Abstract
Soluble forms of the HIV-1 receptor CD4 (sCD4) have been extensively characterized for more than 2 decades as promising inhibitors and components of vaccine immunogens. However, they were mostly based on the first two CD4 domains (D1D2), and numerous attempts to develop functional, high-affinity, stable soluble one-domain sCD4 (D1) have not been successful because of the strong interactions between the two domains. We have hypothesized that combining the power of structure-based design with sequential panning of large D1 mutant libraries against different HIV-1 envelope glycoproteins (Envs) and screening for soluble mutants could not only help solve the fundamental stability problem of isolated D1, but may also allow improvement of D1 affinity while preserving its cross-reactivity. By using this strategy, we identified two stable monomeric D1 mutants, mD1.1 and mD1.2, which were significantly more soluble and bound Env gp120s more strongly (50-fold) than D1D2, neutralized a panel of HIV-1 primary isolates from different clades more potently than D1D2, induced conformational changes in gp120, and sensitized HIV-1 for neutralization by CD4-induced antibodies. mD1.1 and mD1.2 exhibited much lower binding to human blood cell lines than D1D2; moreover, they preserved a β-strand secondary structure and stability against thermally induced unfolding, trypsin digestion, and degradation by human serum. Because of their superior properties, mD1.1 and mD1.2 could be potentially useful as candidate therapeutics, components of vaccine immunogens, and research reagents for exploration of HIV-1 entry and immune responses. Our approach could be applied to other cases where soluble isolated protein domains are needed.
INTRODUCTION
CD4 is a transmembrane glycoprotein expressed on the surfaces of most thymocytes and a subpopulation of mature T cells (CD4+ T cells) (16). It is composed of four immunoglobulin-like extracellular domains, a transmembrane segment, and a cytoplasmic tail noncovalently associated with Lck, a src-family tyrosine kinase. As an important component of the immune system, CD4 functions as a coreceptor of the T-cell receptor (TCR) on the surfaces of CD4+ T cells for stronger association with the class II major histocompatibility complex (MHCII) on antigen-presenting cells (APCs). This association is sufficient to trigger T-cell signaling transduction, resulting in activation of the CD4+ T cells. The crystal structure of human CD4-murine MHCII complex shows that only the first extracellular domain (D1) of CD4 contacts MHCII (37). However, mutational analysis indicates that, in addition to D1, other domains also affect binding to MHCII (27). Moreover, oligomerization of CD4 is required for stable interaction with MHCII and efficient T-cell activation (31).
CD4 is also the primary receptor for HIV-1 (9). HIV-1 entry is initiated by its binding to the viral envelope glycoprotein (Env) gp120. The interaction results in extensive conformational rearrangements of gp120 and subsequently gp41 after engagement of a coreceptor (either CCR5 or CXCR4). The structural rearrangements of Envs and the interplay between Envs and the cellular receptor and coreceptor bring viral and plasma cell membranes within close proximity and eventually cause membrane fusion and entry of the viral genome into cells. Because CD4 plays a key role in HIV-1 infections, recombinant solubly expressed CD4 (sCD4) containing either all four (T4) (10) or the first two (D1D2) (35) extracellular domains is a potent inhibitor of HIV-1 entry and was used for crystallization alone (30, 39) or with gp120 (21). Crystallized CD4 binds similarly to HIV-1 gp120 and MHCII through D1.
We have, therefore, hypothesized that by using protein-engineering techniques it would be possible to generate a smaller version of sCD4 that contains only the first domain, D1, while preserving not only binding activity and specificity, but also other functions, such as induction of conformational changes in HIV-1 gp120. Due to decreased molecular size, D1 could have excellent biological properties, including improved binding kinetics; soluble expression in Escherichia coli; higher solubility, stability, and specificity; lower immunogenicity in animals; and better penetration into tissues, such as the densely packed lymphoid environments (e.g., spleen, lymph node, and gut), where HIV-1 mostly replicates and spreads. In a previous study (34), D1 was generated using a mutational strategy based on single-amino-acid replacement and purified from a sonicated supernatant of E. coli. However, the purified protein was stable only at low pH (4.0), was partially improperly folded, and had affinity severalfold lower than that of D1D2 and T4. In this study, we used the power of library methodologies and generated a large phage-displayed library of D1 by randomizing four hydrophobic residues at the interface with the second domain (D2) and introducing additional mutations into other positions by error-prone PCR. The library was panned sequentially against different HIV-1 Envs. The selected D1 mutants were highly soluble, expressible in E. coli, stable, and monomeric in phosphate-buffered saline (PBS) (pH 7.4), and showed a dramatic increase in affinity compared to D1D2. In addition, they preserved other functions of CD4, such as induction of structural rearrangements in HIV-1 gp120 and possible interactions with other targets.
MATERIALS AND METHODS
Cells, viruses, plasmids, and HIV-1 Env gp140s.
BJAB cells were a gift from Anu Puri (National Cancer Institute, Frederick, MD). We purchased the 293T and SUPT1 cells from ATCC and the 293 freestyle cells from Invitrogen. Other cell lines and plasmids used for expression of various HIV-1 Envs were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (ARRRP). The D1D2 gene was synthesized and cloned in our laboratory by Xiaodong Xiao (National Cancer Institute, Frederick, MD; currently at Medimmune, Gaithersburg, MD). gp140SC and gp140MS were produced in our laboratory as described previously (5). Gp140Con-s (23) and gp140CH12.0544.2 were gifts from Barton F. Haynes (Duke University Medical Center, Durham, NC).
Library construction and panning.
The phage-displayed library of D1 mutants was constructed by random mutagenesis. To randomize positions 5, 76, 96, and 98 of D1, we used the following degenerate primers: D1mF, 5′-CGCTACCGTGGCCCAGGCGGCCAAGAAGGTGGTGNNSGGCAAGAAGGGCGACACC-3′ (sense); D1mR1, 5′-GTGGTGGCCGGCCTGGCCGCCWNNCACWNNCAGCTGCACCTCCTCCTTCTGGTCCTCCACCTCGCAGATGTA-3′ (antisense); and D1mR2, 5′-CTCGCAGATGTAGGTGTCGCTGTCCTCWNNCTTCAGGTTCTTGATGATCAG-3′ (antisense). The D1 gene fragment was first amplified by PCR with a D1D2-encoding plasmid as the template and primers D1mF and D1mR2. The PCR product was gel purified and used as a template for amplification of full-length D1 with primers D1mF and D1mR1. To introduce point mutations in other positions, we performed random DNA mutagenesis with the purified full-length D1 gene as a template, primers D1mF and D1mR1, and the Gene-Morph PCR Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The PCR products were gel purified, digested with SfiI, and gel purified again. The purified fragments were then cloned into phagemid pComb3X linearized by SfiI. A phage library was prepared by electroporation of E. coli strain TG1 electroporation-competent cells (Stratagene, La Jolla, CA) with desalted and concentrated ligation, as described previously (8).
The phage library was used for selection of D1 mutants against HIV-1 antigens used to coat 96-well plates as described previously (13). For sequential panning, 200, 100, and 20 ng of gp140SC, gp140MS, and gp140SC were used in the first, second, and third rounds, respectively. Clones that bound to HIV-1 antigens were identified from the third round of panning using soluble expression-based monoclonal enzyme-linked immunosorbent assay (semELISA) as described previously (7).
Cloning of D1 mutants for mammalian expression and fusion proteins of D1 mutants and D1D2.
The following primers were used: D1-49F, 5′-TGACGCGGCCCAGCCGGCCAAGAAGGTGGTGATCGGC-3′ (sense); D1-49R, 5′-CGGGTTTAAACTCAGTGGTGGTGGTGGTGGTGGCCTAGCACTATCAGCTG-3′ (antisense); D1-53F, 5′-TGACGCGGCCCAGCCGGCCAAGAAGGTGGTGTACGGC-3′ (sense); D1-53R, 5′-CGGGTTTAAACTCAGTGGTGGTGGTGGTGGTGGCCTACCACTACCAGCTG-3′ (antisense); CH2F1, 5′-GCACCTGAACTCCTGGGG-3′ (sense); CH2F2, 5′-TCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCAGCACCTGAACTCCTGGGG-3′ (sense); CH2R, 5′-CGGGTTTAAACTCAGTGGTGGTGGTGGTGGTGTTTGGCTTTGGAGATGGT-3′ (antisense); D1R4, 5′-GAGTTCAGGTGCGCCTAGCACTATCAGCTG-3′ (antisense); D1R5, 5′-AGAGCCACCTCCGCCTGAACCGCCTCCACCGCCTAGCACTATCAGCTG-3′ (antisense); D1-49FcR, 5′-TTTGTCGGGCCCGCCTAGCACTATCAGCTG-3′ (antisense); D1-53FcR, 5′-TTTGTCGGGCCCGCCTACCACTACCAGCTG-3′ (antisense); CD4FcF, 5′-TGGTTTCGCTACCGTGGCCCAGCCGGCCAAGAAGGTGGTGCTGGGC-3′ (sense); and CD4FcR, 5′-GTGAGTTTTGTCGGGCCCGGCCAGCACCACGATGTC-3′ (antisense).
To clone two stable monomeric D1 mutants, mD1.1 and mD1.2, for mammalian expression, the gene fragments were PCR amplified with their bacterial expression plasmids as templates and primer combinations D1-49F/D1-49R and D1-53F/D1-53R, respectively. The PCR products were gel purified, digested with SfiI and PmeI, and cloned into pSecTagB. To construct mD1.1CH2, the mD1.1 fragment was PCR amplified with primers D1-49F and D1R4. The CH2 fragment was amplified with an IgG1-encoding plasmid as a template and primers CH2F1 and CH2R. mD1.1 was joined to CH2 by overlapping PCR performed in a volume of 50 μl by using both templates at the same molarities for 7 cycles in the absence of primers and 15 additional cycles in the presence of 500 pM primers D1-49F and CH2R. mD1.1L3CH2 was constructed in the same way, except for the use of the primer combinations D1-49F/D1R5 and CH2F2/CH2R for amplification of the mD1.1 and CH2 fragments, respectively. The resulting products, with SfiI and PmeI restriction sites appended on both sides, were digested and cloned into pSecTagB. For construction of mD1.1Fc, mD1.2Fc, and D1D2Fc, gene fragments were PCR amplified with primer combinations D1-49F/D1-49FcR, D1-53F/D1-53FcR, and CD4FcF/CD4FcR, respectively; digested with SfiI and ApaI; and cloned into pSecTagB-Fc.
Cloning of gp120SC-mD1.2, gp120SC-D1D2, and gp120SC.
The following primers were used: SCF, 5′-TGACGCGGCCCAGCCGGCCGAGGTGGTGCTGGGCAAC-3′ (sense); SCR, 5′-TGAACCGCCTCCACCGCTTCCTCCTCCTCCGGATCCTCCTCCGCCGGATCCTCCTCCTCCCTCGATCTTCACCACCTT-3′ (antisense); SCD1F, 5′-GGTGGAGGCGGTTCAAAGAAGGTGGTGTACGGC-3′ (sense); D1-53R, 5′-CGGGTTTAAACTCAGTGGTGGTGGTGGTGGTGGCCTACCACTACCAGCTG-3′ (antisense); SCD12F, 5′-GGTGGAGGCGGTTCAAAGAAGGTGGTGCTGGGC-3′ (sense); SCD12R, 5′-CGGGTTTAAACTCAGTGGTGGTGGTGGTGGTGGGCCAGCACCACGATGTC-3′ (antisense); and SCR1, 5′-CGGGTTTAAACTCAGTGGTGGTGGTGGTGGTGCTCGATCTTCACCACCTT-3′ (antisense).
For cloning of gp120SC-mD1.2, the gp120SC and mD1.2 gene fragments were PCR amplified with primer pairs SCF/SCR and SCD1F/D1-53R, respectively. Gp120SC was joined to mD1.2 by overlapping PCR using primers SCF/D1-53R. The resulting product was digested with SfiI and PmeI and cloned into pSecTagB. gp120SC-D1D2 was constructed in the same way, with the substitution of primers SCD12F/SCD12R for amplification of D1D2 and SCF/SCD12R during overlapping PCR. To generate gp120SC, the gene fragment was amplified by PCR with primers SCF/SCR1, digested with SfiI and PmeI, and cloned into pSecTagB.
Protein expression and purification.
The D1 mutants were expressed in E. coli HB2151 or 293 freestyle cells; D1D2 and all fusion proteins were produced in 293 freestyle cells, as described previously (6). The D1 mutants, D1D2, gp120SC, mD1.1-CH2, gp120SC-mD1.2, and gp120SC-D1D2 fusion proteins, which were tagged with hexahistidine at their C termini, were purified from the soluble fraction of HB2151 periplasm or the 293 cell culture supernatants by immobilized metal ion affinity chromatography (IMAC) using Ni-nitrilotriacetic acid (NTA) resin (Qiagen, Valencia, CA) according to the manufacturer's protocols. The Fc fusion proteins were purified from the 293 cell culture supernatants by Protein A Sepharose 4 Fast Flow (GE Healthcare, Piscataway, NJ) column chromatography.
Size exclusion chromatography.
A Superdex75 10/300 GL column (GE Healthcare, Piscataway, NJ) was calibrated with protein molecular mass standards of 14-kDa RNase A, 25-kDa chymotrypsin, 44-kDa ovalbumin, 67-kDa albumin, 158-kDa aldolase, 232-kDa catalase, 440-kDa ferritin, and 669-kDa thyroglobulin. Purified D1 mutants and D1D2 in PBS were loaded onto the preequilibrated column and eluted with PBS at 0.5 ml/min.
ELISA.
ELISA was performed as described previously (6). Bound D1D2, the D1 mutants, and mD1.1-CH2 fusion proteins were detected by horseradish peroxidase (HRP)-conjugated anti-hexahistidine tag antibody (Sigma-Aldrich, St. Louis, MO). Bound antibody-Fc fusion proteins in the absence or presence of sCD4 were detected by HRP-conjugated anti-human IgG (Fc-specific) antibody (Sigma-Aldrich, St. Louis, MO). The half-maximal binding (50% effective concentration [EC50]) was calculated by fitting the data to the Langmuir adsorption isotherm.
SPR analysis.
The binding kinetics of D1D2, the D1 mutants, and their derivatives with HIV-1 gp140 were assessed by surface plasmon resonance (SPR) analysis on Biacore X100 (GE Healthcare) using a single-cycle approach according to the manufacturer's instructions. Briefly, purified HIV-1 gp140 was diluted in sodium acetate (pH 5.0) and immobilized directly onto a CM5 sensor chip by a standard amine-coupling method. The reference cell was injected with N-hydroxysuccinimide/1-ethyl-3-(3-dimethyaminopropy)carbodiimide and ethanolamine without injection of gp140. The proteins were diluted with running buffer HBS-EP (100 mM HEPES, pH 7.4, 1.5 M NaCl, 30 mM EDTA, 0.5% surfactant 20). All analytes were tested at 500, 100, 20, 4, and 0.8 nM concentrations. The kinetic constants were calculated from the sensorgrams fitted with the monovalent binding model of the BiacoreX100 Evaluation software 2.0.
Flow cytometry (FACS).
For detection of the expression of human MHCII (HLA-DR), approximately 106 BJAB or SUPT1 cells in 100 μl PBS containing 0.1% bovine serum albumin (BSA) (PBSA) were mixed at different ratios (vol/vol) with fluorescein isothiocyanate (FITC)-conjugated monoclonal mouse anti-human HLA-DR (Sigma-Aldrich, St. Louis, MO) and incubated for 30 min at room temperature. The cells were washed twice with 200 μl PBSA and then used for fluorescence-activated cell sorter (FACS) analysis. For measurement of the interaction of sCD4, 106 cells in 100 μl PBSA were incubated with sCD4-Fc fusion proteins at final concentrations of 0.2, 2, and 20 μM for 1 h at room temperature. The cells were washed twice and resuspended in 100 μl PBSA, and 0.5 μl Alexa Fluor 488-conjugated mouse anti-human IgG1 (Fc specific; Invitrogen, Carlsbad, CA) was added. Following a 30-min incubation at room temperature, the cells were washed twice and then subjected to FACS.
Pseudovirus neutralization assay.
HIV-1 pseudoviruses were generated, and the neutralization assay was performed as described previously (6).
Solubility measurement.
Proteins in PBS (pH 7.4) were concentrated using Millipore centrifugal filters with a low molecular mass cutoff of 3 kDa and an Eppendorf 5804R centrifuge. The 15-ml filtration devices were centrifuged at 4,000 × g at room temperature until the volume was reduced to about 50 μl. The concentrated proteins were transferred to 1.5-ml Eppendorf tubes and centrifuged at 15,000 × g with an Eppendorf 5417R centrifuge for 10 min at room temperature. The supernatant was collected, and the protein concentration in solution was determined after appropriate dilution by measuring the absorbance at 280 nm. To further concentrate the proteins, Microcon Ultracel YM-3 centrifugal filters with a cutoff of 3 kDa and an Eppendorf 5417R centrifuge were used. When the volumes were reduced to between 5 and 10 μl, samples were transferred to 0.5-ml Eppendorf tubes and centrifuged at 15,000 × g at room temperature for 10 min. The supernatant was collected, and protein concentrations in solution were determined as described above. After storage of the supernatants at 4°C for 5 days, the samples were centrifuged, and the concentrations of proteins in the supernatant were quantified again.
CD.
The secondary structure and thermal stability of D1D2 and D1 mutants were determined by circular dichroism (CD) spectroscopy as described previously (17).
Serum stability measurement.
The proteins in PBS were mixed at a 1:1 ratio (vol/vol) with human serum or PBS as a control to give a final concentration of 8,300 nM in a total volume of 35 μl. After 5, 10, and 15 days of incubation at 37°C, respectively, the reactions were stopped by freezing the samples at −20°C. After all samples were collected, 35 μl of 4% milk in PBS was added to each sample, and the diluted proteins were used in ELISAs with gp140Con-s. Standard curves were generated using the original protein stocks to quantify functional sCD4 surviving different periods of serum incubation.
Proteolysis.
Proteolytic digestion of sCD4 in PBS was performed using trypsin at a protease/substrate ratio of 1:600 (wt/wt). For each reaction, 5 ng of trypsin in 2 μl PBS was added to 3 μg sCD4 in 5.5 μl PBS. Separate samples were incubated for 15, 30, and 60 min. The reactions were stopped by adding 2.5 μl SDS-PAGE gel-loading buffer containing 100 mM dithiothreitol (DTT) to each reaction mixture and boiling the samples for 5 min at 100°C. Samples collected at different time points and stored at −20°C were resolved by SDS-PAGE, followed by staining with Coomassie brilliant blue R250.
RESULTS
Structure-based library design and construction and selection of soluble D1 mutants with high binding and cross-reactivity to HIV-1 gp140s.
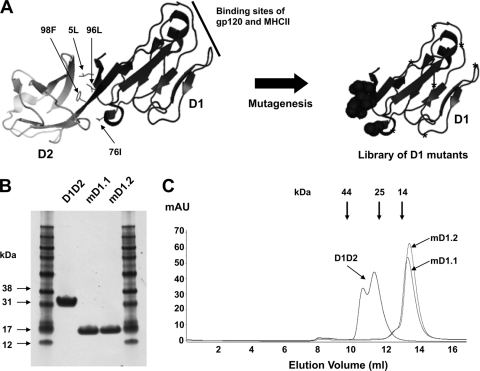
In crystallized D1D2, four hydrophobic residues (5L, 76I, 96L, and 98F) on D1 contact D2 (Fig. 1A). These residues were randomized by using the degenerate codon NNS, which encodes the complete set of standard amino acids, and additional point mutations were introduced into other positions by random mutagenesis through error-prone PCR. The resulting D1 mutants were used to construct a relatively large phage-displayed library with approximately 109 members (Fig. 1A). The library was panned sequentially against two HIV-1 Envs from different clades, gp140SC (clade B) and gp140MS (clade A), to ensure preservation of D1 cross-reactivity and to eventually increase affinity. To identify individual mutants that specifically bind to Envs and are soluble in the E. coli periplasm, clones were randomly selected after three rounds of panning and subjected to screening by semELISA. Sequencing of the 40 highest-affinity binders revealed that they represented 19 different clones (see Fig. S1 in the supplemental material). A majority (89%) of the mutants retained hydrophobic residues in positions 5 and 96, where isoleucine dominated, while 58% and 68% of the mutants contained hydrophobic residues in positions 76 and 98, respectively.
Fig. 1.
Library construction and selection of high-affinity, soluble, stable D1 mutants. (A) Schematic representation of library construction. The X-ray crystal structure of D1D2 (left) was adapted from Protein Data Bank (PDB) 2B4C. The four hydrophobic residues in D1 interacting with D2 are indicated by small arrows. The random-mutagenesis library of D1 is shown on the right, where circles denote randomization of the four hydrophobic residues with the degenerate codon NNS and stars denote randomization by error-prone PCR. (B) Reducing SDS-PAGE of D1D2 and the D1 mutants purified from 293 freestyle cell cultures. (C) Size exclusion chromatography analysis. The thick arrows at the top indicate the elution volumes of molecular mass standards in PBS: RNase A (14 kDa), chymotrypsin (25 kDa), and ovalbumin (44 kDa).
Two clones, designated mD1.1 and mD1.2, were chosen for further characterization because of their high binding to all gp140s tested in semELISA and their relatively high yields of 0.5 and 0.75 mg liter−1, respectively, from the soluble fraction of E. coli periplasm. In order to further characterize these clones and for comparison with D1D2, which had been expressed in E. coli as an insoluble inclusion body protein (15), mD1.1, mD1.2, and D1D2 were cloned into a mammalian expression vector, expressed in 293 freestyle cells, and purified from the cell culture supernatants. mD1.1 and mD1.2 ran on a reducing SDS-PAGE with apparent molecular masses of approximately 16 kDa (Fig. 1B), which were greater than their calculated molecular masses (12.040 and 12.061 kDa, respectively, including the hexahistidine tag). Both mD1.1 and mD1.2 were seen to be monomeric in PBS at pH 7.4 by size exclusion chromatography, with apparent molecular masses similar to their calculated molecular masses (Fig. 1C). D1D2 was also monomeric but did not elute as a single peak.
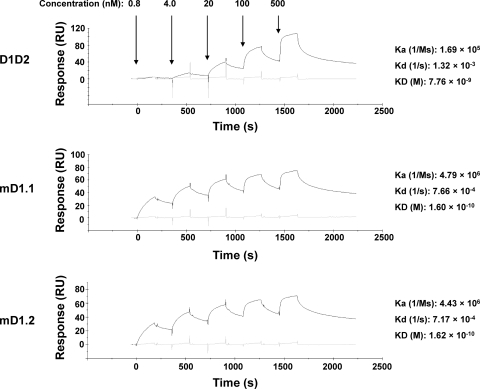
mD1.1 and mD1.2 bound to gp140s with much higher affinity than D1D2 and enhanced binding of CD4-induced (CD4i) antibodies to gp140s.
Binding of mD1.1 and mD1.2 was analyzed by SPR and ELISA. Both D1 mutants bound to gp140Con-s (23), which was a consensus gp140 designed by aligning >1,000 sequences of group M, with picomolar affinity (KD [equilibrium dissociation constant] = 1.60 × 10−10 and 1.62 × 10−10 M, respectively) 48-fold higher than that of D1D2 (KD = 7.76 × 10−9 M) (Fig. 2). They had much higher (28-fold) association rates and lower (2-fold) dissociation rates. To assess cross-reactivity and confirm the high binding affinity of mD1.1 and mD1.2, ELISAs were performed with two additional gp140s (gp140CH12.0544.2 and gp140SC) from clade B isolates. As expected, both mutants were cross-reactive against all three gp140s and in all cases had EC50s about 10-fold lower than those of D1D2 (see Fig. S2 in the supplemental material).
Fig. 2.
Binding kinetics characterization of mD1.1 and mD1.2. SPR analysis was performed on Biacore X100 by using a single-cycle approach according to the manufacturer's instructions. The tested sCD4 concentrations corresponding to each sensorgram are indicated by arrows, and the calculated kinetic constants are shown on the right.
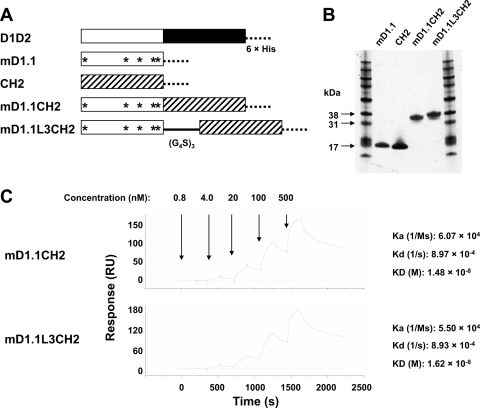
To determine whether the significantly increased affinity of the D1 mutants is due to their decreased molecular size, mutation-induced structural refinement, or both, we made two fusion proteins of mD1.1 with the human IgG1 CH2 domain, mD1.1CH2, without a linker, and mD1.1L3CH2, with a polypeptide linker composed of three repeats of the G4S motif (Fig. 3A and B). As shown in a previous study (17), isolated CH2 is solubly expressed in E. coli with a high yield; it has a mass (approximately 13 kDa) and folding similar to those (mass, approximately 10 kDa, and immunoglobulin-like folding) of D2 (28). Therefore, the fusion proteins could mimic D1D2 in molecular size and shape. The association rate constants (Ka) of mD1.1CH2 and mD1.1L3CH2 (Ka = 6.07 × 104 and 5.50 × 104 M s−1, respectively) were decreased 80-fold compared to that of mD1.1 (Ka = 4.79 × 106 M s−1) as measured by SPR (Fig. 3C). The fusion proteins had similar dissociation rate constants (Kd = 8.97 × 10−4 and 8.93 × 10−4 s−1, respectively) comparable with that of mD1.1 (Kd = 7.66 × 10−4 s−1) and slightly lower than that of D1D2 (Kd = 1.32 × 10−3 s−1). These results suggest that the improved binding kinetics of mD1.1 compared to D1D2 is mostly due to decreased molecular size, although possible structural adjustments induced by the mutations could also be a contributing factor.
Fig. 3.
Effects of molecular size on binding kinetics of mD1.1. (A) Generation of mD1.1-CH2 fusion proteins is schematically represented. The 6×His denotes a hexahistidine tag, the stars denote the mutations in mD1.1 compared to wild-type D1, and (G4S)3 represents a polypeptide linker composed of three repeats of the G4S motif. (B and C) The purified fusion proteins were analyzed on reducing SDS-PAGE (B) and by SPR assay (C).
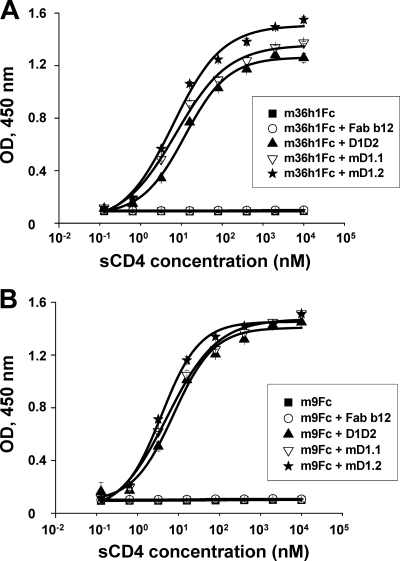
CD4 induces conformational changes in gp120 leading to the exposure of CD4i epitopes. To find out whether the D1 mutants can do likewise, we tested two CD4i antibody-based fusion proteins, m9Fc (41) and m36h1Fc (6), for binding to gp140s in the absence or presence of mD1.1, mD1.2, or D1D2. As expected, binding of the two antibodies at a fixed concentration of 50 nM to gp140Con-s was dramatically enhanced in the presence of the D1 mutants; the increase in binding with mD1.2 was 3-fold more than that seen with D1D2 (Fig. 4). Fab b12 (29), which is a well-characterized broadly neutralizing monoclonal antibody targeting the CD4-binding site on gp120, did not enhance the interaction of the CD4i antibodies with gp140.
Fig. 4.
ELISA binding of CD4i antibodies to gp140s in the absence or presence of the D1 mutants. Two CD4i antibodies, m36h1Fc (A) and m9Fc (B), at a fixed concentration of 50 nM were mixed with different concentrations of mD1.1 or mD1.2 prior to incubation with gp140Con-s used to coat 96-well plates. D1D2 and Fab b12 were used as positive and negative controls, respectively. OD, optical density.
mD1.1 and mD1.2 potently neutralized HIV-1 and sensitized the virus for neutralization by CD4i antibodies.
To determine the potency and breadth of HIV-1 neutralization by the D1 mutants, we used viruses pseudotyped with Envs from R5, X4, and R5X4 HIV-1 isolates representing clades A, B, C, D, and E. Of the 13 isolates tested, 8 were better neutralized (>2-fold) by mD1.1 and mD1.2 than by D1D2, 4 (Bal, JRFL, IIIB, and NL4-3) were neutralized with about the same potency, and only 1 (GXC-44) showed reduced sensitivity to the D1 mutants (Table 1). The D1 mutants had on average 2-fold-lower arithmetic and geometric means of 50% inhibitory concentrations (IC50s) and IC90s than D1D2. The mutants were also more potent than Fab b12, which neutralizes mainly clade B isolates. IgG1 m102.4 (42), a control antibody specific for Nipah and Hendra viruses, did not inhibit any of the viruses.
Table 1.
mD1.1 and mD1.2 potently inhibit infection of HIV-1 pseudotyped with Envs from different clades
| Virus | Clade | Tropism | IgG1 m102.4 |
Fab b12 |
D1D2 |
mD1.1 |
mD1.2 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50a | IC90b | IC50 | IC90 | IC50 | IC90 | IC50 | IC90 | IC50 | IC90 | |||
| 92UG037.8 | A | R5 | −c | − | − | − | 15 ± 2.1 | 63 ± 9.5 | 6.5 ± 0.4 | 38 ± 2.0 | 9.8 ± 3.1 | 40 ± 2.5 |
| Bal | B | R5 | − | − | 4.0 ± 0.2 | 29 ± 1.4 | 2.4 ± 0.7 | 17 ± 4.9 | 2.3 ± 0.9 | 13 ± 1.7 | 1.6 ± 0.2 | 11 ± 0.6 |
| JRFL | B | R5 | − | − | 1.4 ± 0.1 | 13 ± 2.7 | 14 ± 0.8 | 111 ± 7.5 | 12 ± 1.6 | 85 ± 6.4 | 15 ± 2.0 | 103 ± 5.6 |
| JRCSF | B | R5 | − | − | 30 ± 2.2 | 144 ± 23 | 41 ± 2.9 | 160 ± 8.5 | 17 ± 1.3 | 72 ± 5.0 | 11 ± 0.3 | 65 ± 1.4 |
| R2 | B | R5 | − | − | 720 ± 55 | >1,667 | 3.3 ± 0.9 | 45 ± 7.0 | 1.0 ± 0.9 | 8.0 ± 3.5 | 0.6 ± 0.4 | 7.9 ± 1.5 |
| AD8 | B | R5 | − | − | 73 ± 11 | 280 ± 44 | 76 ± 9.1 | 150 ± 22 | 28 ± 4.5 | 136 ± 19 | 30 ± 1.7 | 125 ± 7.6 |
| 92HT | B | R5X4 | − | − | 10 ± 0.6 | 610 ± 39 | 6.0 ± 0.4 | 117 ± 10 | 1.1 ± 0.2 | 12 ± 3.4 | 1.8 ± 0.3 | 32 ± 8.5 |
| IIIB | B | X4 | − | − | <0.2 | 1.1 ± 0.2 | 0.3 ± 0.1 | 1.3 ± 0.2 | <0.2 | 1.1 ± 0.2 | <0.2 | 0.9 ± 0.3 |
| NL4-3 | B | X4 | − | − | 1.2 ± 0.4 | 30 ± 5.8 | <0.2 | 4.5 ± 0.7 | 0.4 ± 0.1 | 5.0 ± 0.9 | 0.3 ± 0.1 | 5.3 ± 1.6 |
| GXC-44 | C | R5 | − | − | − | − | 27 ± 0.3 | 1,000 ± 68 | 155 ± 17 | >1,667 | 50 ± 0.9 | >1,667 |
| Z2Z6 | D | R5 | − | − | 145 ± 18 | >1,667 | 72 ± 5.6 | >1,667 | 12 ± 0.9 | 660 ± 34 | 60 ± 7.7 | 783 ± 15 |
| GXE | E | R5 | − | − | 630 ± 45 | >1,667 | 154 ± 8.9 | 1,080 ± 46 | 54 ± 3.2 | 280 ± 6.3 | 44 ± 1.5 | 150 ± 17 |
| CM243 | E | R5 | − | − | − | − | 43 ± 2.5 | 670 ± 50 | 3.6 ± 0.2 | 150 ± 9.2 | 18 ± 2.1 | 161 ± 4.0 |
| Arithmetic meand | − | − | 161 | 711 | 35 | 417 | 23 | 266 | 19 | 268 | ||
| Geometric meand | − | − | 16 | 138 | 10 | 102 | 5.0 | 50 | 5.6 | 51 | ||
IC50, antibody concentration (nM) resulting in 50% inhibition of virus infection.
IC90, antibody concentration (nM) resulting in 90% inhibition of virus infection.
−, no significant neutralization at the highest antibody concentration (2,000 nM) tested.
Arithmetic and geometric means were calculated for sCD4 constructs and all viruses, including those with values of <0.2 nM, which were assigned a value of 0.1, and those with values of >1,667 nM, which were assigned a value of 2,000. The means for Fab b12 were calculated based on the values with 10 isolates that were significantly neutralized.
Synergistic effects of a combination of sCD4 and CD4i antibodies on HIV-1 neutralization have been described previously (12). The major mechanism of action is that sCD4 enhances the exposure of the antibody epitopes, and therefore, the antibodies can better bind the Envs. To find out whether there is synergy between the D1 mutants and CD4i antibodies, we preincubated Bal with m36h1Fc in the presence of low concentrations of mD1.1, mD1.2, or D1D2. Both m36h1Fc at a concentration of 1,000 nM and IgG1 m102.4 in combination with 2 nM mD1.1 or mD1.2 exhibited very low or no neutralizing activity (see Fig. S3 in the supplemental material). As expected, preincubation of the virus with both m36h1Fc and the D1 mutants resulted in a dramatic increase in neutralization.
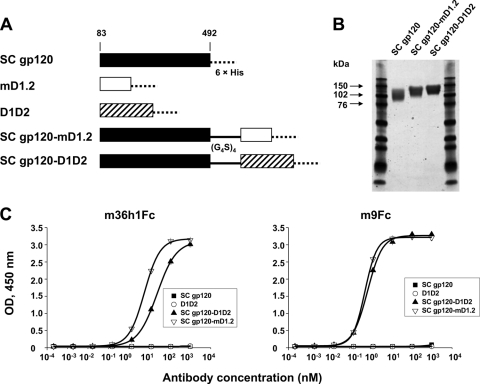
Generation and characterization of a gp120-mD1.2 fusion protein as a candidate vaccine immunogen.
gp120-sCD4 fusion proteins are potentially useful as vaccine immunogens because of the highly conserved neutralizing epitopes on gp120 exposed in the CD4-bound state (11). To assess the degree to which the D1 mutants could stabilize gp120 in this state, we made gp120SC-mD1.2 and two control proteins, gp120SC-D1D2 and gp120SC (Fig. 5A), and measured their binding to CD4i antibodies. The three proteins were expressed in 293 freestyle cells and purified from the cell culture supernatants with yields of ∼2.1, 2.4, and 1.7 mgl−1, respectively. They ran on a reducing SDS-PAGE as relatively broad bands due to glycosylation (Fig. 5B). Notably, m36h1Fc bound gp120SC-mD1.2 more strongly (6-fold) than gp120SC-D1D2; another CD4i antibody, m9Fc, also bound gp120SC-mD1.2 slightly better than gp120SC-D1D2 (Fig. 5C). These results suggest that mD1.2 could induce and stabilize structural rearrangements of gp120 more efficiently than D1D2.
Fig. 5.
Generation and binding to CD4i antibodies of gp120-mD1.2 fusion proteins. (A) Schematic representation of gp120SC-based constructs. The gp120SC sequence used in this study ranges from amino acid residues 83 to 492. 6×His denotes a hexahistidine tag, and (G4S)3 represents a polypeptide linker composed of three repeats of the G4S motif. (B) Reducing SDS-PAGE of the purified proteins. (C) ELISA binding of the purified proteins to two CD4i antibodies, m36h1Fc and m9Fc. gp120SC, gp120SC-mD1.2, gp120SC-D1D2, and D1D2 were used to coat 96-well plates at 2-μg/ml concentration. Binding of the CD4i antibodies was measured as described in Materials and Methods.
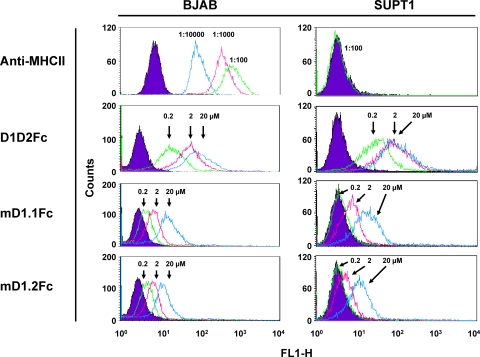
Binding of mD1.1 and mD1.2 to human blood cell lines.
To estimate non-HIV-specific interactions of the D1 mutants in vivo, we measured FACS binding of mD1.1, mD1.2, and D1D2 as Fc fusion proteins to a human B-cell line, BJAB, and a human T-cell line, SUPT1. According to a previous study (3), BJAB cells expressed high-level MHCII, whereas MHCII was not detectable on the surfaces of SUPT1 cells, which was further confirmed in our study with a specific mouse anti-human MHCII monoclonal antibody (Fig. 6). Interestingly, D1D2Fc bound to SUPT1 cells as well as to BJAB cells, suggesting multitarget and/or nonspecific interactions of CD4. mD1.1Fc and mD1.2Fc also bound to both cell lines, but a great decrease in binding strength (>100-fold) was observed. Notably, at 200 nM, which is a similar early antibody concentration in human serum after administration of therapeutic antibodies (a 50-kg person with 4 liters of blood receiving a dose of 3 mg antibodies per kg body weight), the D1-Fc constructs did not interact with SUPT1 cells at all and very weakly with BJAB cells, whereas D1D2Fc still bound strongly.
Fig. 6.
FACS binding of the Fc fusion proteins of mD1.1 and mD1.2 to human blood cell lines. The diagrams for reference cells are in purple. In the anti-MHCII group, the reference cells were not incubated with FITC-conjugated monoclonal mouse anti-human HLA-DR, while the tested cells were mixed with the antibody at different dilutions. In all other groups, the reference cells were not incubated with sCD4-Fc fusion proteins but with the same concentration (1:200 dilution) of Alexa Fluor 488-conjugated mouse anti-human IgG1 (Fc specific) as for the tested cells. Protein concentrations, antibody dilutions, and their corresponding diagrams are indicated by arrows.
mD1.1 and mD1.2 are significantly more soluble than D1D2.
Protein solubility in PBS (pH 7.4) was determined by the ultrafiltration method (40). mD1.1 and mD1.2 were concentrated to 135.2 and 92.6 mg/ml, respectively, without visible precipitation after high-speed centrifugation. Higher concentrations were not tested because of the large amount of protein required. In contrast, precipitation was observed with D1D2, and its concentration in the supernatant after centrifugation was 49.9 mg/ml. The supernatants of the three samples were stored at 4°C for 5 days, but no additional precipitation was observed, suggesting that they remained soluble at those concentrations and under those conditions.
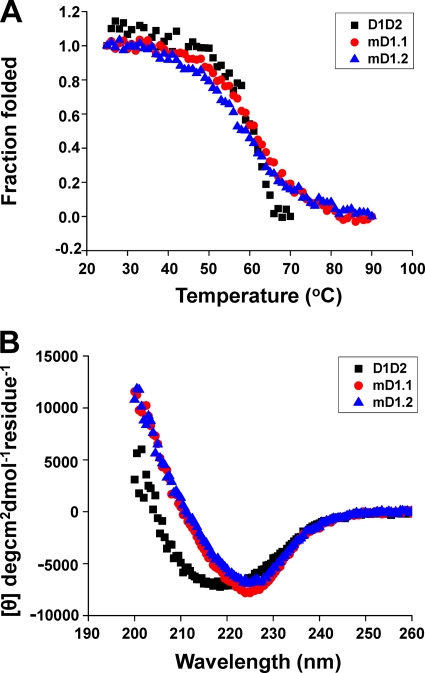
Preservation of mD1.1 and mD1.2 stability and secondary structure.
The thermal stability and secondary structure of mD1.1 and mD1.2 were determined by CD spectroscopy. D1D2 unfolding was observed to begin at 46°C, and the protein was completely unfolded at 67°C, with a temperature of 50% unfolding (midpoint temperature [Tm]) of 58.5°C (Fig. 7A). The measurement was terminated at 70°C, where D1D2 aggregated. A relatively early start of unfolding was also observed with mD1.1 and mD1.2, but about 25% of both remained folded at 67°C and their unfolding was complete at 82°C. The Tms for mD1.1 and mD1.2 were 58.3 and 55.1°C, respectively, which were comparable to that of D1D2. The CD spectra of mD1.1 and mD1.2 were similar to that of D1D2, although there was a shift, suggesting that the D1 mutants still consisted primarily of β strands at 25°C (Fig. 7B).
Fig. 7.
Thermally induced unfolding (A) and secondary structure (B) of mD1.1 and mD1.2 measured by CD. The fraction folded (ff) of the proteins was calculated as follows: ff = ([θ] − [θM])/([θT] − [θM]). [θT] and [θM] are the mean residue ellipticities at 216 nm of the folded state at 25°C and the unfolded state of 90°C (for the D1 mutants) or 70°C (for D1D2). The Tm value from CD was determined by the first derivative [d(fraction folded)/dT] with respect to temperature (T).
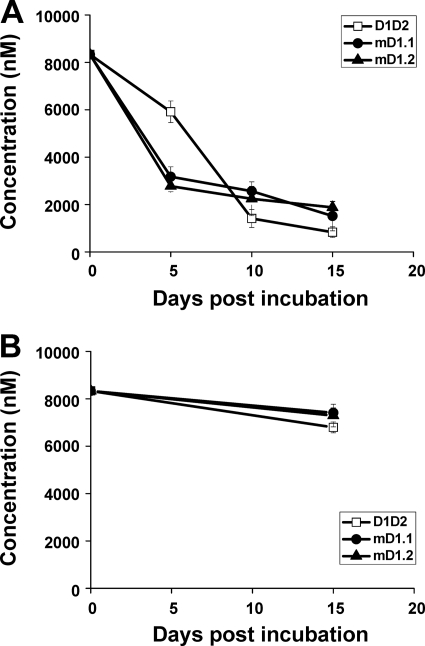
The proteins were further assessed for sensitivity to trypsin digestion and degradation by human serum at 37°C. After 30 min of incubation with trypsin, the majority of D1D2 was digested, while a large percentage of the D1 mutants remained intact and a significant portion of the proteins survived 60-min digestion (see Fig. S4 in the supplemental material). With human serum, D1D2 was degraded slowly within the first 5 days of incubation and then quickly thereafter until 15 days postincubation (p.i.), when less than 1,000 nM protein was left (Fig. 8A). In contrast, the D1 mutants disappeared more rapidly within the first 5 days, but the degradation was slower in the 10 days thereafter, and more than 1,000 nM protein was detected 15 days p.i. In all cases, the degradation was specific to trypsin or human serum, because incubation of the proteins in PBS alone at 37°C for 15 days caused no significant loss (Fig. 8B).
Fig. 8.
Serum stability of mD1.1 and mD1.2. Functional proteins following treatment with (A) or without (B) serum at 37°C for different periods were quantified by using ELISA and standard curves made with untreated proteins, as described in Materials and Methods.
DISCUSSION
D1 and D2 of CD4 pack against each other with a large hydrophobic interface (21, 30). Although all residues directly interacting with gp120 or MHCII are in D1 on a surface opposite to its interface with D2, mutations in D2 and other domains have been shown to affect binding of CD4 to HIV-1 gp120 or MHCII (27), suggesting that there are strong interdomain interactions and that the interactions are important for CD4 functions. As a result, isolated D1 is not expressed in a soluble and functional form. In a previous study (34), five hydrophobic residues identified by crystallographic analysis to significantly interact with D2 were mutated to either alanine or threonine residues. The mutant protein was purified from a sonicated supernatant of E. coli; however, the resulting purified protein was stable only at low pH (4.0) and had an affinity with HIV-1 gp120 severalfold lower than that of D1D2 or T4. The results of size exclusion chromatography suggest that a significant portion of the protein may be improperly folded. These drawbacks may limit the usefulness of the protein as a research reagent, candidate therapeutic, or component of vaccine immunogens. In another study (25), CD4 mimics (CD4M32 and CD4M33) containing 27 amino acid residues were generated by transferring 9 residues of the human CD4 CDR2 loop to a scorpion toxin scaffold and introducing nonstandard (nonproteinogenic) amino acid residues; they exhibited potent antiviral activity, with IC50s in the nanomolar range, comparable to that of human sCD4. Unfortunately, the usefulness of CDM32 and CDM33 as candidate therapeutics or components of vaccine immunogens is limited due to the presence of nonstandard amino acid residues and the use of a nonhuman scaffold with high likelihood of immunogenicity in humans.
In this study, we combined several strategies, including structure-based library design, construction, biopanning, and screening. The selected D1 mutants were highly soluble, expressible in the E. coli periplasm fraction, monomeric, and stable under physiological conditions. They showed a great increase in affinity with HIV-1 gp120 over that of D1D2. While the randomization strategies used in this study create tremendous diversity, sequence analysis of 19 selected D1 mutants showed a large percentage of them with a preference for hydrophobic residues in positions 5 and 96, suggesting that nonhydrophobic residues at these positions may potentially prevent the folding of stable and functional D1 structures. In contrast, positions 76 and 98 were more tolerant of substitution. Based on these results, it is conceivable that other domains, especially D2, could influence the interactions and biological functions of CD4 by interfering with D1 structure.
A previous study (19) showed that increasing antibody molecular size by PEGylation resulted in a marked reduction in affinity observed in kinetic Biacore measurements. This effect was independent of antibody formats and traced to lower association rate constants caused by enlarged antibody hydrodynamic size without significantly perturbing the dissociation rate constants of the antibodies. To better understand the molecular mechanism for the observed increase in affinity of the D1 mutants, we fused mD1.1 to the isolated human IgG1 CH2 domain to mimic D1D2 (Fig. 3A and B). CH2 has a calculated molecular mass only slightly larger than that of D2, and the two independently folded moieties in the mD1.1-CH2 fusion proteins may have a higher level of flexibility than D1D2, where D1 and D2 are tightly associated. We also applied a highly flexible polypeptide linker to one of the fusion proteins generated in order to see whether increased flexibility would have additional effects on binding kinetics. In an SPR-based analysis, the fusion proteins exhibited a dramatic decrease (80-fold) in association rate constants while showing dissociation rate constants comparable to that of mD1.1 (Fig. 3C), in agreement with the previous study (19). The fusion protein mD1.1L3CH2, linked by three repeats of the G4S motif, had even slower association than mD1.1CH2, which contains no linker, most likely due to a further increase in the molecular size and/or flexibility of both moieties. The fusion proteins demonstrated similar kinetics with D1D2, although the former associated more slowly and the latter dissociated more rapidly. These results suggest that most, if not all, of the improved kinetics of the D1 mutants should be attributed to their decreased molecular size (minimized hydrodynamic size).
The D1 mutants have substantially higher affinity for HIV-1 gp120 than D1D2 but neutralized pseudoviruses to a similar degree. A likely explanation is that under the conditions used in our assay, the time allowed for the inhibitors to react with the viruses before exposure to the target cells is sufficient, so that neutralization kinetics is mainly affected by the events occurring thereafter, such as the dissociation of the inhibitors from the viruses.
The D1 mutants described in this study are potentially useful in several respects. First, they could be used as a tool to elucidate the mechanisms of CD4-mediated HIV-1 entry and immune responses. For example, previous studies indicated that CD4 binds to MHCII as a dimer formed either by swapping its D2 domain (24) or through D4 (39). Here, we propose a strategy to explore such possibilities, where the D1 mutants, as a single domain or D1-X-D3-D4 fusion proteins (where X could be any independently folded single domain other than D2 but comparable in molecular size), are displayed on CD4− T-cell surfaces and the activation of these T cells by APCs is studied. This idea appears practical, as in a previous study (24), murine CD4 mutants were successfully presented on human CD4− Jurkat 8.2 T cells and impaired T-cell activation was observed.
Second, the D1 mutants could be superior to D1D2 as a component of HIV-1 gp120-based vaccine immunogens. Binding of CD4 induces extensive conformational changes in gp120, leading to formation and/or exposure of highly conserved structures. These structures are functionally important, and therefore, gp120-sCD4 complexes are potential immunogens capable of eliciting broadly neutralizing antibodies that inhibit viruses from different genetic subtypes. A recent study (11) showed that the antibodies to CD4i epitopes elicited by a gp120-D1D2 fusion protein accounted for the control of SHIV challenge in macaques. When the gp120-D1D2 regimen was evaluated in animal models, however, the resulting broadly neutralizing sera were found to be primarily due to anti-CD4 antibodies (36). It would be of interest to use the smallest version of sCD4, D1, which could possibly minimize the immune responses against CD4 by the gp120-sCD4 combinations. Moreover, the higher affinity of the D1 mutants generated in this study led to more efficient exposure of CD4i epitopes (Fig. 5C); their much weaker interactions with the human blood cell lines (Fig. 6) could generally result in decreased toxicities in humans. One could speculate, therefore, that gp120-D1 fusion proteins could elicit gp120-specific immune responses more efficiently than gp120-D1D2, providing more insights into how the gp120 structures exposed by CD4 binding could elicit neutralizing responses. One should note that the D1 mutants could also be more likely than D1D2 to be immunogenic because of the mutations and the exposed interface with D2. Their possible immunogenicity needs to be assessed in human clinical trials.
Third, the D1 mutants could be more useful than D1D2 to generate candidate HIV-1 therapeutics, not only because of higher potency, but also due to smaller size. The antiviral activities of monomeric (18, 32, 33), dimeric (22, 26), and tetrameric (1, 14) human sCD4 as Fc fusion proteins have been evaluated in animal models and human clinical trials. They were well tolerated by patients with no significant clinical or immunologic toxicities and exhibited significant inhibitory activities, such as reductions in plasma viremia and virus titer. However, their activities were transient and not consistent; viral rebound rapidly occurred, and no significant efficacy was observed in some patients. While the in vitro antiviral potency of the reagents needs to be improved, the short serum half-life problem with monomeric sCD4 in vivo should be addressed, and in contrast, large fusion proteins with Fc, which could have much longer half-lives, may have poor biodistribution in the densely packed lymphoid environment where HIV-1 mostly replicates and spreads. To achieve avidity effects, higher-order oligomers of sCD4 had been previously created. A dodecameric sCD4 molecule generated by placing D1D2 into an IgG-IgA tailpiece context showed more than a 1,000-fold increase in neutralization of HIV-1 primary isolates compared to monomeric sCD4 (2, 20). In other studies (5, 38), sCD4 was fused with CD4i antibodies for synergistic and avidity effects and further linked to Fc for an increase in serum half-life. The resulting chimeric fusion proteins were exceptionally potent and broadly neutralizing. In both cases, however, their large molecular sizes (>650 kDa for the dodecamer and >150 kDa for the CD4i antibody-Fc fusion proteins of sCD4) may create difficulties in producing large quantities of the proteins for clinical use and penetrating into solid tissues in vivo. Therefore, the use of single-domain sCD4 may aid in the generation of smaller fusion proteins that exhibit favorable biodistribution and long half-life in circulation, as well as conjugates with toxic small-molecule drugs in which a short half-life is desirable to reduce toxicity. Another potential application of D1 would be as an addition to the reagents of detection of HIV-1 infection because of its high cross-reactivity and picomolar affinity, which could confer favorable diagnostic sensitivity and accuracy.
Although sCD4 could induce immune responses in humans, there are no indications of such responses or other effects on the immune system. A previous study (4) showed that monomeric sCD4 did not inhibit immune functions in monkeys, suggesting it was unlikely to be immunosuppressive in humans. However, the experiment needs to be repeated in humans with higher-order oligomers, because oligomerization of CD4 is required for interaction with MHCII or, presumably, other targets, and the highly potent sCD4 derivatives described above are exclusively multimeric (1, 14, 22, 26). Although sCD4-Fc fusion proteins have been safely administered to a number of humans in several clinical trials, it was uncertain whether long-term treatment would be safe. Therefore, safety will continue to be one of the major concerns for the use of sCD4 in humans, and strategies to bias sCD4 specificity toward HIV-1 should be strongly pursued. Our results showed that mD1.1Fc and mD1.2Fc exhibited much lower binding to two human blood cell lines than D1D2Fc, although the possibility that they could retain interactions with MHCII was not ruled out. The weaker binding of the D1-Fc constructs is most likely due to slightly altered D1 structures and/or smaller surface area of nonspecific interactions in the absence of D2. An alternative explanation is that D1D2Fc could aggregate more easily than the D1-Fc constructs, with more efficient recognition of the multiple Fc domains of D1D2Fc aggregates by the secondary antibody. Therefore, the D1 mutants selected in this study could lead to safer and more effective sCD4-based HIV-1 inhibitors, although in vivo experimental data are needed to prove it.
Supplementary Material
ACKNOWLEDGMENTS
We thank Barton F. Haynes and Anu Puri for providing reagents and cells and Emily Streaker for helpful comments.
This project was supported by the Intramural AIDS Targeted Antiviral Program of the National Institutes of Health (NIH); by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research; and by the Gates Foundation (D.S.D.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Allaway G. P., et al. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retroviruses 11:533–539 [DOI] [PubMed] [Google Scholar]
- 2. Arthos J., et al. 2002. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: implications for therapeutic and vaccine strategies. J. Biol. Chem. 277:11456–11464 [DOI] [PubMed] [Google Scholar]
- 3. Bosshart H., Jarrett R. F. 1998. Deficient major histocompatibility complex class II antigen presentation in a subset of Hodgkin's disease tumor cells. Blood 92:2252–2259 [PubMed] [Google Scholar]
- 4. Bugelski P. J., Thiem P. A., Truneh A., Morgan D. G. 1991. Recombinant human soluble CD4 does not inhibit immune function in cynomolgus monkeys. Toxicol. Pathol. 19:580–588 [DOI] [PubMed] [Google Scholar]
- 5. Chen W., Xiao X., Wang Y., Zhu Z., Dimitrov D. S. 2010. Bifunctional fusion proteins of the human engineered antibody domain m36 with human soluble CD4 are potent inhibitors of diverse HIV-1 isolates. Antiviral Res. 88:107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen W., Zhu Z., Feng Y., Dimitrov D. S. 2008. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc. Natl. Acad. Sci. U. S. A. 105:17121–17126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen W., Zhu Z., Feng Y., Dimitrov D. S. 2010. A large human domain antibody library combining heavy and light chain CDR3 diversity. Mol. Immunol. 47:912–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen W., Zhu Z., Feng Y., Xiao X., Dimitrov D. S. 2008. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J. Mol. Biol. 382:779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dalgleish A. G., et al. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767 [DOI] [PubMed] [Google Scholar]
- 10. Deen K. C., et al. 1988. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature 331:82–84 [DOI] [PubMed] [Google Scholar]
- 11. DeVico A., et al. 2007. Antibodies to CD4-induced sites in HIV gp120 correlate with the control of SHIV challenge in macaques vaccinated with subunit immunogens. Proc. Natl. Acad. Sci. U. S. A. 104:17477–17482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dey B., Del Castillo C. S., Berger E. A. 2003. Neutralization of human immunodeficiency virus type 1 by sCD4-17b, a single-chain chimeric protein, based on sequential interaction of gp120 with CD4 and coreceptor. J. Virol. 77:2859–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng Y., et al. 2006. Novel human monoclonal antibodies to insulin-like growth factor (IGF)-II that potently inhibit the IGF receptor type I signal transduction function. Mol. Cancer Ther. 5:114–120 [DOI] [PubMed] [Google Scholar]
- 14. Fletcher C. V., et al. 2007. Nonlinear pharmacokinetics of high-dose recombinant fusion protein CD4-IgG2 (PRO 542) observed in HIV-1-infected children. J. Allergy Clin. Immunol. 119:747–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garlick R. L., Kirschner R. J., Eckenrode F. M., Tarpley W. G., Tomich C. S. 1990. Escherichia coli expression, purification, and biological activity of a truncated soluble CD4. AIDS Res. Hum. Retroviruses 6:465–479 [DOI] [PubMed] [Google Scholar]
- 16. Germain R. N. 1997. T-cell signaling: the importance of receptor clustering. Curr. Biol. 7:R640–E644 [DOI] [PubMed] [Google Scholar]
- 17. Gong R., et al. 2009. Engineered human antibody constant domains with increased stability. J. Biol. Chem. 284:14203–14210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kahn J. O., et al. 1990. The safety and pharmacokinetics of recombinant soluble CD4 (rCD4) in subjects with the AIDS and AIDS-related complex. A phase 1 study. Ann. Intern. Med. 112:254–261 [DOI] [PubMed] [Google Scholar]
- 19. Kubetzko S., Sarkar C. A., Pluckthun A. 2005. Protein PEGylation decreases observed target association rates via a dual blocking mechanism. Mol. Pharmacol. 68:1439–1454 [DOI] [PubMed] [Google Scholar]
- 20. Kwong P. D., et al. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 21. Kwong P. D., et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langner K. D., et al. 1993. Antiviral effects of different CD4-immunoglobulin constructs against HIV-1 and SIV: immunological characterization, pharmacokinetic data and in vivo experiments. Arch. Virol. 130:157–170 [DOI] [PubMed] [Google Scholar]
- 23. Liao H. X., et al. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353:268–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maekawa A., Schmidt B., Fazekas de St. Groth B., Sanejouand Y. H., Hogg P. J. 2006. Evidence for a domain-swapped CD4 dimer as the coreceptor for binding to class II MHC. J. Immunol. 176:6873–6878 [DOI] [PubMed] [Google Scholar]
- 25. Martin L., et al. 2003. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat. Biotechnol. 21:71–76 [DOI] [PubMed] [Google Scholar]
- 26. Meng T. C., et al. 1995. Combination therapy with recombinant human soluble CD4-immunoglobulin G and zidovudine in patients with HIV infection: a phase I study. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 8:152–160 [PubMed] [Google Scholar]
- 27. Moebius U., Pallai P., Harrison S. C., Reinherz E. L. 1993. Delineation of an extended surface contact area on human CD4 involved in class II major histocompatibility complex binding. Proc. Natl. Acad. Sci. U. S. A. 90:8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prabakaran P., et al. 2008. Structure of an isolated unglycosylated antibody C(H)2 domain. Acta Crystallogr. D Biol. Crystallogr. 64:1062–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roben P., et al. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ryu S. E., et al. 1990. Crystal structure of an HIV-binding recombinant fragment of human CD4. Nature 348:419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sakihama T., Smolyar A., Reinherz E. L. 1995. Oligomerization of CD4 is required for stable binding to class II major histocompatibility complex proteins but not for interaction with human immunodeficiency virus gp120. Proc. Natl. Acad. Sci. U. S. A. 92:6444–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schacker T., et al. 1995. Phase I study of high-dose, intravenous rsCD4 in subjects with advanced HIV-1 infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:145–152 [PubMed] [Google Scholar]
- 33. Schooley R. T., et al. 1990. Recombinant soluble CD4 therapy in patients with the AIDS and AIDS-related complex. A phase I-II escalating dosage trial. Ann. Intern. Med. 112:247–253 [DOI] [PubMed] [Google Scholar]
- 34. Sharma D., et al. 2005. Protein minimization of the gp120 binding region of human CD4. Biochemistry 44:16192–16202 [DOI] [PubMed] [Google Scholar]
- 35. Traunecker A., Luke W., Karjalainen K. 1988. Soluble CD4 molecules neutralize human immunodeficiency virus type 1. Nature 331:84–86 [DOI] [PubMed] [Google Scholar]
- 36. Varadarajan R., et al. 2005. Characterization of gp120 and its single-chain derivatives, gp120-CD4D12 and gp120-M9: implications for targeting the CD4i epitope in human immunodeficiency virus vaccine design. J. Virol. 79:1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J. H., et al. 2001. Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule. Proc. Natl. Acad. Sci. U. S. A. 98:10799–10804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. West A. P., Jr., et al. 2010. Evaluation of CD4-CD4i antibody architectures yields potent, broadly cross-reactive anti-human immunodeficiency virus reagents. J. Virol. 84:261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu H., Kwong P. D., Hendrickson W. A. 1997. Dimeric association and segmental variability in the structure of human CD4. Nature 387:527–530 [DOI] [PubMed] [Google Scholar]
- 40. Wu S. J., et al. 2010. Structure-based engineering of a monoclonal antibody for improved solubility. Protein Eng. Des. Sel. 23:643–651 [DOI] [PubMed] [Google Scholar]
- 41. Zhang M. Y., et al. 2004. Improved breadth and potency of an HIV-1-neutralizing human single-chain antibody by random mutagenesis and sequential antigen panning. J. Mol. Biol. 335:209–219 [DOI] [PubMed] [Google Scholar]
- 42. Zhu Z., et al. 2008. Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. J. Infect. Dis. 197:846–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.