Abstract
We previously reported that the oncoproteins E6 and E7 from cutaneous human papillomavirus type 38 (HPV38) can immortalize primary human keratinocytes in vitro and sensitize transgenic mice to develop skin cancer in vivo. Immunofluorescence staining revealed that human keratinocytes immortalized by HPV38 E6 and E7 display fewer actin stress fibers than do control primary keratinocyte cells, raising the possibility of a role of the viral oncoproteins in the remodeling of the actin cytoskeleton. In this study, we show that HPV38 E7 induces actin stress fiber disruption and that this phenomenon correlates with its ability to downregulate Rho activity. The downregulation of Rho activity by HPV38 E7 is mediated through the activation of the CK2–MEK–extracellular signal-regulated kinase (ERK) pathway. In addition, HPV38 E7 is able to induce actin fiber disruption by binding directly to eukaryotic elongation factor 1A (eEF1A) and abolishing its effects on actin fiber formation. Finally, we found that the downregulation of Rho activity by HPV38 E7 through the CK2-MEK-ERK pathway facilitates cell growth proliferation. Taken together, our data support the conclusion that HPV38 E7 promotes keratinocyte proliferation in part by negatively regulating actin cytoskeleton fiber formation through the CK2-MEK-ERK-Rho pathway and by binding to eEF1A and inhibiting its effects on actin cytoskeleton remodeling.
INTRODUCTION
Human papillomaviruses (HPVs) are double-stranded small DNA viruses, comprising more than 100 members, that are the causative agents of several human diseases, including cancers (25). According to their tropisms, HPVs are subdivided into mucosal and cutaneous types. High-risk mucosal HPV types, such as HPV type 16 (HPV16) and HPV18, are etiological agents of cervical cancer and other anogenital cancers (25). Their ability to induce cellular immortalization and transformation is attributed primarily to the viral oncoproteins E6 and E7 (20, 25). While E6 prevents apoptosis by inducing the degradation of the tumor suppressor p53 through the proteasome system, E7 disrupts cell cycle regulation by inactivating pRb (20, 25). In addition, HPV16 E6 and E7 alter several other cellular signaling pathways by interacting with a number of cellular proteins, enhancing their carcinogenic properties (20, 38, 39, 41).
A large subgroup of cutaneous HPV types belonging to the beta genus of the HPV phylogenetic tree has been proposed to be involved in the development of nonmelanoma skin cancer (NMSC), since they were isolated for the first time from patients suffering from a rare autosomal recessive genetic disorder called epidermodysplasia verruciformis (EV), characterized by susceptibility to beta HPV infection and NMSC development at sun-exposed anatomical regions (7). Subsequent epidemiological studies have also provided lines of evidence for a possible oncogenic role of beta HPV types in non-EV patients, including the healthy population and immunocompromised individuals, e.g., organ transplant recipients (7, 51).
Our group has previously reported that E6 and E7 from beta HPV type 38 display transforming properties in vitro and in vivo (9, 15). Indeed, similar to HPV16, HPV38 E6 and E7 can efficiently immortalize primary human keratinocytes in vitro (9). Transgenic mice expressing HPV38 E6 and E7 are more susceptible to developing skin cancer induced by carcinogenic chemical agents (15). As with HPV16 E7, HPV38 E7 can associate with and inactivate the tumor suppressor pRb, leading to cell cycle deregulation (9). However, in contrast to HPV16 E6, which induces p53 degradation, thus affecting the control of DNA repair and apoptosis, the loss of p53 function by HPV38 E6 and E7 occurs through the accumulation of its inhibitor ΔNp73 (1). Moreover, the protein sequences of HPV38 E6 and E7 share a similarity of only less than 30% with HPV16 E6 and E7, while mucosal HPV18 E6 and E7 share almost 60% identical amino acid compositions with HPV16 E6 and E7. Additionally, HPV38 E6 and E7 may associate with various cellular proteins affecting other cellular signaling pathways that are important for virally induced cell transformation.
The actin filament, the filamentous form of actin molecules (F-actin), is one of the primary structural components of the eukaryotic cytoskeleton and plays a key role in the regulation of various cellular processes, including cell morphology, migration, cytokinesis, muscle contraction, and transformation (48, 54). Composed of the globular form of actin (G-actin), F-actin can assemble into higher orders of cellular structures, such as stress fibers and filopodia (43). Various environmental stimuli have been shown to be able to rapidly and dynamically regulate the actin filament structure. For instance, during cell transformation, many oncogenes commonly induce the disruption of the actin filament, which is believed to contribute to anchorage-independent proliferation (48–50). However, the role of actin stress fiber disruption by HPV oncoproteins in cell growth transformation has been poorly investigated.
In this study, we showed that HPV38 E7 can induce the disruption of actin stress fibers by lowering the level of F-actin in the cells by two distinct mechanisms. In fact, HPV38 E7 destroys actin stress fibers through the activation of the casein kinase 2 (CK2)–MEK–extracellular signal-regulated kinase (ERK) cascade, leading to the inhibition of Rho. In addition, HPV38 E7 alters actin stress fiber formation by binding to and inhibiting eukaryotic translation elongation factor 1A (eEF1A), which, in addition to its role in protein translation, binds cellular structures, including actin filaments, and increases levels of actin stress fibers (16, 57). Finally, our data demonstrate that the activation of the CK2-MEK-ERK-Rho cascade by HPV38 E7, leading to the disruption of the actin cytoskeleton, contributes to facilitating cell proliferation mediated by HPV38 E6 and E7.
MATERIALS AND METHODS
Cell culture, transfection, and growth curves.
Human epithelial kidney 293 (HEK 293) (adenovirus E1a- and E1b-transformed human embryonic kidney) cells and the human osteosarcoma cell line U2OS were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (61). Transient or stable transfection was performed with Fugene 6 reagent (Roche) according to the manufacturer's instructions. For stable transfection, G418 (800 μg/ml)-resistant colonies were selected and expanded (35). Human keratinocytes were maintained with NIH 3T3 feeder layers in FAD medium (9). Retroviral infection and antibiotic selection were conducted as previously described (9). Lipofectamine LTX and Plus reagent was specifically used to transfect primary and transduced keratinocytes. To determine growth curves, cells were seeded into 6-cm dishes at a density of 1 × 104 cells and cultured until the 11th day. The medium was changed every 48 h, and the number of cells was counted at different time points (days 1, 3, 5, 7, 9, and 11). The experiments were performed three times in triplicate. To determine the effects of kinase inhibitors, cells were treated with the following inhibitors resuspended in dimethyl sulfoxide (DMSO) at different concentrations: U0126 (25 μM), PD98059 (50 μM), LY294002 and API-2 (10 μM), (E)-3-(2,3,4,5-tetrabromophenyl)acrylic acid (TBCA; 10 μM), SP600125 (20 μM), and roscovitine (10 μM).
Plasmids.
pGEX-4T-1-HPV38E7 (glutathione S-transferase [GST]–HPV38 E7); pGEX-4T-1-HPV38E6 (GST-HPV38 E6); pEGFP-HPV38E7 (green fluorescent protein [GFP]-HPV38 E7); pcDNA3-Flag-HPV38E7 (F-HPV38 E7); pcDNA3-Flag-HPV38E7 deletion mutants (residues 1 to 15, 16 to 37, 38 to 101, 1 to 37, and 16 to 101); pcDNA-Flag-HPV1, -17, and -20 E7s; pcDNA3-Flag-human-eEF1A1 (F-eEF1A1); pcDNA3-Flag-human-eEF1A2 tissue (F-eEF1A2); the truncated mutants of pcDNA3-F-eEF1A1 (F-eEF1A1-A, residues 1 to 238; F-eEF1A1-B, residues 239 to 332; F-eEF1A1-C, residues 333 to 462); and additional C-terminally truncated mutants of eEF1A1 (residues 1 to 349, 1 to 364, 1 to 383, 1 to 416, 1 to 429, and 1 to 444) were generated by standard molecular cloning methods. The pRb-binding-defective mutant of HPV38 E7 in the pcDNA3-Flag and pLXSN vectors was generated by incorporating the two mutations of C24G and E26G (27) into the HPV38 E7 wild-type coding sequence by using a QuikChange II mutagenesis kit. pCMV-HA-human-eEF1A1 (hemagglutinin [HA]-eEF1A1) and pCMV-HA-mouse-eEF1A2 (HA-eEF1A2) were kindly provided by Stuart Pitson, Institute of Medical and Veterinary Science, Australia (33). pEGFP-Rho14V was obtained from Pierre Jurdic's laboratory, Ecole Normale Supérieure de Lyon, Lyon, France. The LifeAct-red fluorescent protein (RFP) plasmid was purchased from the Ibidi Company (Germany).
GST pulldown assay and mass spectrometry.
The purification of GST fusion proteins, extractions of cell lysates, and the GST pulldown assay were performed as described previously (2, 61). For mass spectrometry (MS) detection, lysates from 2 × 109 HEK 293 cells were subjected to a pulldown with glutathione-agarose-immobilized GST or GST-HPV38 E7 as described above, and protein precipitates were resolved by preparative SDS-PAGE. Proteins bound to GST-HPV38 E7 but not to GST were visualized by Coomassie brilliant blue staining, excised, and subjected to in-gel reduction, carbamidomethylation, and tryptic digestion. Multiple peptide sequences were determined by mass spectrometry performed by using a Q-Star XL electrospray ionization (ESI) quadrupole-time of flight (TOF) tandem mass spectrometer nanoESI-qQ-TOF-MS/MS (Applied Biosystems, France) coupled to an online nano-liquid chromatography (LC) system (Ultimate Famos Switchos; Dionex, the Netherlands). A 1-s TOF MS survey scan was acquired over 400 to 1,600 atomic mass units (amu), followed by three 3-s production scans over a mass range of 65 to 2,000 amu. The three most intense peptides with a charge state of 2 to 4 above a 30-count threshold were selected for fragmentation and were dynamically excluded for 60 s with 50 millimass units (mmu) of mass tolerance. The collision energy was set by the software according to the charge and mass of the precursor ion. The MS and tandem MS (MS/MS) data were recalibrated by using internal reference ions from a trypsin autolysis peptide at m/z 842.51 [M + H]+ and m/z 421.76 [M + 2H]2+.
Antibodies and reagents.
Antibodies against eEF1A, pRb, ERK1/2, phospho-ERK1/2 (Thr 202/204), Akt, phospho-Akt (Ser 473), cofilin, and phospho-cofilin (Ser 3) were obtained from Cell Signaling. Antibodies against Flag (M5), Flag coupled to beads (M2 beads), and actin were obtained from Sigma. Antibody against HA was obtained from Roche, and Rho-specific antibody was obtained from Millipore. Antibody against the CK2 alpha subunit was obtained from Santa Cruz. GST-specific antibody was a gift of G. Mosialos and E. Kieff (Harvard Medical School, Boston, MA). The kinase inhibitors for MEK (U0126 and PD98059), for Akt (LY294002 and API-2), for CK2 (TBCA), for Jun N-terminal protein kinase (JNK) (SP600125), for Cdk4 and Cdk6 (roscovitine), and for ROCK (Y27632) were obtained from Calbiochem. Latrunculin B and jasplakinolide were purchased from Santa Cruz Biotechnology.
Immunoprecipitation and Immunoblotting.
Immunoprecipitation and immunoblotting were performed as previously described (46). Briefly, cells were lysed in a lysis buffer containing 50 mM Tris-Cl (pH 7.4), 0.5% Nonidet P-40, 150 mM NaCl, 2 mM EDTA, 3% glycerol, and a Halt protease and phosphatase inhibitor cocktail (Pierce Biotechnology). After clearance with Sepharose 6 beads (Sigma), supernatants were added with anti-Flag (M2) beads or anti-eEF1A antibody plus protein A/G beads for immunoprecipitation. Equal amounts of supernatants and immunoprecipitated samples were loaded onto an SDS-PAGE gel, followed by the transfer of proteins onto polyvinylidene difluoride (PVDF) membranes (pore size, 0.45 μm). Immunoblotting was performed by sequentially incubating the membranes with the indicated primary antibodies and secondary antibodies, followed by visualization using an enhanced chemiluminescence detection system (Amersham Biosciences). To detect the small-fragment truncated proteins of Flag-tagged HPV38 E7 mutants (see Fig. 5I), a PVDF membrane with a pore size of 0.2 μm (Millipore) was used for protein transfer (18). For immunoprecipitation to detect F-actin binding, cells were lysed in a specific buffer (10 mM HEPES [pH 6.8], 5 mM MgCl2, 5 mM EGTA, 100 mM KCl, 1 mM dithiothreitol [DTT], 0.05% NP-40, 5 mM ATP) and gently sonicated on ice for 10 s. After sonication and lysis, immunoprecipitation was performed as described above.
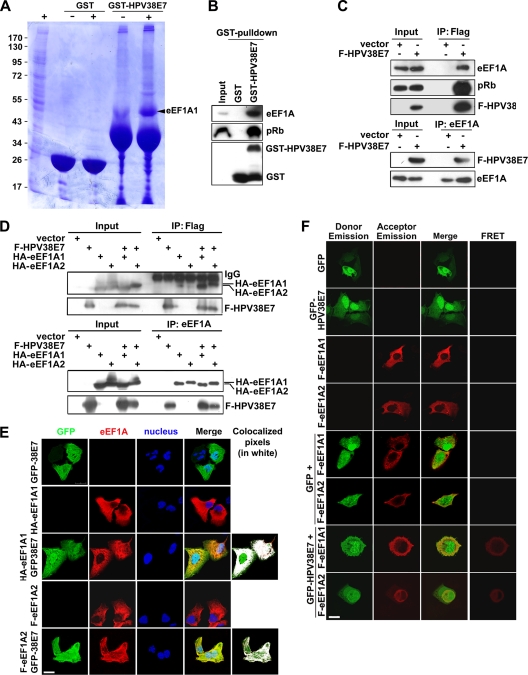
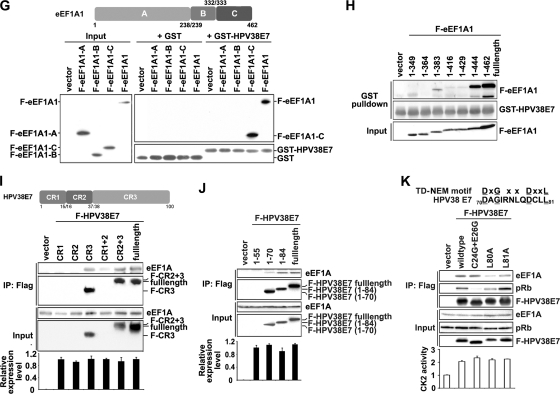
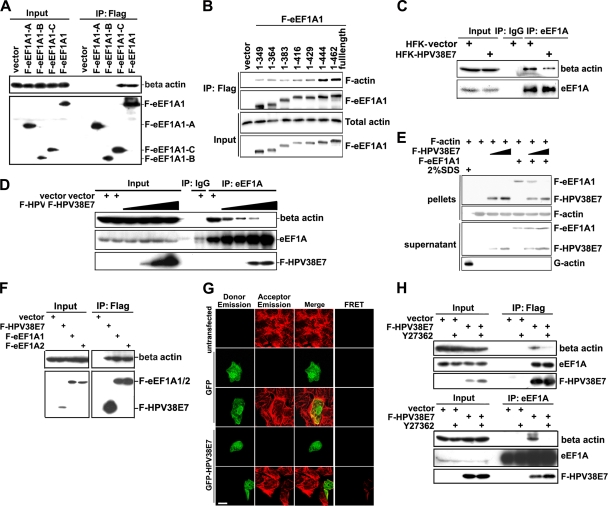
Fig. 5.
HPV38 E7 interacts and colocalizes with eEF1A. (A) HEK 293 cell lysates were incubated with GST or the GST-HPV38 E7 fusion protein bound to glutathione-Sepharose beads, and bound proteins were subjected to SDS-PAGE followed by Coomassie blue staining. The differential bands from GST-HPV38 E7-pulled-down samples were collected for mass spectrometry sequencing. The arrow indicates the eEF1A protein associated with GST-HPV38 E7. + and − indicate whether beads were incubated with or without cell lysates, respectively. (B) GST and the GST-HPV38 E7 fusion protein bound to glutathione-Sepharose beads were incubated with HEK 293 cell lysates. Following a GST pulldown as described in A, beads were used for SDS-PAGE and subjected to immunoblotting by the indicated antibodies. (C) HEK 293 cells were transfected with the expression plasmid for Flag-HPV38E7 (F-HPV38 E7), and 48 h after transfection, cell lysates were immunoprecipitated (IP) with anti-Flag (M2) beads or anti-eEF1A antibody plus protein A/G beads. Total lysates and immunoprecipitated proteins were detected by immunoblotting. (D) HEK 293 cells were transfected with the indicated expression plasmids and processed as described above (C). (E) U2OS cells were transfected with the indicated expression plasmids, and 24 h after transfection, cells were fixed and stained for immunofluorescence. Human HA-eEF1A1 and F-eEF1A2 (red) were stained with anti-HA and anti-Flag, respectively, using Alexa 555, and GFP-HPV38 E7 (green) was revealed by fluorescence. Nuclei were stained with DAPI. White was used to highlight the colocalized pixels using ImageJ 1.41o software. Bar, 25 μm. (F) U2OS cells were transfected and stained as described above (E). A FRET assay was performed to examine the association between HPV38 E7 and eEF1A isoforms. Bar, 25 μm. (G, top) schematic representation of eEF1A1 (462 aa) highlighting the three functional domains A (aa 1 to 238), B (aa 239 to 332), and C (aa 333 to 462). (Bottom) Lysates of HEK 293 cells expressing full-length (FL) Flag-eEF1A1 or Flag-eEF1A1 truncated mutants A, B, and C were incubated with GST and the GST-HPV38 E7 fusion protein bound to glutathione beads. After the pulldown assay, cobound proteins were detected by immunoblotting. (H) GST pulldown assay conducted by incubating the lysates of HEK 293 cells expressing full-length (FL) Flag-eEF1A1 or its C-terminally truncated mutants (aa 1 to 349, aa 1 to 364, aa 1 to 383, aa 1 to 416, aa 1 to 429, and aa 1 to 444) with the GST-HPV38 E7 fusion protein bound to glutathione beads, followed by immunoblotting. (I, top) Schematic representation of HPV38 E7 (101 aa) with its three domains, CR1 (aa 1 to 15), CR2 (aa 16 to 37), and CR3 (aa 38 to 101). (Bottom) The lysates of HEK 293 cells expressing full-length (FL) Flag-HPV38 E7 or its truncated mutants (CR1, CR2, CR3, CR1 plus CR2, and CR2 plus CR3) were immunoprecipitated (IP) with anti-Flag (M2) beads, followed by immunoblotting. CR1, CR2, and CR1 plus CR2 proteins were not detectable due to their small sizes (less than 5 kDa). Quantitative RT-PCR was performed to detect the expression level of full-length Flag-HPV38 E7 and its mutants. (J) Cell lysates of HEK 293 cells expressing full-length (FL) Flag-HPV38 E7 or its C-terminal deletion mutants (aa 1 to 50, aa 1 to 70, and aa 1 to 84) were immunoprecipitated (IP) with anti-Flag (M2) beads, followed by immunoblotting. Quantitative RT-PCR was performed to check the RNA expression level of all the mutants (bottom). (K, top) Alignment of the TD-NEM motif with a similar sequence in the HPV38 E7 protein. The coordinates of the corresponding amino acids are indicated. The conserved residues are underlined. (Middle) HPV38 E7 mutants were transfected into HEK 293 cells, and the lysate was analyzed by immunoprecipitation and immunoblotting. (Bottom) Level of CK2 enzymatic activity determined as described in the legend of Fig. 3A.
Immunofluorescence.
Immunofluorescence was performed as described previously, with minor modifications (46). Briefly, cells on cover slides were fixed in 4% formaldehyde at room temperature for 15 min and then permeabilized with 0.5% Triton X-100 in phosphate-buffered saline (PBS) for 20 min. Blocking was performed with a 5% bovine serum albumin (BSA)–PBS solution, followed by the incubation of the following primary antibodies diluted in blocking solution: mouse anti-HA (1:100) and mouse anti-Flag (M2) (1:400). The secondary antibodies were anti-mouse antibody coupled with Alexa 488, Alexa 555, or Alexa 647 (Invitrogen). The actin cytoskeleton and nuclei were visualized with Alexa 568-labeled phalloidin and DAPI (4′,6-diamidino-2-phenylindole), respectively. Slides were analyzed with the same settings by using the Leica TCS SP5 II spectral confocal system. The colocalization analysis was performed by using ImageJ software.
FRET.
For the detection of the association between GFP-HPV38 E7 and Flag-eEF1A isoforms (F-eEF1A1/2), cells were fixed, permeabilized, and stained with Alexa 555-labeled secondary antibody, as described above. After staining, cells were analyzed by using the Leica TCS SP5 II spectral confocal system. To measure fluorescence resonance energy transfer (FRET), three images were acquired sequentially in the same order through (i) a GFP filter set (excitation at 488 nm, emission at 520 to 580 nm, and filter at 488 nm), (ii) an Alexa 555 filter set (excitation at 516 nm, emission at 580 to 650 nm, and filter at 550 nm), and (iii) a FRET filter set (excitation at 488 nm, emission at 580 to 650 nm, and filter at 550 nm). For the detection of the association between GFP-HPV38 E7 and F-actin, cells were fixed, permeabilized, and stained with phalloidin-Alexa 568, as described above. After staining, cells were analyzed by using the Leica TCS SP5 II spectral confocal system. To measure FRET, three images were acquired sequentially in the same order through (i) a GFP filter set (excitation at 488 nm, emission at 520 to 580 nm, and filter at 488 nm), (ii) an Alexa 568 filter set (excitation at 543 nm, emission at 620 to 700 nm, and filter at 560 nm), and (iii) a FRET filter set (excitation at 488 nm, emission at 620 to 700 nm, and filter at 560 nm), according to data in previous reports (14, 44).
Quantification of F-actin and total actin levels.
Quantifications of F-actin and total actin levels were performed as previously reported (47). For F-actin quantification, cells were fixed in 4% formaldehyde in PBS by a 15-min incubation at room temperature. After being permeabilized with 0.2% Triton X-100 in PBS, cells were stained with Alexa 568-coupled phalloidin (1.5 μM in PBS) for 30 min at room temperature in the dark. Cells were then washed three times with PBS and scraped off into 0.5 ml of 0.1 M NaOH. Fluorescence was measured by using excitation and emission wavelengths of 550 and 600 nm, respectively. The optical density (OD) values were then normalized to the total protein concentration determined by using the BCA assay kit (Interchim) according to the manufacturer's instructions. For the quantification of the total actin level, cells were suspended in lysis buffer containing 10 mM K2HPO4, 100 mM NaF, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.2 mM dithiothreitol, 0.5% Triton X-100, and 1 M sucrose (pH 7.0). The total actin content was measured by reference to a standard curve for the inhibition of DNase I activity, prepared with commercial rabbit muscle actin (Sigma). The lysate protein concentration was also examined to normalize the total actin content. The final results are presented as the relative ratio of F-actin to total actin (averages ± standard deviations [SD]).
Reverse transcription (RT) and quantitative PCR.
Total RNAs were extracted and reverse transcribed by using an Absolutely RNA kit from Stratagene (La Jolla, CA). Quantitative PCR (Q-PCR) was conducted by using a MesaGreen qPCR MasterMix Plus for SYBR assay (Eurogentec, San Diego, CA) according to the manufacturer's instructions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used to normalize RNA inputs. The primers used are listed in the Table S1 in the supplemental material. The relative expression level of HPV38 E7 was calculated by using the comparative threshold cycle (CT) method (2−ΔΔCT) (37).
In vitro F-actin binding assay.
The in vitro F-actin binding assay was performed according to previously described protocols (28), with some modifications. In brief, lyophilized rabbit skeletal muscle actin (Sigma) was solubilized in buffer (5 mM Tris-HCl [pH 8.0], 0.2 mM CaCl2, 0.5 mM DTT, 0.2 mM ATP). After 1 h of incubation on ice, the solution was centrifuged for 1 h at 200,000 × g at 4°C to deplete residual actin polymers. The supernatant was mixed with polymerization stock buffer to produce a final concentration of 12.5 mM KCl, 0.5 mM MgCl2, and 0.25 mM ATP. After 1 h of incubation at room temperature, the solution was centrifuged for 30 min at 430,000 × g at 4°C. The F-actin pellets were washed with buffer (5 mM Tris-HCl [pH 8.0], 0.2 mM CaCl2, 0.5 mM DTT, 0.25 mM ATP, 12.5 mM KCl, 0.5 mM MgCl2) and resuspended in F-actin binding buffer (20 mM Tris-HCl [pH 8.0], 10 mM NaCl, 1 mM MgCl2, 1 mM ATP, and 1 mM dithiothreitol) at an actin concentration of 20 μM. The F-HPV38 E7 and F-eEF1A1 proteins were purified from transfected HEK 293 cells by immunoprecipitation with anti-Flag M2 beads. The F-HPV38 E7 and F-eEF1A1 proteins were then eluted from the beads by using Flag peptide (100 μg/ml; Sigma). Eluted Flag fusion proteins were concentrated with Microcon centrifugal filter columns (Ultracel YM-10 kDa; Millipore), and the concentration was determined. For binding, fusion proteins (5 or 15 μM) were incubated with 20 μM F-actin for 30 min and then centrifuged at 430,000 × g for 30 min to pellet F-actin. SDS (2%) was added to depolymerize F-actin to G-actin. Supernatants and pellets were collected for immunoblotting.
In vitro actin polymerization assay.
Actin polymerization assays were performed according to procedures reported previously (52), with some modifications. The F-HPV38E7 and F-eEF1A1 proteins were purified from transfected HEK 293 cells by immunoprecipitation as described above. G-actin (Sigma) was solubilized in buffer (20 mM HEPES [pH 7.2], 2 mM MgCl2, 2 mM EGTA, 1 mM dithiothreitol, 1 mM ATP, 0.25 mM GDP, and 1 mM phenylmethylsulfonyl fluoride) and precleared by ultracentrifugation (Beckman Optima) (150,000 × g for 30 min at 4°C). G-actin (5 μM) was then incubated with F-HPV38 E7 (0.5 μM), F-eEF1A (0.5 μM), or an equal amount of Flag peptide, in the presence or absence of latrunculin B and jasplakinolide (1 μM) in a total volume of 100 μl at 4°C for overnight. The mixture was then centrifuged at low speed (50,000 × g for 2 min at 4°C) to enrich bundled actins. Supernatants and pellets were analyzed by immunoblotting.
Rho-GTP pulldown assay.
Measurements of GTP-bound Rho levels were performed by using the Rho activation assay kit (Upstate Biotechnology). Briefly, cells were lysed in cold Mg2+ lysis/wash buffer (25 mM HEPES [pH 7.5], 150 mM NaCl, 1% legal CA-630, 10 mM MgCl2, 1 mM EDTA, 2% glycerol, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 25 mM sodium fluoride, and 1 mM sodium orthovanadate). Cell lysates with protein concentrations of above 5 mg/ml were incubated with the GST-tagged RhoA-binding domain of rhotekin-binding agarose beads to precipitate GTP-bound Rho. Precipitated samples were then detected by immunoblotting using the polyclonal anti-Rho (RhoA, -B, and -C) antibody included in the kit.
In vitro CK2 kinase activity assay.
CK2 peptide substrate (Arg-Arg-Arg-Glu-Glu-Glu-Thr-Glu-Glu-Glu; Promega) was used as a substrate for CK2 in a 50-μl reaction mixture under the following conditions: 25 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 200 mM NaCl, 0.1 mM ATP, 1 μCi [γ-32P]ATP, 3 mg of cell lysates, and 2 mg/ml CK2 peptide substrate. The reaction mixture was incubated at 37°C for 5 min, and 40 μl of the reaction mixture was then spotted onto a Whatman phosphocellulose P-81 filter, which was then washed 5 times in 200 ml of a 0.5% H3PO4 solution. The filter was finally rinsed with a small amount of ethanol for dryness and counted with a scintillation counter. Results are presented as relative radioactivities (averages ± SD) from three different samples.
RESULTS
HPV38 E7 disrupts actin stress fibers.
The staining of cells with F-actin dye revealed that primary human keratinocytes transduced with an empty vector contained much more actin stress fibers than did keratinocytes immortalized by HPV38 E6 and E7 (Fig. 1A). Actin stress fibers were clearly present all over the cytoplasm of primary keratinocytes, while such an organization was missing or altered in keratinocytes expressing the HPV38 oncoproteins E6 and E7, and F-actin was localized mainly at the periphery of the cells (Fig. 1A). The F-actin quantification presented in Fig. 1B correlates with the actin fiber loss in keratinocytes immortalized by HPV38 E6 and E7 shown in Fig. 1A.
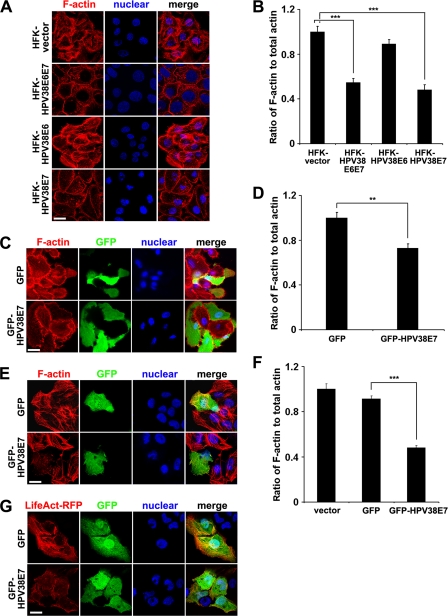
Fig. 1.
HPV38 E7 disrupts actin stress fibers. (A) Human primary forkhead keratinocytes (HFKs) containing the empty pLXSN vector (HFK-vector) or expressing HPV38 E6 and E7 (HFK-HPV38E6E7), HPV38 E6 (HFK-HPV38E6), or HPV38 E7 (HFK-HPV38E7) were visualized for F-actin with phalloidin-Alexa 568, and the nucleus was stained with DAPI. Bar, 25 μm. (B) Quantification of the F-actin/total actin ratio in the indicated human keratinocytes. Data are means ± standard deviations of results from three independent experiments carried out in triplicate. ***, P < 0.001. (C and D) Human primary keratinocytes were transfected with GFP or a GFP-HPV38 E7 plasmid, and 24 h later cells were processed as described above (A and B, respectively). **, P < 0.01. (E and F) U2OS cells were transfected with the indicated expression plasmids and processed as described above (A and B, respectively). ***, P < 0.001. (G) U2OS cells were cotransfected with GFP and LifeAct-RFP or GFP-HPV38 E7 and LifeAct-RFP plasmids. Images were taken to demonstrate actin fiber remodeling. Bar, 25 μm.
To evaluate the role of HPV38 E6 or E7 in the disruption of actin stress fibers, primary human keratinocytes were transduced with a retrovirus expressing E6 or E7, and the F-actin content was evaluated. While E6 had a moderate effect on actin stress fibers, keratinocytes expressing E7 showed a dramatic reduction in the amounts of actin stress fibers and F-actin (Fig. 1A and B). The effect of E7 alone is comparable to that of HPV38 E6 and E7 together, suggesting a major role of E7 in actin stress fiber disruption and in the decrease of the F-actin content. Furthermore, the direct role of HPV38 E7 in disrupting actin stress fibers, which corroborates its decreased F-actin content, was demonstrated by ectopically expressing the oncoprotein in human primary keratinocytes (Fig. 1C and D). Similarly, HPV38 E7 induced stress fiber disruption and a decrease in F-actin content in the heterologous human osteosarcoma cell line U2OS (Fig. 1E and F). Due to the low transfection efficiency in primary keratinocytes, the relative level of the decrease in the F-actin content induced by HPV38 E7 is not as high as that in the U2OS cell line. Since all the events observed with human primary keratinocytes with HPV38 E7 can be reproduced with U2OS cells (see also below), the latter cells were frequently used. To further evaluate the effects of HPV38 E7 on actin stress fiber remodeling, we used the LifeAct-GFP plasmid to monitor actin fiber loss by HPV38 E7. The LifeAct-GFP plasmid is a recently developed approach to visualize F-actin structures in eukaryotic cells and tissues without interfering with actin dynamics in vivo and in vitro (56). As shown in Fig. 1G, HPV38 E7- and LifeAct-RFP-coexpressing U2OS cells displayed the disrupted actin fiber structure stained by LifeAct-RFP. The percentage of the transfected cells expressing HPV38 E7 and LifeAct-RFP with obvious actin structure disruption reached up to 70%, a result from observations by two individuals. Monitoring of the dynamics of actin fibers using LifeAct-RFP confirmed the inhibitory effect of HPV38 E7 on actin stress fiber formation (see Fig. S1 in the supplemental material). Thus, HPV38 E7 is a strong inhibitor of F-actin structure formation, which leads to the loss of stress fibers.
HPV38 E7 disrupts actin stress fibers by activating the MEK-ERK pathway to inhibit Rho activity.
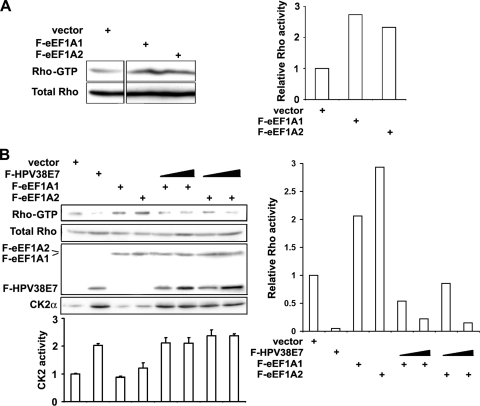
HPV38 E7 may inhibit actin stress fiber formation and remodeling by interacting directly with actin fibers, leading to conformational changes, and/or inhibiting the cellular signaling pathway that promotes fiber formation. The organization of actin filaments has been well established to be controlled by the Rho family members of small GTPases, Rho, Rac, and Cdc42 (6, 24). Among these, Rho regulates the assembly of actin stress fibers and focal contacts through the activation of the downstream effectors mDia and ROCK kinase (3, 64). Therefore, HPV38 E7 likely affects actin stress fiber formation through the inhibition of Rho GTPase. To evaluate this hypothesis, the level of the active form of Rho (Rho-GTP) in cells expressing HPV38 E7 was monitored. Indeed, the level of active Rho-GTP was significantly decreased in HPV38 E7-transduced human keratinocytes compared to the level in vector-transduced keratinocytes (Fig. 2A). Similar results were obtained with HPV38 E6- and E7-immortalized keratinocytes (data not shown) and with U2OS cells ectopically expressing HPV38 E7 (Fig. 2B).
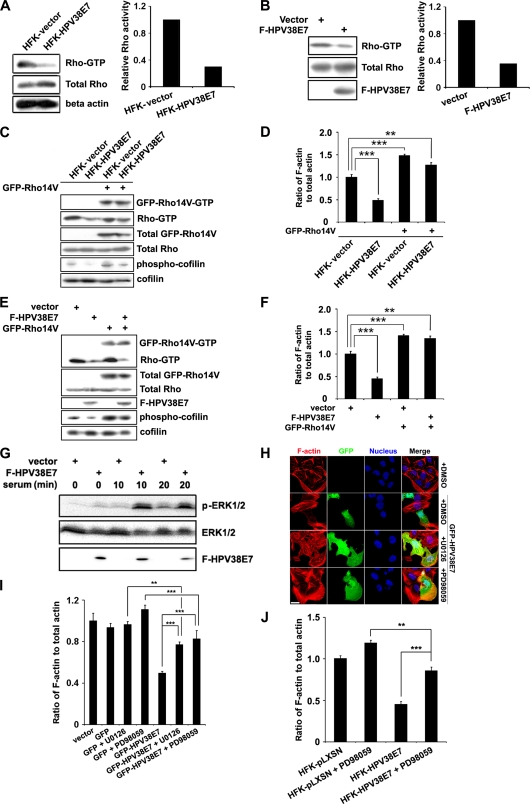
Fig. 2.
Effects of HPV38 E7 on the MEK-ERK-Rho signaling pathway in the regulation of actin stress fiber formation. (A) The Rho-GTP levels in vector-transduced HFKs and HFKs expressing HPV38 E7 detected by immunoblotting (left) and densitometry quantification of Rho-GTP compared to total Rho bands (right). (B) U2OS cells were transfected with expression plasmids for F-HPV38E7, and 48 h after transfection, cells were harvested to detect the Rho-GTP level. (C) Vector-transduced HFKs and HFKs expressing HPV38 E7 were transfected with the constitutively active mutant of Rho (GFP-Rho14V). Proteins were detected by immunoblotting. (D) Quantification of F-actin relative to the total actin from the indicated cells. ***, P < 0.001; **, P < 0.01. (E) U2OS cells were transfected with the indicated plasmids, and protein levels were detected by immunoblotting. (F). Quantification of F-actin relative to the total actin from the indicated cells. ***, P < 0.001; **, P < 0.01. (G) Cells were transfected with the F-HPV38 E7 plasmid or empty vector, and 24 h after transfection, cells were serum starved for 18 h. The level of ERK activation was determined by immunoblotting at the indicated time points after serum restoration at 10%. (H) U2OS cells were transfected with the GFP-HPV38 E7-expressing plasmid, and 24 h later, cells were treated with the MEK inhibitors U0126 and PD98059 or treated with vehicle (DMSO) for 12 h. F-actin and the nucleus were visualized by staining with phalloidin-Alexa 568 and DAPI, respectively. Bar, 25 μm. (I) Quantification of F-actin relative to the total actin in U2OS cells under the indicated conditions. ***, P < 0.001; **, P < 0.01. (J) Vector- or HPV38 E7-transduced human primary keratinocytes were treated with the indicated MEK inhibitors for 12 h. The quantification of F-actin relative to the total actin is presented. ***, P < 0.001; **, P < 0.01.
To further monitor the role of HPV38 E7 in the downregulation of Rho-GTP, the phosphorylation level of cofilin, the downstream effector of activated Rho-GTP and ROCK kinase (6, 24) (see Fig. 10), was evaluated. Interestingly, the low level of activated Rho-GTP in keratinocytes expressing HPV38 E7 correlated with a significant decrease in the level of phospho-cofilin (Fig. 2C). Moreover, the overexpression of the constitutively active mutant of Rho (GFP-Rho14V) in keratinocytes transduced by HPV38 E7 can partially rescue the inhibitory effect of HPV38 E7 on the level of phosphorylated cofilin (Fig. 2C) and F-actin contents (Fig. 2D). These observations were also confirmed with U2OS cells ectopically expressing HPV38 E7 (Fig. 2E and F). Of note, HPV38 E7 has no inhibitory effect on the constitutive activated Rho mutant (Rho14V), in contrast to the endogenous Rho-GTP (Fig. 2A, B, C, and E). Thus, the ability of HPV38 E7 to destroy actin stress fibers correlates with its inhibitory effects on Rho activation.
Fig. 10.
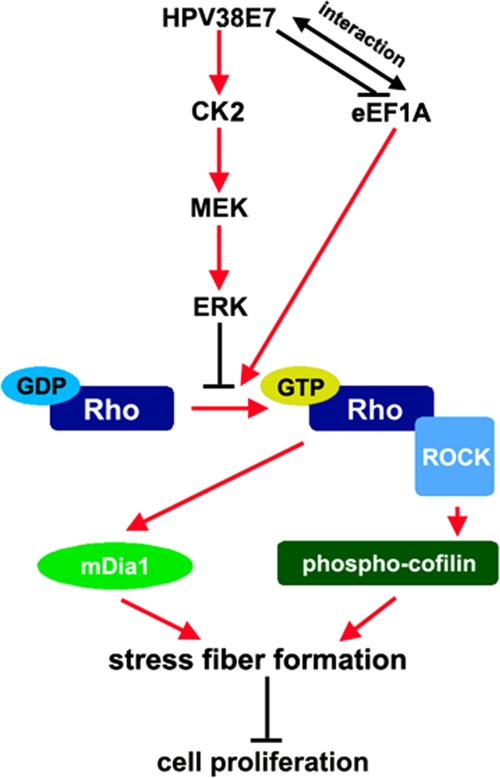
Schematic model of the mechanisms of HPV38 E7-induced actin cytoskeleton disruption in cell growth proliferation. Activated Rho-GTP promotes actin stress fiber formation that inhibits cell proliferation. The activation of ERK inhibits the accumulation of Rho-GTP, therefore preventing actin stress fiber formation and promoting cell proliferation. HPV38 E7 induces the disruption of actin stress fiber formation by activating CK2, the upstream activator of MEK-ERK signaling. eEF1A1 can promote the formation of actin stress fibers and therefore prevents cell proliferation by inhibiting Rho activation. HPV38 E7 can also promote the disruption of actin stress fibers by associating with eEF1A and blocking its effects on Rho-GTP accumulation, thus promoting cell proliferation. HPV38 E7 can also interact directly with F-actin and may induce conformal changes (not highlighted in the figure), which could lead to actin stress fiber remodeling and cell proliferation.
A number of protein kinases, including mitogen-activated protein kinases (MAPKs), regulate various cellular activities and actin cytoskeleton remodeling in response to extracellular stimuli (8, 31). Several viral oncoproteins also activate the Raf-MEK-ERK pathway to induce cellular transformation (21, 53). In addition, the MEK-ERK pathway can inhibit Rho and its downstream kinase ROCK (31, 49), and this contributes to the disruption of the actin stress fibers. HPV38 E7 may then induce actin stress fiber disruptions through the activation of the MEK-ERK pathway and the subsequent inhibition of Rho (Fig. 2A and B). To illustrate this possibility, we first demonstrated that HPV38 E7 is indeed able to activate ERK activity and that this activation is enhanced in the presence of serum stimulation (Fig. 2G). An early independent study conducted with a rat fibroblast cell line showed that HPV16 E7 activates ERK (66). This implies that the activation of the MEK-ERK pathway may be a common event for multiple HPV types. To establish a direct link between HPV38 E7-induced ERK activation and the disruption of actin stress fibers, U2OS cells expressing HPV38 E7 were exposed or not to two different MEK inhibitors, U0126 and PD98059, and examined for the disruption of actin stress fibers. As illustrated in Fig. 2H, MEK inhibitors significantly restored the actin stress fibers destroyed by HPV38 E7. These data were also correlated with a significant restoration of the F-actin content by MEK inhibitors (Fig. 2I). Similar results were obtained with HPV38 E7-expressing keratinocytes exposed to MEK inhibitors (Fig. 2J). Other inhibitors affecting kinases, such as Akt, JNK, or cyclin-dependent kinases (CDKs), had no significant effects on actin fiber remodeling (data not shown), demonstrating the specific and important role of ERK activation in the downregulation of actin stress fiber formation. Thus, the HPV38 E7-induced actin stress fiber disruption is correlated with its ability to activate the MEK-ERK pathway, which then downregulates Rho activity. Moreover, it was shown previously that HPV16 E7 modulates Rho activity but through the Akt pathway (11). This highlights one of the differences between HPV38 E7 and HPV16 E7.
Casein kinase 2 is the upstream regulator of the MEK-ERK-Rho cascade that is activated by HPV38 E7 during actin stress fiber disruption.
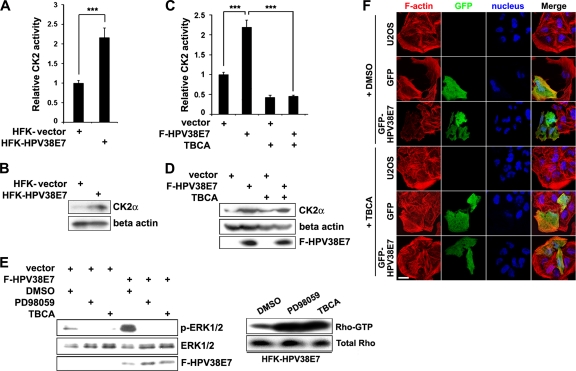
Casein kinase 2 (CK2) is a ubiquitous and constitutively active Ser/Thr protein kinase that is involved in a number of cellular signaling pathways and functions (23, 40). CK2 can activate the Raf-MEK-ERK pathway through the recruitment of the scaffold protein kinase suppressor of Ras (KSR) and plays a role in tumorigenesis (58). It was reported previously that CK2 can phosphorylate the E7 protein of high-risk mucosal HPV types 16 and 18, and this phosphorylation appears to be important for HPV16 and HPV18 to induce cell proliferation (5, 12, 17, 19). In the case of HPV38, we observed that HPV38 E7 is not a CK2 substrate in in vitro kinase assays (data not shown). Since the disruption of actin stress fibers by HPV38 E7 is dependent on the activation of the MEK-ERK pathway (Fig. 2), we hypothesized that HPV38 E7 may target CK2 to induce the disruption of the actin cytoskeleton via this pathway. To test this possibility, we examined whether HPV38 E7 can activate CK2. We first performed in vitro kinase assays and observed that the level of phosphorylation of the CK2-specific substrate peptide was increased more than 2-fold in the lysates of HPV38 E7-transduced keratinocytes (Fig. 3A). The high level of CK2 enzymatic activity correlated with the increased expression level of the CK2 alpha subunit protein in these cells (Fig. 3B). Thus, HPV38 E7 may induce CK2 enzymatic activity by increasing its protein level. Similar results were obtained when HPV38 E7 was ectopically expressed in heterologous U2OS cells (Fig. 3C and D). The specificity of the CK2 activation induced by HPV38 E7 was confirmed by using the CK2 chemical inhibitor TBCA, which significantly inhibited the enzymatic activity of CK2 without affecting its protein level (Fig. 3C and D).
Fig. 3.
HPV38 E7-induced disruption of the actin cytoskeleton is mediated through the CK2-MEK-ERK-Rho signaling pathway. (A) Equal amounts of lysates from vector-transduced HFKs or HFKs expressing HPV38 E7 were incubated with a CK2 substrate peptide in the presence of 32P-labeled ATP for CK2 activity measurements. The level of radioactivity incorporated into the CK2 substrate peptide was measured, and data are means ± standard deviations from three different experiments performed in duplicate. ***, P < 0.001. (B) Immunoblotting was conducted to determine the expression level of the CK2 alpha subunit (CK2α) in the cell lysates used in A. (C) U2OS cells were transfected with empty vector or the F-HPV38 E7 plasmid, and 24 h after transfection, the CK2 activity assay was performed as described above (A). TBCA, a chemical CK2 kinase inhibitor, was used to monitor the specificity of the reaction. ***, P < 0.001. (D) Immunoblotting was conducted to determine the expression level of the indicated proteins in the cell lysates used in C. (E, left) U2OS cells were transfected with empty vector or the F-HPV38 E7 plasmid, and 24 h later, cells were serum starved for 18 h and treated with a MEK inhibitor (PD98059) or CK2 inhibitor (TBCA) for 6 h. Cells were then cultured in normal medium with 10% serum for 20 min, and the level of activated ERK was detected by immunoblotting. (Right) HFKs expressing HPV38 E7 were treated with the indicated inhibitors or not treated for 12 h, and the level of Rho-GTP was determined in cell lysates as described in the legend of Fig. 2A. (F) Cells were transfected with the indicated expression plasmids. Twenty-four hours after transfection, cells were treated with the CK2 inhibitor TBCA for 12 h before staining with phalloidin-Alexa 568 and DAPI for immunofluorescence. Bar, 25 μm.
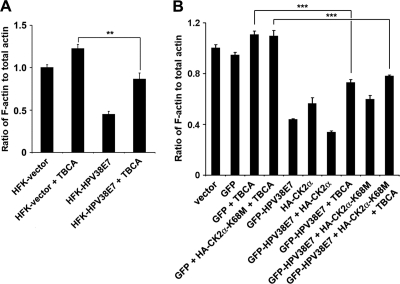
Similar to the MEK inhibitor, the CK2 inhibitor TBCA also abolished ERK activity upregulated by HPV38 E7 (Fig. 3E, left). Importantly, both inhibitors, i.e., CK2 and MEK inhibitors, induced a significant increase in the level of Rho-GTP (Fig. 3E, right), in agreement with the potential implication of the role of CK2 in actin stress fiber remodeling. To establish a direct link between CK2 activation and the disruption of actin stress fibers, HPV38 E7-expressing cells with a decreased level of actin stress fibers were treated with the CK2 inhibitor, and the actin stress fiber content was examined by immunofluorescence. As expected, cells expressing HPV38 E7 had fewer actin stress fibers than did control cells (Fig. 3F, compare the stress fiber contents of the GFP-expressing and the GFP-HPV38 E7-expressing cells). Treatment with the CK2 inhibitor significantly restored the actin fibers in HPV38 E7-expressing cells (Fig. 3F). These observations are also consistent with the F-actin content in primary keratinocytes transduced with HPV38 E7 and treated with the CK2 inhibitor (Fig. 4A). Interestingly, the overexpression of the CK2 alpha subunit resulted in decreased F-actin contents and a disruption of actin fibers, while more fibers were observed for non-CK2-overexpressing cells or for cells expressing the CK2 kinase-dead mutant (Fig. 4B and see Fig. S2 in the supplemental material). However, neither the CK2 inhibitor nor the CK2 kinase-dead mutant, alone or in combination, could completely rescue the loss of actin stress fibers induced by HPV38 E7, suggesting the existence of an additional mechanism(s) (see below).
Fig. 4.
Blocking of CK2 activity cannot completely rescue the actin fiber loss induced by HPV38 E7. (A) Vector-transduced HFKs and HFKs expressing HPV38 E7 were treated with the CK2 inhibitor TBCA for 12 h. The quantification of the F-actin/total actin ratio is presented. **, P < 0.01. (B) U2OS cells were transfected with the indicated expression plasmids in the presence or absence of the CK2 inhibitor TBCA. Twenty-four hours later, cells were lysed to quantify the level of F-actin and total actin. ***, P < 0.001.
HPV38 E7 associates with eEF1A.
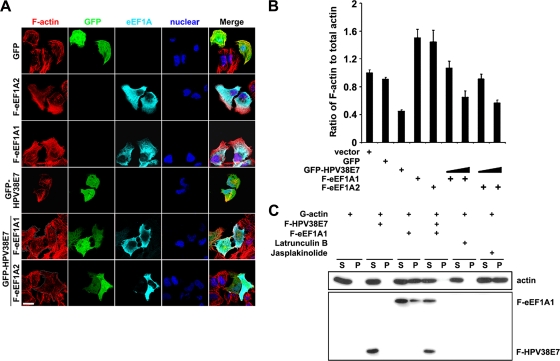
To explore other possible mechanisms by which HPV38 E7 alters actin stress fiber formation, we determined whether HPV38 E7 has the ability to interact with cellular proteins involved in actin stress fiber remodeling. To this end, a bacterially expressed GST-HPV38 E7 protein was used to perform affinity purification followed by mass spectrometry sequencing. Human eukaryotic elongation factor 1A (eEF1A) was identified as one of the cellular proteins that associated with HPV38 E7 (Fig. 5A). eEF1A, as the alpha subunit of the elongation factor 1 complex, is responsible for the enzymatic delivery of aminoacyl-tRNAs to the ribosome (57). In addition to its functions in protein translation elongation, eEF1A was shown previously to associate with some important cellular processes, such as apoptosis, carcinogenesis, and signal transduction (16). These additional eEF1A activities appear to be mediated by its ability to bind cellular structures, including actin filaments, and increase numbers of actin stress fibers. GST pulldown assays followed by immunoblotting showed that HPV38 E7 strongly associated with eEF1A as well as pRb, a well-characterized target of E7 (Fig. 5B) (9). The interaction between HPV38 E7 and endogenous eEF1A was also confirmed with HEK 293 cells expressing Flag epitope-tagged HPV38 E7 (Fig. 5C). Similar data were obtained for keratinocytes ectopically expressing the HPV38 E7 protein (data not shown). Since HPV38 E7 interacts with pRb, it is possible that the formation of the complex between HPV38 E7 and eEF1A requires pRb. To test this hypothesis, we generated an HPV38 E7 mutant defective in binding to pRb and evaluated its ability to associate with eEF1A. We found, as shown in Fig. 5K, that the pRb-binding-defective mutant of HPV38 E7 (C24G/E26G) binds as efficiently to eEF1A as wild-type HPV38. Thus, the interaction between HPV38 E7 and eEF1A is independent of the presence of pRb. E7s from various cutaneous HPVs, including HPV types 1, 7, 10, 13, 17, 20, and 48, which appear to display much weaker transforming activities than those of HPV38 E7, were less efficient in binding to eEF1A and less potent in inhibiting Rho and decreasing the F-actin level (see Fig. S3 in the supplemental material).
Two isoforms of eEF1A have been identified so far. The eEF1A1 isoform is ubiquitously expressed in various tissues and cell lines (32), while the eEF1A2 isoform is limited to the brain, heart, and skeletal muscle (29, 34). HPV38 E7 interacted with both eEF1A isoforms with similar affinities (Fig. 5D). In agreement with the immunoprecipitation data, immunofluorescence staining showed that HPV38 E7 colocalized with eEF1A1 and eEF1A2 (Fig. 5E). ImageJ 1.41o software was used to analyze the red and green channels in colocalization situations. HPV38 E7 was found to exhibit a strong colocalization with eEF1A1 and eEF1A2, a sign of a real interaction, as magnified by the white color, which highlights the colocalized pixels analyzed by ImageJ software (Fig. 5E, right). To further strengthen the interaction between HPV38 E7 and eEF1A1 and eEF1A2, a fluorescence resonance energy transfer (FRET) assay, a well-developed technology for studying protein-protein interactions in cells (14, 44), was performed. The data showed that the excitation of GFP-HPV38 E7, but not of GFP, can resonantly transfer energy to and therefore activate Alexa 555-labeled eEF1A isoforms (Fig. 5F). Thus, the HPV38 E7 protein is spatially associating with eEF1A. Taken together, these results reveal that HPV38 E7 interacts with eEF1A in the cells.
eEF1A interacts with HPV38 E7 through its C-terminal domain.
Human eEF1A1 contains three functional domains: domain A (amino acids [aa] 1 to 238), responsible for the GTP binding of aminoacyl-tRNAs to ribosomes; domain B (aa 239 to 332), involved in binding to charged tRNA; and domain C (aa 333 to 462), for binding to charged tRNA, other elongation factors, and actin filaments (F-actin) (Fig. 5G, top) (42, 45, 60, 63). A bioinformatics analysis using the Pfam database (http://pfam.sanger.ac.uk) showed that eEF1A2 contains similar domain components. GST pulldown experiments showed that HPV38 E7 interacts specifically with domains C of eEF1A1 (Fig. 5G, bottom) and eEF1A2 (data not shown). To further narrow down the interaction domain, various C-terminally truncated eEF1A1 mutants were generated. As shown in Fig. 5H, a mutant eEF1A1 (aa 1 to 444) exhibits an affinity of binding to GST-HPV38 E7 similar to that of full-length eEF1A1. A weaker interaction also occurred with another mutant eEF1A1 (aa 1 to 383). Thus, the eEF1A1 region between aa 430 and 444 in its C terminus contains the major HPV38 E7 interaction domain. Additional specific amino acid mutations in this domain allowed us to obtain the eEF1A(T430A/V431A) mutant, which exhibits a reduced affinity of eEF1A1 for HPV38 E7 (see Fig. S5A in the supplemental material). T430 and V431A were targeted for mutagenesis because they are important for the completeness of the beta sheet in the C domain of eEF1A (59). Taken together, these results indicated that this region (aa 430 to 444), in particular amino acids T430 and V431, is important for eEF1A1 to interact with HPV38 E7.
Identification of the HPV38 E7 domain interacting with eEF1A.
To identify the HPV38 E7 domain(s) required for its interaction with eEF1A, HPV38 E7 was divided into three conserved regions, CR1 (aa 1 to 15), CR2 (aa 16 to 37), and CR3 (aa 38 to 101), in reference to HPV16 E7 (Fig. 5I, top) (39). Flag-tagged proteins of CR1, CR2, CR3, CR1 plus CR2, and CR2 plus CR3 of HPV38 E7 were generated, and a coimmunoprecipitation assay was performed (Fig. 5I, bottom). For technical reasons, CR1, CR2, and CR1 plus CR2 were not detectable due to their small sizes (less than 5 kDa), although Q-PCR showed that all the mutants had RNA expression levels similar to those of full-length HPV38 E7 (Fig. 5I, bottom). The data indicated that HPV38 E7 CR3 and HPV38 E7 CR2 plus CR3 display affinities for endogenous eEF1A similar to those of full-length HPV38 E7 (Fig. 5I, middle). Thus, the CR3 domain of HPV38 E7 plays a major role in the efficient interaction with eEF1A. The possibility that CR1 and CR2 may also contribute to this interaction cannot be completely excluded. The generation of additional C-terminal deletion mutants (aa 1 to 55, 1 to 70, and 1 to 84) of HPV38 E7 indicated that the region in the CR3 domain between aa 70 and 84 mainly contributes to the interaction of HPV38 E7 with eEF1A (Fig. 5J). Interestingly, the region of aa 70 to 84 contains a sequence (70-DAGIRNLQDCLL-81) which shows similarity to the consensus motif of the transcription-dependent nuclear export motif (TD-NEM motif) (DxGxxDxxL) (Fig. 5K, top) (30). The TD-NEM motif mediates the nuclear export of proteins independently of the classical CRM1/nuclear export signal (NES) system and requires ongoing RNA polymerase II (Pol II)-mediated transcription. In this process, eEF1A plays a major role in nuclear exclusion by binding to the TD-NEM motif (30). We reasoned that the likely TD-NEM motif present in HPV38 E7 could mediate its interaction with eEF1A. To test this hypothesis, we generated two mutants of HPV38 E7, HPV38 E7(L80A) and HPV38 E7(L81A), and evaluated their interactions with eEF1A. As shown in Fig. 5K (middle), HPV38 E7(L80A) and HPV38 E7(L81A) exhibited less affinity for eEF1A than wild-type HPV38 E7, while the pRb-binding-defective mutant of HPV38 E7 [HPV38 E7(C24G/E26G)] still efficiently interacted with eEF1A. Therefore, the TD-NEM motif in the CR3 domain of HPV38 E7 plays a major role in its interaction with eEF1A. The possibility that eEF1A can also be involved in HPV38 E7 (a diffuse nuclear/cytoplasmic protein) nuclear exclusion is currently under investigation in our laboratory.
HPV38 E7 interferes with eEF1A binding to actin filaments.
It was previously reported that eEF1A can interact with F-actin (42). Consistent with these findings, we confirmed that eEF1A indeed associates with F-actin through its C domain (Fig. 6A). The use of C-terminally truncated mutants of eEF1A1 allowed us to narrow down the major F-actin-associating domain to aa 430 to 444 (Fig. 6B), the same region required for its binding to HPV38 E7 (Fig. 5H). In addition, the eEF1A1(T430A/V431A) mutant, defective in interactions with HPV38 E7, also exhibits a lower affinity for F-actin (see Fig. S5B in the supplemental material). Thus, HPV38 E7 may interfere with the association between eEF1A and F-actin. Indeed, the expression of eEF1A in the presence of increasing concentrations of HPV38 E7 nearly abolished the interaction of eEF1A with F-actin (Fig. 6D and E). In HPV38 E7-transduced keratinocytes, eEF1A also showed a reduced association with F-actin, compared to control cells (Fig. 6C). HPV38 E7 also interacts with F-actin, and this interaction occurs independently of eEF1A (Fig. 6E, F, and G). Thus, HPV38 E7 associates with F-actin and can block eEF1A binding to F-actin, probably by occupying and saturating the region of the C domain of eEF1A required for its association with F-actin.
Fig. 6.
HPV38 E7 interferes with eEF1A binding to F-actin. (A) U2OS cells were transfected with Flag-eEF1A1 truncated mutants A, B, and C, and 48 h after transfection, cells were lysed for immunoprecipitation using anti-Flag (M2) beads to detect F-actin binding, and proteins were detected by immunoblotting. (B) Cells were transfected with full-length Flag-eEF1A1 (aa 1 to 462) and its C-terminally truncated mutants (aa 1 to 349, aa 1 to 364, aa 1 to 383, aa 1 to 416, aa 1 to 429, and aa 1 to 444), and 48 h after transfection, immunoprecipitation and immunoblotting were performed as described above (A). (C) The lysates of vector-transduced HFKs and HFKs expressing HPV38 E7 in F-actin detection buffer were used for immunoprecipitation with anti-eEF1A. Immunoblotting was performed as described above (A). (D) Cells were transfected with increasing concentrations of the F-HPV38 E7-expressing plasmid, and 48 h after transfection, immunoprecipitation and immunoblotting were performed as described above (A). (E) F-actin was incubated with immunoprecipitated F-HPV38 E7 and/or F-eEF1A1. Supernatants and pellets were separated by centrifugation and analyzed by immunoblotting. F-actin and its binding proteins appear in the pellets, and G-actin and unbound proteins appear in the supernatant. SDS (2%), an actin-depolymerizing agent, was included in the reaction mixture as a control. (F) Cells were transfected with the indicated expression plasmids and processed as described above (A) to determine the association between HPV38 E7 and F-actin. (G) Cells were transfected with the indicated expression plasmids, and 24 h after transfection, F-actin was stained with phalloidin-Alexa 568. A FRET assay was carried out to examine the association between HPV38 E7 and F-actin. Bar, 25 μm. (H) Cells were transfected as indicated and treated with the ROCK inhibitor Y27632 for 12 h before harvesting. Immunoprecipitation and immunoblotting were performed as described above (A), except that anti-Flag M2 beads (top) or anti-eEF1A antibody (bottom) was used for immunoprecipitation.
To determine whether F-actin is necessary for the interaction of HPV38 E7 and eEF1A1, cells were treated with the ROCK kinase inhibitor Y27632, a chemical compound known to destroy actin stress fibers composed of F-actin, and interactions with proteins of interest were monitored. As shown in Fig. 6H, in agreement with previous findings, HPV38 E7 readily associated with eEF1A1 and F-actin. However, in the presence of the ROCK kinase inhibitor, only the complex of HPV38 E7 and eEF1A1 was observed (Fig. 6H, top). Similar results were obtained when the reciprocal immunoprecipitation was performed with eEF1A1 antibody (Fig. 6H, bottom). Taken together, these experiments indicated that HPV38 E7 can interact with eEF1A1 independently of F-actin. In these cells, HPV38 E7 can also independently associate with F-actin and eEF1A1, and the latter interaction blocks the interaction of eEF1A1 with F-actin. However, it is not known at this stage whether the direct interaction of HPV38 E7 with F-actin is also necessary to disrupt stress fibers, as all the E7 mutants generated retained their ability to bind to F-actin (see Fig. S4 in the supplemental material). Further studies are required to delineate the F-actin-binding region in E7 and to clarify whether the direct binding of E7 to actin is required for stress fiber disruption.
eEF1A promotes actin stress fiber formation, and HPV38 E7 inhibits its effects.
In addition to functioning as the basic component of translation elongation machinery, eEF1A can bundle actin and promote the remodeling of actin stress fibers (4, 16). The effects of eEF1A on actin remodeling are probably mediated through its C-terminal domain, required for its binding to F-actin. Since HPV38 E7 interferes with eEF1A to associate with F-actin through the same domain of eEF1A, HPV38 E7 may have an opposite effect on actin stress fiber formation. Thus, in addition to activating CK2 to disrupt actin stress fibers, HPV38 E7 may also interfere with actin fiber remodeling mediated by eEF1A. To test this hypothesis, we first evaluated the effects of eEF1A on actin stress fiber formation. Immunofluorescence analysis performed on U2OS cells showed that both eEF1A1 and eEF1A2 induced an increase in the amounts of actin stress fibers, in comparison to GFP-expressing cells used as a control (Fig. 7A). In contrast, and in agreement with previous observations, cells expressing HPV38 E7 had significantly fewer actin stress fibers than did control GFP- or eEF1A-expressing cells (Fig. 7A). Next, we determined whether HPV38 E7 can reverse the actin stress fiber formation mediated by eEF1A. As shown in Fig. 7A (bottom), cells expressing both eEF1A and HPV38 E7 had fewer actin stress fibers than did cells expressing eEF1A alone. This observation was confirmed by measuring the amount of F-actin relative to the amount of total actin in GFP-, eEF1A-, or HPV38 E7-expressing cells (Fig. 7B). Indeed, a significant increase in the level of F-actin was observed for eEF1A1-expressing cells compared to that of the control, while HPV38 E7 had an opposite effect, i.e., a much lower F-actin level than GFP or eEF1A (Fig. 7B). In agreement with the data shown in Fig. 7A, the level of F-actin induced by eEF1A significantly decreased in the presence of increasing concentrations of HPV38 E7 (Fig. 7B). To further confirm the inhibitory effect of HPV38 E7, we used an in vitro assay to evaluate its ability to interfere with actin polymerization mediated by eEF1A. The results indicated that in contrast to eEF1A, HPV38 E7 does not promote in vitro G-actin polymerization; moreover, it inhibits the actin polymerization induced by eEF1A (Fig. 7C, compare lanes S and P). Jasplakinolide (26), known as an actin linker agent, induced actin polymerization similarly to eEF1A, while the actin fiber disruptor latrunculin B (26) did not promote actin polymerization, as with HPV38 E7 (Fig. 7C).
Fig. 7.
eEF1A induces actin stress fiber formation, and HPV38 E7 inhibits its effects. (A) U2OS cells were transfected with the indicated expression plasmids, and 24 h later cells were visualized for F-actin with phalloidin-Alexa 568, GFP-HPV38 E7 by fluorescence, HA-eEF1A1 (anti-HA), F-eEF1A1 (anti-Flag), and nucleus (DAPI). Bar, 25 μm. The dotted line delineates the HPV38 E7-expressing cells. (B) Cells were transfected with the indicated expression plasmids, and 24 h later cells were lysed to quantify the level of F-actin, the total actin, and the ratio of F-actin/total actin. Results are means ± standard deviations of data from three independent experiments carried out in triplicate. (C) G-actin was incubated with immunoprecipitated F-HPV38 E7 and/or F-eEF1A1 at 4°C overnight. The supernatant (S) (nonpolymerized actin) and pellet (P) (polymerized actin) were separated by centrifugation and analyzed by immunoblotting. Latrunculin B (actin fiber disruptor) and jasplakinolide (actin-polymerizing agent) were used as controls.
Taken together, these data show that eEF1A and HPV38 E7 have opposite effects on the regulation of actin polymerization and that HPV38 E7 can inhibit the effects of eEF1A on actin stress fiber formation and remodeling.
HPV38 E7 blocks the Rho-GTPase activity induced by eEF1A.
In contrast to HPV38 E7, which destroys actin stress fibers and inhibits Rho-GTPase activity, eEF1A may favor actin stress fiber formation through the activation of Rho. To evaluate this assumption, the level of the active form of Rho (Rho-GTP) in cells expressing eEF1A was monitored. The Rho-GTP level was significantly increased in eEF1A-expressing cells compared to control cells (Fig. 8A). In addition, the eEF1A1 defective mutant in its binding to HPV38 E7 has lost its ability to activate Rho (see Fig. S5C in the supplemental material). Interestingly, HPV38 E7 inhibited the activation of Rho induced by eEF1A (Fig. 8B). As expected, HPV38 E7 increased the CK2 activity, which is associated with the decreased Rho-GTP level (Fig. 8B). The presence of eEF1A alone was not associated with a decreased level of CK2 activity, suggesting that eEF1A may induce Rho-GTP by a mechanism independent of CK2, and HPV38 E7 may have an inhibitory effect on this pathway as well. Moreover, the dose-dependent inhibition of the Rho-GTP level by the increasing amount of HPV38 E7 is not due simply to CK2 activation, since the CK2 activity was not changed with an increased amount of HPV38 E7 in the presence of eEF1A1 (Fig. 8B). Thus, the ability of eEF1A to promote actin stress fiber formation correlates with its effects to activate Rho, which is not associated with inhibiting CK2. In addition, HPV38 E7 has a potent inhibitory effect on eEF1A-induced Rho activation in the actin stress fiber-remodeling process. Taken together, HPV38 E7 can abrogate actin stress fiber formation by at least two independent mechanisms, involving the activation of the CK2-MEK-ERK-Rho pathway and the inhibition of eEF1A-mediated actin stress fiber remodeling.
Fig. 8.
HPV38 E7 inhibits eEF1A-mediated Rho activation. (A) U2OS cells were transfected with expression plasmids for eEF1A isoforms, and 48 h after transfection, cells were harvested to detect the Rho-GTP level by using a Rho activation assay kit. Activated Rho-GTP detected by immunoblotting (left) and the densitometric quantification of the ratio of Rho-GTP to total Rho bands (right) are shown. (B) Cells were transfected with the indicated expression plasmids, and the levels of the CK2 alpha subunit and its kinase activity were determined (bottom). The quantification of the amount of Rho-GTP compared to total Rho bands is shown on the right.
HPV38 E7-induced disruption of the actin cytoskeleton is important for HPV38 E6E7-induced cell proliferation.
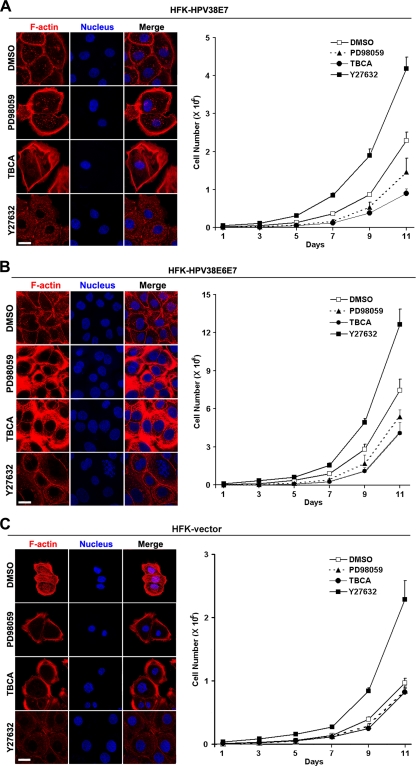
HPV38 E6 and E7 induce cellular immortalization in part by disrupting or activating some cellular signaling pathways affecting cell proliferation. The disruption of actin fibers may also be important for HPV38 E6- and E7-induced cell proliferation and immortalization. To investigate this possibility, we treated HPV38 E7-expressing human keratinocytes with different inhibitors affecting the CK2-MEK-ERK axis activated by HPV38 E7 and then examined the pattern of actin fibers in relation to cell proliferation. The exposure of cells to the MEK inhibitor PD98059 resulted, as expected, in an increase in the numbers of actin fibers (Fig. 9A, left). This event is associated with a significant reduction in cell proliferation (Fig. 9A, right). Similarly, more actin stress fibers were observed when cells were treated with the CK2 inhibitor TBCA (Fig. 9A, left), also with a great reduction in cell proliferation (Fig. 9A, right). In contrast, the inhibition of ROCK kinase (a downstream effector of Rho) with Y27632 is accompanied by a strong reduction in numbers of actin stress fibers and an increased level of cell proliferation (Fig. 9A). Similar observations were obtained with HEK 293 cells constitutively expressing HPV38 E7 (see Fig. S6 in the supplemental material) or immortalized human keratinocytes expressing HPV38 E6 and E7 (Fig. 9B) but to a lesser extent with vector-containing HEK 293 cells (Fig. S6A) or vector-transduced primary keratinocytes (Fig. 9C). In both cases, the MEK inhibitor PD98059 and the CK2 inhibitor TBCA did not affect cell proliferation and actin fibers significantly as in HPV38 E7-transduced or HPV38 E6E7-immortalized keratinocytes. However, as reported previously by Chapman et al. (10), the ROCK inhibitor Y27632 has a positive effect on cell proliferation in primary keratinocytes, which is further enhanced by HPV38 E7 as a result of increased stress fiber disruption.
Fig. 9.
Disruption of the actin cytoskeleton by HPV38 E7 correlates with cell proliferation. (A, left) HPV38 E7-expressing human keratinocytes (HFK-HPV38E7) were not treated or treated with different kinase inhibitors of MEK (PD98059), CK2 (TBCA), and ROCK kinase (Y27632) for 12 h, and cells were examined for their F-actin contents by immunofluorescence. (Right) Cells were exposed to kinase inhibitors for the indicated time courses and monitored for proliferation. Bar, 25 μm. (B) HPV38 E6- and E7-expressing human keratinocytes (HFK-HPV38E6E7) were treated and monitored as described above (A). Bar, 25 μm. (C) Vector-transduced HFKs were treated and monitored as described above (A). Bar, 25 μm.
Other inhibitors affecting the kinases Akt and JNK had no significant effect on actin fiber restoration or cell proliferation (data not shown). Thus, the inhibition of actin stress fibers is important for HPV38 E6 and E7 to promote cell proliferation in the process of the immortalization of primary keratinocytes.
DISCUSSION
Viral oncoproteins induce cell transformation by altering a number of normal cellular functions, leading to uncontrolled cell proliferation. The remodeling of the actin cytoskeleton is a key event accompanying the cell transformation process (48, 55, 62). It is now well established that highly oncogenic mucosal HPVs use their oncoproteins E6 and E7 to induce neoplastic transformation by altering signaling pathways regulating cell cycle progression and apoptosis (25, 41). Evidence is now accumulating that cutaneous beta HPVs also display oncogenic properties through their oncoproteins E6 and E7 by affecting similar cellular signaling pathways albeit by different mechanisms (7, 51). In this study, we show for the first time that cutaneous HPV38 E7 can alter actin stress fiber formation, and this event is associated with enhanced cell proliferation, suggesting a key role of actin filament remodeling during cell transformation induced by HPV38 E6 and E7. Immortalized human keratinocytes expressing HPV38 E6 and E7 display a strong reduction in the F-actin content and fewer actin stress fibers than in nonimmortalized primary human keratinocytes. The disruption of actin stress fibers was attributed mainly to HPV38 E7, which had an effect similar to that of the combination of E6 and E7 of HPV38 (Fig. 1). As Rho plays an important role in stress fiber formation, HPV38 E7 may induce actin stress fiber disruption by inhibiting Rho-GTP. Indeed, the expression of HPV38 E7 leads to a significant decrease in Rho-GTP levels and a subsequent diminution of the level of phosphorylated cofilin, a downstream effector of Rho-GTP and ROCK kinase (Fig. 2). This event occurred not only in cells ectopically expressing HPV38 E7 but also in human keratinocytes in an experimental model that closely resembles the natural situation (Fig. 2). Interestingly, the constitutive active Rho mutant can rescue the inhibitory effects of HPV38 E7 on cofilin phosphorylation and, more importantly, on actin stress fiber formation (Fig. 2C, D, E, and F). This result furthermore supports the key role of HPV38 E7-mediated Rho inactivation in actin stress fiber remodeling. Analysis of the additional effectors upstream of Rho signaling led to the identification of the MEK-ERK axis as a target of HPV38 E7 and the negative regulator of Rho. Indeed, we showed that the HPV38 E7-induced inhibition of actin stress fibers is associated with its ability to activate the MEK-ERK pathway (Fig. 2G). This observation was corroborated by the fact that cells exposed to MEK inhibitors showed a strong accumulation of Rho-GTP (Fig. 3E) and a significant increase in numbers of actin stress fibers (Fig. 2H, I, and J). One of the upstream activators of MEK-ERK signaling is the CK2 kinase, which is involved in several cellular functions (58). Thus, HPV38 E7 may target CK2 to induce the disruption of the actin cytoskeleton through the MEK-ERK pathway. Indeed, the level of the kinase activity of CK2 is significantly increased in cells ectopically expressing HPV38 E7 or in human keratinocytes transduced with HPV38 E7, associated with a decrease of the F-actin content, and those effects are sensitive to the CK2 inhibitor TBCA (Fig. 3 and 4). Interestingly, both the MEK inhibitor and the CK2 inhibitor induced a robust accumulation of Rho-GTP in cells expressing HPV38 E7 and an increased level of actin stress fibers (Fig. 3), demonstrating a clear link between HPV38 E7, CK2, MEK-ERK, Rho, and actin cytoskeleton remodeling. However, our data do not elucidate the mechanisms by which HPV38 E7 activates CK2. HPV38 E7 may bind to CK2 and induce its kinase activity, but immunoprecipitation experiments indicated that HPV38 E7 did not bind to endogenous CK2 as well as the overexpressed CK2 protein (data not shown). Instead, we consistently observed that the level of the CK2 alpha subunit was significantly increased in cells expressing HPV38 E7 (Fig. 3B and D). Thus, HPV38 E7 could increase the expression level of CK2 alpha mRNA and/or indirectly mediate the stability of the protein, which may lead to the increase in the level of CK2 kinase activity.
Further investigations revealed that the inhibition of the CK2 activity induced by HPV38 E7 was unable to fully rescue the loss of actin stress fibers by HPV38 E7, suggesting additional mechanisms for HPV38 E7 to disrupt the actin cytoskeleton; for example, HPV38 E7 could act through a cellular partner that may play a role in actin remodeling. Using a GST pulldown assay combined with mass spectrometry protein sequencing, we identified the human eukaryotic elongation factor eEF1A as a partner of HPV38 E7. Further studies showed that HPV38 E7 binds to the two isoforms of eEF1A (eEF1A1 and eEF1A2) in vitro and in the cells (Fig. 5). The colocalization in the cell is not incidental, as the physical interaction of the two proteins was confirmed by a FRET assay (Fig. 5F). The region of eEF1A located between aa 430 and 444 in its C domain appears to be important for this interaction (Fig. 5H). Indeed, a specific mutation in this region decreases the affinity of eEF1A for HPV38 E7 (see Fig. S5A in the supplemental material). It was shown previously that the eukaryotic elongation factor eEF1A is not only a highly conserved major translational factor but also a functionally pleiotropic protein (16). In addition to its role in signal transduction related to translational regulation and multiple cellular processes, eEF1A has also been shown to promote actin stress fiber formation and remodeling by association with actin filaments (16). However, as previously reported (22, 36, 52), the ability of eEF1A to regulate the cytoskeletal organization is distinct from its translation elongation function. We confirmed this finding by using specific inhibitors in the absence or presence of HPV38 E7 (data not shown). We also discovered that the binding of eEF1A to F-actin occurred through its C-terminal fragment (aa 430 to 444), the same region involved in its interaction with HPV38 E7 (Fig. 6). Thus, HPV38 E7 could interfere with the association of eEF1A with F-actin, suggesting a potential role of HPV38 E7 in actin remodeling and in regulating the effects of eEF1A. Indeed, the CR3 domain at the C terminus of HPV38 E7 plays a major role in its interaction with eEF1A (Fig. 5I to K). Interestingly, a domain similar to the TD-NEM motif involved in non-CRM1-mediated protein nuclear export (30) and present in the CR3 domain appears to be important for this interaction (Fig. 5K). In this regard, the possibility that eEF1A is involved in HPV38 E7 (a diffuse nuclear/cytoplasmic protein) nuclear exclusion is currently under investigation in our laboratory. pRb, a protein known to interact with HPV38 E7, is not absolutely required for HPV38 E7 to associate with eEF1A (Fig. 5K), and HPV38 E7 can bind to F-actin independently of eEF1A (Fig. 6H). Taken together, HPV38 E7 associates with eEF1A and blocks its interaction with F-actin. The expression of an HPV38 E7-binding-defective mutant of eEF1A1 [eEF1A(T430A/V431A)] has an inhibitory effect on cell proliferation, while the truncated mutants (Fig. 5H) have a toxic effect (data not shown). Thus, it is likely that the interaction of HPV38 E7 and eEF1A is important for cell proliferation. However, due to the various functions attributed to eEF1A (including protein translation), we cannot exclude that those mutants may also be defective in other functions of eEF1A important for cell proliferation.
In contrast to HPV38 E7, which destroys actin stress fibers, eEF1A promotes actin stress fibers by increasing the F-actin content, and HPV38 E7 abrogates the actin stress fiber remodeling mediated by eEF1A (Fig. 1 and 7). Since HPV38 E7 activates CK2 and MEK-ERK to abolish actin stress fibers, we may expect that eEF1A, which promotes actin stress fiber formation and induces Rho-GTP accumulation, inhibits ERK and CK2 activation. However, eEF1A had no effect on CK2 activity (data not shown) or on ERK activation (see Fig. S8 in the supplemental material). Thus, eEF1A activates Rho independently of the MEK-ERK axis. Taken together, we can conclude that HPV38 E7 blocks actin stress fiber formation by at least two independent mechanisms. The first mechanism involves the activation of the CK2-MEK-ERK pathway leading to Rho inhibition, and the second mechanism is through the binding and inhibition of eEF1A-mediated actin stress fiber formation. Since the actin-bundling effect of eEF1A is dependent on its binding to F-actin, we conclude that HPV38 E7 inhibits the formation of actin fibers by eEF1A, through the blocking of eEF1A binding to F-actin, independently of the activation of CK2-induced HPV38 E7. At present, it is not known whether the binding of HPV38 E7 to actin can independently induce actin stress fiber remodeling.
As E7, together with E6, plays a major role in HPV38-induced human keratinocyte immortalization (1), we raised the possibility that its ability to activate CK2 and ERK in the process of disrupting actin stress fibers could be related to its oncogenic activity. Indeed, the inhibition of MEK-ERK or CK2 in keratinocytes expressing HPV38 E7 or HPV38 E6 and E7 leads to an increase in stress fiber formation and a significant inhibition of cell proliferation. In contrast, the inhibition of Rho in the same cells was associated with robust cell proliferation (Fig. 9), which is consistent with a recent report by Chapman et al., where they reported that the inhibition of Rho can promote keratinocyte immortalization (10). Similar observations were noted for heterologous HEK 293 cells expressing HPV38 E7 (see Fig. S6B in the supplemental material). This is consistent with the role of CK2 as an oncogenic agent in the development of tumors (58). Thus, the disruption of the actin cytoskeleton is important for HPV38 E7 to facilitate cell proliferation in the process of keratinocyte immortalization. CK2 also contributes to the S-phase entry of high-risk mucosal HPV16 and HPV18 into human primary keratinocytes through the phosphorylation of E7 from these viruses (5, 12, 17, 19). HPV38 E7, not being a substrate of CK2, increases its kinase activity in the process of cell proliferation. In addition, HPV38 E7, as a multifunctional protein, is involved in keratinocyte immortalization by altering several cellular pathways, including pRb inactivation (9). An HPV38 E7 mutant (C24G plus E26G), defective for binding to pRb, is less potent in promoting cell proliferation than wild-type HPV38 E7 (see Fig. S7 in the supplemental material), suggesting complementary effects of pRb inhibition and actin stress fiber remodeling on the process of cell proliferation by HPV38 E7.
Various viral proteins have been reported to bind to eEF1A. For example, the human immunodeficiency virus type 1 Gag polyprotein interacts with eEF1A through tRNA, which is implicated in viral RNA maturation (13). The NS5A proteins of bovine viral diarrhea virus and tobacco mosaic virus RNA-dependent RNA polymerase were shown previously to interact with eEF1A, indicating a potential role of eEF1A in the virus replication complex (65). Our study demonstrated that the viral oncoprotein E7 of HPV38 can also associate with eEF1A, an event that could be important for HPV38-induced keratinocyte immortalization. Indeed, we tested the binding of E7s from various cutaneous HPVs, including HPV types 1, 7, 10, 13, 17, 20, and 48. Among all these HPVs, only E7 of HPV38, which has the ability to immortalize human keratinocytes, showed a significant efficiency of binding with eEF1A (see Fig. S4 in the supplemental material). Interestingly, the E7s of HPV types 1, 17, and 20 affect neither Rho activities nor F-actin contents (Fig. S4B and S4C), suggesting a correlation between the ability of the virus to disturb actin stress fibers and the ability to induce cellular immortalization.
The alteration of the cellular actin cytoskeleton, which is related to both anchorage-independent growth and cellular tumorigenesis, is a hallmark of several oncoproteins (48). Our study has revealed that HPV38 oncoprotein E7 can disrupt actin fibers by activating the CK2-MEK-ERK-Rho cascade, binding to the eukaryotic translation factor eEF1A, and inhibiting its effects on actin stress fiber remodeling. The inhibition of actin stress fiber formation contributes to the enhanced cell proliferation induced by HPV38 E6 and E7, which may be important for HPV38-mediated keratinocyte immortalization (Fig. 10).
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Mosialos, E. Kieff, and S. Pitson for reagents and J. Daniel for editing. We are grateful to all the members of our laboratory for their comments and cooperation.
The study was partially supported by grants from the Association pour la Recherche Contre le Cancer and La Ligue Contre le Cancer du Comité du Rhône and Comité de la Drôme (to B.S.S. and M.T.), by a grant from the Association for Research on Cancer (to P.J.), and by the IARC fellowship program (to J.Y., R.S., and I.H.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Accardi R., et al. 2006. Skin human papillomavirus type 38 alters p53 functions by accumulation of deltaNp73. EMBO Rep. 7:334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alunni-Fabbroni M., et al. 2000. Induction of S phase and apoptosis by the human papillomavirus type 16 E7 protein are separable events in immortalized rodent fibroblasts. Oncogene 19:2277–2285 [DOI] [PubMed] [Google Scholar]
- 3. Amano M., et al. 1997. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science 275:1308–1311 [DOI] [PubMed] [Google Scholar]
- 4. Amiri A., et al. 2007. eEF1A2 activates Akt and stimulates Akt-dependent actin remodeling, invasion and migration. Oncogene 26:3027–3040 [DOI] [PubMed] [Google Scholar]
- 5. Barbosa M. S., et al. 1990. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and Sv40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation. EMBO J. 9:153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bishop A. L., Hall A. 2000. Rho GTPases and their effector proteins. Biochem. J. 348(Pt. 2):241–255 [PMC free article] [PubMed] [Google Scholar]
- 7. Bouvard V., Gabet A. S., Accardi R., Sylla B. S., Tommasino M. 2006. The cutaneous human papillomavirus types and non-melanoma skin cancer, p. 269–277 In Campo S. (ed.), Papillomavirus research: from natural history to vaccines and beyond. Caister Academic Press, Hethersett, Norwich, United Kingdom [Google Scholar]
- 8. Cain R. J., Ridley A. J. 2009. Phosphoinositide 3-kinases in cell migration. Biol. Cell 101:13–29 [DOI] [PubMed] [Google Scholar]
- 9. Caldeira S., et al. 2003. The E6 and E7 proteins of the cutaneous human papillomavirus type 38 display transforming properties. J. Virol. 77:2195–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chapman S., Liu X., Meyers C., Schlegel R., McBride A. A. 2010. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Invest. 120:2619–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Charette S. T., McCance D. J. 2007. The E7 protein from human papillomavirus type 16 enhances keratinocyte migration in an Akt-dependent manner. Oncogene 26:7386–7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chien W. M., Parker J. N., Schmidt-Grimminger D. C., Broker T. R., Chow L. T. 2000. Casein kinase II phosphorylation of the human papillomavirus-18 E7 protein is critical for promoting S-phase entry. Cell Growth Differ. 11:425–435 [PubMed] [Google Scholar]
- 13. Cimarelli A., Luban J. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 73:5388–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clegg R. M. 1992. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 211:353–388 [DOI] [PubMed] [Google Scholar]
- 15. Dong W., et al. 2005. Skin hyperproliferation and susceptibility to chemical carcinogenesis in transgenic mice expressing E6 and E7 of human papillomavirus type 38. J. Virol. 79:14899–14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ejiri S. 2002. Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci. Biotechnol. Biochem. 66:1–21 [DOI] [PubMed] [Google Scholar]
- 17. Firzlaff J. M., Luscher B., Eisenman R. N. 1991. Negative charge at the casein kinase II phosphorylation site is important for transformation but not for Rb protein binding by the E7 protein of human papillomavirus type 16. Proc. Natl. Acad. Sci. U. S. A. 88:5187–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallagher S. R. 2006. One-dimensional SDS gel electrophoresis of proteins, unit 10 2A, p. 1–37 In Ausubel F. M., et al. (ed.), Current protocols in molecular biology. Wiley Interscience, New York, NY: [DOI] [PubMed] [Google Scholar]
- 19. Genovese N. J., Banerjee N. S., Broker T. R., Chow L. T. 2008. Casein kinase II motif-dependent phosphorylation of human papillomavirus E7 protein promotes p130 degradation and S-phase induction in differentiated human keratinocytes. J. Virol. 82:4862–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghittoni R., et al. 2010. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 40:1–13 [DOI] [PubMed] [Google Scholar]
- 21. Grammatikakis N., et al. 2001. Simian virus 40 large tumor antigen modulates the Raf signaling pathway. J. Biol. Chem. 276:27840–27845 [DOI] [PubMed] [Google Scholar]
- 22. Gross S. R., Kinzy T. G. 2005. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12:772–778 [DOI] [PubMed] [Google Scholar]
- 23. Gyenis L., Litchfield D. W. 2008. The emerging CK2 interactome: insights into the regulation and functions of CK2. Mol. Cell. Biochem. 316:5–14 [DOI] [PubMed] [Google Scholar]
- 24. Hall A. 2005. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33:891–895 [DOI] [PubMed] [Google Scholar]
- 25. Howley P. M., Lowy D. R. 2007. Polyomaviruses, p. 229–2354 In Knipe D. M., et al. (ed.), Fields virology, 5th ed Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 26. Iyer S. S., Kusner D. J. 2009. Coordinate regulation of sphingosine kinase and actin dynamics. Methods Mol. Biol. 531:347–361 [DOI] [PubMed] [Google Scholar]
- 27. Jewers R. J., Hildebrandt P., Ludlow J. W., Kell B., McCance D. J. 1992. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J. Virol. 66:1329–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson H. W., Schell M. J. 2009. Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol. Biol. Cell 20:5166–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kahns S., et al. 1998. The elongation factor 1 A-2 isoform from rabbit: cloning of the cDNA and characterization of the protein. Nucleic Acids Res. 26:1884–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khacho M., et al. 2008. eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol. Biol. Cell 19:5296–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klein R. M., Spofford L. S., Abel E. V., Ortiz A., Aplin A. E. 2008. B-RAF regulation of Rnd3 participates in actin cytoskeletal and focal adhesion organization. Mol. Biol. Cell 19:498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Knudsen S. M., Frydenberg J., Clark B. F., Leffers H. 1993. Tissue-dependent variation in the expression of elongation factor-1 alpha isoforms: isolation and characterisation of a cDNA encoding a novel variant of human elongation-factor 1 alpha. Eur. J. Biochem. 215:549–554 [DOI] [PubMed] [Google Scholar]
- 33. Leclercq T. M., Moretti P. A., Vadas M. A., Pitson S. M. 2008. Eukaryotic elongation factor 1A interacts with sphingosine kinase and directly enhances its catalytic activity. J. Biol. Chem. 283:9606–9614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee J. M. 2003. The role of protein elongation factor eEF1A2 in ovarian cancer. Reprod. Biol. Endocrinol. 1:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee S., et al. 2004. Frameshift mutation in the Dok1 gene in chronic lymphocytic leukemia. Oncogene 23:2287–2297 [DOI] [PubMed] [Google Scholar]
- 36. Liu G., et al. 1996. F-actin sequesters elongation factor 1alpha from interaction with aminoacyl-tRNA in a pH-dependent reaction. J. Cell Biol. 135:953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 38. Mantovani F., Banks L. 2001. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20:7874–7887 [DOI] [PubMed] [Google Scholar]
- 39. McLaughlin-Drubin M. E., Munger K. 2009. The human papillomavirus E7 oncoprotein. Virology 384:335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meggio F., Pinna L. A. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349–368 [DOI] [PubMed] [Google Scholar]
- 41. Moody C. A., Laimins L. A. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer 10:550–560 [DOI] [PubMed] [Google Scholar]
- 42. Morita K., Bunai F., Numata O. 2008. Roles of three domains of Tetrahymena eEF1A in bundling F-actin. Zoolog. Sci. 25:22–29 [DOI] [PubMed] [Google Scholar]
- 43. Naumanen P., Lappalainen P., Hotulainen P. 2008. Mechanisms of actin stress fibre assembly. J. Microsc. 231:446–454 [DOI] [PubMed] [Google Scholar]
- 44. Nieminen J., Kuno A., Hirabayashi J., Sato S. 2007. Visualization of galectin-3 oligomerization on the surface of neutrophils and endothelial cells using fluorescence resonance energy transfer. J. Biol. Chem. 282:1374–1383 [DOI] [PubMed] [Google Scholar]
- 45. Nissen P., et al. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270:1464–1472 [DOI] [PubMed] [Google Scholar]
- 46. Niu Y., et al. 2006. A nuclear export signal and phosphorylation regulate Dok1 subcellular localization and functions. Mol. Cell. Biol. 26:4288–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Papakonstanti E. A., Stournaras C. 2007. Actin cytoskeleton architecture and signaling in osmosensing. Methods Enzymol. 428:227–240 [DOI] [PubMed] [Google Scholar]
- 48. Pawlak G., Helfman D. M. 2001. Cytoskeletal changes in cell transformation and tumorigenesis. Curr. Opin. Genet. Dev. 11:41–47 [DOI] [PubMed] [Google Scholar]
- 49. Pawlak G., Helfman D. M. 2002. MEK mediates v-Src-induced disruption of the actin cytoskeleton via inactivation of the Rho-ROCK-LIM kinase pathway. J. Biol. Chem. 277:26927–26933 [DOI] [PubMed] [Google Scholar]
- 50. Pawlak G., Helfman D. M. 2002. Post-transcriptional down-regulation of ROCKI/Rho-kinase through an MEK-dependent pathway leads to cytoskeleton disruption in Ras-transformed fibroblasts. Mol. Biol. Cell 13:336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pfister H. 2003. Human papillomavirus and skin cancer. J. Natl. Cancer Inst. Monogr. 2003:52–56 [DOI] [PubMed] [Google Scholar]
- 52. Pittman Y. R., Kandl K., Lewis M., Valente L., Kinzy T. G. 2009. Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Balpha. J. Biol. Chem. 284:4739–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pleschka S. 2008. RNA viruses and the mitogenic Raf/MEK/ERK signal transduction cascade. Biol. Chem. 389:1273–1282 [DOI] [PubMed] [Google Scholar]
- 54. Pollard T. D., Cooper J. A. 2009. Actin, a central player in cell shape and movement. Science 326:1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rao J., Li N. 2004. Microfilament actin remodeling as a potential target for cancer drug development. Curr. Cancer Drug Targets 4:345–354 [DOI] [PubMed] [Google Scholar]
- 56. Riedl J., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5:605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Riis B., Rattan S. I., Clark B. F., Merrick W. C. 1990. Eukaryotic protein elongation factors. Trends Biochem. Sci. 15:420–424 [DOI] [PubMed] [Google Scholar]
- 58. Ritt D. A., et al. 2007. CK2 is a component of the KSR1 scaffold complex that contributes to Raf kinase activation. Curr. Biol. 17:179–184 [DOI] [PubMed] [Google Scholar]
- 59. Soares D. C., Barlow P. N., Newbery H. J., Porteous D. J., Abbott C. M. 2009. Structural models of human eEF1A1 and eEF1A2 reveal two distinct surface clusters of sequence variation and potential differences in phosphorylation. PLoS One 4:e6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stark H., et al. 1997. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389:403–406 [DOI] [PubMed] [Google Scholar]
- 61. Sylla B. S., et al. 2000. The X-linked lymphoproliferative syndrome gene product SH2D1A associates with p62dok (Dok1) and activates NF-kappa B. Proc. Natl. Acad. Sci. U. S. A. 97:7470–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vasiliev J. M. 2004. Cytoskeletal mechanisms responsible for invasive migration of neoplastic cells. Int. J. Dev. Biol. 48:425–439 [DOI] [PubMed] [Google Scholar]
- 63. Wang Y., Jiang Y., Meyering-Voss M., Sprinzl M., Sigler P. B. 1997. Crystal structure of the EF-Tu.EF-Ts complex from Thermus thermophilus. Nat. Struct. Biol. 4:650–656 [DOI] [PubMed] [Google Scholar]
- 64. Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1:136–143 [DOI] [PubMed] [Google Scholar]
- 65. Yamaji Y., et al. 2006. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 347:100–108 [DOI] [PubMed] [Google Scholar]
- 66. Yuan H., et al. 2009. Human papillomavirus type 16 oncoprotein E7 suppresses cadherin-mediated cell adhesion via ERK and AP-1 signaling. Int. J. Oncol. 35:309–314 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.