Abstract
Susceptibility to norovirus (NoV), a major pathogen of epidemic gastroenteritis, is associated with histo-blood group antigens (HBGAs), which are also cell attachment factors for this virus. GII.4 NoV strains are predominantly associated with worldwide NoV epidemics with a periodic emergence of new variants. The sequence variations in the surface-exposed P domain of the capsid protein resulting in differential HBGA binding patterns and antigenicity are suggested to drive GII.4 epochal evolution. To understand how temporal sequence variations affect the P domain structure and contribute to epochal evolution, we determined the P domain structure of a 2004 variant with ABH and secretor Lewis HBGAs and compared it with the previously determined structure of a 1996 variant. We show that temporal sequence variations do not affect the binding of monofucosyl ABH HBGAs but that they can modulate the binding strength of difucosyl Lewis HBGAs and thus could contribute to epochal evolution by the potentiated targeting of new variants to Lewis-positive, secretor-positive individuals. The temporal variations also result in significant differences in the electrostatic landscapes, likely reflecting antigenic variations. The proximity of some of these changes to the HBGA binding sites suggests the possibility of a coordinated interplay between antigenicity and HBGA binding in epochal evolution. From the observation that the regions involved in the formation of the HBGA binding sites can be conformationally flexible, we suggest a plausible mechanism for how norovirus disassociates from salivary mucin-linked HBGA before reassociating with HBGAs linked to intestinal epithelial cells during its passage through the gastrointestinal tract.
INTRODUCTION
Noroviruses (NoVs) are highly contagious human pathogens that cause both sporadic and epidemic gastroenteritis. A recent estimate suggests that NoVs are responsible for 1 million hospitalizations and up to 200,000 deaths of children under the age of 5 years worldwide annually (31). NoVs constitute one of the four major genera in the family Caliciviridae (15). They are phylogenetically divided into five genogroups (genogroup I [GI] to GV). GI, GII, and GIV contain human pathogens, and each genogroup is further divided into several genotypes (44). NoVs belonging to genogroup II and genotype 4 (GII.4) are the most prevalent, accounting for 70 to 80% of the norovirus outbreaks worldwide (20). Since at least 1995, the emergence of new variants has been associated with worldwide norovirus epidemics (37). The GII.4 viruses have been suggested to undergo epochal evolution, with the emergence of a new GII.4 variant coinciding with a new epidemic peak that is typically followed by a period of stasis (10, 36). Our understanding of the factors that drive this punctuated evolution of GII.4 viruses has been limited mainly because of the lack of a cell culture system or a readily available animal model for NoVs.
Several studies have indicated that histo-blood group antigens (HBGAs), which are genetically determined glycans found in mucosal secretions and on epithelial cells, are susceptibility factors and cell attachment factors for NoVs (16, 17, 22, 25). HBGAs are oligosaccharides with various carbohydrate compositions and linkages between them (24). They are synthesized by the linkage-specific sequential addition of a monosaccharide to the terminal disaccharide of a precursor oligosaccharide by various glycosyltransferases, resulting in the ABH and Lewis families of HBGAs (see Fig. S1 in the supplemental material). Depending upon the linkage and carbohydrate composition of the precursor disaccharide, these HBGAs are further classified into different types. Previous binding studies using recombinant virus-like particles (VLPs) of NoVs with saliva, red blood cells, and synthetic carbohydrates have demonstrated direct interactions between VLPs and HBGAs (16, 18, 23, 35, 38). Subsequent crystallographic studies using the recombinant P domain of the capsid protein have shown that HBGAs bind to the surface-exposed P2 subdomain and that NoVs recognize various HBGAs in a distinct strain-dependent manner (4–6).
In addition to variations in HBGA binding patterns between genogroups (16, 34, 39), variations are also observed within a genotype. This is particularly highlighted in the case of periodically evolving GII.4 viruses (10, 23). Based on phylogenetic analyses, several evolutionary patterns have been observed for GII.4 variants, which include major variants such as GII.4-pre-1995 (Camberwell), GII.4-1996 (Grimsby), GII.4 2002 (Farmington Hills), GII.4-2004 (Hunter), GII.4 2006a (Laurens), and GII.4 2006b (Minerva) and minor variants such as GII.4 2001Japan, GII.4 2001Henry (Houston virus), and GII.4 2003Asia (Sakai) (10, 23, 37). Recently, two more variants have been identified, which include GII.4 2008 (Apeldoorn) and GII.4 2010 (New Orleans). Most of the sequence variations among the GII.4 variants occur in the P2 subdomain. Antigenic drift and differential HBGA binding specificities, and therefore susceptibility, resulting from these sequence changes were suggested previously to drive the observed epochal evolution of GII.4 variants (10). However, there are conflicting reports regarding the role of HBGA in epochal evolution and also the evolving HBGA patterns in the GII.4 variants (23, 39, 41). In the case of 2004-2005 variants, there have been conflicting reports regarding HBGA binding. One study of VLPs representing GII.4 2004-2005 variants showed that these variants do not bind to any known carbohydrates, and these variants were suggested to have acquired novel receptors or novel carbohydrate ligands (10). Another study using recombinant P particles, higher-order oligomers of the P domain obtained by using a cysteine-linked peptide at one of the termini, indicated that these variants bind to all of the secretor HBGAs although to a lesser extent than those of other variants (43). Finally, most recently, de Rougemont et al., using a variety of binding studies, including surface plasmon resonance with well-characterized VLP preparations, have shown that the 2004-2005 variants bind to both ABH and Lewis secretor HBGAs (9).
The only GII.4 structure available is that of the P domain of VA387, a 1996 GII.4 variant belonging to the Grimsby cluster (5). Crystallographic studies of the P domain of this variant in complex with A- and B-type trisaccharides provided the first structural details of HBGA interactions and showed that both trisaccharides bind to the same site, referred to as site 1, in the P domain with only the terminal α-fucose involved in the direct interactions. However, several questions remain regarding HBGA recognition in GII.4 variants. Given that site 1 (Fig. 1) is highly conserved across the GII.4 variants and also that only the nondiscriminatory α-fucose is primarily involved in HBGA binding, how the temporal sequence variations modulate HBGA binding preferences in GII.4 variants is not clear. In the crystal structures, the central β-galactose (β-Gal) of the trisaccharides, which would attach to the proximal residues in a longer oligosaccharide, points toward a nearby loop (amino acids [aa] 390 to 395) and makes solvent-mediated interactions. Based on this observation, it was hypothesized that this loop provides further stability to longer oligosaccharides (5). In subsequent studies using evolutionary analysis of the GII.4 sequences, computer modeling, and mutational analyses, this loop structure, referred to as site 2 (Fig. 1), which exhibits temporal sequence variability, was suggested to be involved in modulating the HBGA binding patterns in GII.4 variants (10, 23). However, the direct involvement of site 2 with longer oligosaccharides and how the sequence variations in this site affect HBGA binding in GII.4 variants are not directly established. Several biochemical studies have shown the binding of GII.4 NoVs to secretor difucosyl Lewis HBGA (9, 16, 23, 34). How these HBGAs with two terminal fucose moieties, in contrast to monofucosyl ABH HBGAs, interact with the P domain is not structurally characterized.
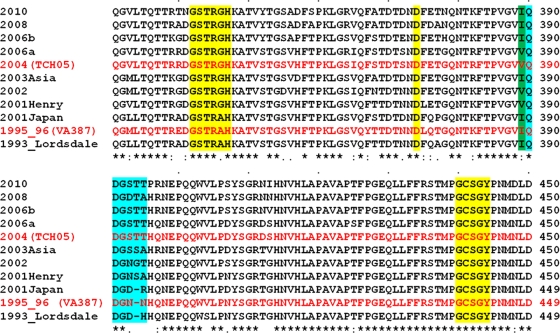
Fig. 1.
Multiple-sequence alignment of representative GII.4 variants from 1995 to 2010. Only residues from aa 331 to 450 in the P domain of the capsid protein are shown. Amino acid residues in site 1 are highlighted in yellow. Site 2 with temporally evolving residues is highlighted in cyan. The I-to-V changes at position 389 are highlighted in green. The amino acid sequences of 1996 (VA387) and 2004 (TCH05) GII.4 variants are shown in red. Every 10th residue is denoted by a dot above the sequences. A sequence similarity index is shown below the sequences according to ClustalW conventions (asterisk, invariant; colon, highly similar; dot, similar). GenBank accession numbers for the various sequences represented here are as follows: X86557 for Lordsdale, AJ004864 for 1995_96, AB294779 for 2001Japan, EU310927 for 2001Henry, AY485642 for 2002; AB220922 for 2003Asia, JF827296 for 2004, EF126963 for 2006a, EF126965 for 2006b, AB445395 for 2008 Apeldoorn 317/2007, and GU445325 for 2010 New Orleans 1805/2009.
To characterize the structural properties of another epidemic GII.4 variant and identify the structural determinants that could modulate HBGA binding specificities, we studied a 2004 variant (TCH05) because it not only is temporally distant from the 1996 epidemic variant (VA387) for which the P domain structure is already determined but also has prominent sequence changes in the proposed site 2. Our crystallographic studies of this 2004 variant reported herein provide a more complete understanding of GII.4 NoV interactions with different HBGAs, including monofucosyl ABH and secretor difucosyl Lewis HBGAs, and have revealed novel features that have implications for the epochal evolution of the GII.4 strain. A remarkable observation from our studies is the conformational flexibility in the regions involved in the P domain dimerization and formation of the HBGA binding site, which may be an important factor for the dissociation and reassociation of HBGA during virus passage through the gastrointestinal tract.
MATERIALS AND METHODS
Expression, purification, and crystallization of the P domain.
The P domain (amino acids [aa] 221 to 531) construct of the 2004 GII.4 variant (TCH05) with an N-terminal His6-MBP (maltose binding protein) tag was cloned into an expression vector, pMal-C2E (New England BioLabs), with a TEV (tobacco etch virus) protease (19) cleavage site between MBP and the P domain. The recombinant protein was expressed in Escherichia coli BL-21(DE3) (Novagen) cells. The His-MBP-tagged P domain was first purified over a Ni-nitrilotriacetic acid (NTA) (Qiagen) column, and the His-MBP tag was removed by using TEV protease. The P domain was then separated from His-MBP by rerunning the mixture through the Ni-NTA column and was purified to homogeneity by size exclusion chromatography (Superdex S75p). The purified P domain was concentrated and stored in 20 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl, 1 mM dithiothreitol (DTT), and 5 mM MgCl2. Crystal screening was done by using the Mosquito nanoliter handling system (TTP LabTech), and crystals were visualized by using a Rock Imager (Formulatrix). Crystals of the unliganded and liganded P domains of the 2004 variant were obtained by the hanging-drop vapor diffusion method at 20°C. The unliganded protein (∼3.5 mg/ml) was crystallized by using a solution containing 0.2 M zinc acetate, 0.1 M morpholineethanesulfonic acid (MES) (pH 6.0), and 15% (vol/vol) ethanol (C2 space group) and by using a solution containing 0.2 M sodium citrate, 0.1 M HEPES (pH 7.5), and 20% isopropanol (C2221 space group), and the liganded protein was crystallized by using a solution containing 0.8 M potassium sodium tartrate, 100 mM Tris (pH 8.5), and 0.5% (wt/vol) polyethylene glycol (PEG) monomethyl ether 5000 in a 1:1 ratio. The P domain was cocrystallized in the presence of each of the following ligands: A-type trisaccharide, H type 1 pentasaccharide, and Leb hexasaccharide (Dextra Laboratories), with a 1:60 excess molar ratio of ligand. Prior to data collection, crystals were flash-frozen using either 20% glycerol or 20% ethylene glycol as a cryoprotectant.
Diffraction, data collection, structure determination, and refinement.
Diffraction data for the unliganded crystal were collected at the Baylor College of Medicine home source by using Rigaku FR-E+ (Rigaku), and for liganded P domain crystals, data were collected on the SBC-CAT 19ID beamline at the Argonne National Laboratory, Chicago, IL, and on the 5.0.2 beamline at Advance Light Source, Berkeley, CA. Diffraction data were processed by using either HKL2000 (29), D*TREK (32), or IMOSFLM as implemented in the CCP4 suite (7). The space group in either crystal form was confirmed by using POINTLESS (13). The structure of the unliganded P domain in the C2 space group, with one molecule in the asymmetric unit, at a ∼2-Å resolution, was determined first. The initial electron density map was obtained by molecular replacement (MR) using the previously reported VA387 1996 variant P domain structure (Protein Data Bank [PDB] accession no. 20BR) (5) as the phasing model using PHASER (26). After automated model building and solvent addition using ARP/wARP (27), the structure was refined by using Refmac (28). This unliganded structure of the P domain of the 2004 variant was subsequently used as a phasing model for the structure determination of the native P domain and its complexes with different HBGAs in the C2221 space group with 10 molecules in the crystallographic asymmetric unit. For these structures, model building was carried out by using COOT (12) software, followed by iterative cycles of refinement and model building using Refmac (28) or the PHENIX suite (1). During the refinement, both translation/libration/screw (TLS) parameters (30) and NCS (noncrystallographic symmetry) constraints were included. The oligosaccharide moieties of the HBGAs were generated by using the SWEET2 package (2) of the Glycosciences.de server (http://www.glycosciences.de/), modeled into the electron density by using COOT, and validated by computing simulated annealing omit maps using PHENIX (1). Following each cycle of refinement, the model was corrected based on the Fo-Fc maps, and the stereochemistry of the structures was checked by using COOT modules and PROCHECK (21). The stereochemistry of the oligosaccharides, including the allowed conformational angles, was checked by using the CARP package at the Glycosciences.de server (http://www.glycosciences.de/). Data collection and refinement statistics are provided in Table 1. The electrostatic potential surface was calculated by using the vacuum electrostatics feature of the PyMOL software package (http://www.pymol.org/). The same software package was also used for the generation of the final figures. The superposition of the various structures was carried out by using either COOT or PyMOL. Ligand interactions were analyzed by using COOT and LIGPLOT (42), with donor-to-acceptor distances of between 2.6 Å and 3.3 Å for hydrogen-bonding interactions and C-C distances of between 3.4 Å and 4.5 Å for hydrophobic interactions.
Table 1.
Data processing and refinement statistics
| Parameter | Value for P domain 2004 variant forma |
||||
|---|---|---|---|---|---|
| Monomer | Unliganded | Complex with A type | Complex with H type | Complex with Leb | |
| Space group | C2 | C2221 | C2221 | C2221 | C2221 |
| Cell dimensions | |||||
| a, b, c (Å) | 89.0, 72.9, 55.6 | 243.9, 339.1, 125.1 | 244.5, 341.5, 124.8 | 242.2, 339.0, 124.3 | 242.7, 339.8, 125.0 |
| α, β, γ (°) | 90, 98, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–2.0 (2.05–2.0) | 27.5–3.2 (3.4–3.2) | 50–2.67 (2.72–2.67) | 40–2.85 (2.9–2.85) | 50–3.0 (3.1–3.0) |
| Wavelength (Å) | 1.54 | 1.54 | 0.98 | 0.98 | 0.98 |
| No. of reflections | 68,635 | 799,199 | 782,703 | 2,878,040 | 718,946 |
| No. of unique reflections | 23,043 | 79,184 | 143,411 | 119,306 | 98,994 |
| R merge (%) | 5.4 (30.8) | 17.1 (36.1) | 10.6 (40.4) | 12.2 (89.6) | 13.5 (51.7) |
| I/σI | 19.2 (3.8) | 7.8 (3.9) | 12.7 (2.4) | 15.9 (2.2) | 10.3 (2.6) |
| Completeness (%) | 97.2 (95.4) | 94.2 (93.7) | 99.4 (99.4) | 99.4 (99.2) | 100 (100) |
| Redundancy | 3.0 (2.8) | 3.9 (4.2) | 5.5 (5.4) | 6.7 (6.7) | 7.3 (6.9) |
| Rwork/Rfree (%) | 19.8/24.2 | 20.0/24.4 | 18.0/23.0 | 18.6/23.1 | 16.6/22.3 |
| Bond length (Å) | 0.007 | 0.011 | 0.020 | 0.020 | 0.018 |
| Bond angle (°) | 1.02 | 1.32 | 1.98 | 1.91 | 1.81 |
Values in parentheses are for the highest-resolution shell.
Protein structure accession numbers.
Coordinates and structure factors of the structures discussed in this work have been deposited in the Protein Data Bank under accession numbers 3SJP (native, monomer), 3SKB (native, unliganded), 3SLD (A-type complex), 3SLN (H-type complex), and 3SEJ (Leb complex).
RESULTS
The TCH05 variant was isolated from a 1-year-old boy in 2005 at the Texas Children's Hospital, Houston, TX, and phylogenetic analysis (data not shown) of the capsid protein sequence (GenBank accession no. JF827296) confirmed that it is a GII.4 2004 variant belonging to the Hunter cluster. The recombinant P domain (aa 221 to 531) of this GII.4 2004 variant was expressed in E. coli cells and purified to homogeneity. It crystallized into two space groups: orthorhombic space group C2221, with 4 dimers and 2 monomers in the asymmetric unit (see Fig. S2 in the supplemental material), and the monoclinic space group C2, with one monomer in the asymmetric unit. The structure determination of the 2004 P domain in each of these space groups was carried out as described in Materials and Methods. We first discuss the P domain dimer structure in the C2221 space group, because of its relevance to HBGA binding, followed by the description of the monomeric structures in both space groups.
Structure of the P domain dimer of the 2004 variant.
The overall structure of the P domain dimer of the 2004 variant in the C2221 space group is similar to the previously observed P domain dimer structures with a well-defined P2 subdomain (aa 275 to 417) inserted between the N- and C-terminal parts of the P1 subdomain (aa 221 to 274 and aa 418 to 531) in each subunit. The P1 subdomain consists of a twisted antiparallel β-sheet and a single α-helix, and the P2 subdomain consists of a six-stranded antiparallel β-barrel with elaborate loops connecting the β-strands. Analysis of the GII.4 2004 P domain dimer using the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) showed that the dimer interface in the P domain structure of the 2004 variant is extensive, with a total buried surface area of ∼3,010 Å2, and the majority of the dimeric interactions involve the P2 subdomain. The four P domain dimers in the asymmetric units are very similar to one another, with an average root mean square deviation (RMSD) of ∼0.5 Å.
Structural comparison between 2004 and 1996 GII.4 P domain dimers.
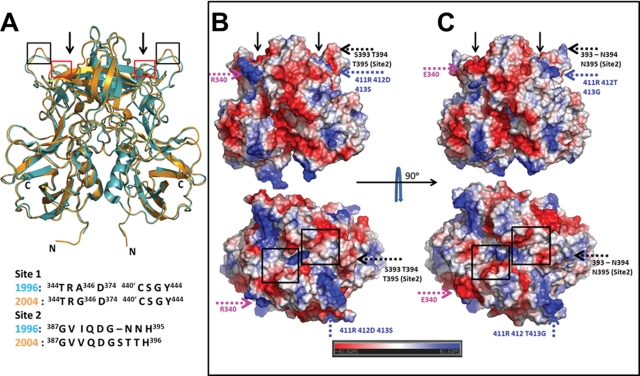
To examine if the P domain structure of the 2004 variant had undergone any structural changes from that of the 1996 variant (PDB accession no. 2OBS) (5), we superimposed both P domain structures (Fig. 2A). The structures superimposed with an RMSD of 0.7 Å, indicating that the overall structure of the P domain dimer is evolutionarily conserved from 1996 to 2004. The primary HBGA binding site 1, which has been shown to bind to the α-fucose in the 1996 variant, is structurally conserved despite some amino acid changes in the 2004 variant (E340R, A346G, and L375F) (Fig. 1). The most significant difference is found in the orientation of the loop that consists of site 2 residues (aa 390 to 395) (Fig. 2A and see Fig. S3A in the supplemental material). Site 2, which is suggested to provide further stabilizing interactions for HBGA binding and also to modulate the binding specificity, is weakly conserved, with an insertion in the post-2002 variants (Fig. 1).
Fig. 2.
Structural comparison of P domain dimers of 2004 and 1996 GII.4 variants. (A) Superposition of the 2004 (orange) and 1996 (cyan) P domain dimer structures. The N and C termini of the dimeric subunits in the 2004 P domain structure are indicated. HBGA binding sites observed for the 1996 variant structure are indicated by arrows. Red and black boxes indicate locations of site 1 and site 2, respectively, in the P domain dimer. A conformational change in the site 2 loop (black box) between the two variants is clearly evident. The alignment of residues in site 1 and site 2 of the 1996 and 2004 GII.4 variants is shown below. (B and C) Side-by-side comparison of the electrostatic potential surfaces of the 2004 and 1996 P domain dimers. (B) Side (above) and top (below) views of the 2004 variant. (C) Side (above) and top (below) views of the 1996 variant. The electrostatic potential variation from negative (red) to positive (blue) is indicated by a bar at the bottom. Significant differences in the electrostatic potential surfaces between the variants are indicated by colored dashed arrows, with each color representing a different region. The temporally varying residues that contribute to these changes are also indicated. For reference, the locations of the HBGA binding sites in the P domain dimer are indicated by black arrows in the side views and by black boxes in the top views. Residues 340 (pink), 393 to 395 (black), and 412 (blue) have all been identified as hot spots for GII.4 epochal evolution.
Although the overall structure is mostly conserved between the two variants, the temporal sequence variations result in significant differences in the electrostatic potential surfaces of the two dimers, particularly in the surface-exposed regions of the P2 subdomain (Fig. 2B and C). In some regions, there is a pronounced reversal from a positively to a negatively charged surface and vice versa. The differences are likely indicative of antigenic variations. However, some of these changes are in close proximity to HBGA binding sites, particularly site 2, and thus could potentially affect HBGA binding in the 2004 variant. Regarding HBGA binding in the 2004 variants, there have been conflicting reports (9, 10, 43). To examine if the changes observed for the 2004 P domain structure influence the HBGA binding preference of the 2004 variant, we carried out cocrystallization experiments with both monofucosyl ABH HBGAs and difucosyl secretor Lewis HBGA, for which the structure in complex with GII.4 has not been previously reported.
Structure of the GII.4 2004 P domain–A-type trisaccharide complex.
Analysis of the diffraction data from the 2004 P domain crystals obtained by cocrystallization with A-type trisaccharide showed these crystals also belonged to the C2221 space group, with 4 dimers and 2 monomers in the asymmetric unit. The structure was determined by using MR as described in Materials and Methods. The Fo-Fc map showed density due to the bound ligand. The carbohydrate density was observed exclusively in association with the dimers, consistent with the previous finding that the HBGA binding pocket is composed of residues from both subunits in the dimer. The density due to the terminal fucose moiety is clearly represented at a contour level of ∼3σ in the simulated annealing omit difference map (see Fig. S3B in the supplemental material). The densities for the other two carbohydrate residues, however, were progressively weaker, requiring a lowering of the contour level to ∼2.5σ, likely indicating conformational flexibility in this region of the trisaccharide due to a lack of constraining interactions with the P domain. A similar weakening of the density for the proximal saccharide moieties was also reported in previous crystallographic studies with the 1996 variant P domain (5).
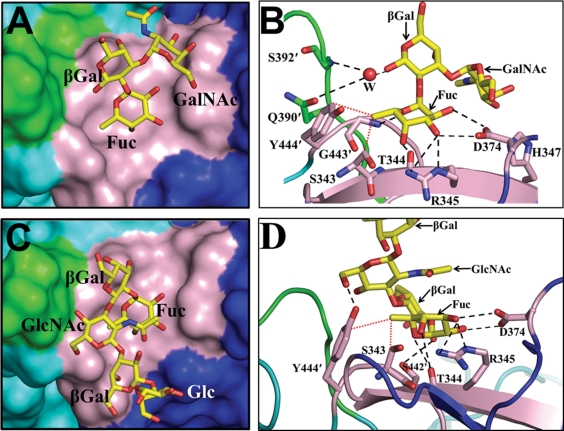
The binding of the A-type trisaccharide to the 2004 variant is similar to that observed for the 1996 variant. As in the case of the 1996 variant, the HBGA binding site in the 2004 variant is located at the surface-exposed dimeric interface, involving residues from the P2 subdomain. It is formed by residues S343, T344, R345, H347, and D374 from one subunit in the dimer and by residues S442′, G443′, and Y444′ from the other subunit (indicated by the ′ hereafter) (Fig. 3A and B). The terminal α-fucose moiety of the A-type trisaccharide is firmly anchored in the binding site by an extensive network of hydrogen bond interactions involving its exocyclic carboxyl groups and residues D374, R345, T344, and G443′ (Fig. 3B). In addition to these hydrogen-bonding interactions, the methyl group at position 6 of the fucose makes strong hydrophobic interactions with the side chain Cε atom of residue Y444′ and the Cβ atom of residue S343. The other two carbohydrate residues do not make any direct interactions. However, the O1 of the central β-Gal makes water-mediated hydrogen bond interactions with residues G392′ and Q390′ in site 2. This water is placed firmly into its position by hydrogen-bonding interactions involving the main-chain amide of residue G392′ and the main-chain carbonyl group of residue Q390′ (Fig. 3B). This is in contrast to the water-mediated interactions between β-Gal and site 2 observed in the case of the 1996 variant. In the 1996 variant, because the site 2 loop is oriented differently from that in the 2004 variant, these interactions mediated by two water molecules involve residue D391′ in site 2. In both variant structures, the C1 atom of the central β-Gal, which would link to proximal saccharides (through a 1-3 linkage) in a longer oligosaccharide, is positioned pointing toward site 2. It was this observation, as mentioned above, that provided a basis for Cao et al. (5), and later on for others (11), to suggest the possible involvement of site 2 in providing further stability to longer oligosaccharides in modulating the HBGA binding specificity.
Fig. 3.
Binding of monofucosyl ABH family HBGAs to the 2004 P domain dimer. (A) Surface representation (top view) showing a close-up view of one of the two binding sites in the 2004 P domain dimer bound to an A-type trisaccharide. Identical interactions with the trisaccharide were observed for the other binding site of the dimer and are not shown. Amino acid residues in site 1 and site 2 are shown in pink and green, respectively, with other residues from the dimeric subunits in the vicinity shown as blue and cyan. The HBGAs, here and in all subsequent figures, are shown as yellow sticks, with nitrogen atoms and oxygen atoms shown in blue and red, respectively. The individual saccharide moieties in the A-type trisaccharide are labeled as Fuc (fucose), βGal (galactose), and GalNAc (N-acetylgalactosamine). (B) Detailed interactions of the A-type trisaccharide with the 2004 P domain dimer. P domain residues participating in the hydrogen-bonding (black dashed lines) and hydrophobic (red dotted lines) interactions with the trisaccharide are labeled and shown as pink sticks, with nitrogen and oxygen atoms in blue and red, respectively; the rest of the dimer is shown as a cartoon representation. The individual moieties of the trisaccharide are labeled as described above (A). The water molecule involved in solvent-mediated interactions with site 2 is labeled W and is shown as a red sphere. (C) Surface representation (top view) showing a close-up view of one of the two binding sites in the 2004 P domain dimer bound to the H-type pentasaccharide. Amino acid residues in site 1 and site 2 are colored as described above (A). The H-type pentasaccharide is shown as stick model with individual moieties labeled as Fuc (fucose), βGal (galactose), GlcNAc (N-acetylglucosamine), and Glc (glucose). (D) Detailed interactions of the H-type pentasaccharide with the 2004 P domain. P domain residues participating in the hydrogen-bonding (black dashed lines) and hydrophobic (red dotted lines) interactions with the pentasaccharide (individual moieties labeled as described above) are labeled and shown as pink sticks, with nitrogen and oxygen atoms in blue and red, respectively. The individual moieties of the pentasaccharide are labeled as described above (C).
Structure of the P domain-H type 1 complex.
Although the cocrystallization of the 1996 P domain with the H-type complex was attempted, the bound ligand could not be visualized in the P domain structure (5). Thus, it was of interest to examine if the bound H-type HBGA could be visualized with the 2004 P domain. The cocrystallization of the 2004 P domain with the H type 1 complex also yielded crystals in the same space group, C2221, as described above, with 4 dimers and 2 monomers in the asymmetric unit. In this experiment, instead of a trisaccharide, as was attempted with the 1996 P domain, a longer pentasaccharide of the H type was used so that the proposed role of site 2 in stabilizing the longer oligosaccharides could be evaluated. The structure determination was carried out as described above for the P domain–A-type complex. The different density in the simulated annealing omit map for the first three carbohydrate residues (α-Fuc–β-Gal–GlcNAc) of the pentasaccharide was clear (contour level, ∼3σ), and for the proximal two carbohydrate residues, it was significantly lower (see Fig. S3C in the supplemental material). The H-type pentasaccharide binds at the same location as the A-type trisaccharide at the dimeric interface of the P2 subdomain (Fig. 3C). As in the A-type trisaccharide, the terminal α-fucose of the H-type pentasaccharide is primarily involved in the interactions with site 1, making similar hydrogen bonds with residues T344, R345, D374, and G443′ and van der Waals contacts with residues Y444′ and S343 (Fig. 3D). However, in the case of the H-type pentasaccharide, β-Gal, which is linked to the terminal α-fucose, is oriented differently than the β-Gal in the A-type trisaccharide, and as a result, the carboxyl group at position C3 hydrogen bonds with the side-chain hydroxyl group of S442′. In addition, the O6 of GlcNAc hydrogen bonds with the side-chain hydroxyl group of residue Y444′. Thus, in the case of the H-type pentasaccharide, residue Y444′ is involved in both the hydrogen bond and hydrophobic interactions, in contrast to the A-type trisaccharide, in which this residue participate only in the hydrophobic interactions. The other two distal sugar residues in the pentasaccharide are positioned away from the HBGA binding site. Despite the longer oligosaccharide used in this study, site 2 residues are not involved in the interactions with HBGA.
Structure of the P domain-Leb complex.
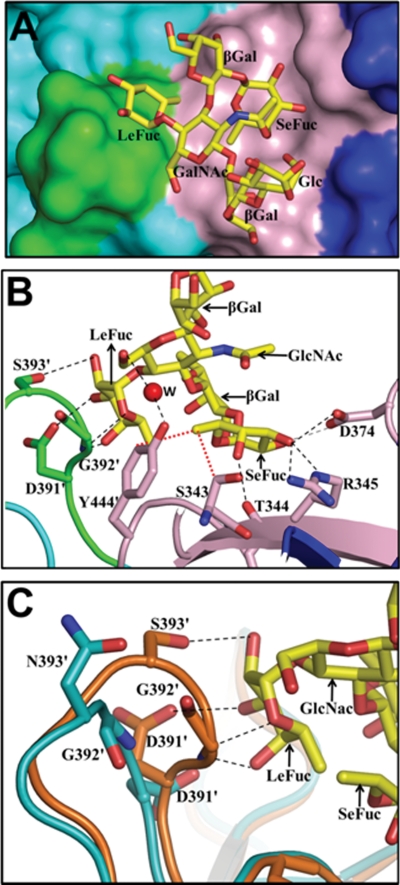
Although various biochemical studies have indicated the binding of secretor Lewis HBGAs to GII.4 NoVs (9, 16, 23, 34), there has been no structural characterizations of GII.4 NoVs binding to Lewis HBGAs. In contrast to ABH HBGAs, which have one fucose moiety (referred to as secretor fucose) added by the FUT2 enzyme, the secretor Lewis HBGAs have an additional fucose moiety (referred to as Lewis fucose) linked to the precursor GlcNAc (type 1), which is added by the FUT3 enzyme (see Fig. S1 in the supplemental material). To structurally characterize the binding of difucosyl Lewis HBGAs, we cocrystallized the 2004 P domain with a Leb hexasaccharide under the same conditions as those used for the other crystals discussed above. Analysis of the diffraction data from these crystals indicated the same C2221 space group, with 4 dimers and 2 monomers in the asymmetric unit. Based on the simulated annealing Fo-Fc omit map (Fig. S3D), we could unambiguously model the terminal four saccharide moieties of the Leb hexasaccharide. In contrast to the binding of monofucosyl ABH HBGAs, as described above, which involves primarily site 1 residues, the binding of the difucosyl Leb prominently involves residues from both site 1 and site 2 (Fig. 4A). The secretor fucose of Leb binds to site 1, making hydrogen bonds and van der Waals contacts similar to those observed in the case of monofucosyl A and H types, involving residues T344, R345, D374, Y444′, and S343 (Fig. 4B). The Lewis fucose of Leb, with its exocyclic carboxyl groups facing oxygen-rich site 2, interacts extensively with this site, making a total of four hydrogen bonds involving residues D391′, G392′, and S393′. In addition, the O6 of GlcNAc, which is proximally linked to the Lewis fucose, interacts with the side-chain hydroxyl group of Y444′ through water-mediated hydrogen bonds.
Fig. 4.
Binding of difucosyl Lewis HBGA to the 2004 P domain dimer. (A) Surface representation (top view) showing a close-up view of one of the two binding sites in the 2004 P domain dimer bound to Leb hexasaccharide. The coloring scheme for the amino acid residues in site 1 and site 2 is the same as that described in the legend of Fig. 3A. The individual saccharide moieties in the Leb hexasaccharide are labeled SeFuc (secretor fucose), LeFuc (Lewis fucose), βGal (galactose), GlcNAc (N-acetylglucosamine), and Glc (glucose). (B) Detailed interactions of the Leb hexasaccharide with the 2004 P domain. P domain residues participating in the hydrogen-bonding (black dashed lines) and hydrophobic (red dotted lines) interactions with Leb (individual moieties denoted as described above) are labeled and shown as pink (site 1) and green (site 2) sticks, with nitrogen and oxygen atoms in blue and red, respectively. The water molecule involved in solvent-mediated interactions with residue Y444′ and O6 of GlcNAc is labeled W and is shown as a red sphere. (C) Close-up view of the superposition of site 2 from the 1996 variant (cyan) with the Leb-bound 2004 variant (orange). Dashed lines represent hydrogen-bonding interactions between site 2 residues of the 2004 variant and the Lewis fucose of Leb. The site 2 residues in the 2004 variant participating in hydrogen-bonding interactions are shown as orange sticks, with nitrogen and oxygen atoms labeled in blue and red, respectively. Their counterparts in the 1996 strain are shown as cyan sticks. Because of the structural alterations in site 2 of the 1996 variant, similar hydrogen-bonding interactions with Leb involving site 2 residues are not possible.
Although the structure of the 1996 variant with secretor Lewis HBGAs has not been determined, a relevant question is whether the1996 P domain structure allows similar interactions with this Leb. The superposition of the P domain structures of the 2004 and 1996 variants clearly shows that the site 2 loop in the 1996 variant is positioned slightly away from site 1 compared to that in the 2004 P domain structure. As a result, the site 2 residues in the 1996 P domain are not in a position to make similar hydrogen bond interactions with the Lewis fucose moiety of Leb (Fig. 4C). Of the four hydrogen bonds observed between the Lewis fucose and site 2 residues in the 2004 P domain, the conformation of site 2 in the 1996 P domain structure allows for one rather weak hydrogen bond (∼3.3 Å) involving residue D391′.
P domain dimer and monomer structures and conformational flexibility.
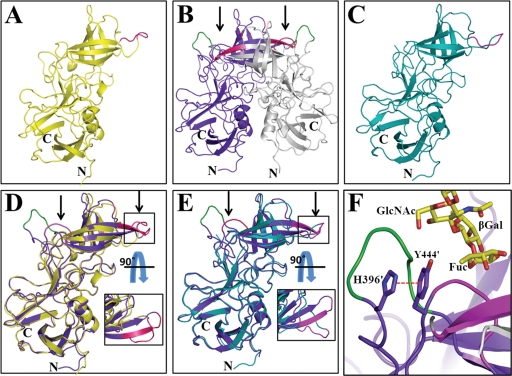
As mentioned above, the asymmetric unit in the C2221 crystals of the GII.4 2004 P domain, in addition to four dimers, consists of two monomers. Analysis of the crystal packing confirmed that these monomers do not pair with other monomers using any of the symmetry operations in the space group. Furthermore, the GII.4 2004 P domain also crystallized in the C2 space group as a monomer alone under different crystallization conditions (Fig. 5A). This provided us with a unique opportunity to examine how the P domain in the monomeric state differed from the subunits in the dimeric form, particularly in the regions that are involved in the dimerization and formation of the HBGA binding site. As described above, HBGA binding is observed exclusively with dimers, as the primary HBGA binding site (site 1) is composed of residues from both subunits in the dimer. The superimposition of the monomers in C2221 and in C2 with the subunits in the C2221 dimers revealed interesting conformational changes that may have implications for dimer formation and, consequently, for the formation of the HBGA binding site (Fig. 5B to E). Most significant conformational changes are particularly in some of the loop regions. The largest conformational change is observed in the loop containing residues 334 to 347, which shows a movement of ∼13 Å in the Cα atom position at maximum divergence (Fig. 5E). This loop contains some of the residues, as a part of site 1, which contribute to HBGA binding. It adopts three different conformations between the monomers in C2 and C2221 and the individual subunits in the dimer. In both C2 and C2221 monomers, this region is essentially a random coil; however, upon dimerization the N- and C-terminal portions of this loop undergo a transition to β-strands, forming an antiparallel β-sheet. Such a transition brings residues involved in HBGA binding into the appropriate orientation. Without this rearrangement, this loop region would sterically hinder dimer formation. In addition to this loop, two other regions, aa 439 to 447, which include some of the residues that constitute site 1, and aa 389 to 400, which consist of residues in site 2, also exhibit significant changes. In the C2 and C2221 monomer structures, the electron densities in these two regions are weak or absent, indicating conformational flexibility in these two regions, whereas in the dimer, both of these regions adopt well-defined conformations, thereby allowing the proper formation of the HBGA binding site.
Fig. 5.
Comparison of P domain monomers and dimers. (A) Cartoon representation of the P domain monomer in the C2 crystal form (yellow). Site 1 residues are shown in magenta for reference. (B) Cartoon representation of the P domain dimer in the C2221 crystal form. The dimeric subunits are indicated in purple and gray, and black arrows indicate the location of the HBGA binding sites. HBGA binding sites 1 and 2 are indicated in magenta and green, respectively. (C) Cartoon representation of the P domain monomer in the C2221 crystal form (cyan). Site 1 residues are shown in magenta for reference. (D) Superposition of the C2 monomer (yellow) and one of the dimer subunits (purple) in the C2221 crystal form. Regions with conformational flexibility, which include site 1, are shown inside a box (side view). The inset shows the top view of this boxed loop region. Site 2 (green) is seen only in the dimer subunit (purple); this region in the C2 monomer is disordered. Arrows indicate the HBGA binding sites. (E) Superposition of the C2221 monomer (cyan) and a subunit from the dimer (purple) in the same C2221 crystal form. Regions with conformational flexibility, which include site 1, are shown inside a box (side view). The top view of this boxed region is shown in the inset. As in D, site 2 (green) is seen only in the dimer subunit (purple), and this region in the C2221 monomer is disordered. Arrows indicate the HBGA binding region. (F) Cation-π interaction between residues H396 and Y444 in the P domain of the 2004 variant with a face-to-face distance of ∼3.8 Å (indicated by a red dashed line). These two highly conserved residues are in close proximity to the dimeric interface, HBGA binding sites 1 (magenta) and 2 (green). Bound H-type HBGA is shown (yellow) for reference. The cation-π interaction is observed only for the dimeric subunits and is not observed for the monomeric state, as the loops containing residues H396 and Y444 in this state are disordered.
One interesting feature in the P domain dimeric conformation of the 2004 variant is the stabilizing face-to-face stacking interaction observed between the positively charged imidazolium ring of residue H396 and the phenyl ring of residue Y444. With a distance of ∼3.8 Å between the rings, this interaction is clearly indicative of a cation-π interaction (Fig. 5F). Such an interaction is absent in the monomeric state, as the loops at aa 389 to 400 and 439 to 447 are disordered. An identical cation-π interaction between residues H395 and Y443 was also observed in the P domain dimer of the 1996 variant, and these two residues are highly conserved in all the GII.4 variants. Their close proximity to the dimeric interface and the HBGA binding sites with residue Y444 participating in the direct interactions with HBGAs suggest that these two residues and the cation-π interaction play important roles in both stabilizing the dimeric conformation and HBGA binding. In the case of the 1996 variant, it was shown previously that a Y443A mutation abolishes HBGA binding (40).
DISCUSSION
Although many factors may be involved, previous studies suggested that in addition to antigenic drift, alterations in HBGA binding patterns resulting from temporal sequence changes in the surface-exposed P2 subdomain contribute to GII.4 epochal evolution. The main focus of our studies was to characterize how such temporal sequence variations in the GII.4 variants influence the P domain structure and to determine if they affect HBGA binding. In these studies we analyzed a 2004 epidemic variant that is temporally distant from a 1996 GII.4 variant for which the P domain structure with A- and B-type trisaccharides was already known (5).
Temporal sequence variations do not affect binding of monofucosyl HBGA.
Our structural studies of the P domain of a 2004 variant indicate that temporal sequence variations do not affect the binding of monofucosyl ABH HBGAs. The binding of these HBGAs predominantly involves interactions between the terminal fucose of the HBGA and the residues in site 1. Despite two amino acid changes in site 1 of the 2004 variant, the conformation of this site is unaltered compared to that in the 1996 variant. The binding of the A-type trisaccharide is essentially the same for both of these variants. Although the structure of the 1996 variant with the H type is not available for direct comparison, our studies with the H-type pentasaccharide indicate that it is very likely that the 1996 variant binds to the H type in a similar manner. This is because the binding of the H-type pentasaccharide in the 2004 variant involves a set of interactions between the terminal fucose and the site 1 residues similar to that of the A-type trisaccharide. The additional hydrogen bond interactions observed, involving residues S442 and Y444, because of the longer H-type oligosaccharide used in our studies, are also possible in the case of the 1996 variant, as both of these residues are conserved in the 1996 variant (residue S441), and they superimpose perfectly. All the residues that participate in direct interactions with monofucosyl ABH HBGAs, including those that contribute to hydrophobic interactions, such as residues Y444 and S343, are highly conserved in all the GII.4 variants. Despite the longer oligosaccharide used in our studies with the H-type HBGA, contrary to previous suggestions, we did not observe an involvement of site 2, which exhibits significant temporal variations. Thus, the binding of ABH HBGAs, which may contribute to the higher prevalence of GII.4 NoV, is unlikely to be a major factor in GII.4 epochal evolution. However, the possibility that subtle variations in the binding affinities for various ABH HBGAs could contribute to GII.4 evolution cannot be discounted. These variations could indeed occur in the context of the multivalent binding of the virions driven by avidity effects, which cannot be assessed directly from our crystallographic studies with the P domain alone. Although further studies are required to assess how the binding of ABH HBGAs contributes to GII.4 evolution, de Rougemont et al. indeed observed a stronger binding of 2004 GII.4 VLPs to ABH HBGAs than the pre-2002 GII.4 strains (9).
Recently, based on saliva binding studies using P particles of various GII.4 variants, an interesting observation was made regarding the binding of the A-type HBGA (43). From those studies it was suggested that an amino acid change at position 389, which is close to site 2, may alter the binding preference for the A type in the GII.4 variant. GII.4 variants prior to 2002 have isoleucine at this position, whereas in the GII.4 variants from between 2002 and 2006, it is mutated to a valine, and in the subsequent 2006-2008 variants, it reverted back to isoleucine (Fig. 1). According to these studies, “V389” strains have a reduced affinity for the A type compared to “I389” phenotypes. Our studies clearly show that the P domain of the 2004 variant, a V389 phenotype, binds to the A-type trisaccharide in a manner similar to that of the P domain of the 1996 variant, an I389 phenotype, making very similar hydrogen-bonding interactions. The I389 residue in the 1996 variant, except for the terminal methyl groups of the isoleucine, superimposes exactly with the V389 residue in the 2004 variant. In both structures, these residues do not make any contact with the A-type HBGA, and the closest distance to the HBGA is ∼6.5 Å, rather far for it to be exerting any influence on the binding of the A type. Thus, from our structural observations, it is unclear as to how an amino acid change at residue 389 could affect the binding preference for A-type HBGA in GII.4 variants.
Based on mutational studies using GII.4 VLPs, some of the residues in site 2, particularly those under positive selection, were also suggested to alter the binding affinity/specificity for ABH HBGAs. A mutation of residue D393, which is found in the GII.4 1987 variant, to a glycine, which was observed for the 1997 variant, was reported previously to promote B-type binding in the 1987 variant (23). More recently, it was shown that the deletion of residue T395 in a GII.4 2004 variant resulted in a markedly lower level of binding to A and B types than the wild type (9). Except for water-mediated hydrogen-bonding interactions with the A type, our studies show that residues in site 2 do not participate directly in the binding of ABH HBGAs. It is possible that these mutations cause alterations in the solvent structure near site 2, thereby causing subtle differences in the relative binding affinities/specificities of ABH HBGAs. Alternatively, it is possible that these mutations cause other conformational changes. Further crystallographic studies are required to address these possibilities.
Interactions with difucosyl secretor Lewis HBGA involves site 2 with temporal variations.
In contrast to the binding of ABH HBGA, the binding of difucosyl Leb HBGA to the 2004 variant involves prominently both site 1, which is highly conserved, and site 2, which exhibits significant temporal sequence variability. Strong hydrogen bond interactions involving both fucosyl moieties of Leb indicate significantly increased binding stability for Lewis HBGA compared to ABH HBGAs. Because of the involvement of the conserved site 1 in interacting with secretor fucose in Leb, it is very likely that all the GII.4 variants show binding to Leb; however, their binding strength may be significantly altered because of the sequence variability in site 2. For instance, in the 1996 variant, the site 2 loop is not positioned as favorably as that in the 2004 variant to make similar hydrogen-bonding interactions with the Lewis fucose of Leb (Fig. 4C). With the loss of these hydrogen-bonding interactions, the affinity for Leb would be significantly reduced in the 1996 variant, although it is unlikely to be completely eliminated, because the secretor fucose of Leb can still make hydrogen-bonding interactions with site 1.
The noticeable structural shift that was observed for the position of the site 2 loop between 1996 and 2004 variants was most likely caused by an insertion following residue 393 in the latter. Interestingly, such an insertion is consistently observed in all post-2002 variants, suggesting that this region is under positive selection (Fig. 1). However, the amino acid compositions immediately following this position (aa 394 to 395) vary considerably in these post-2002 variants. One possibility is that these variations may be involved in modulating the specificity/affinity for different secretor Lewis HBGAs, such as Leb, ALeb, and BLeb, derived from the type 1 precursor disaccharide, and their counterparts, derived from other precursor disaccharide types, such as Ley, ALey, and BLey (type 2). In all the post-2002 variants, the interactions between site 1 and secretor fucose in these Lewis HBGAs are likely to be the same. However, because of the differences in the terminal nonfucosyl saccharide moieties and also the linkage differences in these HBGAs (see Fig. S1 in the supplemental material), the orientation of the Lewis fucose moiety that interacts with site 2 is likely to differ, and accordingly, the amino acid changes in site 2 may provide a selectivity filter to specifically modulate the affinity for a particular (secretor) Lewis HBGA or a particular combination of these HBGAs. Thus, the temporal variations in site 2 resulting in the differential binding of secretor Lewis family HBGAs are likely an important contributing factor in GII.4 epochal evolution.
Temporal variations result in significant differences in the electrostatic landscapes of the P domains.
Although the sequence variations between the P domains of the 1996 and 2004 variants, except those in site 2, cause minimal structural alterations elsewhere, they result in significant differences in the electrostatic potential surfaces of the two dimers, particularly in the surface-exposed regions of the P2 subdomain (Fig. 2B and C). These regions include site 2, which we have shown to be important for the binding of Leb in the 2004 variant; residue R340; and residues 410 to 412. Although residue R340 does not participate in any direct interactions with HBGA, it is indeed close to site 1, whereas residues 410 to 412 are not involved in HBGA binding. Based on the evolutionary analysis of the GII.4 sequences, the residues in site 2, aa 393 to 395, and also aa 340 have been identified as “hot spots” for the evolution of GII.4 variants (3, 11). The differences in the electrostatic landscapes are likely indicative of antigenic variation, and the proximity of some of these changes to HBGA binding sites may suggest an interplay between HBGA binding and antigenic variation in the epochal evolution of GII.4 variants. The changes in the surface electrostatic potential, resulting from immunological pressure, could affect the manner by which NoVs interact with the cell surface receptors through avidity effects. The modulation of cell binding due to alterations of the surface charge was noted previously in the cases of influenza virus (14) and HIV (8). This may explain the results of elegant surrogate HBGA blockade assays, used in lieu of neutralization assays, that showed different binding activities of pre- and postepidemic antisera (23). However, because of the current lack of data describing the neutralization epitopes for any of the human NoVs, a more definitive correlation between HBGA binding and how it would be affected by neutralization antibodies is not possible.
Local flexibility in the P2 subdomain and possible functional implications.
An interesting observation from our crystallographic studies is that the P domain of the 2004 variant exists as both monomers and dimers within the asymmetric unit of the same crystal. A comparative analysis of the monomeric and dimeric forms points to an inherent flexibility in the loop regions of the P2 subdomain that participate in the dimerization and formation of the HBGA binding site. Such a flexibility in the P2 subdomain suggests the interesting possibility that the dimeric interactions and, consequently, the HBGA binding site can be “locally” disrupted and reformed under certain conditions. NoV infection occurring through the oral route to ultimately target intestinal epithelial cells raises the puzzling question of how the bound salivary mucin-linked HBGA is disassociated to enable the virus to interact with HBGAs linked to epithelial cells for entry. We hypothesize that the local flexibility in the P2 subdomain could allow the virus to disassociate itself from the salivary HBGA in the changing microenvironment (pH, for example) of the gut to subsequently interact with intestinal epithelial cell HBGAs for cell entry.
Although this is an interesting possibility, it remains to be shown whether such a conformational flexibility is a common feature of the GII NoVs and whether bound HBGA can be disassociated under various conditions, such as pH. One structural interaction that could be susceptible to such changes in the microenvironment is the observed cation-π interaction between highly conserved residues H396 and Y444, which is present only upon dimerization (Fig. 5F). The importance of the Y444 residue, and, consequently, that of the cation-π interaction, in HBGA binding is strongly supported by data from previous mutational analyses of the case of the 1996 variant, wherein it was shown that a Y443A mutation abolishes HBGA binding (40). The destabilization of the cation-π interaction could be a pH-sensitive trigger to initiate the partial unraveling of the dimeric interface. Such a process should then exist for other NoVs as well. In the case of GI NoVs, such as Norwalk virus (NV) (GI.1), in which the HBGA binding site, in contrast to GII NoVs, is composed of residues from the same subunit, a highly conserved His-Trp pair involved in cation-π interactions was shown previously to be critical for HBGA binding (6). The local destabilization of this cation-π interaction in the GI NoVs may provide a pH-sensitive mechanism for the disassociation and reassociation of HBGAs. It is interesting that His-Trp cation-π interactions are implicated in the pH-sensitive gating of the M2 ion channel in influenza virus (33).
In conclusion, in addition to providing the first structural details of GII.4 NoV interactions with H-type and secretor Lewis HBGAs, our studies have revealed several novel observations that have implications for our understanding of the epochal evolution of GII.4 variants. The observation that interactions with both ABH and secretor HBGAs involve the highly conserved site 1 suggests that all GII.4 variants bind to these HBGAs with similar affinities. However, subtle variations in the binding affinities for these HBGAs could occur in the context of the multivalent binding of the virion and thus could contribute to GII.4 evolution. The observation that the binding of Lewis HBGAs also involves site 2, which is susceptible to conformational alterations because of temporally evolving residues, suggests that this site could function as a selectivity filter to more directly modulate the affinities for secretor Lewis family HBGAs and thus could contribute to epochal evolution by the potentiated targeting of Lewis-positive, secretor-positive individuals. Further epidemiological studies are required to examine if such individuals are differentially targeted during epidemic peaks. From the observation of distinct differences in the electrostatic landscapes of the two temporally distant GII.4 variants, we have suggested that these changes are indicative of antigenic variation, and the proximity of some of these changes to HBGA binding sites leads to a coordinated interplay between HBGA binding and antigenic variation in the epochal evolution of GII.4 variants. Another novel observation is that the regions involved in P domain dimerization and the formation of the HBGA binding site can be conformationally flexible. This has led us to postulate that such flexibility may play an important role in the dissociation and reassociation of HBGA during virus passage through the gastrointestinal tract.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the use of the synchrotron beamlines at Advanced Light Source (5.0.1), Berkeley, CA, and Argonne National Laboratories (SBC-CAT 19ID), Chicago, IL, for diffraction data collection. We thank their staff for excellent help.
This work was supported by grants from the NIH (PO1 AI057788 to M.K.E., R.L.A., and B.V.V.P. and P30DK5638 to M.K.E.) and the Robert Welch Foundation (Q1292 to B.V.V.P.). The Berkeley Center for Structural Biology is supported in part by the NIH, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. SBC-CAT 19ID at Advanced Photon Source is supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Adams P. D., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bohne A., Lang E., von der Lieth C. W. 1999. SWEET—WWW-based rapid 3D construction of oligo- and polysaccharides. Bioinformatics 15:767–768 [DOI] [PubMed] [Google Scholar]
- 3. Bok K., et al. 2009. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83:11890–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bu W., et al. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 82:5340–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao S., et al. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi J. M., Hutson A. M., Estes M. K., Prasad B. V. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175–9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 8. Cornelissen M., Hogervorst E., Zorgdrager F., Hartman S., Goudsmit J. 1995. Maintenance of syncytium-inducing phenotype of HIV type 1 is associated with positively charged residues in the HIV type 1 gp120 V2 domain without fixed positions, elongation, or relocated N-linked glycosylation sites. AIDS Res. Hum. Retroviruses 11:1169–1175 [DOI] [PubMed] [Google Scholar]
- 9. de Rougemont A., et al. 2011. Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens. J. Virol. 85:4057–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Donaldson E. F., Lindesmith L. C., Lobue A. D., Baric R. S. 2008. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 225:190–211 [DOI] [PubMed] [Google Scholar]
- 11. Donaldson E. F., Lindesmith L. C., Lobue A. D., Baric R. S. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. Microbiol. 8:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emsley P., Cowtan K. 2004. COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 13. Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72–82 [DOI] [PubMed] [Google Scholar]
- 14. Gambaryan A. S., Matrosovich M. N., Bender C. A., Kilbourne E. D. 1998. Differences in the biological phenotype of low-yielding (L) and high-yielding (H) variants of swine influenza virus A/NJ/11/76 are associated with their different receptor-binding activity. Virology 247:223–231 [DOI] [PubMed] [Google Scholar]
- 15. Green K. Y. 2007. Caliciviridae: the noroviruses, p. 949–979 In Knipe D. M., et al. (ed.), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 16. Huang P., et al. 2005. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 79:6714–6722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hutson A. M., Atmar R. L., Graham D. Y., Estes M. K. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 185:1335–1337 [DOI] [PubMed] [Google Scholar]
- 18. Hutson A. M., Atmar R. L., Marcus D. M., Estes M. K. 2003. Norwalk virus-like particle hemagglutination by binding to H histo-blood group antigens. J. Virol. 77:405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kapust R. B., et al. 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14:993–1000 [DOI] [PubMed] [Google Scholar]
- 20. Kroneman A., et al. 2008. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe network from 1 July 2001 to 30 June 2006. J. Clin. Microbiol. 46:2959–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 22. Lindesmith L., et al. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548–553 [DOI] [PubMed] [Google Scholar]
- 23. Lindesmith L. C., et al. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marionneau S., et al. 2001. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 83:565–573 [DOI] [PubMed] [Google Scholar]
- 25. Marionneau S., et al. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. 2007. PHASER crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morris R. J., Perrakis A., Lamzin V. S. 2003. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374:229–244 [DOI] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 30. Painter J., Merritt E. A. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450 [DOI] [PubMed] [Google Scholar]
- 31. Patel M. M., et al. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pflugrath J. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55:1718–1725 [DOI] [PubMed] [Google Scholar]
- 33. Sharma M., et al. 2010. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science 330:509–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shirato H., et al. 2008. Noroviruses distinguish between type 1 and type 2 histo-blood group antigens for binding. J. Virol. 82:10756–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shirato-Horikoshi H., Ogawa S., Wakita T., Takeda N., Hansman G. S. 2007. Binding activity of norovirus and sapovirus to histo-blood group antigens. Arch. Virol. 152:457–461 [DOI] [PubMed] [Google Scholar]
- 36. Siebenga J. J., et al. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siebenga J. J., et al. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 200:802–812 [DOI] [PubMed] [Google Scholar]
- 38. Tan M., Jiang X. 2005. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 13:285–293 [DOI] [PubMed] [Google Scholar]
- 39. Tan M., Jiang X. 2010. Norovirus gastroenteritis, carbohydrate receptors, and animal models. PLoS Pathog. 6:pii=1000983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tan M., et al. 2008. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology 379:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tan M., et al. 2009. Conservation of carbohydrate binding interfaces: evidence of human HBGA selection in norovirus evolution. PLoS One 4:e5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wallace A. C., Laskowski R. A., Thornton J. M. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134 [DOI] [PubMed] [Google Scholar]
- 43. Yang Y., et al. 2010. Genetic and phenotypic characterization of GII-4 noroviruses that circulated during 1987 to 2008. J. Virol. 84:9595–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng D. P., et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.