Abstract
The UL16 tegument protein of herpes simplex virus (HSV) is conserved throughout all of the herpesvirus families. Previous studies have shown that the binding of HSV to heparan sulfate molecules on the host cell triggers the release of UL16 from the capsid, but the mechanism by which the signal is sent from the virion surface into the tegument is unknown. Here, we report that a glutathione S-transferase chimera bearing the cytoplasmic tail of viral glycoprotein E (gE) is capable of binding to UL16 in lysates of eukaryotic cells or purified from bacteria. Moreover, mass spectrometry studies of native-UL16 complexes purified from infected cells also revealed the presence of gE. Proof that UL16-gE can interact within cells required the fortuitous discovery of a mutant possessing only the first 155 residues of UL16. Confocal microscopy of cotransfected cells revealed that this mutant colocalized with gE in the cytoplasm, whereas it was found throughout the cytoplasm and nucleus when expressed alone. In contrast, the full-length UL16 molecule was very poorly capable of finding gE. Moreover, membrane flotation assays showed that UL16(1-155) was able to float to the top of sucrose step gradients when coexpressed with gE, whereas full-length UL16 was not. Thus, the discovery of the UL16(1-155) mutant confirmed the specific in vitro interaction with gE and provides evidence that a binding domain at the N terminus of UL16 may be controlled by a regulatory domain within the C terminus. These findings suggest the possibility that the UL16-gE interaction may play roles in the tegument signaling mechanism, virus budding, and the gE-mediated mechanism of cell-to-cell spread.
INTRODUCTION
Open reading frame number 16 within the unique-long segment of the herpes simplex virus (HSV) genome encodes a 373-amino-acid (aa) protein whose function is almost entirely unknown, as is the case for the homologs found in all families (alpha, beta, and gamma) of herpesviruses (25, 36, 48, 50, 56, 60, 78). The UL16 protein is expressed late in the infection and initially accumulates in the nucleus, but at later times is found primarily in the cytoplasm (48, 56). When virions bud into cytoplasmic membranes, UL16 is packaged into the tegument—the layer of the virion situated between the capsid and the viral envelope (50, 51). Mutants that do not express UL16 are viable but produce only ∼10% the number of infectious virions compared to the wild type in cell cultures (3). Thus, this protein plays an augmenting role in the replication cycle; one that is highly conserved.
Previous studies have suggested two potential functions for UL16. First, it may provide one of the bridging functions that link capsids to membranes during the envelopment process within the cytoplasm. In support of this hypothesis, a population of UL16 molecules has been found that is associated with cytoplasmic capsids (48), and there is a strong interaction between UL16 and UL11 (43, 81), a small tegument protein that is peripherally bound to membranes via two covalently attached fatty acids, myristate and palmitate (6, 42). Like UL16, UL11 is needed for efficient envelopment and is conserved among all of the herpesviruses (4, 9, 23, 36, 39, 64).
The second potential function for UL16 comes from studies of extracellular virions. These showed that binding of the virus to attachment receptors (heparan sulfate), either on the surface of host cells or immobilized on agarose beads, causes a signal to be sent into the tegument to trigger the release UL16 from the capsid (49). The purpose of this rearrangement in the tegument is unknown, but it could be important for uncoating of the capsid and/or activation of the fusion apparatus prior to virus entry. In any case, it is clear from studies of UL16 that the assembly of the tegument creates machinery with moving parts that respond to signals detected on the outside of the virion.
To understand how the tegument machine is assembled and activated, a thorough understanding is needed of the network of interactions in which UL16 operates. Prior to the experiments described here, three interactions were known. One is the interaction with UL11, and within that protein, UL16 specifically recognizes a cluster of acidic residues (43, 81). Attempts to map the part of UL16 involved in this interaction were not successful, but modification of its free cysteines with N-ethylmaleimide (NEM) blocked binding to UL11 (81). A second protein with which UL16 interacts is UL21, a tegument protein that is conserved among alphaherpesviruses and is also bound to capsids (2, 15, 28, 72). This interaction involves a domain in the second half of UL21 (28), but again, the part of UL16 that participates in binding remains unknown. The site must be distinct from that used for UL11 because binding to UL21 is not blocked with NEM (28). Evidence for a third protein in the interaction network comes from the discovery that UL21 is not required for binding of UL16 to capsids (47). The unidentified protein must be a part of the capsid structure or another tegument protein that is bound to capsids. Although the UL16-capsid interaction is destabilized upon binding of the virion to its attachment receptors (49), other changes in the network remain to be elucidated.
To receive the signal from outside the virion, the UL16-interaction network must interface in some manner with viral glycoproteins on the surface. The only clear linkage to the membrane is via UL11; however, because that protein does not span the membrane, another network connection with a viral glycoprotein must exist. There is one report of a possible interaction between UL11 and the tails of glycoprotein D (gD) and gE (22), but the data are limited and have been suggested to be the result of nonspecific binding (32). The companion paper (26a) shows that the gE-UL11 interaction indeed occurs and is biologically significant; however, neither gE nor gD binds to attachment receptors. That function is provided by gB and gC (10, 66). There is some evidence that gB and gC reside in a complex with gD and gE on the surface of the virion (27, 30), and thus signals might be sent between glycoproteins prior to being routed into the tegument. Clearly, a much better understanding of tegument-glycoprotein “wiring” is needed.
The experiments described below began with the question of whether UL16 interacts directly with the cytoplasmic tail of gB, one of the glycoproteins known to bind attachment receptors (66). The data quickly ruled against that hypothesis. Instead, converging lines of evidence suggested that UL16 interacts with gE, a glycoprotein that has long been known to be required for the spread of HSV in a cell-to-cell manner (33, 45). Rigorous proof that the interaction can occur within cells required the fortuitous discovery of a particular UL16 mutant, one that also provides evidence that the interaction with gE is regulated.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Vero and human melanoma (A7) cell lines were maintained in Dulbecco modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS), penicillin (65 μg/ml), and streptomycin (131 μg/ml). The KOS strain of HSV-1 (65) was used for these studies, along with a recombinant that expresses UL16-TAP (see below). In all cases, infected cells were grown in DMEM supplemented with 2% FBS, 25 mM HEPES buffer, glutamine (0.3 μg/ml), penicillin, and streptomycin. A recombinant baculovirus BV.UL16-GFP and Sf21 insect cells were used and maintained as described previously (81). The UL16- and green fluorescent protein (GFP)-specific antibodies produced in rabbits (Cocalico Biologicals, Inc.) were raised against GST-UL16 and His6-GFP antigens, and these have been described previously (6, 43, 48). The His6-GFP-specific antiserum (diluted 1:3,000) recognizes both GFP and the His6 tag, which was fused to the N terminus of GFP and UL16 for purification from Escherichia coli (81). UL16 antibodies used in the coimmunoprecipitation and membrane flotation assays specifically recognize a sequence near the N terminus of UL16 (residues 21 to 32 plus a C-terminal cysteine to enable conjugation to a carrier protein) and were produced in rabbits (Cocalico Biologicals, Inc.) after cross-linking the peptide to purified keyhole limpet hemocyanin. The rabbit polyclonal antibody against VP5 was kindly provided by Richard J. Courtney (Pennsylvania State University). The polyclonal gE antibody (UP1725), kindly provided by Harvey M. Friedman (University of Pennsylvania), was produced in rabbits, using baculovirus-expressed gE aa 24 to 409 as the antigen (40). The monoclonal antibody 3114, which is specific for gE (13, 46) and was used in the immunofluorescence assays, was kindly provided by David C. Johnson (Oregon Health and Science University).
E. coli expression constructs.
A plasmid encoding GST-UL11 was described previously (43). A plasmid encoding the cytoplasmic tail of gE fused to glutathione S-transferase (GST-gEtail) was kindly provided by David C. Johnson and has been described previously (58). For expression of GST-gEtail derivatives, an acidic cluster (DEED/As) and deletion mutants were created by QuikChange site-directed mutagenesis. Codons for four acidic amino acids (two aspartates and two glutamates within the sequence SSDSEGERDQV) in the gE tail were replaced with ones for alanine by using a forward primer (5′-GAC TTG AGC TCG GCC AGC GCG GGA GCA CGC GCC CAG GTC CCG TG-3′) and a primer containing the reverse-complementary sequence. A plasmid encoding the cytoplasmic tail of gB (GST-gBtail) was provided by Richard J. Courtney (Pennsylvania State University). A plasmid encoding the cytoplasmic tail of gD (GST-gDtail) was kindly provided by Duncan W. Wilson (Albert Einstein College of Medicine) (12). All GST fusion proteins were purified from E. coli on glutathione beads according to the standard methods described by the manufacturer (GE Healthcare). A plasmid encoding His6-tagged UL16 was generated previously (81). A clone expressing the first 155 aa of UL16, followed by a frameshift sequence of 70 aa, was a result of a random frameshift mutation in His6-UL16 and is referred to as His6-UL16(FS). The plasmid encoding only first 155 aa of UL16 [referred as His6-UL16(1-155)] was generated by inserting a stop codon immediately after codon 155 in the His6-UL16 construct by QuikChange mutagenesis with the following primers: ATA CGG GCG GCC ACC CCC CCG TAA AGC GGC CGC ACT CGA GCA CC (forward) and the reverse complement of the forward sequence. UL16(1-155)-GFP was created by deletion mutagenesis of pCMV.UL16-GFP (43) with the use of primer 5′-ATA CGG GCG GCC ACC CCC CCG TGG ATC CAC CGG CCG GTC GCC-3′ (forward) and the reverse complement of the forward sequence. A plasmid encoding HSV-1 glycoprotein E (pCMV.gE) was kindly provided by Harvey M. Friedman (University of Pennsylvania).
GST pulldown assay.
To analyze the interaction of UL16 with gE, pCMV. UL16-GFP was transfected into A7 cells by means of the calcium phosphate precipitation method. At 20 h posttransfection, the cells were harvested in NP-40 lysis buffer (0.5% NP-40, 150 mM NaCl, 50 mM Tris-HCl [pH 8.0]) and precleared with glutathione-Sepharose-4B beads for 4 h at room temperature. Samples (2 μl) of the bead-bound GST fusion proteins were added to the precleared lysates, followed by incubation at room temperature for 5 h. The beads were pelleted and washed three times with NP-40 lysis buffer for 10 min each time and mixed with sample buffer, and the bound proteins were separated by SDS-PAGE. The proteins were then transferred to nitrocellulose membranes and analyzed by enhanced chemiluminescence (ECL)-based immunoblotting (GE Healthcare). The same protocol was used to analyze the interaction of GST-gEtail with UL16 produced in Vero cells infected with wild-type HSV or insect cells infected with BV.UL16-GFP, all produced in 60-mm plates with a multiplicity of infection (MOI) of 1. To ascertain whether UL16 can interact directly with the tail of gE, purified proteins were prepared and analyzed as previously described (81). Briefly, the GST-gEtail protein and its corresponding mutants were purified on glutathione beads. His6-UL16 and its corresponding derivatives were purified with nickel beads, washed, eluted with imidazole, and dialyzed against 20 mM Tris-HCl (pH 7.9). The bead-bound GST-fusion proteins were mixed with soluble His6-UL16 proteins in 1 ml of 0.5% NP-40 lysis buffer and incubated at room temperature for 5 h. All proteins bound to the beads were collected by centrifugation, washed, separated by SDS-PAGE, and subjected to immunoblotting to detect the presence of His6-UL16.
NEM treatment.
To determine whether the UL16-gE interaction is prevented when free cysteines are blocked, BV.UL16-GFP or HSV-infected cells were treated with 10 mM NEM to covalently modify free cysteines in UL16 before lysing the cells with NP-40, essentially as described previously (81). For in vitro binding assays, His6-UL16 proteins made in bacteria were purified on nickel beads (as described above), untreated or treated with NEM before elution as described previously (81). Eluted proteins were dialyzed before being added to in vitro binding assays.
Construction of KOS.UL16-TAP.
To construct a recombinant virus that expresses UL16 fused to a TAP tag, a bacterial artificial chromosome (BAC) containing the HSV-1 KOS strain genome was used (KOS BAC, generously provided by David A. Leib [24]). Recombinant HSV-1 clones were made using a galK-positive/negative selection-based recombineering strategy, as described previously (75). Briefly, a galK expression cassette was inserted at the 3′ terminus of the UL16 open reading frame in the KOS BAC. Next, the galK cassette was replaced with a DNA fragment that encodes the TAP tag sequence (pCAGGS. TAP tag C cassette, kindly provided by J. C. de la Torre, Scripps Research Institute). The resulting BAC was then transfected into A7 cells by means of the calcium phosphate precipitation method. After 5 to 6 days, transfected cells were harvested when showing cytopathic effects and used to infect fresh Vero cells to produce a viral stock. Confirmation that the desired virus was obtained was provided by PCR analyses using primers that flank the UL16-TAP-coding sequence (yielding a larger product than untagged UL16) and the failure to express wild-type UL16 (as determined by immunoblotting).
Single-step viral growth.
Plates (60 mm) of Vero cells (3.2 × 106/plate) were infected with the specified viruses at an MOI of 5 at 37°C. After 1 h of incubation, cells were washed with phosphate-buffered saline (PBS) plus FBS and then overlaid with 4 ml of DMEM containing 2% FBS. At the indicated times postinfection, the medium and cells from each plate were harvested and treated as follows. The medium was cleared of cells by centrifugation at 3,000 rpm for 5 min and frozen at −80°C. Cells were scraped into PBS, washed three times with PBS, and freeze-thawed three times to release cell-associated viruses. The virus titer of each sample was measured on Vero cells in standard plaque assays.
Tandem affinity purification and mass spectrometry.
Vero cells infected with KOS.UL16-TAP at an MOI of 5 were lysed in NP-40 buffer at 20 h postinfection. The cell lysates were first incubated with IgG-Sepharose beads (GE Healthcare) overnight at 4°C. The IgG beads were then washed with TEV protease cleavage buffer (0.1% NP-40, 150 mM NaCl, 1 mM dithiothreitol, 10 mM Tris-HCl [pH 8.0]) and incubated with TEV protease (generously provided by John M. Flanagan, Pennsylvania State University) at room temperature for 4 h. The cleavage product (UL16-CBP) and its associated proteins were harvested on calmodulin resin (GE Healthcare) in calmodulin-binding buffer (0.1% NP-40, 150 mM NaCl, 10 mM β-mercaptoethanol, 10 mM Tris-HCl [pH 8.0], 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2). The collected proteins from separate experiments were analyzed with two different approaches in two different laboratories, as follows.
In the first mass spectrometry experiment (conducted at Eastern Virginia Medical School), proteins bound to the calmodulin beads were eluted in sample buffer, resolved by SDS-PAGE, and visualized using SYPRO Ruby protein gel stain according to the manufacturer's instructions (Invitrogen). The major protein bands were excised from SDS-PAGE gels, cut into 1- to 2-mm cubes, washed three times with 500 μl of water, and incubated in 100% acetonitrile for 45 min. The samples were further processed and subjected to mass spectrometry, essentially as described before (28). Sequence analysis was performed with MASCOT (Matrix Sciences, London, United Kingdom) using an indexed viral and human subset database of the nonredundant protein database from National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/).
In the second mass spectrometry experiment (provided as a service of the Pennsylvania State University College of Medicine Mass Spectrometry and Proteomics Core Facility), proteins bound to the calmodulin beads were eluted with 2 mM EGTA, and the entire protein mixture was digested with sequencing-grade trypsin. The resulting peptides were separated in a reversed-phase Nanoflow C18 high-pressure liquid chromatography column, and eluted fractions were automatically spotted onto a stainless steel matrix-assisted laser desorption ionization target plate every 6 s (0.6 μl per spot), for a total of 370 spots. MS spectra were then acquired from each spot (400 laser shots per spot). A plate-wide interpretation was then automatically performed, choosing the highest peak of each observed m/z value for subsequent MS-MS analysis. Sequence analysis was performed using the Paragon and ProGroup algorithm as implemented in the ProteinPilot 2.0 software (MDS Sciex/Applied Biosystems).
Coimmunoprecipitation from HSV-infected cells.
Confluent monolayers of Vero cells were infected with wild-type HSV at an MOI of 10. At 20 h postinfection, cells were scraped into PBS, pelleted by centrifugation at 1,000 × g for 5 min, and resuspended into NP-40 lysis buffer with protease inhibitors (Sigma catalog no. P8340). The nuclei were removed by centrifugation for 5 min at 1,000 × g, and the supernatant was further cleared by spinning at 18,000 × g for 10 min. Lysates were precleared for 1 h with protein A-agarose beads (Roche) at 4°C and subsequently incubated with UL16- or gE-specific antibodies for 1 h. To capture antibody-protein complexes, lysates were incubated with protein A-agarose beads for an additional 3 h or overnight, and the beads were washed thoroughly with NP-40 lysis buffer and resuspended in sample buffer. Bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with UL16- or gE-specific primary antibodies and True-Blot horseradish peroxidase (HRP)-conjugated secondary antibody (eBioscience). True-Blot HRP preferentially binds to native rabbit IgG and thereby reduces background from the denatured heavy and light chains of the primary antibodies used for the immunoprecipitations.
Immunofluorescence and confocal microscopy.
To determine the subcellular localization of UL16-GFP and UL16(1-155)-GFP in the presence or absence of gE, confocal immunofluorescence microscopy was performed. Vero cells were grown in 35-mm dishes on coverslips and transfected with plasmids encoding UL16-GFP, UL16(1-155)-GFP, or gE via Lipofectamine 2000 (Invitrogen). For cotransfections, 1:2 ratios of the UL16 and gE DNAs were used (respectively). After 16 to 20 h posttransfection, the cells were washed three times with PBS and fixed with 3.7% paraformaldehyde in PBS for 10 min. After three 5-min washes with PBS, the cells were permeabilized for 15 min with 0.1% Triton X-100 in blocking buffer (2% bovine serum albumin in PBS). The cells were then incubated in blocking buffer for 30 min, followed by staining with monoclonal antibody against gE (1:4,000) for 1 h. After three washes with PBS, the cells were incubated with goat anti-mouse secondary antibody conjugated with Alexa Fluor 568 (Invitrogen) for 1 h and washed three times with PBS before mounting the coverslips on slides. Cells were imaged using the GFP and Alexa Fluor 568 channels and a ×60 oil immersion objective lens of a Leica TCS SP2 AOBS confocal microscope.
Membrane flotation assay.
Vero cells grown to 70% confluence in 100-mm dishes were either transfected or cotransfected with GFP-tagged UL16 constructs and/or gE, as described above. At 16 to 20 h posttransfection, the cells were harvested, washed twice with cold NTE buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA), and then resuspended in 300 μl of hypotonic lysis buffer (10 mM Tris-HCl [pH 7.4], 0.2 mM MgCl2) on ice for 20 min. Swollen cells were lysed on ice by 35 strokes with a Dounce homogenizer and then centrifuged at low speed to remove unbroken cells and nuclei. Postnuclear supernatants (∼300 μl) were mixed with 1.7 ml of 65% sucrose (wt/wt), placed at the bottom of a Beckman SW55Ti tube, and sequentially overlaid with 2.5 ml of 45% and 0.5 ml of 2.5% sucrose. All sucrose solutions were made in NTE buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA). The samples were centrifuged for 20 h at 200,000 × g and 4°C in a Beckman ultracentrifuge, and six equal-volume fractions (∼833 μl of each) were collected from the top. The membranes in each fraction were solubilized with 200 μl of 5× radioimmunoprecipitation assay (RIPA) buffer and immunoprecipitated with either anti-GFP antibodies [to precipitate UL16-GFP and mutant UL16(1-155)-GFP] or anti-gE antibodies. Immunocomplexes were captured with protein A-agarose beads (Roche), washed thrice with RIPA buffer, and lysed in sample buffer. Samples were separated by SDS-PAGE, followed by Western blot analysis with the respective primary antibodies and True-Blot HRP-conjugated secondary antibody.
RESULTS
Having discovered that the interaction of UL16 with the capsid is destabilized when HSV binds to attachment receptors (49), we considered the possibility that this tegument protein is directly linked to one of the two viral glycoproteins that interact with heparan sulfate (66). Binding to the tail of gB seemed more likely because it is longer (109 aa) than that of gC (only 14 aa). Inconsistent with this was the observation that the signal sent into the virion does not require gB in the heparin bead-binding assay (49); however, since viral glycoproteins exist in complexes on the surface of the virion, we proceeded to test the hypothesis in a simple in vitro assay.
Interaction between UL16 and the cytoplasmic tail of gE in GST-pulldown assays.
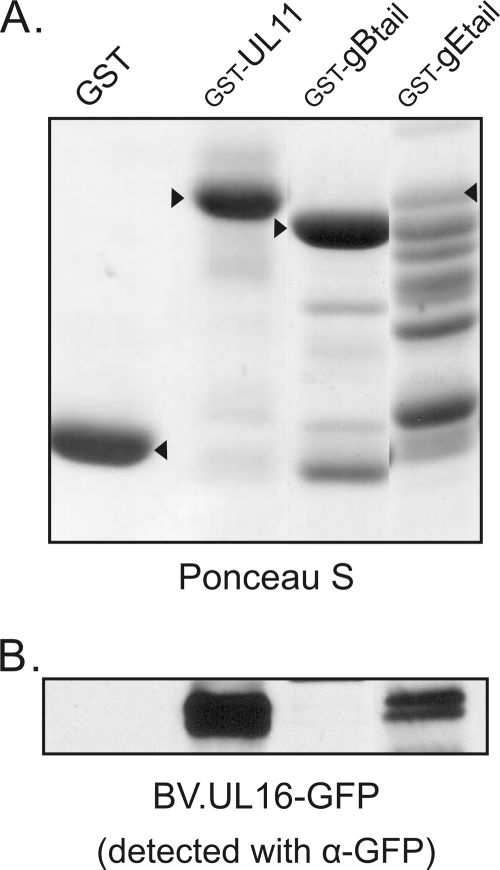
GST fusion proteins possessing the tails of gB, gD, or gE were purified from E. coli. As a positive control for UL16 binding, GST-UL11 was also isolated (Fig. 1A). The molecular masses of the fusion proteins were as expected with the exception of GST-gDtail, which appeared to have lost the foreign sequence (data not shown). Also, the GST-gEtail fusion protein exhibited extensive degradation in spite of the use of protease inhibitors and the construction of a codon-optimized sequence (data not shown). To look for binding, a recombinant baculovirus that expresses high levels of UL16-GFP was used to infect Sf21 cells, and a cell lysate was prepared with buffer containing NP-40 detergent. Glutathione beads bound with GST or the individual GST fusion proteins were incubated with the lysate, washed, and then analyzed by immunoblotting with antibodies specific for GFP. As expected, GST-UL11 readily pulled down UL16-GFP, while the GST-only construct did not (Fig. 1B). Surprisingly, the tail of gE (106 aa), but not that of gB, was capable of pulling down UL16 in spite of the extensive degradation of the former and the high abundance of the latter. This result was highly reproducible and indicated that the UL16-gEtail interaction does not require other viral or mammalian proteins. The use of insect cells for expression of UL16-GFP was not an influencing factor since this protein could also be pulled out of lysates of transfected mammalian cells (Fig. 2). However, it was quite possible that all of these findings were an artifact of the in vitro binding assay, and the two proteins might not interact at all within infected cells. Indeed, UL16 is known to interact with the acidic cluster of UL11 (43), and the tail of gE happens to contain such a sequence (Fig. 2A).
Fig. 1.
Interaction of UL16 with the gE tail. (A) Approximately equal amounts of glutathione bead-bound GST constructs, as indicated by arrowheads on a Ponceau S-stained gel, were mixed with lysates of insect cells infected with a recombinant baculovirus that expresses UL16-GFP. (B) Proteins bound to the GST constructs were loaded onto SDS-PAGE gels and analyzed by immunoblotting for GFP. The GST-UL11 construct was used as a positive control.
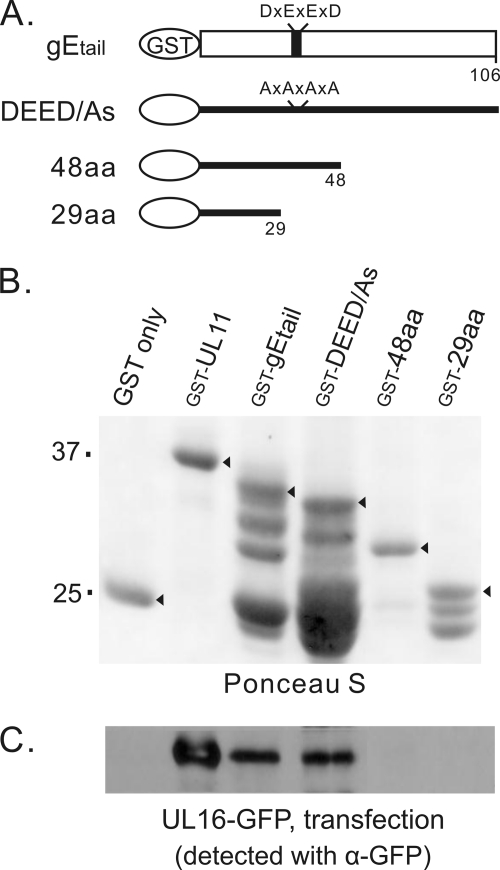
Fig. 2.
Interactions of gEtail mutants with UL16-GFP from mammalian cells. (A) Diagrams of GST-gEtail wild-type and mutant constructs. For mutant DEED/As, codons for four acidic amino acids (D, aspartate; E, glutamate) were replaced with codons for alanine. (B) Approximately equal amounts of glutathione bead-bound GST constructs, as indicated by arrowheads on a Ponceau S-stained gel, were mixed with lysates from UL16-GFP-transfected mammalian cells. (C) Proteins bound to the GST constructs were loaded onto SDS-PAGE gels and analyzed by immunoblotting for GFP.
To obtain a better understanding of the specificity of the interaction, several alterations were made to the GST-gEtail construct. First, to eliminate the acidic cluster, four alanine substitutions were inserted to create mutant DEED/As (Fig. 2A and B), but these changes had no effect on binding (Fig. 2C). This suggests that UL16 interacts with gE in a manner that is different from the UL16-UL11 interaction (i.e., there are two distinct binding sites within UL16). Reasoning that the region of interaction in gE might reside in the more conserved, membrane-proximal portion of the tail (1), two C-terminal deletion mutants were made (Fig. 2A and B), but neither was able to bind UL16-GFP (Fig. 2C). Two additional mutants were made that lack either the first 29 or the first 48 residues of the tail, but these were also unable to bind UL16-GFP (data not shown). Based on these data, it appears that the binding site within the tail may be conformationally dependent.
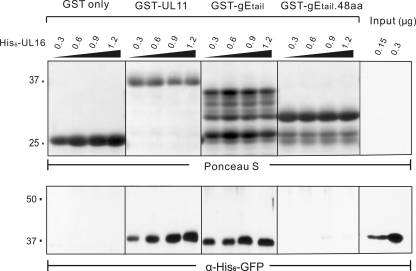
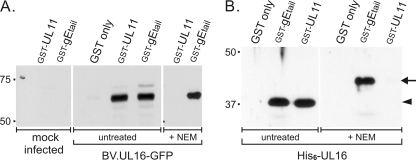
To ascertain whether the interaction is direct or indirect, binding assays were performed using proteins purified from E. coli. Similar to GST-UL11 (81), purified GST-gEtail was able to pull down purified His6-UL16; however, the interaction was lost once again when only the first 48 aa of the gE tail were used (Fig. 3). These data show that UL16 is able to directly bind to the gE tail. In contrast to the UL11 interaction (81), NEM modification of the free cysteines in UL16 did not reduce binding to gE, whether the source of UL16 was insect cells (Fig. 4A) or E. coli (Fig. 4B). This provides a second line of evidence to suggest that UL16 contains distinct sites for binding these two partners, at least in these in vitro assays.
Fig. 3.
Direct-interaction assays with UL16 and gEtail. The indicated amounts of purified, soluble His6-UL16 protein were mixed with the indicated GST fusion proteins immobilized on glutathione beads and incubated to allow binding. The beads were washed to remove unbound His6-UL16, and the samples were subjected to SDS-PAGE. Two samples of input His6-UL16 were included to show the efficiency of binding (rightmost lanes). The separated proteins were transferred to a nitrocellulose membrane, and the GST-fusion proteins were visualized by Ponceau S staining (upper panel). Bound and input His6-UL16 proteins were detected by immunoblotting with an antibody that recognizes the His6 tag (bottom panel).
Fig. 4.
Insensitivity of the UL16-gEtail interaction to NEM treatment. (A) Insect cells infected with a recombinant baculovirus expressing UL16-GFP were either untreated or treated with NEM prior to detergent lysis. The lysates were incubated with approximately equal amounts of the indicated GST constructs (determined by Ponceau S staining [data not shown]) attached to glutathione beads. The bound proteins were collected by centrifugation, separated by SDS-PAGE, and analyzed by immunoblotting for GFP. (B) Equal aliquots of purified His6-UL16 protein (1.2 μg) were incubated with approximately equal amounts of the indicated GST constructs as estimated from a Ponceau S-stained gel (data not shown), and bound proteins were analyzed by immunoblotting with antibody specific for the His6 tag. The positions of the unmodified and NEM-modified forms of UL16 are indicated (arrowhead and arrow, respectively). The shift in migration is less obvious in panel A because of the larger mass of the UL16-GFP chimera.
Detection of gE in native complexes with UL16 from infected cells.
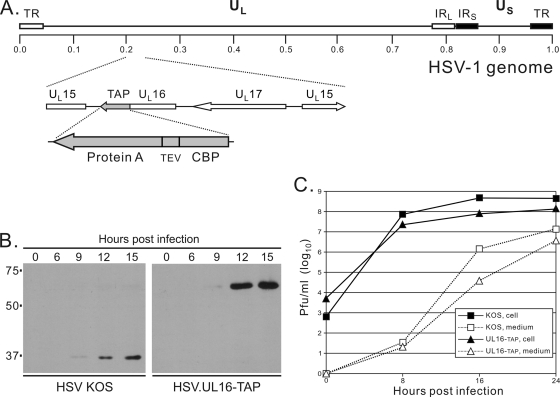
In a concurrent study within our laboratory, native UL16 complexes were being isolated from cells infected with a virus that expresses UL16-TAP, a chimera that contains two C-terminal tags for affinity purification (Fig. 5A). This recombinant virus was found to replicate substantially slower than the WT-KOS strain at 16 h postinfection, but by 24 h the difference was only half a log (Fig. 5C). This may be due to interference of the TAP tag; nevertheless, we proceeded to use this virus to test whether any UL16-containing complexes could be captured by tandem affinity purification. Vero cells infected for 20 h were lysed in NP-40 buffer, and complexes containing UL16 were purified via the TAP tag. Briefly, IgG beads were incubated with the lysates to allow binding of the protein A tag, washed, and then treated with tobacco etch virus (TEV) protease to release the complexes from the beads. Because of the high specificity of TEV protease, it is unlikely that cellular proteins present in the complex would be cleaved; moreover, there are no identifiable TEV cleavage sites within any of the HSV-encoded proteins (based on published sequences, data not shown). Next, the released UL16 complexes were collected on calmodulin resin and processed in two different ways for analysis by mass spectrometry at two separate facilities (see Materials and Methods). In both cases, gE was found to be present (16 and 15% coverage of 550 aa from methods 1 and 2, respectively; data not shown). These results were striking in light of the in vitro binding assays described above.
Fig. 5.
Characterization of the HSV.UL16-TAP recombinant. (A) The diagram shows the position of the TAP-tag-coding sequence at the 3′ end of the UL16 open reading frame. Starting from the position closest to UL16, the tag consists of calmodulin-binding peptide (CBP), a TEV protease cleavage site, and protein A. The wild-type and HSV.UL16-TAP viruses were compared with regard to their kinetics of UL16 expression (B) and production of cell-associated and extracellular infectious virions in single-step growth curves (C).
In retrospect, one concern with these tandem-affinity-purification experiments is that gE, within a heterodimer with glycoprotein I, has a high-affinity binding site for the Fc domain of IgG, which was present on the beads used in the first purification step. When these experiments were designed, the idea that UL16 might interact with gE had not yet emerged, but even so, any gE that was bound to the beads independently of UL16-TAP should not have been released by digestion with TEV protease, and any IgG-gE complexes that happened to come off would not be expected to bind to calmodulin beads in the final step. Thus, the detection of gE in two independent mass spectrometry experiments is consistent with the pull-down results (Fig. 1).
Coimmunoprecipitation of gE and UL16.
In an attempt to validate the putative UL16-gE interaction, coimmunoprecipitation assays were performed under various conditions with extracts of HSV-infected cells. The results were discouraging. Although antibodies specific for UL16 also precipitated gE under all conditions (Fig. 6), this was not necessarily due to an interaction between these two proteins because gE also came down in the same assay when cells were infected with a UL16-null mutant (data not shown). This is to be expected since all IgG molecules have two binding sites within their Fc region (31, 68, 69), one of which can bind gE/gI, while the other binds to the protein A beads used to collect the antigen-antibody complexes (i.e., binding to an antigen is not needed). In the other half of the experiment, antibodies against gE did not coimmunoprecipitate UL16 either at 4°C or at room temperature (Fig. 6A and B). UL16 was detected when the immunoprecipitation was done at 37°C overnight, but in this case, the complexes probably formed after lysis of the infected cells because UL16 was not seen when gE was collected for only 3 h at this temperature (Fig. 6C). This result shows that UL16 can bind to the tail of gE, but it does not prove that the interaction takes place in intact cells.
Fig. 6.
Coimmunoprecipitation of UL16 and gE from HSV-infected cell lysates. Vero cells were mock infected or infected with HSV at an MOI of 10. At 20 h postinfection, the cells were harvested, washed with PBS, and lysed with NP-40 buffer. Equal amounts of infected-cell lysate (based on the expression levels of UL16, gE, and VP5; data not shown) were subjected to immunoprecipitation (IP) using rabbit polyclonal antibodies against UL16 or gE for the indicated times and at three different temperatures: 4°C (A), room temperature (RT) (B), and 37°C (C). The collected molecules were analyzed by SDS-PAGE and immunoblotting with antisera specific for UL16 or gE.
Three possible explanations were considered for the discrepancy between the coimmunoprecipitation experiments and all of the in vitro binding assays. First, it was possible that UL16 interacts with gE in a transient manner that is negatively regulated by other viral proteins during an infection. If so, then it might be possible to coimmunoprecipitation complexes from transfected cells that express only UL16 and gE, but this was not the case (data not shown). Second, it was possible that NP-40 disrupts the UL16-gE interaction as it does the UL16-capsid interaction in virions (48). If so, then it might be possible to detect complexes by osmotically disrupting the transfected cells and floating membranes containing gE to the top of a sucrose step gradient, but UL16 remained at the bottom whether gE was present or not. Likewise, subcellular localization of UL16-GFP, as measured by confocal microscopy, was not altered by coexpressing gE. In light of all of these failed attempts (confirmed by multiple individuals), we began to favor the third possibility: binding of UL16 to the tail of gE could be an in vitro artifact.
Fortuitous discovery of a UL16 mutant that binds gE within cells.
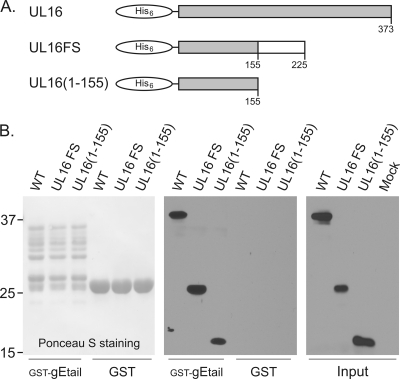
During a screen of a library of point mutations scattered throughout the UL16-coding sequence, an unintended frameshift mutant was found that expresses only the first 155 aa (Fig. 7A) and yet retains the ability to interact with the tail of gE in the in vitro binding assay (Fig. 7B). This finding was unexpected because the only portion of UL16 that had ever been found to be dispensable for any of its binding functions was the first 40, nonconserved residues (81). To ascertain whether binding was due to the foreign sequence that had been introduced as a result of the frameshift, a stop codon was inserted immediately after residue 155 (Fig. 7A), and the truncated protein was still able to recognize the GST-gEtail construct (Fig. 7B).
Fig. 7.
Discovery of a UL16-deletion mutant that retains the ability to bind gE. (A) Diagrams showing His6-UL16, the fortuitous frameshift mutant, and the construct containing only the first 155 residues. (B) Lysates from E. coli expressing wild-type (WT) or mutant His6-UL16 proteins were mixed with the indicated GST fusion proteins on glutathione beads and incubated to allow binding. The beads were washed to remove unbound proteins, and the samples were subjected to SDS-PAGE. The separated proteins were transferred to a nitrocellulose membrane, and the GST fusion proteins were visualized by Ponceau S staining. Bound and input His6-UL16 proteins were detected by immunoblotting with an antibody that recognizes the His6 tag.
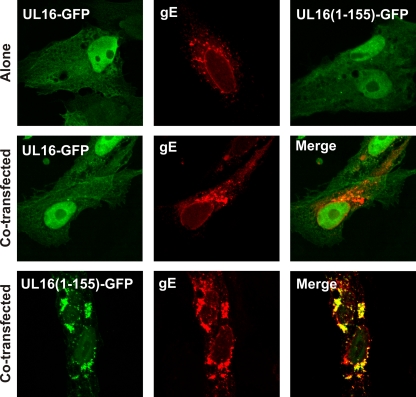
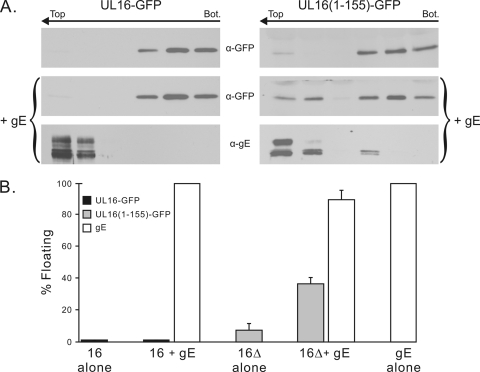
The binding properties of the mutant led us to consider the possibility that UL16 binds to gEtail in a regulated manner. That is, a binding domain at the N terminus might be controlled by a regulatory domain at the C terminus. According to this hypothesis, wild-type UL16 present in NP-40 lysates happens to be in an “open” state and is able to bind to gEtail in vitro, but within transfected cells (and in the absence of other binding partners) the full-length molecule exists in a “closed” state. This model predicts that UL16(1-155) will interact with gE in cotransfection experiments, and this was found to be the case in two different assays. First, confocal microscopy revealed that UL16(1-155) relocalizes to gE in the cytoplasm (Fig. 8, bottom row), whereas by itself, this truncated molecule was found throughout the cell, including the nucleus (top right). In contrast (as mentioned earlier), the full-length UL16 molecule lacks the ability to find gE and remains primarily in the nucleus (Fig. 8, middle row and top left). Second, when cells were osmotically lysed and membranes were placed at the bottom of a sucrose step gradient, UL16(1-155) was able to float to the top when coexpressed with gE (Fig. 9A, right panels), whereas wild-type UL16 was not (Fig. 9A, left panels). Moreover, a clear and reproducible alteration of the flotation properties of gE was observed such that a portion of these molecules were retained closer to the bottom of the gradient and in the less highly glycosylated, immature forms. This alteration in processing suggests that UL16 can bind to and inhibit transport of gE through the secretory pathway. Measuring only the amounts of the UL16 mutant that appeared in the top fractions, coexpression of gE resulted in an ∼4-fold increase in membrane binding (Fig. 9B). Collectively, these data strongly argue that UL16 contains a N-terminal domain that is capable of binding the tail of gE, and it will be interesting to learn more about the regulation and purpose of this binding event (see Discussion).
Fig. 8.
Relocalization of UL16(1-155)-GFP when coexpressed with gE. Vero cells were transfected with plasmids encoding UL16-GFP, UL16(1-155)-GFP or gE alone (top panels) or cotransfected with the indicated UL16-GFP constructs and gE (middle and bottom panels). At 16 to 20 h posttransfection the cells were fixed, stained with gE antibodies, and visualized by confocal microscopy.
Fig. 9.
Flotation analysis of UL16 in the presence or absence of gE. (A) Vero cells expressing UL16-GFP or deletion mutant UL16(1-155)-GFP, either alone or together with gE, were osmotically disrupted, and the ability of the proteins to float to the upper fractions of sucrose step gradients during centrifugation was examined. Representative immunoblots are shown. The tops and bottoms of the gradients are indicated, along with the direction of flotation (arrows). (B) Densitometric analysis was used to quantitate the immunoblots, and the results are shown as the percentage of floating protein (top three fractions) relative to the total protein (all fractions). The averages of three experiments are shown, along with the standard deviations.
DISCUSSION
The experiments described here have shown that HSV tegument protein UL16 interacts directly with the cytoplasmic tail of gE, both in vitro and within cells. For gE, this expands its network of known binding partners to at least three: gI, VP22, and UL16 (Fig. 10). The companion paper (26a) definitively shows that UL11 is another, as previously suggested (22, 32). It also remains to be shown that VP22 binds directly to gE; nevertheless, it is clear that this interaction does not require any other viral proteins (58). Whether UL16 and VP22 compete for binding to the tail of gE or assist one another for some common purpose is unknown and should be explored. It is conceivable that UL16 and VP22 directly interact with each other, but no evidence for this has been reported.
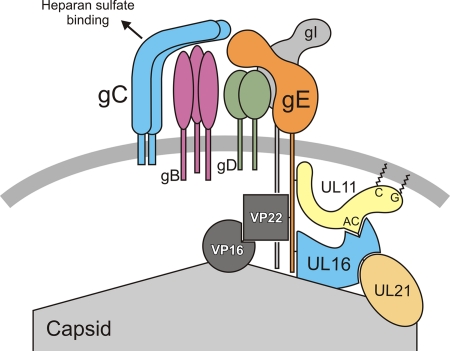
Fig. 10.
Model for the UL16-interaction network within virions. gE exists as a heterodimer with gI within a larger complex containing other viral glycoproteins. Binding of the virus to heparan sulfate via gC sends a signal that destabilizes the UL16-capsid interaction. That signal may be passed through gE, which has UL16 bound to its tail. UL16 also binds to an acidic cluster (AC) motif within UL11, which is peripherally associated with the membrane via an N-terminal glycine (G) modification with myristate and cysteine (C) modifications with palmitate. Moreover, UL11 may be associated with the tail of gE. For further details of this and other possible models, see the Discussion.
For UL16, its network of direct-binding partners has been expanded to four, the other members being UL11, UL21, and the unknown protein that enables UL16 to associate with capsids (Fig. 10). There are likely to be other members of this direct-interaction network. In particular, UL16 travels into the nucleus of infected cells, but nothing is known about its function there, and insight would likely be provided by the identification of nuclear-binding partners. However, care is needed when searching for these because of the presence of many free cysteines within UL16. For example, when virion proteins are separated in gels that lack reducing agent, very little monomeric UL16 can be detected, indicating the presence of extensive disulfide bonding with other proteins (49). However, these bonds seem to be irrelevant products—ones that arise only after lysis—because virions that have been pretreated with NEM (to block free, reactive cysteines) yield only monomers of UL16 in nonreducing gels (49). This concern does not apply to the interaction with gE because it takes place even after UL16 is modified with NEM, as shown here. The same is true for the UL16-UL21 interaction and the UL16-capsid interaction (28, 48, 49). In contrast, the UL16-UL11 interaction is blocked by NEM treatment, even though the interaction itself does not involve disulfide bonds (81).
Significance of the UL16-gE interaction.
Binding of UL16 to gE is likely important for at least three different events in the HSV replication cycle. Beginning with the infectious virion, it is possible that UL16 interacts with gE within virions (as illustrated in Fig. 10), although this is a question that remains to be addressed, not only for this pair of proteins but for virtually all of the interactions that have been ascribed to tegument proteins. Nevertheless, this model provides a framework for thinking about how binding of the virus to heparan sulfate (on cells or beads) might send a signal into the tegument to trigger the release of UL16 from the capsid. In support of the model, we have shown that the signal goes through gC rather than gB, and it requires clustering of glycoproteins on the virion surface; that is, exposure of virions to soluble heparan sulfate does not affect UL16 (49). Others have reported that gC and gE/gI are in a complex that blocks neutralizing antibodies directed at gB and gD (30). Thus, binding to immobilized heparan sulfate presumably rearranges the glycoprotein complex on the exterior of the virion to cause a mechanical signal to be sent into the tegument to alter the conformation of UL16 and its interaction with the capsid. Almost all of the molecular details of these complicated events remain to be elucidated, but none of that can be pursued without a clear understanding of which proteins are in the interaction network.
A second step in the replication cycle for which binding of UL16 to gE is likely to be important is cytoplasmic envelopment, where all of the components of the virus are assembled into their final structures. From a naive point of view, the UL16-gE interaction might be thought of as just another, redundant connection that helps “glue” the components of the tegument to the tails of glycoproteins in the envelope. After all, capsid-bound UL16 is also known to interact with capsid-bound UL21 and membrane-bound UL11 (28, 37, 41, 43, 81), while VP22 is also known to interact with the tail of gE (22, 58, 59, 70) and to capsid-bound VP16 (26, 38, 58). Thinking of these various interactions as being more or less equal might seem reasonable since mutants that lack any one of the four proteins in the UL16-direct interaction network still produce infectious virions at merely reduced levels (2–4, 7, 8, 15–18, 23, 25, 37, 39, 73). However, the assembly of an infectious virion involves much more than simply placing an envelope around the capsid. It has become increasingly clear that a machine is created within the tegument as the virion is assembled, and the absence of a single “gear” may have a huge effect on that machinery, even though some virions can still be released that are infectious in cell cultures. Indeed, numerous studies have shown that the absence of a single protein can affect the overall composition of the tegument (5, 21, 52–54, 82), much as the composition of a mechanical clock is altered when a single gear is removed and others spring loose. How the gears of the tegument fit, move, and work together will require a great deal of investigation.
The third event in which the UL16-gE interaction is likely to participate is cell-to-cell spread. It has long been known that gE plays a critical role in enabling virions produced in one cell to avoid neutralizing antibodies as they infect adjacent cells that are in direct contact. There are two commonly proposed mechanisms for this, both of which are built on the observation that in polarized epithelial cells, virions contained in transport vesicles are delivered to, and released from, lateral cell junctions (16–18, 32, 34, 80). In the first, the virions can only infect adjacent cells when gE/gI—either on the surface of the virion or the surface of the infected cell—interacts with a hypothetical receptor on the target cell (61, 79). It is not obvious why such an interaction would be required because the HSV fusion machinery (consisting of gB, gD, and gH/gL) is well known to be sufficient for virus entry in all other situations and is also known to be required for cell-to-cell spread (33, 55, 66, 67, 71, 74). This model is based largely on loss-of-function data (61, 63), and it may be that some alterations of the extracellular domain affect a more important function in the cytoplasmic domain. Nevertheless, it remains quite possible that the exposed portion of gE/gI has a function beyond that of being an Fc receptor because mutants have been found that fail to bind IgG but retain the ability to spread laterally (76), and antibodies against gE can block cell-cell fusion of HSV syncytial strains (11, 14). In the second model, the only parts of gE/gI that are needed for cell-to-cell spread are the cytoplasmic tails, which face into the cytoplasm on transport vesicles (32), where they help direct the enclosed virion to the lateral cell junction (61). It is easy to imagine that the interactions of UL16 and other tegument proteins with gE might be needed for such a mechanism. Indeed, it has been suggested that VP22 plays a role in cell-to-cell spread (19–21, 35, 62). With this model in mind, it is intriguing that one of our two mass spectrometry analyses of UL16-TAP complexes yielded Rich1 (7% coverage, data not shown). This host-encoded GTPase-activating protein has been proposed to be part of a sorting mechanism that interacts with Cdc42 to enable the transport of vesicles from recycling endosomes to tight junctions (29, 44, 57, 77). Further experiments will be needed to test the hypothesis that UL16 or one of its direct or indirect binding partners plays a role in recruiting Rich1 for the purpose of moving virion-containing vesicles to lateral membranes.
There may be other UL16-containing complexes that have different locations, different protein compositions, different dynamic properties, and different functions in the virion or infected cell. However, it is already clear that targeting of this conserved tegument protein is likely to affect multiple events in the replication cycle, not just for HSV but also for distantly related herpesviruses.
New insights on the structure and regulation of UL16-binding domains.
Perhaps the most exciting finding of the present study is the identification of subdomains within UL16. Previously, we showed that the first 40 aa are dispensable for binding to UL11 and UL21 (28, 81), but these residues are the least conserved and in fact are absent in most homologs. All of the other UL16 mutants in those studies (and others we made in unpublished mapping attempts) failed to reveal subdomains. Now, it is clear that the first 155 residues of UL16 are capable of binding to the tail of gE, both in vitro and within cotransfected cells. Although it is conceivable that the C-terminal deletion results in the creation of a totally new and irrelevant binding activity within the remaining N-terminal sequence of UL16, we think this is very unlikely because of the high efficiency with which the truncated protein found gE within the vast expanses of the cell, even leading to its exclusion from the nucleus (Fig. 8, bottom). Also, full-length UL16 occasionally was seen to colocalize with gE (yellow dot near the nucleus in Fig. 8, rightmost-middle panel), although this was not robust or stable enough to detect in the flotation experiments. Combined with all of the detailed in vitro binding data, which demonstrated a specific interaction between full-length UL16 and the tail of gE, the simplest interpretation of this deletion mutant is that it has lost a regulatory domain. Efforts are under way to examine the ability of the N- and C-terminal domains to interact in trans and to find single-residue substitutions in the C-terminal domain that activate binding of full-length UL16 to gE. We hypothesize that interaction with another binding partner (e.g., UL11 or UL21) would normally activate UL16 for binding to gE within cells. We further hypothesize that disruption of cells with NP-40 switches some UL16 molecules into an “open” state, enabling them to bind GST-gEtail in pulldown assays. This proposed activation is based in part on the observation that NP-40 artificially triggers the release of UL16 from capsids (48), a molecular rearrangement that is normally induced by the binding of virions to heparan sulfate and is blocked by prior NEM treatment (49). Precisely how UL16 is regulated, and whether its highly conserved cysteine residues are involved, will be fundamental to understanding how this tegument protein works.
Now that the gE-binding domain has been identified, much more work is needed to elucidate which parts of UL16 interact with UL11, UL21, and the capsid. Thus far, it is clear that the interaction with UL21 is not sensitive to NEM (28), as is the case for the gE interaction (shown here), but these limited observations are insufficient for predicting that the UL21-binding site will also be found in the N-terminal domain of UL16. With regard to UL11, it might be tempting to predict that its binding site will be found in the C-terminal domain because that portion of UL16 contains 15 of the 20 cysteine residues (including all of the conserved ones), and this interaction is sensitive to NEM (81). Caution is needed in considering this hypothesis because nothing is known about which cysteines in UL16 are free and which reside in disulfide bonds. Moreover, modification of the putative C-terminal regulatory domain with NEM might lock UL16 in a conformation that hides a UL11-binding site in the N-terminal domain. It will be interesting to learn whether mutant UL16(1-155) will be found to associate with UL11 in confocal microscopy and membrane-flotation experiments. Ironically, a nearly identical UL16 deletion mutant containing residues 41 to 155 was made during a previous search for the UL11-binding domain (81), but it failed to bind in GST pulldown experiments. Perhaps a different result might be found in the absence of NP-40. In any case, the large number of binding partners and evidence for multiple, regulated conformational states for UL16 make this conserved tegument protein both interesting and challenging to study.
ACKNOWLEDGMENTS
We thank our coworkers Jacob A. Marsh, Jason L. Starkey, and Carol B. Wilson for helpful discussions and encouragement. We especially thank J. C. de la Torre for the plasmid containing the TAP tag and David Leib for providing protocols and reagents for HSV-1 BAC recombineering.
This study was supported by National Institutes of Health (NIH) grants to J.W.W. (RO1 AI071286) and O.J.S. (RO1 CA76595). D.G.M. was supported by a training grant from the NIH (T32 CA60395).
Footnotes
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Alconada A., Bauer U., Sodeik B., Hoflack B. 1999. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J. Virol. 73:377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baines J. D., Koyama A. H., Huang T., Roizman B. 1994. The UL21 gene products of herpes simplex virus 1 are dispensable for growth in cultured cells. J. Virol. 68:2929–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baines J. D., Roizman B. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baines J. D., Roizman B. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baird N. L., Starkey J. L., Hughes D. J., Wills J. W. 2010. Myristylation and palmitylation of HSV-1 UL11 are not essential for its function. Virology 397:80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baird N. L., Yeh P. C., Courtney R. J., Wills J. W. 2008. Sequences in the UL11 tegument protein of herpes simplex virus that control association with detergent-resistant membranes. Virology 374:315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balan P., et al. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI, or the putative gJ. J. Gen. Virol. 75:1245–1258 [DOI] [PubMed] [Google Scholar]
- 8. Berarducci B., et al. 2006. Essential functions of the unique N-terminal region of the varicella-zoster virus glycoprotein E ectodomain in viral replication and in the pathogenesis of skin infection. J. Virol. 80:9481–9496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Britt W. J., Jarvis M., Seo J. Y., Drummond D., Nelson J. 2004. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai W. H., Gu B., Person S. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chatterjee S., Koga J., Whitley R. J. 1989. A role for herpes simplex virus type 1 glycoprotein E in induction of cell fusion. J. Gen. Virol. 70:2157. [DOI] [PubMed] [Google Scholar]
- 12. Chi J. H., Harley C. A., Mukhopadhyay A., Wilson D. W. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253–261 [DOI] [PubMed] [Google Scholar]
- 13. Cross A. M., Hope R. G., Marsden H. S. 1987. Generation and properties of the glycoprotein E-related 32K/34K/35K and 55K/57K polypeptides encoded by herpes simplex virus type 1. J. Gen. Virol. 68:2093. [DOI] [PubMed] [Google Scholar]
- 14. Davis-Poynter N., Bell S., Minson T., Browne H. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J. Virol. 68:7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Wind N., Wagenaar F., Pol J., Kimman T., Berns A. 1992. The pseudorabies virus homology of the herpes simplex virus UL21 gene product is a capsid protein which is involved in capsid maturation. J. Virol. 66:7096–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dingwell K. S., et al. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dingwell K. S., Doering L. C., Johnson D. C. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 69:7087–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dingwell K. S., Johnson D. C. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933–8942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dorange F., Tischer B. K., Vautherot J. F., Osterrieder N. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duffy C., et al. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J. Virol. 80:8664–8675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elliott G., Hafezi W., Whiteley A., Bernard E. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735–9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farnsworth A., Wisner T. W., Johnson D. C. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81:319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fulmer P. A., Melancon J. M., Baines J. D., Kousoulas K. G. 2007. UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J. Virol. 81:3097–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gierasch W. W., et al. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197–206 [DOI] [PubMed] [Google Scholar]
- 25. Guo H., Wang L., Peng L., Zhou Z. H., Deng H. 2009. Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J. Virol. 83:10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hafezi W., Bernard E., Cook R., Elliott G. 2005. Herpes simplex virus tegument protein VP22 contains an internal VP16 interaction domain and a C-terminal domain that are both required for VP22 assembly into the virus particle. J. Virol. 79:13082–13093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a. Han J., Chadha P., Meckes D. G., Jr., Baird N. L., Wills J. W. 2011. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J. Virol. 85:9437–9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Handler C. G., Eisenberg R. J., Cohen G. H. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J. Virol. 70:6067–6070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harper A. L., et al. 2010. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J. Virol. 84:2963–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harris K. P., Tepass U. 2010. Cdc42 and vesicle trafficking in polarized cells. Traffic 11:1272–1279 [DOI] [PubMed] [Google Scholar]
- 30. Hook L. M., Huang J., Jiang M., Hodinka R., Friedman H. M. 2008. Blocking antibody access to neutralizing domains on glycoproteins involved in entry as a novel mechanism of immune evasion by herpes simplex virus type 1 glycoproteins C and E. J. Virol. 82:6935–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johansson P. J., Nardella F. A., Sjöquist J., Schröder A. K., Christensen P. 1989. Herpes simplex type 1-induced Fc receptor binds to the Cgamma2-Cgamma3 interface region of IgG in the area that binds staphylococcal protein A. Immunology 66:8–13 [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson D. C., Baines J. D. 2011. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9:382–394 [DOI] [PubMed] [Google Scholar]
- 33. Johnson D. C., Huber M. T. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson D. C., Webb M., Wisner T. W., Brunetti C. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75:821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalthoff D., Granzow H., Trapp S., Beer M. 2008. The UL49 gene product of BoHV-1: a major factor in efficient cell-to-cell spread. J. Gen. Virol. 89:2269–2274 [DOI] [PubMed] [Google Scholar]
- 36. Kelly B. J., Fraefel C., Cunningham A. L., Diefenbach R. J. 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145:173–186 [DOI] [PubMed] [Google Scholar]
- 37. Klupp B. G., Bottcher S., Granzow H., Kopp M., Mettenleiter T. C. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J. Virol. 79:1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ko D. H., Cunningham A. L., Diefenbach R. J. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J. Virol. 84:1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kopp M., et al. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin X., Lubinski J. M., Friedman H. M. 2004. Immunization strategies to block the herpes simplex virus type 1 immunoglobulin G Fc receptor. J. Virol. 78:2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y., et al. 2009. The tegument protein UL94 of human cytomegalovirus as a binding partner for tegument protein pp28 identified by intracellular imaging. Virology 388:68–77 [DOI] [PubMed] [Google Scholar]
- 42. Loomis J. S., Bowzard J. B., Courtney R. J., Wills J. W. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209–12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Loomis J. S., Courtney R. J., Wills J. W. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417–11424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Macara I. G., Spang A. 2006. Closing the GAP between polarity and vesicle transport. Cell 125:419–421 [DOI] [PubMed] [Google Scholar]
- 45. McGraw H. M., Awasthi S., Wojcechowskyj J. A., Friedman H. M. 2009. Anterograde spread of herpes simplex virus type 1 requires glycoprotein E and glycoprotein I but not Us9. J. Virol. 83:8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McMillan T. N., Johnson D. C. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meckes D. G., Jr., Marsh J. A., Wills J. W. 2010. Complex mechanisms for the packaging of the UL16 tegument protein into herpes simplex virus. Virology 398:208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meckes D. G., Jr., Wills J. W. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 81:13028–13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meckes D. G., Jr., Wills J. W. 2008. Structural rearrangement within an enveloped virus upon binding to the host cell. J. Virol. 82:10429–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mettenleiter T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167–180 [DOI] [PubMed] [Google Scholar]
- 51. Mettenleiter T. C., Klupp B. G., Granzow H. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423–429 [DOI] [PubMed] [Google Scholar]
- 52. Michael K., Bottcher S., Klupp B. G., Karger A., Mettenleiter T. C. 2006. Pseudorabies virus particles lacking tegument proteins pUL11 or pUL16 incorporate less full-length pUL36 than wild-type virus, but specifically accumulate a pUL36 N-terminal fragment. J. Gen. Virol. 87:3503–3507 [DOI] [PubMed] [Google Scholar]
- 53. Michael K., Klupp B. G., Karger A., Mettenleiter T. C. 2007. Efficient incorporation of tegument proteins pUL46, pUL49, and pUS3 into pseudorabies virus particles depends on the presence of pUL21. J. Virol. 81:1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Michael K., Klupp B. G., Mettenleiter T. C., Karger A. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Muggeridge M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH, and gL in transfected cells. J. Gen. Virol. 81:2017–2027 [DOI] [PubMed] [Google Scholar]
- 56. Nalwanga D., Rempel S., Roizman B., Baines J. D. 1996. The UL16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226:236–242 [DOI] [PubMed] [Google Scholar]
- 57. Nelson W. J. 2009. Remodeling epithelial cell organization: transitions between front-rear and apical-basal polarity. Cold Spring Harbor Perspect. Biol. 1:a000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. O'Regan K. J., Bucks M. A., Murphy M. A., Wills J. W., Courtney R. J. 2007. A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358:192–200 [DOI] [PubMed] [Google Scholar]
- 59. O'Regan K. J., Murphy M. A., Bucks M. A., Wills J. W., Courtney R. J. 2007. Incorporation of the herpes simplex virus type 1 tegument protein VP22 into the virus particle is independent of interaction with VP16. Virology 369:263–280 [DOI] [PubMed] [Google Scholar]
- 60. Oshima S., et al. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch. Virol. 143:863–880 [DOI] [PubMed] [Google Scholar]
- 61. Polcicova K., Goldsmith K., Rainish B. L., Wisner T. W., Johnson D. C. 2005. The extracellular domain of herpes simplex virus gE is indispensable for efficient cell-to-cell spread: evidence for gE/gI receptors. J. Virol. 79:11990–12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pomeranz L. E., Blaho J. A. 2000. Assembly of infectious Herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041–10054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Saldanha C. E., et al. 2000. Herpes simplex virus type 1 glycoprotein E domains involved in virus spread and disease. J. Virol. 74:6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Silva M. C., Yu Q. C., Enquist L., Shenk T. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594–10605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smith K. O. 1964. Relationship between the envelope and the infectivity of herpes simplex virus. Proc. Soc. Exp. Biol. Med. 115:814–816 [DOI] [PubMed] [Google Scholar]
- 66. Spear P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol. 6:401–410 [DOI] [PubMed] [Google Scholar]
- 67. Spear P. G., Longnecker R. 2003. Herpesvirus entry: an update. J. Virol. 77:10179–10185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sprague E. R., Martin W. L., Bjorkman P. J. 2004. pH dependence and stoichiometry of binding to the Fc region of IgG by the herpes simplex virus Fc receptor gE-gI. J. Biol. Chem. 279:14184–14193 [DOI] [PubMed] [Google Scholar]
- 69. Sprague E. R., Wang C., Baker D., Bjorkman P. J. 2006. Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging. PLoS Biol. 4:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stylianou J., Maringer K., Cook R., Bernard E., Elliott G. 2009. Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J. Virol. 83:5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Subramanian R. P., Geraghty R. J. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. U. S. A. 104:2903–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Takakuwa H., et al. 2001. Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in overexpressing cells. Genes Cells 6:955–966 [DOI] [PubMed] [Google Scholar]
- 73. Tirabassi R. S., Townley R. A., Eldridge M. G., Enquist L. W. 1997. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J. Virol. 71:6455–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Turner A., Bruun B., Minson T., Browne H. 1998. Glycoproteins gB, gD, and gH/gL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weeks B. S., Sundaresan P., Nagashunmugam T., Kang E., Friedman H. M. 1997. The herpes simplex virus-1 glycoprotein E (gE) mediates IgG binding and cell-to-cell spread through distinct gE domains. Biochem. Biophys. Res. Commun. 235:31–35 [DOI] [PubMed] [Google Scholar]
- 77. Wells C. D., et al. 2006. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell 125:535–548 [DOI] [PubMed] [Google Scholar]
- 78. Wing B. A., Lee G. C., Huang E. S. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 70:3339–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wisner T., Brunetti C., Dingwell K., Johnson D. C. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J. Virol. 74:2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wisner T. W., Johnson D. C. 2004. Redistribution of cellular and herpes simplex virus proteins from the trans-Golgi network to cell junctions without enveloped capsids. J. Virol. 78:11519–11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yeh P. C., Meckes D. G., Jr., Wills J. W. 2008. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J. Virol. 82:10693–10700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang Y., McKnight J. L. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 67:1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]