during an in vivo infection. Although it is possible that the difference in D13L transcription could contribute to the observed phenotype of D14L KO MPXV-Z, the major difference between the parental and recombinant virus is the lack of MOPICE expression by D14L KO MPXV-Z, which is therefore most likely to be responsible for any major phenotypic variations observed between these viruses.
To assess the role of MOPICE in disease progression and the development of the adaptive immune response in vivo, RM were infected intrabronchially with either WT MPXV-Z or D14L KO MPXV-Z. Our results show that this model closely recapitulates MPXV disease in humans, with the development of fever, widespread lesions, and respiratory symptoms in animals infected with WT MPXV-Z. Results of the WT MPXV-Z infection studies in RM also parallel recent findings in a study of i.b. infection of cynomolgus macaques, with the development of similar patterns of disease progression in both model systems (16). Further, utilizing an extensive array of immunological tools available in the RM model, we demonstrate that WT MPXV-Z-infected animals generate a humoral and cellular immune response against the virus, as evidenced by an increase in the number of proliferating memory B cells, the production of MPXV-specific IgG, an increase in the number of proliferating memory T cells, and the generation of IFN-γ-/TNF-α-secreting T cells specific to orthopoxvirus antigens. The onset of the adaptive immune response correlated with the increase in viral loads detected in infected animals, and it closely paralleled the control and resolution of the MPXV infection.
MOPICE is capable of inhibiting complement activity in rhesus macaque serum in vitro (data not shown), and we initially hypothesized that the absence of this protein would result in increased levels of complement activation, viral opsonization, infected cell lysis, and anaphylatoxin production in D14L KO MPXV-Z-infected animals, thereby leading to lower levels of viral replication and a less severe disease than that found in WT MPXV-Z-infected animals. However, although the deletion of MOPICE had no effect on the in vitro growth properties of MPXV-Z, infection of RM with D14L KO MPXV-Z resulted in more severe disease than WT MPXV-Z infections. This was evidenced by the fact that while all animals infected with WT MPXV-Z survived, one of the four animals infected with D14L KO MPXV-Z succumbed to disease at 17 dpi, and two additional D14L KO MPXV-Z-infected animals displayed larger lesions with delayed scabbing and healing compared to those in WT MPXV-Z infection. We also observed higher peak viral loads in D14L KO MPXV-Z-infected animals than in those with WT MPXV-Z infections. Specifically, viral loads in BAL fluid and whole blood in D14LKO MPXV-Z-infected animals were generally higher than those observed in WT MPXV-Z-infected animals, with the highest viral load of any infection being detected in the single D14L KO MPXV-Z-infected animal that succumbed to MPXV-related disease complications. These observations are similar to those made in studies of an IMP knockout cowpox virus in mice, in which footpad swelling and localized tissue damage were increased in the absence of the viral complement inhibitory protein (26). Taken together, these observations suggest that the deletion of MOPICE might result in increased localized inflammation and tissue damage in pox lesions, thereby rendering D14LKO MPXV-Z more virulent.
We also observed a delayed and reduced adaptive immune response in D14L KO MPXV-Z-infected animals. Although the MPXV-specific IgG response appeared with similar kinetics in both WT and D14L KO MPXV-Z-infected animals (14 dpi), initially the endpoint IgG titer was approximately a log lower in the D14L KO-infected animals and the animals did not achieve the same titer as WT-infected animals until 35 dpi. Further, the proliferative burst, as well as the frequency, of MPXV-specific CD4 and CD8 T-cell subsets was reduced in D14L KO MPXV-Z-infected animals. It is very likely that the delayed and reduced adaptive immune response contributed to the increased viral loads seen in some D14L KO MPXV-Z-infected animals, although it is also possible that the initial increase in viral loads could have resulted in T-cell exhaustion in a manner analogous to what has been observed in some models of chronic viral infection (2). Also of interest was the biphasic T-cell response observed in the WT MPXV-infected animals. As described in Results, the decreased frequency of proliferating T cells correlated with a decrease in white blood cell and lymphocyte counts in these animals, suggesting that MPXV infection might result in extensive lymphocyte death. This possibility is supported by the observation that the majority of the IFN-γ-producing, MPXV-specific T cells are detected 21 dpi at the earliest, which corresponds to the second proliferative peak.
Taken together, our data indicate that deletion of MOPICE does not render MPXV-Z less virulent, and that lack of this protein may actually result in higher viral loads and more severe disease outcomes in vivo. This suggests that increased complement activation in the absence of MOPICE expression in vivo may not play a significant role in enhancing the adaptive response to MPXV-Z but, rather, that it may actually be inhibitory to the adaptive response in some fashion. It has been recently found, for example, that despite its known proinflammatory properties, the anaphylatoxin C5a may also possess immunosuppressive properties and that, in certain contexts, including aerosol exposure to inhaled allergens, it can inhibit the adaptive immune response to antigen (19, 37). In addition, C5a has also been shown to suppress anti-tumor CD8 T-cell responses (24). Thus, one possibility in our model is that increased complement activation and anaphylatoxin production during i.b. infection of RM with D14L KO MPXV-Z actually result in a decreased adaptive immune response against the virus, compared to findings with WT MPXV-Z infection. However, exactly how MOPICE modulates the adaptive immune response is still unclear at this point and will be the subject of future studies.
Overall, our findings do not rule out some role for MOPICE in pathogenic differences between MPXV clades, but they do suggest that other viral products also significantly contribute to observed differences in mortality and disease severity. Indeed, it is unlikely that a single protein is fully responsible for pathogenic differences between MPXV clades, but, rather, it is more reasonable to expect that multiple differences between viruses, either small or large, have an additive effect that culminates in varied behavior of different MPXV strains. Other factors such as the route of infection and general disease severity may also have an impact on the development of immune responses against MPXV in vivo and may modulate the extent to which a particular viral protein is involved in pathogenesis in any given context. Further analysis of the activities of MOPICE during MPXV infection in vivo will be necessary in order to provide a better understanding of the overall importance of complement in regulation of the adaptive immune response against MPXV, as well as other poxviruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joanne Strussenberg, Susan Clary, and Gwynn Daniels for their assistance.
Portions of this work were supported by Public Health Service grant RR00163, National Institute of Allergy and Infectious Diseases (NIH/DHHS through interagency agreement Y1-AI-4894-01), and the United States Department of Defense (W81XWH-05-1-0046). The following reagent was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: polyclonal anti-vaccinia virus (WR) A33R protein (antiserum, rabbit), NR-628.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Amanna I. J., Slifka M. K., Crotty S. 2006. Immunity and immunological memory following smallpox vaccination. Immunol. Rev. 211:320–337 [DOI] [PubMed] [Google Scholar]
- 2. Blackburn S. D., et al. 2009. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breman J. G., et al. 1980. Human monkeypox, 1970–79. Bull. World Health Organ. 58:165–182 [PMC free article] [PubMed] [Google Scholar]
- 4. Brodie R., Smith A. J., Roper R. L., Tcherepanov V., Upton C. 2004. Base-By-Base: single nucleotide-level analysis of whole viral genome alignments. BMC Bioinformatics 5:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cameron C. M., Barrett J. W., Mann M., Lucas A., McFadden G. 2005. Myxoma virus M128L is expressed as a cell surface CD47-like virulence factor that contributes to the downregulation of macrophage activation in vivo. Virology 337:55–67 [DOI] [PubMed] [Google Scholar]
- 6. Carroll M. C. 2004. The complement system in regulation of adaptive immunity. Nat. Immunol. 5:981–986 [DOI] [PubMed] [Google Scholar]
- 7. Chen N., et al. 2005. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology 340:46–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ciulla E., Emery A., Konz D., Krushkal J. 2005. Evolutionary history of orthopoxvirus proteins similar to human complement regulators. Gene 355:40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Di Giulio D. B., Eckburg P. B. 2004. Human monkeypox: an emerging zoonosis. Lancet Infect. Dis. 4:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Girgis N. M., et al. 2011. The vaccinia virus complement control protein modulates adaptive immune responses during infection. J. Virol. 85:2547–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hammarlund E., et al. 2003. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 9:1131–1137 [DOI] [PubMed] [Google Scholar]
- 12. Heymann D. L., Szczeniowski M., Esteves K. 1998. Re-emergence of monkeypox in Africa: a review of the past six years. Br. Med. Bull. 54:693–702 [DOI] [PubMed] [Google Scholar]
- 13. Hutin Y. J., et al. 2001. Outbreak of human monkeypox, Democratic Republic of Congo, 1996 to 1997. Emerg. Infect. Dis. 7:434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hutson C. L., et al. 2009. A prairie dog animal model of systemic orthopoxvirus disease using West African and Congo Basin strains of monkeypox virus. J. Gen. Virol. 90:323–333 [DOI] [PubMed] [Google Scholar]
- 15. Isaacs S. N., Kotwal G. J., Moss B. 1992. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc. Natl. Acad. Sci. U. S. A. 89:628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson R. F., et al. 2011. Comparative analysis of monkeypox virus infection of cynomolgus macaques by the intravenous or intrabronchial inoculation route. J. Virol. 85:2112–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khodakevich L., Jezek Z., Messinger D. 1988. Monkeypox virus: ecology and public health significance. Bull. World Health Organ. 66:747–752 [PMC free article] [PubMed] [Google Scholar]
- 18. Khodakevich L., et al. 1987. The role of squirrels in sustaining monkeypox virus transmission. Trop. Geogr. Med. 39:115–122 [PubMed] [Google Scholar]
- 19. Kohl J., Wills-Karp M. 2007. Complement regulates inhalation tolerance at the dendritic cell/T cell interface. Mol. Immunol. 44:44–56 [DOI] [PubMed] [Google Scholar]
- 20. Lambris J. D., Ricklin D., Geisbrecht B. V. 2008. Complement evasion by human pathogens. Nat. Rev. Microbiol. 6:132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Likos A. M., et al. 2005. A tale of two clades: monkeypox viruses. J. Gen. Virol. 86:2661–2672 [DOI] [PubMed] [Google Scholar]
- 22. Liszewski M. K., et al. 2006. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J. Immunol. 176:3725–3734 [DOI] [PubMed] [Google Scholar]
- 23. Manes N. P., et al. 2008. Comparative proteomics of human monkeypox and vaccinia intracellular mature and extracellular enveloped virions. J. Proteome Res. 7:960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Markiewski M. M., et al. 2008. Modulation of the antitumor immune response by complement. Nat. Immunol. 9:1225–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Messaoudi I., et al. 2009. Simian varicella virus infection of rhesus macaques recapitulates essential features of varicella zoster virus infection in humans. PLoS Pathog. 5:e1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller C. G., Shchelkunov S. N., Kotwal G. J. 1997. The cowpox virus-encoded homolog of the vaccinia virus complement control protein is an inflammation modulatory protein. Virology 229:126–133 [DOI] [PubMed] [Google Scholar]
- 27. Pitcher C. J., et al. 2002. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol. 168:29–43 [DOI] [PubMed] [Google Scholar]
- 28. Reed K. D., et al. 2004. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350:342–350 [DOI] [PubMed] [Google Scholar]
- 29. Saijo M., et al. 2009. Virulence and pathophysiology of the Congo Basin and West African strains of monkeypox virus in non-human primates. J. Gen. Virol. 90:2266–2271 [DOI] [PubMed] [Google Scholar]
- 30. Sbrana E., Xiao S. Y., Newman P. C., Tesh R. B. 2007. Comparative pathology of North American and central African strains of monkeypox virus in a ground squirrel model of the disease. Am. J. Trop. Med. Hyg. 76:155–164 [PubMed] [Google Scholar]
- 31. Shchelkunov S. N. 2003. Immunomodulatory proteins of orthopoxviruses. Mol. Biol. 37:37–48(In Russian.) [PubMed] [Google Scholar]
- 32. Shchelkunov S. N., et al. 2002. Analysis of the monkeypox virus genome. Virology 297:172–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stittelaar K. J., et al. 2005. Modified vaccinia virus Ankara protects macaques against respiratory challenge with monkeypox virus. J. Virol. 79:7845–7851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tcherepanov V., Ehlers A., Upton C. 2006. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Upton C., Hogg D., Perrin D., Boone M., Harris N. L. 2000. Viral genome organizer: a system for analyzing complete viral genomes. Virus Res. 70:55–64 [DOI] [PubMed] [Google Scholar]
- 36. Weaver J. R., Isaacs S. N. 2008. Monkeypox virus and insights into its immunomodulatory proteins. Immunol. Rev. 225:96–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wills-Karp M. 2007. Complement activation pathways: a bridge between innate and adaptive immune responses in asthma. Proc. Am. Thorac. Soc. 4:247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zaucha G. M., Jahrling P. B., Geisbert T. W., Swearengen J. R., Hensley L. 2001. The pathology of experimental aerosolized monkeypox virus infection in cynomolgus monkeys (Macaca fascicularis). Lab. Invest. 81:1581–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.