Abstract
High-mobility group box 1 (HMGB1), an abundant nuclear protein that triggers host immune responses, is an endogenous danger signal involved in the pathogenesis of various infectious agents. However, its role in hepatitis C virus (HCV) infection is not known. Here, we show that HMGB1 protein is translocated from the nucleus to cytoplasm and subsequently is released into the extracellular milieu by HCV infection. Secreted HMGB1 triggers antiviral responses and blocks HCV infection, a mechanism that may limit HCV propagation in HCV patients. Secreted HMGB1 also may have a role in liver cirrhosis, which is a common comorbidity in HCV patients. Further investigations into the roles of HMGB1 in the diseases caused by HCV infection will shed light on and potentially help prevent these serious and prevalent HCV-related diseases.
INTRODUCTION
Hepatitis C virus (HCV) is one of the major causative agents of hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC) (17, 30). More than 170 million people are estimated to suffer from HCV infection worldwide (17). Chronic and persistent infection is a characteristic feature of HCV pathogenesis (30). During chronic infection, the production of virus particles is limited, and a restricted number of liver cells are infected. As a result, the viral dose in patients' blood generally is lower than that of other hepatitis-causing viruses, such as hepatitis B virus (HBV) (4). Moreover, a large portion of hepatocytes often remains uninfected by the virus even after long-term infection (28). These phenomena indicate the existence of balance between the HCV infection process and host mechanisms that protect against HCV infection. We speculate that innate and adaptive immunities contribute to the balance between infection and protection.
High-mobility group box 1 (HMGB1) protein is a highly conserved nuclear protein that participates in DNA organization and the regulation of transcription. In addition to its nuclear function, HMGB1 plays an important role as a cytokine, mediating the responses to infection, injury, and inflammation (1, 2, 29, 42). HMGB1 is released passively from necrotic cells and is actively secreted from activated immune cells, such as macrophages, natural killer cells, and mature dendritic cells (2). The functionality of actively secreted HMGB1 is known to be modulated by posttranslational modifications, such as oxidation (2, 36). Extracellular HMGB1 can function by itself and/or in association with other molecules, including CpG DNA, lipopolysaccharide (LPS), and interleukin-1 (IL-1) (5). HMGB1 induces a variety of cellular responses that contribute to innate immunity, tissue repair, and cell migration through interactions with various receptors that activate multiple signal transduction responses. The Toll-like receptors (e.g., TLR2, TLR4, and TLR9) and the receptor for advanced glycation end products (RAGE) are known receptors for the cytokine functions of HMGB1 (2). TLR4, the major component of the LPS recognition receptor complex, engages in downstream signaling through MyD88 and the Toll-like adapter protein TRIF to produce proinflammatory cytokines and type I interferons (IFNs), which potentially participate in blocking virus infections (34).
No role of HMGB1 in HCV infection has been demonstrated yet. However, HMGB1 is known to be an indicator of human liver injury (19), and HMGB1 levels in the sera of patients with chronic hepatitis and cirrhosis are significantly elevated (8). The source of this elevated serum HMGB1 and the molecular mechanism responsible for the secretion of HMGB1 from cells are not known. One possible mechanism underlying elevated HMGB1 secretion is increased reactive oxygen species (ROS) in HCV-infected cells. Two HCV-encoded proteins, core and NS5A, induce oxidative stress in infected cells (15, 25–27), and it has been reported that ROS induces nuclear-to-cytoplasmic translocation and the subsequent secretion of HMGB1 from cells (36). However, the localization of HMGB1 in HCV-infected cells and the effect of HMGB1 on HCV infectivity remain to be elucidated.
Here, we investigated the localization and secretion of HMGB1 upon HCV infection. We found that some HMGB1 protein was translocated from the nucleus to the cytoplasm and secreted into the medium before virus production was observed. This suggests that HCV-infected cells sense the HCV infection and propagate a warning signal to uninfected cells. We tested the potential role of secreted HMGB1 in HCV infection using an antibody against HMGB1 and purified HMGB1 protein. The treatment of HCV culture medium with the anti-HMGB1 antibody increased the infectivity of HCV. Conversely, the pretreatment of cells with purified HMGB1 protein reduced the infectivity of HCV, indicating that HMGB1 secreted from infected cells blocked the infection of neighboring cells by HCV. Our investigation of the molecular basis of this protection further revealed that TLR4, a potential HMGB1 receptor, plays an important role in the antiviral effect via the activation of interferon signaling.
MATERIALS AND METHODS
Cell culture and HCV production.
The human hepatoma cell line Huh7.5.1 was kindly provided by Francis V. Chisari (Scripps Research Institute). Huh7.5.1 and HEK293FT (human kidney carcinoma) cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; HyClone), penicillin, and streptomycin. The in vitro synthesis of HCV RNA (derived from JFH1 and JFH5a-Rluc vectors) and electroporation were performed as described previously (22). Infectious HCV in cell culture (HCVcc) was produced by the transfection of Huh7.5.1 cells with in vitro-transcribed HCV RNA. The virus titer was determined as described elsewhere (11) and stored at −80°C.
Reagents.
The antibodies against HMGB1, TLR4, and isotype control IgG were purchased from Abcam. The antibody against HCV core for immunocytochemistry was obtained from Affinity Bioreagents. The antibodies to PARP [poly(ADP-ribose) polymerase], β-actin, and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) were obtained from Santa Cruz Biotechnology, Sigma, and AbD Serotec, respectively. The secondary antibodies for immunostaining were purchased from Invitrogen and Jackson ImmunoResearch. The antibody against HCV core for Western blotting was a gift from Ralf Bartenschlager (University of Heidelberg). LPS and Hoechst 33258 were obtained from Sigma.
Patient tissue.
Informed consent to use tissue specimens for research purposes was obtained from all patients, and the utilization of specimens for this research was authorized by the Institutional Review Boards of the College of Medicine, Ewha Womans University (case numbers S10-6410 and S09-2974), Yonsei University (case number T6), and Catholic University (case number 14266130).
RT-PCR.
Total RNA was purified with TRI reagent, and cDNA was synthesized using reverse transcription reagents (Promega). Reverse transcription-PCR (RT-PCR) was performed using a LightCycler 480 SYBR green I master system (Roche). Primer sequences for reverse transcription, PCR, and real-time PCR were the following: IFNβ1, 5′-CAG CAA TTT TCA GTG TCA GAA GC-3′ and 5′-TCA TCC TGT CCT TGA GGC AGT-3′; interferon-stimulated gene 15 (ISG15), 5′-ACT CAT CTT TGC CAG TAC AGG AG-3′ and 5′-CAG CAT CTT CAC CGT CAG GTC-3′; GAPDH, 5′-TGC ACC ACC AAC TGC TTA G-3′ and 5′-GAG GCA GGG ATG ATG TTC-3′; and β-actin, 5′-CTG GAA CGG TGA AGG TGACA-3′ and 5′-AAG GGA CTT CCT GTA ACA ATG CA-3′.
Plasmid constructs and mutagenesis.
The plasmid pHMGB1-eGFPN1 was provided by J. S. Shin (Yonsei University, South Korea). To create pcDNA3.1-Flag-HMGB1, we first constructed the plasmid pcDNA3.1-Flag by inserting annealed oligonucleotides encoding Flag sequences (5′-CTA GCC GCC ACC ATG GAC TAC AAA GAC GAT GAC GGT GAT TAT AAA GAT GAT GAC ATC GAT TAC AAG GAT GAC GAT GAC A-3′ and 5′-AGC TTG TCA TCG TCA TCC TTG TAA TCG ATG TCA TCA TCT TTA TAA TCA CCG TCA TCG TCT TTG TAG TCC ATG GTG GCG G-3′) into pcDNA3.1(+)-Hyg (Invitrogen) treated with NheI and HindIII. A pHMGB1-eGFPN1 PCR product amplified using the primers 5′-CCC AAG CTT GGC AAA GGA GAT CCT AAG AAG C-3′ and 5′-GCT GAT ATC TTA TTC ATC ATC ATC ATC TTC TTC TTC ATC-3′ was digested with HindIII and EcoRV and then subcloned into pcDNA3.1-Flag to generate pcDNA3.1-Flag-HMGB1. The mutant construct pcDNA3.1-Flag-HMGB1-C106S was generated by DpnI-mediated site-directed mutagenesis (14) using the primers 5′-GCC TTC TTC CTC TTC TCC TCT GAG TAT CGC-3′ and 5′-GCG ATA CTC AGA GGA GAA GAG GAA GAA GGC-3′. All clones were confirmed by sequencing. The plasmid pcDNA3/myc-hTLR4 was provided by Yoomi Kim (Postech, South Korea).
Purification of HMGB1 and HMGB1-C106S proteins.
Flag-HMGB1 and Flag-HMGB1-C106S proteins were purified using an anti-FLAG M2 affinity gel (Sigma) according to the manufacturer's instructions, with minor modifications. Briefly, HEK293FT cells, grown on 30 150-mm plates, were transfected with 8 μg/plate of pcDNA3.1-Flag-HMGB1 or pcDNA3.1-Flag-HMGB1-C106S using the calcium phosphate transfection method and further cultivated for 3 days. The cells were washed twice with ice-cold phosphate-buffered saline (PBS) and then harvested. The cell pellets were resuspended in 45 ml of lysis buffer (50 mM Tris-Cl, pH 7.4, 1% Triton X-100, 350 mM NaCl, 10 mM mercaptoethanol, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride), sonicated, and then centrifuged at 12,000 × g for 15 min at 4°C. The supernatant was collected, mixed with anti-FLAG M2 affinity gel (200 μl), gently rotated for 2 h at 4°C, and washed five times with lysis buffer. FLAG-HMGB1 was eluted using lysis buffer (100 μl) containing the FLAG peptide (30 μg/ml) and dialyzed with H100 buffer (20 mM HEPES-KOH, pH 7.5, 100 mM KCl, 1 mM MgCl2, 1 mM mercaptoethanol, and 0.1 mM EDTA) at 4°C overnight. Protein concentration was determined by comparing the intensity of Coomassie-stained purified protein bands with those of standard bovine serum albumin (BSA) protein (Sigma).
Cytoplasmic/nuclear protein fractionation.
Cytoplasmic/nuclear protein fractionation was performed as described by Kim et al. (24).
Immunocytochemistry and immunohistochemistry.
Immunocytochemistry was performed as previously described (22). For immunohistochemistry, paraffin-embedded liver tissues from HCV patients were deparaffinized and rehydrated with xylene and ethanol. Antigenic epitopes were exposed by treating samples with 10 mM citrate buffer and heating in a microwave oven. Samples were sliced and incubated in blocking solution (5% horse serum and 0.02% Triton X-100 in PBS) at room temperature for 4 h, followed by an additional overnight incubation with primary antibodies. After washing with blocking buffer, the samples were incubated with secondary antibodies. The subcellular localization of HMGB1 and core proteins was observed and analyzed using an Olympus inverted confocal microscope (FV1000) and accompanying software. Quantitative analyses of fluorescence intensities were performed using Metamorph and ImageJ softwares.
Transfection of DNA and siRNA.
Cells seeded on 12-well plates (4 × 104 cells/well) were transfected with siRNAs (20 nM final concentration) and DNAs (1 μg/well) using Oligofectamine (Invitrogen) and Fugene HD (Roche), respectively, according to the manufacturer's instructions. The small interfering RNA (siRNA) targeting human TLR4 mRNA (5′-CUG CAU AAA GUA UGG UAG A-3′) and a control siRNA were purchased from Bioneer Inc. (South Korea).
Luciferase assay.
Luciferase assays were performed using a luciferase assay kit (Promega) according to the manufacturer's instructions. Luciferase activity in cells was normalized to protein concentrations determined by Bradford assays.
Statistical analysis.
Results are expressed as means ± standard deviations. Comparisons between two groups were performed using Student's t test. Differences with P values of <0.05 were considered significant.
RESULTS
HCV infection promotes nuclear-to-cytoplasmic translocation of HMGB1 in virus-infected cells.
The subcellular localization of HMGB1 changes depending on environmental conditions. For instance, oxidative stress induces HMGB1 translocation from the nucleus to the cytoplasm (37, 38). Because HCV infection induces ROS production (9, 40), we assessed HMGB1 dynamics during HCV infection using an immunocytochemical method with confocal immunofluorescence microscopy detection. Huh7.5.1 cells were mock infected or infected with HCV (JFH; multiplicity of infection of 1), grown on glass coverslips, fixed with paraformaldehyde, and processed for indirect immunofluorescence with anti-core and anti-HMGB1 antibodies. One day postinfection (1 dpi), HCV core proteins were observed mainly around lipid droplets (Fig. 1A). Interestingly, a portion of HMGB1 protein was translocated from the nucleus to the cytoplasm in HCV-infected cells within 1 dpi, when the viral protein core could first be detected by Western blotting, although a majority of the protein remained in the nucleus (Fig. 1A and B). Consistently with this, the amount of HMGB1 in the cytoplasm increased more than 3-fold at 1 and 2 dpi, as monitored by the fractionation of cell extracts and Western blotting (Fig. 1B). Moreover, the secretion of HMGB1 into the medium after HCV infection was detected at 1 and 2 dpi by Western blotting, even though no cytopathic effects were observed at these time points (Fig. 1C and D). These results indicate that HCV infection triggers the translocation and secretion of HMGB1 proteins in virus-infected cells.
Fig. 1.
Translocation of HMGB1 in HCV-infected cells. (A) Huh7.5.1 cells infected with JFH or mock infected were analyzed immunocytochemically 1 day (1d) or 2 days (2d) after infection using an anti-HCV core (green) or anti-HMGB1 (red) antibody. Representative fluorescence micrographic images (HMGB1 and Core) and merged images (Merged) are shown. Combined merged and differential interference contrast microscopic (DIC) images (Merged + DIC) also are shown. Scale bar, 5 μm. (B) Cell extracts from Huh7.5.1 cells, mock infected or HCV infected for 1 or 2 days, were divided into cytosolic (Cyt) and nuclear (Nu) fractions. The fractionation efficiency was confirmed by Western blotting for PARP and GAPDH, which are representative nuclear and cytoplasmic proteins, respectively. The infection of HCV was confirmed by the production of HCV core protein (Core). The levels of nuclear and cytoplasmic HMGB1 protein were analyzed by Western blotting (HMGB1). The intensities of HMGB1 protein bands were quantified densitometrically using ImageJ software. Band intensities of nuclear and cytoplasmic HMGB1 were normalized to those of GAPDH and PARP, respectively. Band intensity values relative to those in mock-infected cells are depicted in the bottom panel. The mean values and standard deviations are depicted as numbers and bars, respectively (*, P < 0.05 versus the mock-infected group). (C) The amount of HMGB1 protein secreted by HCV-infected and uninfected cells was analyzed by Western blotting with an anti-HMGB1 antibody. (D) The viability of Huh7.5.1 cells before and 1 and 2 days after HCV infection was determined by measuring the release of adenylate kinase (AK) from damaged cells into the culture medium using the ToxiLight BioAssay kit (Cambrex) according to the manufacturer's instructions. The means and standard deviations are depicted as numbers and bars, respectively.
Cytoplasmic HMGB1 is observed in HCV-infected cells of HCV patients.
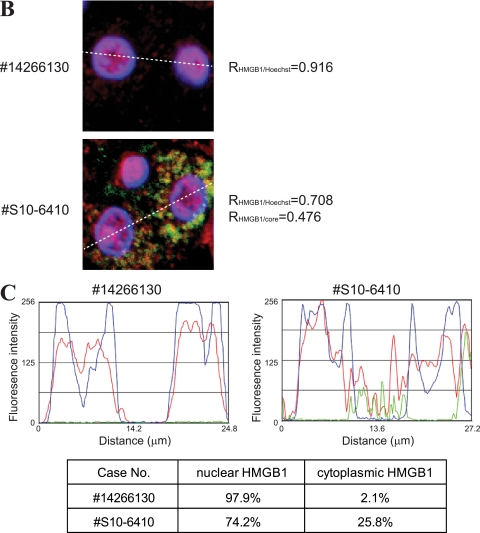
To confirm the physiological relevance of the translocation of HMGB1 in HCV-infected cells, we monitored the subcellular localization of HMGB1 in the livers of HCV patients. HMGB1 proteins were detected mainly in the nuclei of normal liver cells (Fig. 2A, first row) and uninfected liver cells (defined by the absence of HCV core protein) of an HCV patient (Fig. 2A, asterisked cells in the second row), whereas a large portion of HMGB1 proteins was observed in the cytoplasm of HCV-infected liver cells from the same patient (Fig. 2A, second row). A similar redistribution of HMGB1 protein also was observed in liver cancer tissues from patients infected with HCV (Fig. 2A, third and fourth rows). The redistribution of HMGB1 protein was confirmed by quantitative analyses of fluorescence intensities corresponding to HMGB1, HCV core, and Hoechst (Fig. 2B and C). A more than 10-fold increase of cytoplasmic HMGB1 was observed in the HCV-infected cells (Fig. 2C). Patient clinical data are summarized in Table 1. As noted above, elevated levels of HMGB1 have been observed in the sera of patients with liver diseases, such as chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (8). Taken together with this previous report, these data suggest that secreted HMGB1 is involved in the pathogenesis of HCV.
Fig. 2.
Subcellular localization of HMGB1 in the livers of HCV-infected patients. (A) Liver tissues from an uninfected cohort (number 14266130) and HCV patients (numbers S10-6410, S09-2974, and T6) were immunostained with antibodies against HCV core (green) and HMGB1 (red) and stained with Hoechst 33342 (blue). Representative fluorescence micrographic images (HMGB1 and Core), Hoechst-stained images (Hoechst), and merged images (Merged) are shown. Combined merged and DIC images (Merged + DIC) also are shown. Scale bar, 20 μm. Asterisks in the center panels indicate cells lacking HCV core protein (i.e., uninfected cells). (B) Magnified pictures of boxes in panel A. Correlation coefficients for the colocalization of these proteins (R) are depicted by using ImageJ software. (C) Fluorescence intensities of core (green), HMGB1 (red), and Hoechst (blue) corresponding to the white dashed lines in panel B are plotted by using the linescan function of Metamorph software. The proportion of cytoplasmic HMGB1 to nuclear HMGB1 in the HCV-infected cells and in the uninfected cells is depicted in the bottom panel.
Table 1.
Clinical background of HCV patients
| Case no. | Age (yr) | Gendera | Tumor diagnosis | Tumor size | Tumor grade | % Necrosis | Etiology | Pathology |
|---|---|---|---|---|---|---|---|---|
| 14266130 | 53 | F | 0 | Anti-HCV negative | Within normal limits | |||
| S10-6410 | 63 | F | 0 | HCV (2a) | Cirrhosis | |||
| S09-2974 | 61 | M | Hepatocellular carcinoma | 4 × 3 | 2∼3 | 0 | HCV (2b) | Cirrhosis |
| T6 | 61 | M | Hepatocellular carcinoma | 6.4 × 6.5 | 1∼2 | 0 | HCV | Fibrosis |
F, female; M, male.
Extracellular HMGB1 blocks HCV infection.
Because various cytokine functions of HMGB1 have been reported, we next investigated the effect of secreted HMGB1 on HCV infection. First, the effect of an anti-HMGB1 antibody on HCV infection was monitored by adding the antibody to culture medium after HCV (JFH/5aRluc) infection (22). This virus contains the Renilla luciferase gene inserted into the NS5A region and allows the quantitative measurement of HCV infectivity (22). We measured luciferase activity in Huh7.5.1 cells at 3 dpi, a time that corresponds to the first and second rounds of HCV infection. Under these conditions, this experiment monitors the autocrine and/or paracrine effects of HMGB1. We found that the HMGB1 antibody, but not the control antibody, augmented HCV infection up to 176% in a dose-dependent manner (Fig. 3A), indicating that secreted HMGB1 has an antiviral effect.
Fig. 3.
Effects of secreted HMGB1 protein on HCV infection. (A) Huh7.5.1 cells were inoculated with JFH/5aRluc virus for 4 h and then cultivated further in fresh medium containing different amounts of anti-HMGB1 or control IgG antibodies for 3 days. HCV-infected cells were lysed with passive lysis buffer, and luciferase assays were performed to monitor the proliferation of HCV. Luciferase activity in lysates (RLU values) was normalized to protein levels measured by Bradford assays. The mean values and standard deviations are depicted as numbers and bars, respectively (*, P < 0.05; **, P < 0.01 versus the mock-infected group). (B) Huh7.5.1 cells were treated with purified wild-type HMGB1 (30 or 300 ng/ml) or mutant HMGB1-C106S (30 or 300 ng/ml) proteins for 18 h and then infected with JFH/5aRluc virus for 3 days. HCV-infected cells were lysed with passive lysis buffer, and luciferase assays were performed to monitor the proliferation of HCV. RLU values were normalized to protein levels measured by Bradford assays. The mean values and standard deviations are depicted as numbers and bars, respectively (*, P < 0.05 versus the mock-infected group).
The anti-HCV effect of HMGB1 was further investigated using recombinant HMGB1 protein expressed in and purified from HEK293FT cells. As a negative control, we also generated and purified a mutant HMGB1 protein (HMGB1-C106S) containing a cysteine-to-serine substitution at residue 106, which abrogates HMGB1 activity by hampering its interaction with TLR4 (41). The pretreatment of Huh7.5.1 cells with wild-type HMGB1 for 18 h blocked HCV infection in a dose-dependent manner (Fig. 3B). The mutant HMGB1-C106S protein, on the other hand, showed no effect on HCV infection. These data strongly support the interpretation that HMGB1 proteins secreted from HCV-infected cells at least partially block the rapid propagation of HCV infection.
The anti-HCV activity of HMGB1 is mediated by TLR4.
To understand the molecular basis of the anti-HCV effect of HMGB1, we sought to identify the cellular receptor of HMGB1 that mediates HMGB1 signaling. We speculated that TLR4 was the most likely receptor candidate, since it not only triggers proinflammatory responses through NF-κB activation like other potential receptors (RAGE, TLR2, and TLR9) but also mediates antiviral responses through the activation of IFN response factor 3 (IRF-3) (2). To test this supposition, we knocked down TLR4 with siRNA and examined HCV infectivity. siRNA-mediated TLR4 knockdown (Fig. 4B) augmented HCV infection by about 40% (Fig. 4A). Conversely, the overexpression of TLR4 (Fig. 4D) reduced HCV infection by about 25% compared to that of mock-transfected cells (Fig. 4C). To further confirm the TLR4-mediated HMGB1 signaling, we investigated whether the knockdown of TLR4 renders HCV replication resistant to exogenous HMGB1 treatment. HCV production in the presence of exogenous HMGB1 was increased by 80% in TLR4 knockdown cells (Fig. 4E), and the inhibitory effect of the exogenous HMGB1 on HCV production in TLR4 knockdown cells was reduced significantly (from 44 to 28%) compared to that of control siRNA-transfected cells (Fig. 4E). The remaining inhibitory effect of HMGB1 in the siRNA-treated cells likely is attributable to the residual TLR4 proteins on the siRNA-treated cells and/or to other putative HMGB1 receptors, such as TLR2, TLR9, and RAGE, that might exist on the hepatocyte cells. These data indicate that TLR4 is responsible, at least in part, for mediating the anti-HCV activity of HMGB1.
Fig. 4.
Effects of TLR4 knockdown and overexpression on HCV infection. (A) Huh7.5.1 cells were transfected with an siRNA against TLR4 or a control siRNA using Oligofectamine. After 1 day of cultivation, siRNA-transfected cells were infected with JFH/5aRluc virus and further cultivated for 3 days. HCV-infected cells were lysed with passive lysis buffer, and luciferase assays were performed to monitor the proliferation of HCV. Luciferase activity in lysates was normalized to protein levels measured by Bradford assays. The mean values and standard deviations are depicted as numbers and bars, respectively (*, P < 0.05 versus the mock-infected group). (B) The Huh7.5.1 cells described in panel A were subjected to Western blotting with anti-TLR4 and anti-β-actin (control) antibodies. (C) Huh7.5.1 cells were transfected with pcDNA3.1 (negative control) or pcDNA3/myc-hTLR4 using Fugene HD. After 1 day of cultivation, transfected cells were infected with JFH/5aRluc virus and further cultivated for 3 days. Virus infectivity was monitored as described for panel A. (D) The Huh7.5.1 cells described for panel C were subjected to Western blotting with anti-TLR4 and anti-β-actin (control) antibodies. (E) Huh7.5.1 cells were transfected with an siRNA against TLR4 or a control siRNA using Oligofectamine. After 1 day of cultivation, siRNA-transfected cells were treated with purified wild-type HMGB1 (300 ng/ml) or mutant HMGB1-C106S (300 ng/ml) for 18 h and then infected with JFH/5aRluc virus for 3 days. Virus infectivity was monitored as described for panel A. (F) The Huh7.5.1 cells described in panel E were subjected to Western blotting with anti-TLR4 and anti-β-actin (control) antibodies.
HMGB1 triggers an interferon response.
We further analyzed the molecular basis of the antiviral activity of HMGB1 by testing the induction of the antiviral protein beta interferon (IFN-β), which blocks HCV proliferation (13), and IFN-stimulated gene 15 (ISG15), which is a good marker for the induction of interferon-responsive genes (33). Both IFN-β and ISG15 were greatly induced in Huh7.5.1 cells, the same cells used in HCV infection studies, by treatment with wild-type HMGB1 for 48 h (Fig. 5A). LPS, a positive control that triggers TLR4 signaling, also induced the expression of interferon-related genes (Fig. 5A). In contrast, treatment with mutant HMGB1-C106S produced a very limited induction of IFN-β and ISG15 (Fig. 5A). No reagents induced the expression of the negative control, GAPDH (Fig. 5A). To further validate the role of TLR4 in HMGB1-mediated signaling, we investigated the induction levels of IFN-β and ISG15 in TLR4 knockdown cells by treating wild-type HMGB1 for 48 h. As shown in Fig. 5B, the induction of IFN-β and ISG15 with wild-type HMGB1 treatment in the siRNA against TLR4-transfected cells was reduced by 63 and 39%, respectively, compared to that of the control siRNA-transfected cells. These data strongly suggest that secreted HMGB1 exerts anti-HCV activity by triggering an interferon response in HCV-infected cells and neighboring cells.
Fig. 5.
Induction of interferon-related genes by HMGB1 protein. (A) The mRNA levels of the interferon-related genes, IFN-β and ISG15, and the control gene, GAPDH, in Huh7.5.1 cells were monitored by quantitative RT-PCR after treatment with HMGB1 (300 ng/ml), HMGB1-C106S (300 ng/ml), or LPS (2 μg/ml) for the indicated times. The changes in mRNA concentration, normalized to the level of control mRNA (β-actin) 3 h after treatment, are depicted. The values presented in the graphs are means and standard deviations. (B) Huh7.5.1 cells were transfected with an siRNA against TLR4 or a control siRNA using Oligofectamine. After 1 day of cultivation, siRNA-transfected cells were treated with purified wild-type HMGB1 (300 ng/ml) or mutant HMGB1-C106S (300 ng/ml) for 48 h. The mRNA levels of IFN-β, ISG15, and β-actin were measured as described for panel A (*, P < 0.05 versus the control siRNA-transfected group).
DISCUSSION
Here, we provide the first demonstration that HCV infection triggers the translocation of a portion of HMGB1 protein from the nucleus to the cytoplasm and induces HMGB1 secretion into the extracellular milieu (Fig. 1 and 2). The release of HMGB1 protein from cells infected by various viruses has been reported previously. For instance, both RNA virus (West Nile virus, Dengue virus, and HIV-1 [3, 7, 10, 20]) and DNA virus (herpes simplex virus type 2 [6]) infections have been shown to result in the secretion of HMGB1 through apoptosis and/or necrosis. It has been further suggested that HMGB1 protein secreted in response to viral infections contributes to the pathogenesis of these infections (39). The elevation of HMGB1 levels in the plasma of HCV patients with chronic hepatitis, liver cirrhosis, and HCC (8) likely is attributable to both the active secretion of HMGB1 and cytopathic effects of HCV infection, even though we showed that the secretion of HMGB1 can occur without killing the HCV-infected cells from an in vitro HCV culture system (Fig. 1D).
In the current study, we focused on the function of secreted HMGB1 protein. Using an anti-HMGB1 antibody and purified HMGB1 protein, we showed that secreted HMGB1 partially blocks HCV infection (Fig. 3). This is the first report demonstrating an anti-HCV effect of HMGB1. By monitoring the effects of TLR4 knockdown and overexpression on HCV infection, we also showed that the anti-HCV effect of HMGB1 is mediated, at least in part, by TLR4 (Fig. 4). Moreover, we showed that HMGB1 induces the expression of interferon-related genes in the hepatocellular carcinoma cell line (Huh7.5.1) used in HCV infection studies (Fig. 5). These data lead us to conclude that HCV infection induces the secretion of HMGB1 protein into the extracellular milieu, and secreted HMGB1 triggers the production of antiviral proteins through TLR4-mediated interferon responses. We speculate that the anti-HCV effect of HMGB1 at least partly contributes to limiting the propagation of HCV in HCV patients. Moreover, secreted HMGB1 is likely to trigger inflammatory responses (16) by activating neighboring immune cells, such as Kupffer cells and hepatic dendritic cells, to release more proinflammatory cytokines. Therefore, HMGB1 by itself or in combination with other proinflammatory cytokines also may contribute to the chronic inflammation of liver cells (hepatitis) in HCV patients. Because TLR4 enhances the signaling of TGF-β, which is the cytokine best known to trigger liver cirrhosis and promote hepatic fibrosis in quiescent hepatic stellate cells (31), secreted HMGB1 also may be associated with liver cirrhosis, another pathological symptom caused by HCV infection. The involvement of TLR4 in liver cirrhosis also was strongly supported by a recent study indicating that a single-nucleotide polymorphism of TLR4 is the key factor in significantly reducing the risk of developing cirrhosis in patients with chronic HCV infection (18). These observations support the view that secreted HMGB1 functions as a double-edged sword: it protects hepatocytes from infection while at the same time contributing to triggering and maintaining a pathological status (e.g., in hepatitis and liver cirrhosis). Confirming the roles of HMGB1 in HCV pathology and the physiological activities of HMGB1 during HCV infection will require further investigation using appropriate animal model systems.
The molecular basis of the translocation of HMGB1 during HCV infection is not fully understood. However, the most likely mechanism involves ROS generated in HCV-infected cells. HCV proteins are known to cause endoplasmic reticulum (ER) stress, which results in the release of calcium from the ER. The released calcium, in turn, reduces mitochondrial membrane potential and increases ROS production (27). For example, HCV core and NS5A proteins induce oxidative stress in HCV-infected cells (15, 25, 26). In addition, it is known that HMGB1 proteins are translocated from the nucleus to cytoplasm or even secreted into the extracellular milieu under conditions in which intracellular ROS levels are elevated (36). We confirmed that ROS was elevated in HCV-infected cells (data not shown) and demonstrated that HMGB1 was secreted from cells treated with H2O2, which increased intracellular ROS levels (data not shown). These observations suggest that the elevated levels of ROS in HCV-infected cells contribute, at least in part, to the translocation of HMGB1.
Although this study did not focus on the role of cytoplasmic HMGB1, it is worth emphasizing the potential significance of the translocation of HMGB1 from the nucleus to the cytoplasm (Fig. 1A and B and 2). One possibility is that cytoplasmic HMGB1 is involved in the induction of autophagy in HCV-infected cells. Autophagy is known to play an important role in host immunity against virus infections (23). On the other hand, autophagy is known to be required for HCV replication (12), and the knockdown of proteins involved in autophagy enhances the innate immune response in HCV-infected hepatocytes (21, 32). A relationship between autophagy and HMGB1 has been reported recently (35). The authors of this study showed that cytoplasmic HMGB1 enhances autophagic flux by directly interacting with the autophagy protein Beclin1 and displacing Bcl-2 (35). Studies on the role of cytoplasmic HMGB1 in HCV pathogenesis are under way in our laboratory.
In conclusion, HMGB1 protein mobilized by HCV infection appears to play an important role in the pathogenesis of HCV-related diseases. Further investigations into the roles of HMGB1 in the diseases caused by HCV infection will shed light on and potentially help prevent these serious and prevalent HCV-related diseases.
ACKNOWLEDGMENTS
We are grateful to Ralf Bartenschlager (University of Heidelberg) for the core antibodies and to Francis Chisari (Scripps Research Institute) for the Huh7.5.1 cell line.
This work was supported in part by grants from the 21C Frontier Functional Proteomics Project (FPR08B1-220), Bio R&D Program (2010-0018167), World Class University Program (R31-10105), NCRC (2010-0028447), and BRL (2010-0019706) through the National Research Foundation funded by the Ministry of Education, Science and Technology.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Andersson U., Erlandsson-Harris H., Yang H., Tracey K. J. 2002. HMGB1 as a DNA-binding cytokine. J. Leukoc. Biol. 72:1084–1091 [PubMed] [Google Scholar]
- 2. Andersson U., Tracey K. J. 2010. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu. Rev. Immunol. 29:139–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barqasho B., Nowak P., Abdurahman S., Walther-Jallow L., Sonnerborg A. 2010. Implications of the release of high-mobility group box 1 protein from dying cells during human immunodeficiency virus type 1 infection in vitro. J. Gen. Virol. 91:1800–1809 [DOI] [PubMed] [Google Scholar]
- 4. Bertoletti A., Ferrari C. 2003. Kinetics of the immune response during HBV and HCV infection. Hepatology 38:4–13 [DOI] [PubMed] [Google Scholar]
- 5. Bianchi M. E. 2009. HMGB1 loves company. J. Leukoc. Biol. 86:573–576 [DOI] [PubMed] [Google Scholar]
- 6. Borde C., et al. 2011. Stepwise release of biologically active HMGB1 during HSV-2 infection. PLoS One 6:e16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L. C., Yeh T. M., Wu H. N., Lin Y. Y., Shyu H. W. 2008. Dengue virus infection induces passive release of high mobility group box 1 protein by epithelial cells. J. Infect. 56:143–150 [DOI] [PubMed] [Google Scholar]
- 8. Cheng B. Q., et al. 2008. Serum high mobility group box chromosomal protein 1 is associated with clinicopathologic features in patients with hepatocellular carcinoma. Dig. Liver Dis. 40:446–452 [DOI] [PubMed] [Google Scholar]
- 9. Choi J., Ou J. H. 2006. Mechanisms of liver injury. III. Oxidative stress in the pathogenesis of hepatitis C virus. Am. J. Physiol. Gastrointest. Liver Physiol. 290:G847–G851 [DOI] [PubMed] [Google Scholar]
- 10. Chu J. J., Ng M. L. 2003. The mechanism of cell death during West Nile virus infection is dependent on initial infectious dose. J. Gen. Virol. 84:3305–3314 [DOI] [PubMed] [Google Scholar]
- 11. Diamond D. L., et al. 2010. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 6:e1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dreux M., Gastaminza P., Wieland S. F., Chisari F. V. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Festi D., et al. 2004. Safety of interferon beta treatment for chronic HCV hepatitis. World J. Gastroenterol. 10:12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fisher C. L., Pei G. K. 1997. Modification of a PCR-based site-directed mutagenesis method. Biotechniques 23:570–574 [DOI] [PubMed] [Google Scholar]
- 15. Gong G., Waris G., Tanveer R., Siddiqui A. 2001. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc. Natl. Acad. Sci. U. S. A. 98:9599–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gremion C., Cerny A. 2005. Hepatitis C virus and the immune system: a concise review. Rev. Med. Virol. 15:235–268 [DOI] [PubMed] [Google Scholar]
- 17. Hoofnagle J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21–S29 [DOI] [PubMed] [Google Scholar]
- 18. Huang H., et al. 2007. A 7 gene signature identifies the risk of developing cirrhosis in patients with chronic hepatitis C. Hepatology 46:297–306 [DOI] [PubMed] [Google Scholar]
- 19. Ilmakunnas M., et al. 2008. High mobility group box 1 protein as a marker of hepatocellular injury in human liver transplantation. Liver Transpl. 14:1517–1525 [DOI] [PubMed] [Google Scholar]
- 20. Kamau E., et al. 2009. Dengue virus infection promotes translocation of high mobility group box 1 protein from the nucleus to the cytosol in dendritic cells, upregulates cytokine production and modulates virus replication. J. Gen. Virol. 90:1827–1835 [DOI] [PubMed] [Google Scholar]
- 21. Ke P. Y., Chen S. S. 2010. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J. Clin. Investig. 121:37–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim C. S., Jung J. H., Wakita T., Yoon S. K., Jang S. K. 2007. Monitoring the antiviral effect of alpha interferon on individual cells. J. Virol. 81:8814–8820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim H. J., Lee S., Jung J. U. 2010. When autophagy meets viruses: a double-edged sword with functions in defense and offense. Semin. Immunopathol. 32:323–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim J. H., et al. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23:708–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Korenaga M., et al. 2005. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J. Biol. Chem. 280:37481–37488 [DOI] [PubMed] [Google Scholar]
- 26. Okuda M., et al. 2002. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology 122:366–375 [DOI] [PubMed] [Google Scholar]
- 27. Piccoli C., et al. 2007. Hepatitis C virus protein expression causes calcium-mediated mitochondrial bioenergetic dysfunction and nitro-oxidative stress. Hepatology 46:58–65 [DOI] [PubMed] [Google Scholar]
- 28. Rullier A., et al. 2001. Immunohistochemical detection of HCV in cirrhosis, dysplastic nodules, and hepatocellular carcinomas with parallel-tissue quantitative RT-PCR. Mod. Pathol. 14:496–505 [DOI] [PubMed] [Google Scholar]
- 29. Scaffidi P., Misteli T., Bianchi M. E. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418:191–195 [DOI] [PubMed] [Google Scholar]
- 30. Seeff L. B. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35–S46 [DOI] [PubMed] [Google Scholar]
- 31. Seki E., et al. 2007. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat. Med. 13:1324–1332 [DOI] [PubMed] [Google Scholar]
- 32. Shrivastava S., Raychoudhuri A., Steele R., Ray R., Ray R. B. 2010. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology 53:406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Skaug B., Chen Z. J. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143:187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takeuchi O., Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820 [DOI] [PubMed] [Google Scholar]
- 35. Tang D., et al. 2010. Endogenous HMGB1 regulates autophagy. J. Cell Biol. 190:881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang D., Kang R., Zeh H. J., Lotze M. T. 2010. HMGB1, oxidative stress, and disease. Antioxid. Redox Signal. 14:1315–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang D., et al. 2007. Hydrogen peroxide stimulates macrophages and monocytes to actively release HMGB1. J. Leukoc. Biol. 81:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsung A., et al. 2007. HMGB1 release induced by liver ischemia involves Toll-like receptor 4 dependent reactive oxygen species production and calcium-mediated signaling. J. Exp. Med. 204:2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H., Ward M. F., Fan X. G., Sama A. E., Li W. 2006. Potential role of high mobility group box 1 in viral infectious diseases. Viral Immunol. 19:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Woodhouse S. D., et al. 2010. Transcriptome sequencing, microarray, and proteomic analyses reveal cellular and metabolic impact of hepatitis C virus infection in vitro. Hepatology 52:443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang H., et al. 2010. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc. Natl. Acad. Sci. U. S. A. 107:11942–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang H., Wang H., Tracey K. J. 2001. HMG-1 rediscovered as a cytokine. Shock 15:247–253 [DOI] [PubMed] [Google Scholar]