Abstract
It has recently been shown that polymorphism at the rhesus macaque TRIM5 locus can affect simian immunodeficiency virus (SIV) replication. Here we show that TRIM5 alleles can also affect acquisition of SIVsmE660. Animals coexpressing the TRIM5TFP and TRIM5CypA alleles took significantly longer to become infected with SIVsmE660, but not SIVmac239, after repeated limiting-dose intrarectal challenge than did animals expressing other TRIM5 allele combinations. Our results indicate that the TRIM5 alleles can be a barrier to productive infection and that this should be taken into account when designing acquisition studies using SIVsmE660 or related viruses.
TEXT
Lentiviruses and their hosts have been interacting and adapting to each other for millions of years. Humans and nonhuman primates, like other mammals, express cellular restriction factors to inhibit viral replication (6). These restriction factors offer protection against a broad range of viruses and represent an important barrier to zoonotic transmissions. Recently, three major classes of retroviral restriction factors have been identified: APOBEC3, tripartite motif protein 5 alpha (TRIM5α), and tetherin (12, 16, 19, 20). Each of these factors targets a different step in the virus life cycle to suppress viral replication. To counteract these restriction factors, primate lentiviruses have developed a series of accessory proteins that, in part, circumvent their actions and allow viruses to efficiently replicate (11).
Polymorphic alleles at the rhesus macaque TRIM5 locus correlate with a 1.3- to 3-log reduction in simian immunodeficiency virus (SIV) replication (7, 10). The mechanism by which TRIM5α affects viral replication is unclear, but it appears to act by binding incoming viral capsids and mediating premature viral uncoating (18). A recent study also suggests a role for TRIM5 mediating the early, innate immune response to retroviral infection (14). Rhesus macaque TRIM5 is polymorphic, and the encoded protein includes a TRIM5-cyclophilin A (CypA) chimera in which CypA replaced the TRIM5α binding domain (7, 10, 13, 17, 19). While there are multiple TRIM5 alleles, they can be categorized into three functional allelic classes based on variations in their C-terminal domains and their relative effects on SIVsmE543-3 replication: TRIM5CypA, TRIM5TFP, and TRIM5Q (7). Combinations of these alleles result in six possible functional TRIM5 genotypes. A previous study showed that cell lines expressing TRIM5TFP or TRIM5CypA alleles were resistant to infection with the molecular clone SIVsmE543-3 but not with SIVmac239 (7). Cells expressing TRIM5Q alleles were susceptible to both viruses. These in vitro results were recapitulated in vivo in a cohort of animals infected with SIVsmE543-3. Animals with two resistance-conferring TRIM5 alleles (TRIM5TFP/TFP or TRIM5TFP/CypA) had a 2.0- to 3.0-log reduction in chronic-phase plasma virus replication in comparison to animals with the TRIM5Q/Q genotype. Furthermore, animals with one resistance-conferring allele (TRIM5TFP/Q or TRIM5Q/CypA) had intermediate plasma viral loads. A less dramatic effect on plasma virus replication was also seen in a cohort of SIVmac251-infected animals by using similar TRIM5 groupings (10).
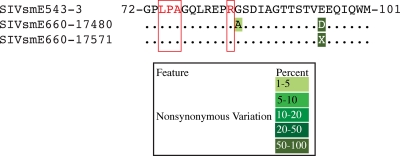
We first investigated whether the biological isolate SIVsmE660 might harbor TRIM5α-resistant variants. Previous studies identified amino acids in Gag p27CA affecting TRIM5-mediated restriction (2, 7, 9). Since two different stocks of SIVsmE660 were used in our studies, we employed ultradeep pyrosequencing as previously described (1) to determine whether there existed any variation that might result in TRIM5α-resistant quasispecies. Briefly, we used four overlapping reverse transcription-PCR amplicons of approximately 2.5 kb spanning the entire SIVsmE660 genome. The reverse transcription-PCR products were then randomly fragmented using modified transposons (Nextera; Epicentre Biotechnology) and pyrosequenced. At average nucleotide depths of 493 for SIVsmE660 stock 17480 and 819 for SIVsmE660 stock 17571, we found no nonsynonymous variation above 1% from the SIVsmE543-3 regions previously identified to affect rhesus macaque TRIM5-mediated restriction (Fig. 1) (7). This suggested that an animal's TRIM5 genotype could impose a bottleneck for productive infection following limiting-dose intrarectal SIVsmE660 challenge.
Fig. 1.
Sequence of SIVsmE660 Gag p27CA known to affect TRIM5-mediated restriction. Partial sequences of the core nucleocapsid protein amino acids 72 to 101 of the two stocks of SIVsmE660 analyzed in this study are aligned against that of the closely related molecular clone SIVsmE543-3. The putative amino acids affecting restriction mediated by TRIM5TFP (74LPA76) and TRIM5CypA (R83) are highlighted in red. Nonsynonymous variations from the SIVsmE543-3 sequence are color coded by frequency. X indicates a mixed amino acid sequence.
Only a few human immunodeficiency virus (HIV) strains cross mucosal barriers to establish infection in humans (4). Investigators have been using repeated limiting-dose SIV challenge of rhesus macaques to mimic HIV transmission and have shown that only one to three viral species similarly initiate infection in monkeys (5, 21). Under these conditions, it is possible that the TRIM5 genotype of an animal might play a role in whether an animal becomes productively infected with SIV. To address this issue, we retrospectively analyzed a cohort of 62 Indian rhesus macaques at the Wisconsin National Primate Research Center (WNPRC) that were not vaccinated with SIV antigens to determine if TRIM5 genotype affected the rate at which they became productively infected with SIVsmE660. We genotyped our animals based on the following three allelic classes: TRIM5CypA, TRIM5TFP, and TRIM5Q. We established the TRIM5 genotypes of the animals either by sequencing genomic DNA as previously described (7) or by sequence-specific priming PCR (PCR-SSP) assays. To distinguish TRIM5TFP and TRIM5Q alleles by PCR-SSP, we took advantage of unique polymorphisms in the TRIM5 SPRY/B30.2 domain and used previously published thermal cycling conditions and reagents (3). The TRIM5TFP and TRIM5Q PCR-SSP reactions share a forward primer (5′-AGA GAG CTA ACA GAT GCC CG-3′) designed to anneal to both of the TRIM5 variants, while the reverse primers targeted unique sites within the TRIM5TFP and TRIM5Q alleles to confer specificity (TFP allele reverse primer, 5′-ACA ATG AAA GGA GCA AAA GGA GTA TGT G-3′; Q allele reverse primer, 5′-CAC AAT GAA AGG AGC AAA AGG AGT ATG TA-3′). Primers were used at a 0.3 μM working concentration. The TRIM5CypA allele was genotyped using a previously published protocol which utilized PCR followed by NsiI digestion (13). To confirm that the consistency of the PCR-SSP genotyping was equivalent to that of genomic DNA sequencing, we tested a subset of animals by both methods and obtained identical results.
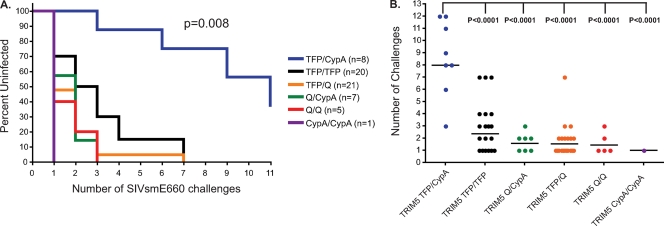
We next investigated whether the TRIM5 genotype affected acquisition of SIVsmE660. Rhesus macaques used in this analysis were intrarectally inoculated on a weekly or biweekly basis with initial limiting doses ranging from 225 to 450 50% tissue culture infective doses (TCID50) for SIVsmE660-17571 (52 animals) and 800 TCID50 for SIVsmE660-17480 (10 animals) as previously described (15, 21). We performed a Kaplan-Meier analysis to test whether any of the TRIM5 genotypes differed in the number of challenges required to achieve productive infection. We discovered that animals having the TRIM5TFP/CypA genotype took significantly more exposures to get infected than animals expressing the other TRIM5 genotypes (log rank test; chi-square = 15.74; df = 5; P = 0.008) (analyses performed with GraphPad Prism version 4.0c) (Fig. 2A). Of note, four animals with the TRIM5TFP/CypA genotype were the only animals to remain uninfected at the conclusion of intrarectal challenges, two animals after the seventh challenge and two after the eleventh challenge. We therefore further explored the resistance to infection with SIVsmE660 of the TRIM5TFP/CypA animals in comparison to the other genotypes. We compared the numbers of challenges needed to achieve productive infection per TRIM5 genotype by using the parametric generalized gamma model because it was determined to be the best model in comparison to nonparametric models by Akaike information criterion scores for examining dosages that increased over time. For this analysis, we designated the inoculation after intrarectal challenges were stopped for the uninfected animals as the infecting dose (e.g., we designated the animals remaining uninfected after 11 inoculations as being infected with the 12th inoculation). Animals with the TRIM5TFP/CypA genotype took significantly longer to productively infect with limiting doses of SIVsmE660 than each of the remaining TRIM5 genotypes (P < 0.0001) (Fig. 2B).
Fig. 2.
Acquisition of SIVsmE660 infection after repeated limiting-dose challenge is affected by TRIM5 genotype. (A) Kaplan-Meier curve analysis of the effects different TRIM5 genotypes have on the rate of acquisition of SIVsmE660 infection after repeated limiting-dose intrarectal challenge. The statistical significance of the rates of infection of the different TRIM5 genotypes was determined by log rank test. (B) A comparison of the numbers of challenges needed to productively infect animals expressing the different TRIM5 genotypes with SIVsmE660. Significance between the rates of infection of animals having the TRIM5TFP/CypA genotype and the other TRIM5 genotypes was determined by parametric generalized gamma model.
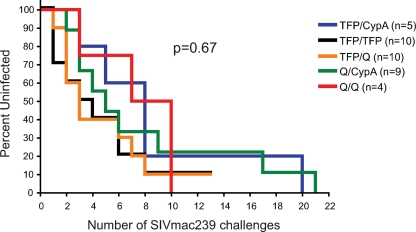
Since the TRIM5 genotype affects acquisition of infection with SIVsmE660, we sought to determine if it has the same effect on another SIV strain commonly used in challenge studies, the molecularly cloned SIVmac239. Kirmaier et al. previously established that SIVmac239, unlike SIVsmE543-3, is resistant to all of the TRIM5 allelic classes in vitro, which is likely due to amino acid substitutions in the viral capsid that prevent or inhibit TRIM5-mediated restriction (7). However, it is still possible in the setting of a limiting-dose challenge in which only a few viruses likely cross the mucosal barrier and establish infection that TRIM5α might have an effect on acquisition. We therefore examined a cohort of 38 naïve animals repeatedly challenged with limiting doses of SIVmac239 (produced in vitro with rhesus macaque peripheral blood mononuclear cells [PBMC]) housed at the WNPRC and at the University of Pittsburgh. Of note, a few animals included in this cohort were repeatedly challenged with either 30 or 300 TCID50 SIVmac239, which in retrospect was probably an extremely limiting dose of the virus, because it required between 8 and 21 challenges to achieve infection. We also had two animals with either the TRIM5TFP/TFP or TRIM5TFP/Q genotype that did not become infected after 13 limiting-dose challenges. Despite this, we detected no significant difference in the rates of acquisition of SIVmac239 for the different TRIM5 genotypes when we performed the Kaplan-Meier analysis (P = 0.67) (Fig. 3). However, due to the relatively small number of animals in this analysis, it is possible that TRIM5 has a more limited effect on acquisition of SIVmac239 that cannot be discerned in our study.
Fig. 3.
Acquisition of SIVmac239 infection after repeated limiting-dose intrarectal challenge is not affected by TRIM5 genotype. The Kaplan-Meier curve analysis revealed no significant differences in the acquisition of SIVmac239 infection after repeated limiting-dose intrarectal challenge among the TRIM5 genotypes. We performed a power analysis and determined that our cohort of SIVmac239 challenged animals had 73% power to detect an effect by the TRIM5TFP/CypA genotype on acquisition of infection similar to that observed for the cohort of SIVsmE660 challenged animals.
Our study highlights the complex nature of virus-host interactions and the difficulty of modeling HIV transmission in rhesus macaques. Our analysis also expands upon a recent report showing that TRIM5α alleles can affect the acquisition of SIVsmE660 infection by naïve and vaccinated rhesus macaques (8). In this study, however, TRIM5TFP and TRIM5CypA were grouped together as “restrictive” alleles, and homologous expression of these alleles resulted in delayed infection with SIVsmE660. In our cohort of animals, we further subdivided the genotypes to include TRIM5CypA as a separate allele and only the heterozygous TRIM5TFP/CypA genotype made a significant impact on the rate of acquisition of SIVsmE660 infection. It is possible that since animals with the TRIM5TFP/CypA genotype have two TRIM5α molecules that bind distinct regions of the viral capsid, they eliminate incoming virus more effectively than animals having TRIM5α molecules targeting only one region of the viral capsid, even for animals homozygous for resistance-conferring TRIM5 alleles. It is also possible that the TRIM5TFP/TFP and TRIM5CypA/CypA genotypes have a less dramatic effect on the acquisition of infection, which would become apparent in a larger cohort of animals. Our study also indicates that under physiological conditions in which only a few viruses are crossing the mucosal barrier, TRIM5α can decrease the probability of productive infection occurring. However, since restriction by TRIM5α is known to be saturable, this effect may be subtle and overcome by high-dose mucosal or intravenous exposures (19). In this regard, the previous studies identifying that TRIM5 genotypes differentially affects SIVsmE543-3 and SIVmac251 replication did not note an effect on acquisition of infection after intravenous inoculation (7, 10). Our results suggest that animals entering repeated limiting-dose SIV challenge studies should be typed for their TRIM5 genotype, especially if SIVsmE660 or a related virus is used. When possible, TRIM5TFP/CypA animals should be either excluded from the study or balanced among test groups in a manner similar to that for animals expressing protective major histocompatibility complex (MHC) class I alleles.
Acknowledgments
We thank the veterinary staff at the Wisconsin National Primate Research Center (WNPRC) for their assistance. We also thank Ben Burwitz, Ben Bimber, Simon Lank, and David O'Connor for their guidance in pyrosequencing and analyzing the SIVsmE660 stocks.
This research was supported by funds from the International AIDS Vaccine Initiative, National Institutes of Health (NIH) grants R01 AI049120, R01 AI076114, R01 AI076114, R24 RR016038, R24 RR015371, and R37 AI076114 and contract HHSN266200400088C to D.I.W., grant R01 AI083118 to W.E.J., grant P51 RR000167 to the WNPRC, and grant RR00168 to the New England Primate Research Center (NEPRC) from the National Center for Research Resources (NCRR), a component of the NIH. This research was also supported by Pfizer Inc.-sponsored research agreement 140-N-2076312 with J.B.S. Additionally, this research was conducted at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01.
This publication's contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
We have no conflicting financial interests.
Footnotes
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Bimber B. N., et al. 2010. Whole-genome characterization of human and simian immunodeficiency virus intrahost diversity by ultradeep pyrosequencing. J. Virol. 84:12087–12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hatziioannou T., Cowan S., Von Schwedler U. K., Sundquist W. I., Bieniasz P. D. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaizu M., et al. 2007. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics 59:693–703 [DOI] [PubMed] [Google Scholar]
- 4. Keele B. F., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keele B. F., et al. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55–67 [DOI] [PubMed] [Google Scholar]
- 7. Kirmaier A., et al. 2010. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8: pii: e1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Letvin N. L., et al. 2011. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci. Transl. Med. 3:81ra36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y., Li X., Stremlau M., Lee M., Sodroski J. 2006. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J. Virol. 80:6738–6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lim S. Y., et al. 2010. TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 6:e1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malim M. H., Emerman M. 2008. HIV-1 accessory proteins-ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398 [DOI] [PubMed] [Google Scholar]
- 12. Neil S. J., Zang T., Bieniasz P. D. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 13. Newman R. M., et al. 2008. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 4:e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pertel T., et al. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reynolds M. R., et al. 2010. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J. Virol. 84:9190–9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 17. Song B., et al. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J. Virol. 79:6111–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strebel K., Luban J., Jeang K. T. 2009. Human cellular restriction factors that target HIV-1 replication. BMC Med. 7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stremlau M., et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 20. Van Damme N., et al. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilson N. A., et al. 2009. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J. Virol. 83:6508–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]