Abstract
The herpes simplex virus (HSV) tegument protein VP1-2 is essential for virus entry and assembly. VP1-2 also contains a highly conserved ubiquitin-specific protease (USP) domain within its N-terminal region. Despite conservation of the USP and the demonstration that it can act on artificial substrates such as polyubiquitin chains, identification of the relevance of the USP in vivo to levels or function of any substrate remains limited. Here we show that HSV VP1-2 USP can act on itself and is important for stability. VP1-2 N-terminal variants encompassing the core USP domain itself were not affected by mutation of the catalytic cysteine residue (C65). However, extending the N-terminal region resulted in protein species requiring USP activity for accumulation. In this context, C65A mutation resulted in a drastic reduction in protein levels which could be stabilized by proteosomal inhibition or by the presence of normal C65. The functional USP domain could increase abundance of unstable variants, indicating action at least in part, in trans. Interestingly, full-length variants containing the inactive USP, although unstable when expressed in isolation, were stabilized by virus infection. The catalytically inactive VP1-2 retained complementation activity of a VP1-2-negative virus. Furthermore, a recombinant virus expressing a C65A mutant VP1-2 exhibited little difference in single-step growth curves and the kinetics and abundance of VP1-2 or a number of test proteins. Despite the absence of a phenotype for these replication parameters, the USP activity of VP1-2 may be required for function, including its own stability, under certain circumstances.
INTRODUCTION
The posttranslational modification of proteins by mono-, multi-, and polyubiquitination is a major regulatory pathway controlling protein stability, trafficking, and function and as a result plays diverse roles in many aspects of cellular regulation and organization (5, 19). Ubiquitination is therefore also involved in numerous aspects of virus-host interactions, from the perspective of cellular response mechanisms, such as immune regulation, but also from the perspective of virus control of ubiquitination pathways (8, 20). Many viruses modulate ubiquitination of cellular and virus components both by indirect mechanisms wherein cellular conjugation and deconjugation processes are altered and direct mechanisms wherein viruses encode specific ubiquitin ligases or ubiquitin-specific proteases (20).
A unique ubiquitin-specific protease (USP) has now been identified, embedded within the N-terminal region of the structural protein VP1-2 of herpes simplex virus (HSV) (22). VP1-2, the product of the UL36 gene in HSV, is a large multifunctional protein and one of a subset of tegument proteins which is both essential for virus replication and conserved across the entire herpesvirus family (11, 14, 25, 26, 28). Moreover, the N-terminal USP domain within VP1-2, while relatively poorly conserved in overall primary sequence, contains the core catalytic residues including C65 (in HSV) which are completely conserved and required for enzymatic activity of a cysteine protease (22, 35). The USP activity of HSV VP1-2 was originally observed using a ubiquitin suicide substrate which revealed a product of ca. 57 kDa in infected cells, representing the N-terminal region of VP1-2 coupled to the substrate (22). A purified protein comprising the N-terminal 533 residues was shown to specifically bind to ubiquitin and not to ubiquitin-like proteins such as Sumo1, Nedd8, or ISG15. This USP protein was also shown to cleave lysine 48-linked polyubiquitin chains, an activity completely dependent upon the conserved C65 core catalytic residue. USP activity has now been demonstrated for the VP1-2 homologues of a number of herpesviruses across the alpha, beta, and gamma families including human cytomegalovirus (HCMV) (23, 41), mouse cytomegalovirus (MCMV) (36), Epstein-Barr virus (EBV) (36, 42), Marek's disease virus (MDV) (21), and Kaposi's sarcoma-associated herpesvirus (KSHV) (16). Analysis of structural features of the USP domain of MCMV indicates that the herpesvirus class of USPs adopts a papain-like fold that is structurally distinct from what is seen for other deubiquitinating enzymes (36). To date, most analysis of USP activity has been performed with ubiquitin probes or with polyubiquitin branched chains linked by lysine 48 (K48) or lysine 63 (K63) linkages. Although these are artificial substrates, such analyses demonstrate at the least the ability to cleave polyubiquitin and may be indicative of type of function, since K48- and K63-branched polyubiquitin are associated respectively with protein degradation or protein trafficking (32). The purified HSV USP was reported to exhibit significant specificity toward K48-linked polyubiquitin (22). This was also reported for analysis of the virion-associated USP activity of the homologue (pUL48) in HCMV (41). However, in a later study of the purified HCMV and HSV USP domains, both K48- and K63-linked chains were cleaved, with if anything more efficient activity on K63-linked chains for both USPs (23). In other studies, the MCMV USP domain cleaved both K48- and K63-linked chains, though with 5-fold greater activity on K48-linked chains. The purified EBV and KSHV USP domains were reported to cleave both types of chains with approximately equal efficiencies (16, 42).
While it is clear that VP1-2 is absolutely essential for virus replication and plays important roles at different phases of the virus life cycle (2, 11, 14, 25, 26, 28, 34), the precise mechanistic role of the USP activity within VP1-2 remains poorly understood. Studies in HCMV provided the first demonstration for an involvement in virus replication where HCMV mutants containing substitutions within the catalytic triad of the pUL48 USP domain (C24I or H162A in this case) replicated more slowly with lower yields of extracellular virus in culture (41). These studies indicated that an active USP was not essential for virus replication but that inactivity abrogated the infection process at some stage. Consistent with this, studies in pseudorabies virus (PRV) demonstrated that a virus with a substitution in the proposed catalytic cysteine of the PRV VP1-2 USP (C26S) resulted in a 20- to 30-fold reduction in virus yields in single-step growth curves, a reduction in plaque size (30%), and a delayed onset of pathogenic features after intranasal infection of mice (6). Moreover, in ultrastructural analysis of infection in culture, accumulations in nonenveloped cytoplasmic capsids were observed for the mutant virus, consistent with a defect in the known role of VP1-2 in virus assembly (6). In studies of a similar PRV mutant containing a C26A mutation, virus yields in culture were delayed and lower (10- to 20-fold), and a very significant defect was observed in animal models of neuroinvasion from peripheral tissues (27). In the MDV VP1-2 homologue, it was reported that a single substitution in the active site of the enzyme abolished its ubiquitin cleavage activity in vitro, and while the corresponding mutation in the context of a recombinant virus had a modest though detectable effect on replication (4-fold lower yields and up to 50% reduction ion plaque size), it had a profound effect on in vivo pathogenesis, with a 1,000- to 10,000-fold reduction in viremic levels and a drastic reduction in T cell lymphomas in infected chickens (21).
Notwithstanding the complete conservation of the USP catalytic core residues and demonstration of ubiquitin binding and polyubiquitin chain cleavage, there has been no evidence of VP1-2 USP involvement in ubiquitination in relation to protein degradation and no identification of potential substrates. One recent report of the VP1-2 homologue of EBV (BPLF1), originating in a broad yeast 2-hybrid screen of interacting proteins, indicated that the BPLF1 USP domain expressed in isolation could interact with the EBV RNase reductase (RR) large subunit. Overexpressed RR exhibited minor amounts of a ubiquitin species, which were reduced further when coexpressed with the BPLF1 USP domain, although overall levels appeared to be unaffected (42). Moreover, although the prototype in which the USP activity was first discovered, there has been little analysis of the HSV VP1-2 USP. Here we report the first demonstration that the VP1-2 USP can act as a USP relevant to removing ubiquitin which would otherwise target for degradation. The core USP of HSV VP1-2 was expressed efficiently without a requirement for its intrinsic catalytic activity. However, N-terminal variants extending into the body of VP1-2, and encompassing other functional determinants, were critically dependent upon the USP for accumulation. Surprisingly, unstable variants containing an inactive USP were stabilized not only by proteosome inhibition but also by virus infection. An HSV recombinant containing a mutant VP1-2 with a catalytically inactive USP exhibited little defect in culture, and the VP1-2 was expressed at similar levels as the wild-type (wt) protein. Together, these results provide the first evidence in vivo for VP1-2 USP activity in relation to reversing ubiquitination of a protein which would be targeted for degradation, an important conclusion given the observation that the herpesvirus USPs comprise a unique, evolutionarily distinct class (12, 22, 36). We also discuss a framework for understanding the pronounced conservation of an activity which can be inactivated with minimal consequences in experimental systems. It is possible that the involvement of the USP is obviated by additional mechanisms operating in virus-infected cells and that it remains important (and conserved) at select phases where these mechanisms may be uncoupled.
MATERIALS AND METHODS
Cells and viruses.
Vero HS30 cells, a derivative of Vero cells containing the UL36 gene (11), and COS 7 cells were grown in Dulbecco's modified minimal essential medium (DMEM, from Gibco) containing 10% newborn calf serum (NCS) and penicillin-streptomycin. RSC (rabbit skin cell) and RSC-HAUL36 cells were grown in DMEM containing 10% fetal calf serum (FCS) supplemented with nonessential amino acids. HeLa Tet-on Advanced and HEK 293 Tet-on Advanced cells (Clontech) were grown in DMEM with 10% Tet system approved fetal bovine serum, penicillin-streptomycin, and Geneticin at a concentration of 100 μg/ml. In an experiment examining expression levels under conditions of proteosomal inhibition, MG132 was added to a final concentration of 20 μM 4 h before final harvesting. For virus infection at a high multiplicity of infection (MOI), cells were plated in 12-well cluster dishes at 3 × 105 cells/well, infected with appropriate viruses (see below) at different multiplicities in serum-free medium, and incubated for 1 to 2 h, after which time the inoculum was removed, any extracellular virus was inactivated with a low-pH wash (40 mM citric acid, 135 mM NaCl, 10 mM KCl, pH 3.0), and cultures were incubated further in medium containing 2% FCS for plaque formation assays (3 to 4 days). Plaque assays were routinely performed with the addition of 1% pooled human serum to the medium. For growth curves and protein analysis, infections of duplicate sets of monolayers were performed 12 h apart to facilitate collection over a 2- to 24-h time frame. HSV-1 KΔUL36 (11), a deletion mutant lacking intact UL36, was propagated by standard methods on HS30 cells. HSV-1.ΔARUL36 (34), an independent mutant lacking the complete gene for UL36, was propagated on RSC-HAUL36 cells.
Generation of doxycycline (Dox)-inducible cell lines.
Cell lines for the inducible expression of VP1-2 N-terminal variants were generated using HEK 293 Tet-on Advanced cells. The cells were cotransfected with pTRE Tight vectors containing the VP1-2 variants (see below) together with a linear fragment containing a puromycin resistance gene at a 10:1 ratio using the GeneJammer transfection reagent (Stratagene) at a 3:1 ratio. Positive colonies were selected by treatment with puromycin, expanded, and tested for induction of the appropriate protein.
Transient transfections.
For expression studies, cells were transfected by standard methods with 1 μg plasmid DNA using the GeneJammer at a 3:1 ratio or at a 3:2 ratio for cotransfections.
Plasmid constructs of VP1-2 mutants.
To analyze the effect of deletion or mutation within the USP domain a number of derivatives were first made in the context of N-terminal variants. pcDNASV5-NT1(1–287), pcDNASV5-NT2(1–498), pcDNASV5-NT3(1–705), and pcDNASV5-NT5(486–705) have been previously described (2).
Mutant versions pcDNASV5-NT1.C65A and pcDNASV5-NT2.C65A were generated by direct mutagenesis of the corresponding vectors using the primers listed in Table 1. pcDNASV5-NT3.C65A (pMB15) was generated by insertion of the HindIII-BamHI fragment from pcDNASV5-NT2.C65A into pcDNASV5-NT3. pcDNASV5-NT6(1-1875) (pMB20) was generated by insertion of a 1.4-kb SnaBI and BamHI fragment of pcDNASV5-NT3 into the 8.1-kb fragment of ΔCT1 (2). The mutant version, pcDNASV5-NT6.C65A (pMB21), was then generated by ligation of the 1.2-kb BamHI-HindIII fragment of pcDNASV5-NT2.C65A with the 8.5-kb BamHI-HindIII fragment of pMB20. pcDNASV5-NT3.C65A.K666R (pMB22) was generated by mutagenesis using pcDNASV5-NT3.C65A (pMB15) as a template with appropriate primers (Table 1). First, we generated PCR product A, using primer pairs F1 and 1R, and PCR product B, using primer pairs 2F and R1. To generate the final PCR product incorporating the mutation, products A and B were used as templates and amplified using primer pairs F1 and R1. This final PCR product was purified and ligated into the backbone of pcDNASV5-NT3.C65A by using HindIII and EcoRI.
Table 1.
Primers used in this study
| Primer(s) | Sequencea |
|---|---|
| NT1.C65A and | |
| NT2.C65A | 5′-GGG TCG GTA TCG GCG ATG CGC TCG TCG CTG TCC-3′ |
| 3′-GGA CAG CGA CGA GCG CAT CGC CGA TAC CGA CCC-5′ | |
| pMB22 | F1: 5′-GGG GGG AAG CTT ATG GGA AAG CCG ATC CCA AAC-3′ |
| R1: 5′-GGG GGG GAA TTC TTA GCC GAG GCG TGT ATA CAG-3′ | |
| 2F: 5′-TCG CCT GGC GCT GGC GAG GCT GAT CCT TGT GGC TAG GGA TGT CAT TC-3′ | |
| 1R: 5′-TAG CCA CAA GGA TCA GCC TCG CCA G-3′ |
F, forward; R, reverse.
To incorporate the single substitution of the cysteine of the USP catalytic triad (C65) in the context of full-length VP1-2, the 1-kb HindIII and RsrII fragment from pcDNASV5-NT2.C65A fragment was ligated with the similarly digested 13.3-kb fragment from pcDNA3SV5-UL36fl to create pcDNASV5-UL36.C65A (pMB3).
To delete the nuclear localization signal (NLS) in the context of the C65A mutant, the double mutant, pcDNASV5-UL36C65A.ΔNLS (pMB4), was constructed by digesting both pcDNA3SV5-UL36ΔNLS (2) and pcDNASV5-NT2.C65A with HindIII and RsrII. The 13.3-kb pcDNA3SV5-UL36ΔNLS was then ligated with the 1-kb pcDNASV5-NT2.C65A fragment.
To generate doxycycline-inducible expression vectors for VP1-2, the following constructs were generated. Construct pTightSV5-NT1 (pMB8) was constructed by ligating the 2.6-kb HindIII-EcoRV fragment from pTRE-Tight (Clontech) with the 1-kb fragment of pcDNASV5-NT1. Construct pTightSV5-NT1.C65A (pMB6) was generated similarly from pcDNASV5-NT1.C65A. Additional constructs pTightSV5-NT2 (pMB9), pTightSV5-NT2.C65A (pMB10), and pTightSV5-NT3 (pMB11) were generated similarly by insertion of the appropriate HindIII-EcoRV fragments into the 2.6-kb fragment of pTRE-Tight. pTightSV5-UL36fl and pTightSV5-UL36.C65A were generated by insertion of the HindIII-HpaI fragment from the corresponding constructs into similarly digested pTRE-Tight. All the VP1-2-expressing constructs were detected by virtue of the N-terminal epitope V5, by using anti-V5 monoclonal antibody.
Complementation assay of VP1-2-defective virus.
Cos cells were seeded in 12-well plates and transfected, in four replicates, with 1 μg of the following plasmids: pcDNASV5-UL36fl(pTD3), pcDNASV5-UL36.C65A(pMB3), pcDNASV5-UL36ΔNLS (pTD3ΔNLS), pcDNASV5-UL36.C65AΔNLS (pMB4), and pEGFP. Transfected cells were infected 24 h later with HSV-1 KΔUL36 (11) at an MOI of 5. After 1 h, unabsorbed virus was briefly inactivated by an acid wash (40 mM citric acid, 135 mM NaCl, 10 mM KCl, pH 3.0), and the infected monolayers were then incubated in DMEM–2% newborn calf serum (NBCS). Sixteen hours postinfection, one replicate was harvested in SDS buffer for examination of expression levels by Western blotting. The infected cultures from the other three replicates and medium were harvested and lysed by freeze-thawing, and total virus yields were determined in triplicate by plaque assay on HS30 cells.
Construction of viruses containing VP1-2 or VP1-2.C65A.
A mutant virus containing VP1-2 C65A was constructed in the background of HSV-1 strain KOS previously cloned in a bacterial artificial chromosome (15) The bacmid was grown in Escherichia coli strain GS1783. The strategy involved recombinatorial insertion of a cassette for kanamycin resistance adjacent to the mutated sequence and selection for kanamycin resistance, followed by recombination excision and counter selection. PCR was carried out with the following primers by using pEPKan-S (38) as a template: COL416 (forward primer for UL36 C65A mutation in KOS bacterial artificial chromosome (BAC) [CCAGTTCGCGCCCGACCTGGAGCCAGGGGGGTCGGTATCGGCCATGCGCTCGTCGCTGTCCTAGGATGACGACGATAAGTAGGG]) and COL417 (reverse primer for UL36 C65A mutation in KOS BAC [ATCAAATATGAGGCTGAGAAAGGACAGCGACGAGCGCATGGCCGATACCGACCCCCCTGGCTCAACCAATTAACCAATTCTGATTAG]). Template DNA was digested by incubating the PCR mix with 20 U DpnI for 1 h at 37°C, and the PCR product was purified using a QIAQuick gel extraction kit (Qiagen) after separation on a 0.8% agarose gel.
The PCR product (100 ng) was electroporated into bacteria containing the KOS bacmid (BAC) and selected for resistance to chloramphenicol and kanamycin. Resistant colonies were screened by restriction digest. Kanamycin- and chloramphenicol-resistant colonies were grown, 2% (wt/vol) arabinose was added to induce the expression of I-SceI, and the culture was heat shocked at 42°C for 30 min. Cultures were then grown at 30°C and dilutions plated on agar plates containing chloramphenicol and 1% (wt/vol) arabinose. Candidate colonies that had lost the kanamycin resistance gene were identified by replica plating. BAC DNA from kanamycin-sensitive colonies was isolated and examined by restriction digestion. BAC DNA (100 μg) from the appropriate colonies, together with 1 μg pGS403 encoding Cre recombinase, was then transfected into Vero cells using Fugene 6 (Roche) in six-well dishes to reconstitute HSV-1 and excise the BAC backbone from the viral genome. Cell monolayers were monitored for plaque formation. After 3 days of incubation at 37°C, cells were harvested and sonicated and samples were stored at −70°C. The efficiency of BAC backbone excision and the virus titers were determined by performing a plaque assay on Vero cells followed by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining. Virus stocks were generated in Vero cells (0.1 to 0.01 PFU/cell) and harvested after 2 to 3 days. The C65A substitution or wild-type C65 was confirmed by sequencing the appropriate region from purified DNA stocks of mutant or parental virus isolates.
Expression and purification of N-terminal USP.
The plasmid encoding the inducible GST-NT1 fusion protein (pGEX-6P-NT1) was used as described previously (2). Briefly, bacterial cultures induced for the expression of GST-NT1 were lysed and clarified, and supernatants were incubated with glutathione-Sepharose 4B beads. Binding and purification were monitored analytically by SDS-PAGE and total protein staining with Coomassie brilliant blue. GST-NT1 purified protein was cleaved from the beads using PreScission protease (Amersham), eluted, and concentrated in Centricon tubes (cutoff, 10 kDa). The resulting sample was processed by fast protein liquid chromatography (FPLC) anion-exchange chromatography on a Mono-Q column equilibrated in the following buffer: 1 mM EDTA (pH 7.5), 20 mM Tris-HCl (pH 7.0), 50 mM NaCl, 1 mM dithiothreitol (DTT). NT1 was then purified with a salt gradient in the same buffer, eluting at approximately 400 mM NaCl. The peak fractions of NT1 (approximately 95% pure) were pooled, desalted in phosphate-buffered saline (PBS), and used for in vitro assays.
In vitro assays of VP1-2 USP activity.
Isopeptidase assays of ubiquitin peptide conjugates were performed as previously described (22). Briefly, the ubiquitin peptide consisting of K48- or K63-linked polyubiquitin branched chains (final concentration, 250 nM) was added to 6 μl of reaction buffer (50 mM Tris [pH 7.4], and 2 mM DTT) and made to a final volume of 11 μl with 50 mM Tris, pH 7.4. Reactions containing 5 to 10 ng of purified protein NT1(VP1-2 USP), VP22 expressing residues from 159 to 301 [VP22(159-301)], or USP5 were incubated at 37°C for 8 h. All reactions were terminated by the addition of reducing sample buffer and separated by 12.5% SDS-PAGE. Proteins were observed by silver staining (Pierce). USP5 and K48- and K63-linked polyubiquitin were obtained from Axxora Ltd. (United Kingdom).
SDS-PAGE and Western blotting.
Transfected or infected cell monolayers were harvested in standard SDS sample buffer. Samples were lysed either using a sonicator bath or by needle shearing using a 25-G needle and boiled for 2 min prior to electrophoresis. Equal cell samples were analyzed by separation on 3 to 8% gradient Tris-acetate gels or 10% Tris-glycine gels and transferred onto Immobilon-P (Millipore) membranes. Primary and secondary antibodies for immunodetection were diluted in PBST (PBS plus 0.1% Tween 20 containing 5% dried milk). Target proteins were visualized by enhanced chemiluminescence (ECL; Pierce). Primary antibodies were used as follows: anti-ICP4 (1/1,000 [Virusys Corp.]); anti-VP5 (1/3,000 [East Coast Bio]), anti-V5 (1/10,000 [Invitrogen]), anti-UL37 (1/10,000 [a kind gift of P. Desai]), anti-UL25 (1/1,000 [a kind gift of V. Preston]), LP1, anti-VP16 (1/4,000 [a kind gift of T. Minson]), anti-VP1-2 (1/2,000 [anti-NT1]), used for detection of VP1-2 during virus infection, FK2 (1/2,000 [Biomol]). Secondary antibodies were anti-mouse (1/2,000 [Bio-Rad]) or anti-rabbit (1/3,000 [Bio-Rad]).
RESULTS
N-terminal variants incorporating an active USP domain.
The observation that VP1-2 encodes a ubiquitin-specific protease was first made by virtue of the incorporation of a ubiquitin suicide substrate into a novel peptide in HSV-infected cells (22). The peptide corresponded to an N-terminal product of VP1-2 of approximately 47 kDa, although incorporation of the ubiquitin substrate into full-length VP1-2 was not observed. A bacterially expressed purified N-terminal polypeptide comprising residues 1 to 533 was shown to have activity on the ubiquitin substrate with a strict requirement for C65 for activity. The USP was also reported to cleave K48-linked polyubiquitin chains but not K63-linked chains (22). A subsequent study examining the activity of the same peptide in vitro observed cleavage with both K48- and K63-linked polyubiquitin (23). Based on sequence alignments with VP1-2 homologues, it was speculated that the minimal region necessary for catalysis would be contained in the first 280 residues of HSV VP1-2 and corresponding peptides from the VP1-2 proteins of EBV and MCMV were shown to have activity in vitro on a ubiquitin suicide substrate (35).
Considering results of this study (see below), it is first pertinent to consider alignment analysis of VP1-2. Our comparisons within the alphaherpesviruses and across the family herpesviridae (see Fig. S1 in the supplemental material) indicate an overall organization whereby the N-terminal USP is linked via a poorly conserved region, of varying lengths, to the central body of the protein (see Fig. S1) (Fig. 1). Within this N-terminal linker region there is a notable exception to the general lack of conservation, i.e., the presence of a highly basic region which is well conserved in all VP1-2 species. In HSV VP1-2, we have shown that this basic region (pink shading) encompasses a functional nuclear localization signal (2). The central region of VP1-2 (gray shading) is reasonably well conserved and fluctuates in the degree of conservation, though distinct N- and C-terminal boundaries of this region are clearly discernible. The total lengths of this central conserved region are similar between the family members, even though, e.g., the total length of HSV VP1-2 is almost 900 residues longer than the HCMV UL48 homologue. This difference in the overall lengths of VP1-2 homologues is partly due to the difference in the linker region between the USP domain and the N-terminal boundary of the central region but mainly due to the differences in length of a poorly conserved region between the C-terminal boundary of the central region and a very highly conserved region at the extreme C terminus of the protein itself (blue shading). In HSV, the C-terminal linker region is approximately 700 residues in length and rich in prolines and glutamines, while in HCMV this linker region is fewer than 100 residues.
Fig. 1.
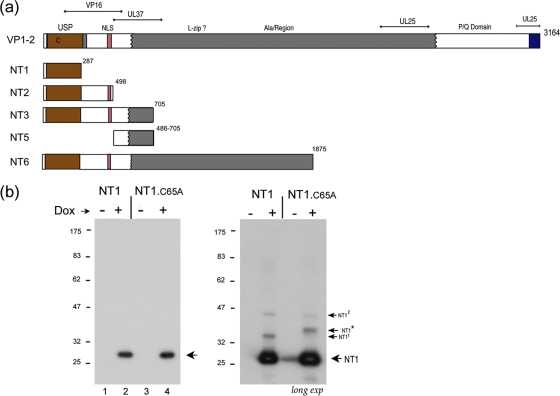
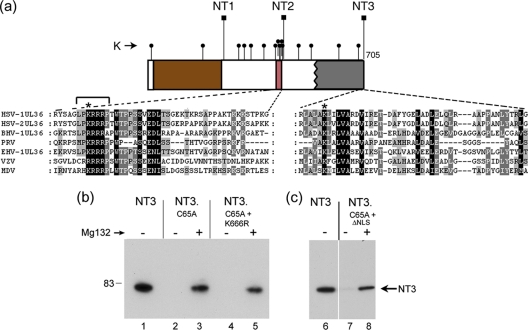
Doxycycline-inducible expression of the VP1-2 USP domain. (a) Summary of various N-terminal constructs of VP1-2, NT1, NT2, NT3, NT5, and NT6 in relation to general features of VP1-2 organization. Numbering refers to the final residue in each construct. The N-terminal core USP is indicated by brown shading, the NLS in pink shading, the central core of VP1-2 in gray, and the C-terminal highly conserved region in blue. Various alignments within each of the alpha, beta, and gamma classes indicate this overall organization, as summarized in Fig. S1 in the supplemental material. Previously characterized regions involved in UL37, VP16, and UL25 binding are indicated (7, 14, 24, 26, 30, 31, 40). (b) Induction of NT1 and NT1.C65A after transfection in 293-Tet On cells. Cells were transfected with the appropriate vectors, Dox was added 24 h later, and cells were harvested 48 h after Dox addition. The primary NT1 species are indicated by an arrow. The panel in the right hand side shows a longer exposure revealing additional lower-abundance species and the presence of NT1* in NT1.C65A, a species never observed for NT1.
We initially examined an N-terminal product NT1 (residues 1 to 287) that encompasses the conserved core USP domain. We expressed and purified NT1 in bacteria as previously described (2) and tested its cleavage activity in vitro on K48- or K63-linked polyubiquitin chains (see Fig. S2 in the supplemental material). The results demonstrated cleavage of both substrates, with additional dose-response experiments indicating a modest preference for K48-linked chains. Further work in this report concerns analysis of activity in vivo.
We generated constructs for expression and analysis in mammalian cells of a series of N-terminal variants of VP1-2 (as summarized in Fig. 1a), for the USP domain only (NT1), or progressively extending to include the nuclear localization sequence [NT2(1-498)], to further encompass determinants previously identified for binding UL37 (14, 24) and VP16 (30, 40) [NT3(1-705)], to extend into the central region encoding almost 60% of the protein [NT6(1-1875)], or the full-length protein. We first made constructs for the inducible expression of NT1 and a variant of NT1 containing a mutation in the conserved catalytic cysteine previously shown to be required for HSV USP activity (22). In transiently transfected cells, expression of NT1 and NT1.C65A was tightly regulated by the addition of doxycycline (Dox), resulting in proteins of the expected sizes and synthesized in equivalent amounts (Fig. 1b, lanes 1 to 4, arrow). Upon longer exposure of the gels (Fig. 1b, long exp), we observed several low-abundance species migrating more slowly (labeled NT11 and NT12) but in addition a species uniquely observed for NT1.C65A (labeled NT1*). The origins of NT11 and NT12 are unclear but could be due to several factors, including SDS resistance multimers or posttranslational modification. However, NT1* was never observed for NT1 expression even after considerable overexposure of the gels (see Fig. 5). The singular presence of NT1* after NT1.C65A expression indicated to us that it may be related to ubiquitination of NT1 itself. An explanation consistent with these observations was that NT1 was subject to ubiquitination but could deubiquitinate itself, while the mutant, or at least a population of it, retained a ubiquitin conjugate.
Fig. 5.
Active USP is required for stabilization in trans. (a) Cells were transfected as for Fig. 4b with constructs for constitutive expression of NT3 (lanes 1, 2, and 5, 6) or NT3.C65A (lanes 3, 4 and 7, 8), together with vectors for the Dox-inducible expression of NT1 or NT1.C65A as indicated. Twenty-four hours after transfection, Dox was added to one set of duplicate transfections and cells were incubated for a further 24 h. Samples were then probed simultaneously for expression of the various species. Dox induction of NT1 resulted in expression of NT3.C65A (lanes 3 and 4), while no increase was observed after induction of NT1.C65A (cf. lanes 7 and 8). (b) Exactly as for panel a, but comparing the effect of NT2 versus NT2.C65A on NT3 and NT3.C65A levels. (c) Cellular protein subject to proteosome-mediated turnover shows little increase upon induction of NT1. Cells were transfected as for panel a, with vectors for expression of NT3.C65A or CREBHΔTMC and processed as described. Despite significant increase in NT3.C65A (lanes 1 and 2), little effect was observed for CREBHΔTMC (lanes 3 and 4).
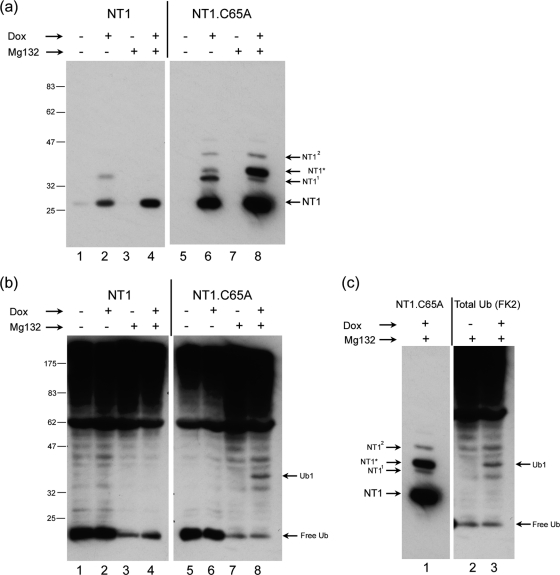
To examine this further, we established inducible cell lines for NT1 and NT1.C65A and examined expression in the absence and presence of the proteosome inhibitor MG132 (Fig. 2a). In the cell lines, in contrast to transient transfections, NT1.C65A was induced to levels higher than those for NT1. We do not know the reason for this, and even though the promoter driving expression enabled tight Dox regulation it is possible that background expression in the uninduced state selected for lines with overall lower levels of NT1. (This was observed in several independent clones [data not shown]). Nevertheless, it was clear that the NT1* species again was observed only for NT1.C65A (Fig. 2a, lane 6) and very long overexposures failed to reveal any NT1* in the wt NT1 (Fig. 2a, lane 2; also data not shown). Moreover, upon proteosome inhibition, NT1* abundance increased markedly for NT1.C65A (Fig. 2a, lane 8), while again this species was not observed for NT1 (Fig. 2a, lane 4). In parallel, we probed the same samples with an antibody (FK2) against endogenous ubiquitin. We did not observe any major changes in the profile on ubiquitinated species upon Dox induction of NT1, though proteosome inhibition reduced the level of free ubiquitin. However, we did observe a novel ubiquitin band, specifically induced when NT1.C65A was induced by Dox addition (Fig. 2b, lane 8, Ub1). Moreover, when the blots for detection of NT1.C65A and for ubiquitin were aligned, it was evident that NT1* comigrated with the induced ubiquitin band (Fig. 2c). These data provide strong support for the conclusions first that NT1* represents a ubiquitinated form of NT1.C65A and second that the parental NT1 is able to remove ubiquitin from itself. This, notwithstanding the mutation of C65A, did not result in lower levels of NT1.C65A, particularly in direct comparison after transient expression.
Fig. 2.
The USP domain can deubiquitinate itself. (a) Cell lines containing inducible NT1 (lanes 1 to 4) and NT1.C65A (lanes 5 to 8) were established and accumulation measured after Dox induction with or without the addition of MG132 (added for 4 h before harvesting) as indicated. The species NT11 and NT12 were readily observed for NT1.C65A and could be seen for NT1 (Fig. 1). NT1* was specific for NT1.C65A and its accumulation was significantly increased by MG132 addition. (b) The same samples as in panel a were probed for the presence of total ubiquitinated species using the anti-ubiquitin antibody FK2. We did not observe major changes in the total ubiquitinated species upon NT1 (or NT1.C65A) induction. However, MG132 treatment resulted in the loss of free ubiquitin and a general increase in higher-molecular-weight species. A novel ubiquitinated species appeared upon Dox induction of NT1.C65A. This species labeled Ub1 (lane 8) was not observed for NT1 induction. (c) Alignment of the same blots as in panels a and b. The Dox-induced NT1.C65A band NT1* (lane 1) comigrates with the novel Dox-induced ubiquitinylated band, Ub1 (lane 3).
An N-terminal variant whose abundance is dependent upon functional USP activity.
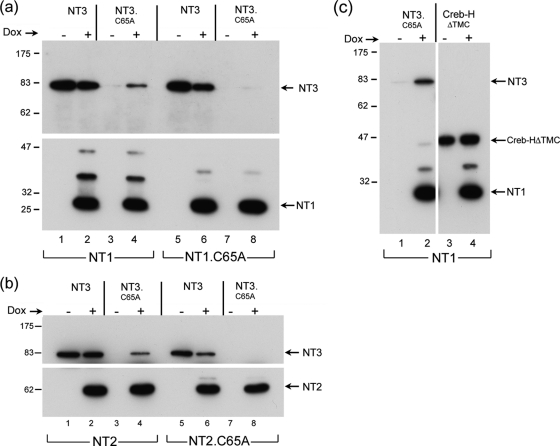
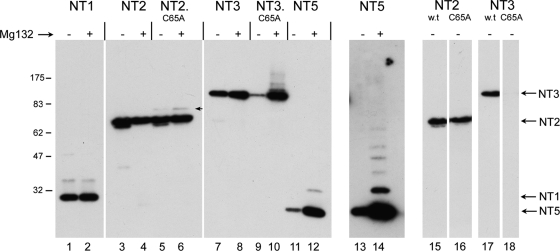
We next examined a series of N-terminal variants (Fig. 1a) which included progressively more of the protein, in particular extending beyond the limits of the core USP to incorporate the NLS (NT2) and then beyond the N-terminal boundary of the central conserved region to incorporate determinants known to be involved in UL37 binding (NT3). In this case, expression was examined in the absence and presence of proteosome inhibition after transient expression (Fig. 3). For NT2, we again noticed a novel band for NT2.C65A, migrating slightly above the main NT2 band (Fig. 3, lane 6). This NT2.C65A-specific band was modestly increased in the presence of MG132 though as for NT1, no higher-molecular-weight species were observed, and the abundances of NT2 and NT2.C65A were similar. In contrast, for NT3, we noticed a marked effect of the C65A substitution. In this case, levels of NT3.C65A were drastically reduced compared to those for NT3 (Fig. 3, cf. lanes 7 and 9; also Fig. 4 to 6). Indeed, in a comparison of shorter exposures of the blot of NT2 and NT3 and the corresponding C65A mutation, the marked difference in the effects of the mutation can be readily seen (Fig. 3, cf. NT2, lanes 15 and 16, to NT3, lanes 17 and 18). In effect, NT3 depended upon the presence of C65 for normal levels of synthesis. Moreover, in contrast to NT1.C65A or NT2.C65A, where proteosome inhibition had at most a relatively modest effect, for NT3.C65A proteosome inhibition resulted in a large increase in levels of the protein and the appearance of a smear of higher-molecular-weight species (Fig. 3, lane 10). We also examined a further extension, NT6, which encompassed residues 1 to 1875. As for NT3, NT6 was expressed and detected as a band with the appropriate size, while NT6.C65A was virtually undetectable (Fig. 4a, lanes 4 and 5). Again, proteosome inhibition restored NT6.C65A levels to almost the same as those of the parental NT6 (Fig. 4a, lanes 5 and 6).
Fig. 3.
Dependence of N-terminal variants on USP activity for inhibition of ubiquitination and degradation. N-terminal variants of VP1-2 containing wt C65 or the C65A substitution as indicated (Fig. 1) were transfected into Cos cells and treated without or with addition of MG132 (added 4 h prior to harvesting). Mutation in the active site of the USP domain in NT2 resulted in a novel low-abundance species (arrowed lane 6) but had very little overall effect on NT2 levels (cf. lanes 3 and 5). Proteosome inhibition had little effect on total levels of either NT2 or NT2.C65A (lanes 3 to 6). In contrast, mutation in the USP active site resulted in a profound reduction in NT3 levels (cf. lanes 7 and 9 to shorter-exposure lanes 17 and 18). Moreover, proteosome inhibition restored levels of NT3.C65A and induced a series of poorly resolved higher-molecular-weight species (cf. lanes 9 and 10). NT5 was also poorly expressed with proteosome inhibition inducing a significant increase in levels and higher-molecular-weight species (cf. lanes 11 and 12) and longer exposure (lanes 13 and 14).
Fig. 4.
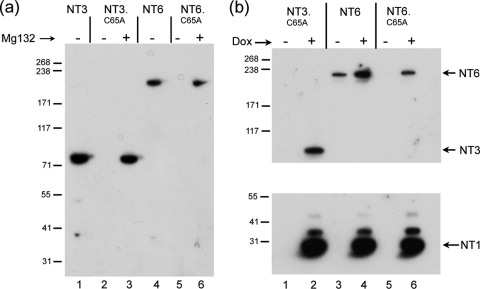
The USP domain expressed in trans stabilizes VP1-2 C65A mutants. (a) Variant encoding approximately 60% of the protein NT6 was expressed in comparison to NT3, each containing C65 or C65A as indicated. As for NT3, substitution of C65A in NT6 resulted in a profound decrease in levels of the protein (cf. lanes 4 and 5) which could be stabilized by proteosome inhibition (cf. lanes 5 and 6). (b) Cells were transfected with constructs for constitutive expression of NT3.C65A, NT6, or NT6.C65A as indicated, together with the vector for the Dox-inducible expression of NT1. Twenty-four hours after transfection, Dox was added to one set of duplicate transfections and cells were incubated for a further 24 h. Samples were then probed simultaneously for expression of the various species. Dox induction resulted in expression of NT1 and a concomitant increase in NT3.C65A and NT6.C65A. An increase was seen also for NT6 itself.
The difference in NT2 and NT3 is in the extension of an approximately 200-amino-acid segment from residue 498 to 703. Interestingly, this region itself was expressed relatively poorly but stabilized by proteosome inhibition with the appearance of a ladder of higher-molecular-weight species (Fig. 3, lanes 11 and 12, and longer-exposure lanes 13 and 14).
We next wished to examine whether the USP domain could act in trans to increase the steady-state levels of C65 mutant variants of itself. We transfected vectors for the constitutive expression of NT3.C65A or NT6.C65A together with Dox-regulatable vectors for expression of parental NT1 (Fig. 4b). Levels of the mutant variants were then examined with or without NT1 induction. NT3.C65A and NT6.C65A were again virtually undetectable (Fig. 4b, lanes 1 and 5). Upon the induction of NT1 by Dox addition (Fig. 4b, lower panel), levels of NT3.C65A and NT6.C65A were increased significantly (Fig. 4b, corresponding upper panel of the same samples, lanes 2 and 6).
These results indicated that the core USP domain NT1 was able to act in trans on the N-terminal extended, unstable C65A mutant versions. To confirm this, we repeated these experiments comparing the effect of NT1 or the NT1.C65A (Fig. 5a). Again, levels of NT3.C65A were increased upon induction of NT1 (Fig. 5a, lanes 3 and 4). Interestingly, NT3 itself was not increased, and if anything modestly lower levels were observed upon NT1 induction (lanes 1 and 2). However, in parallel assays, NT3.C65A levels were not increased after induction of NT1.C65A (lanes 7 and 8). Similar results were obtained in assays for trans stabilization of NT3.C65A when using NT2 or NT2.C65A (Fig. 5b). Induction of NT2 resulted in increased levels of NT3.C65A (Fig. 5b, lanes 3 and 4), while induction of NT2.C65A had no significant effect (Fig. 5b, lanes 7 and 8).
Thus, levels of the unstable NT3.C65A could be increased by proteosome inhibition or by coexpression of the functional USP in NT1 or NT2. Although we found that levels of parental NT3 itself were not significantly affected by coexpression of NT1, we wished to examine whether any relatively unstable protein would be increased by NT1 in this coexpression system. To this end, we coexpressed NT1 with a test cellular protein, CREBHΔTMC, which we have shown to be a short-lived protein subject to ubiquitination and rapid degradation (3). In parallel, we examined the effect of expression of NT1 on levels of NT3.C65A or CREBHΔTMC. As before, while levels of NT3.C65A were significantly increased (Fig. 5c, lanes 1 and 2), levels of CREBHΔTMC were largely unaffected (Fig. 5c, lanes 3 and 4). While this does not demonstrate specificity, the increased levels of NT3.C65A or NT6.C65A do not appear to be due to generalized action of the NT1 USP. Taken together, these results provide strong support for the following proposals: first, that the N-terminal USP domain is functional in vivo as a domain that can deubiquitinate itself; second, that within the region from 498 to 703 reside determinants which destabilize the protein leading to ubiquitination and proteosomal degradation; third, that this can be reversed by the N-terminal USP itself; and finally that while this action may be in cis, the USP can function at least in part in trans to deubiquitinate itself, leading to decreased proteosomal degradation and increased protein levels.
The C65A mutation had little effect on overall levels of NT1 or NT2 but a major effect in the context of NT3. This indicated to us that determinants within the region between the boundaries of NT2 and NT3 may be the target of ubiquitination, which without deubiquitination would destabilize the protein. Interestingly, while there are numerous lysine residues between the boundaries of NT1 and NT2 (Fig. 6a), as indicated these had no effect on the requirement for C65. However, there are 4 additional lysines between NT2 and NT3, and one of these (K666) is both within the N-terminal boundary of the core of VP1-2 and conserved in all members of the alphaherpesvirus family. We wished to examine whether ubiquitination at this lysine might be the explanation for the instability of NT3.C65A mutants and reasoned if this were the case its mutation may have a stabilizing effect on the protein. We therefore made the substitution K666R in the context of NT3.C65A and examined expression with or without proteosomal inhibition (Fig. 6). As described above, levels of NT3.C65A were drastically reduced compared to what was seen for NT3 and virtually undetectable in the absence of proteosomal inhibition, which resulted in significant restoration (Fig. 6, lanes 2 and 3). The NT3.C65A.K666R mutant behaved identically, with expression levels remaining very low, no detectable increase over NT3.C65A and while levels were increased substantially by proteosomal inhibition, these were to the same levels as NT3.C65A. Although precise mapping of the determinants involved in destabilizing NT3.C65A is beyond the scope of the current work, one other possibility was pursued. It remained possible that while the extension from NT2 to NT3 resulted in ubiquitination leading to proteosomal degradation, the site of such ubiquitination could be elsewhere, possibly within the NT2 region itself. We have previously described a highly basic region in the N terminus of VP1-2 (2) which falls within the NT2 region (Fig. 6a, boxed red). This encompasses a nuclear localization signal and numerous lysine residues, one of which is strongly conserved. We made a short 7-residue deletion (Fig. 6a, bracketed) encompassing this conserved lysine in the context of NT3.C65A (NT3.C65A.ΔNLS) but again observed no significant increase in levels compared to NT3.C65A, with the protein requiring proteosomal inhibition for detection (Fig. 6b, lanes 6 to 8).
Fig. 6.
Single conserved lysine residues within the VP1-2 N terminus are not sufficient to mediate degradation of catalytically inactive variants. (a) Schematic of the N-terminal constructs NT1, NT2, and NT3, whose C-terminal endpoints are indicated (square lollipops). Lysine residues are indicated by small round lollipops. Sequence alignments of the N terminus of VP1-2 reveal a highly conserved lysine residue within the boundary between NT2 and NT3, indicated by an asterisk above the expanded section of sequence. Numerous lysine residues are present between the boundaries of NT1 and NT2, which also includes a demonstrated functional NLS (2). Within the NLS is located a conserved lysine, as indicated by an asterisk above the expanded sequence. (b) Substitution of these individual lysines does not result in significant stabilization of NT3.C65A. NT3, NT3.C65A, variants of NT3.C65A containing K666R, and a 7-residue deletion at the NLS (indicated by bracket above sequence alignment) were analyzed in the absence or presence of MG132 (added 8 h before harvesting). The double mutants behaved like NT3.C65A, being virtually undetectable in the absence of proteosome inhibition and stabilized by proteosome inhibition to approximately similar levels.
Stabilization of VP1-2 during virus infection.
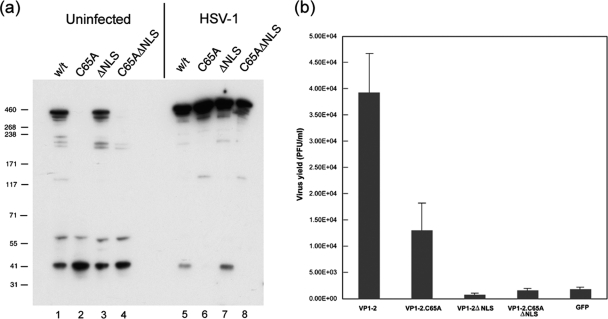
We next examined the effect of the C65 mutation in the context of full-length VP1-2 (Fig. 7a) with results which indicated that the C65A substitution had a significant effect on levels on VP1-2 and on another full-length VP1-2 variant (2) which lacks the consensus NLS motif (Fig. 7, lanes 1 to 4). We generally found that the effect of the C65A substitution on protein levels was less in the context of the full-length protein than in the N-terminal variants NT6 or NT3. We also note that, as in other studies, we observe breakdown products of VP1-2. Considering the tag for detection of VP1-2 is at the N terminus, it was noticeable that levels of these N-terminal products were less affected by the C65A mutation. While there may be other explanations, this could be consistent with the results above, since these products, migrating around 44 and 58 kDa, were smaller than NT2, whose levels were not affected by C65A. Somewhat surprisingly, however, we observed that levels of full-length VP1-2, including the C65A mutant, were increased to overall similar levels in transfected cells that were subsequently superinfected with HSV mutant K.ΔUL36 (11), which lacks intact UL36 (Fig. 7, lanes 5 to 8). We considered the possibility that the stabilization of VP1-2.C65A in cells superinfected with K.ΔUL36 could somehow be related the residual expression of an N-terminal fragment incorporating the USP domain itself. Although it is unclear whether it exhibits any activity, this fragment has been observed in K.ΔUL36-infected cells (38). To rule this possibility out, we performed experiments identical to those above, in this case superinfecting the transfected cells with HSV-1.ARΔUl36, a virus with a complete deletion of UL36. We observed the same overall increase in both VP1-2 and VP1-2 C65A (see Fig. S3 in the supplemental material). These experiments, performed as a control for expression levels in virus complementation assays (see below), indicates that processes that ultimately destabilize variants with an inactive USP are somehow counteracted in the context of virus-infected cells. There are several possibilities to explain this effect (see Discussion), including protein interactions in infected cells, the folding of VP1-2 itself, alterations in translation, or proteosome activity. While the precise explanation is beyond the scope of the current investigation, these observations influence the interpretation of attempts described in the next section to examine the effect of inactivation of the USP in the context of virus infection.
Fig. 7.
Relative instability of full-length VP1-2.C65A is rescued by replication-defective virus. (a and b) Cells were transiently transfected with expression vectors for full-length VP1-2 or mutant versions as indicated. Duplicate sets were either mock infected or superinfected with the VP1-2-negative deletion mutant, KΔUL36 (MOI 5), and further incubated for 16 h, after which time cells were harvested and processed for expression levels of VP1-2 (a) or recovery of virus growth by plaque titration in HS30 cells (b). Virus infection resulted in a significant increase in all species of VP1-2 but in particular altered the ratio of the C65A mutants, whose levels were now overall similar to the wt VP1-2, unlike what was seen for uninfected cells. As described previously, VP1-2 substantially complemented growth of KΔUL36. Background levels seen with the control vector expressing green fluorescent protein (GFP) are due to the low level of revertant virus obtained in KΔUL36 preparation (11). VP1-2.ΔNLS showed no complementation above background level. However, VP1-2.C65A exhibited significant levels of complementation above background and only 2- to 3-fold lower than that for wt VP1-2.
An active USP domain is not required for complementation of a VP1-2-defective virus.
We previously demonstrated the ability of exogenously expressed VP1-2 to complement the growth of the UL36 deletion mutant KΔUL36 and used this assay system to identify a critical basic motif required for VP1-2 function (2). To examine whether substitution of C65A in full-length VP1-2 affected function in virus replication, we first examined the effect on the ability to complement KΔUL36. Cells were transfected with various vectors: VP1-2, VP1-2.C65A, VP1-2.ΔNLS (2), VP1-2.C65A.ΔNLS, or a vector expressing GFP as a control. The cells were then superinfected with KΔUL36 and harvested 16 h later, and yields of virus were measured by titration in the complementing cell line HS30 (11). As shown previously, VP1-2.ΔNLS was completely debilitated in its ability to complement with yields at approximately the same levels as the control GFP vector (Fig. 7b). VP1-2.C65A.ΔNLS was as anticipated similarly defective. In contrast, expression of VP1-2.C65A resulted in significant levels of complementation, with absolute levels only 2- to 3-fold lower than wt VP1-2. Given the potential instability of the C65A mutant, we had anticipated a much lower level of complementation whether due to loss of function of the USP activity or overall levels of the protein. As indicated above, for these experiments we examined levels of the different proteins in the transfected cells, with and without superinfection (Fig. 7a). In infected cells, the comparable levels of the VP1-2.C65A presumably explains the reasonable levels of complementation and indicates that the activity of the USP domain of VP1-2 plays a minor role in this context.
Construction of a recombinant HSV variant expression VP1-2.C65A.
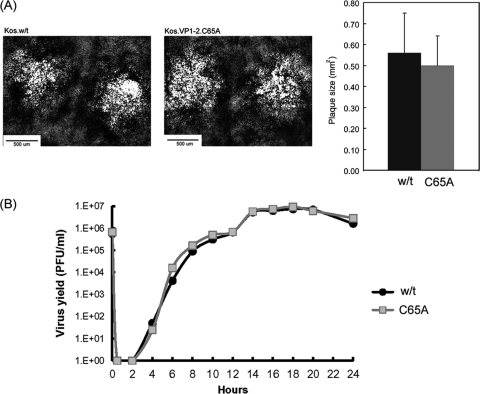
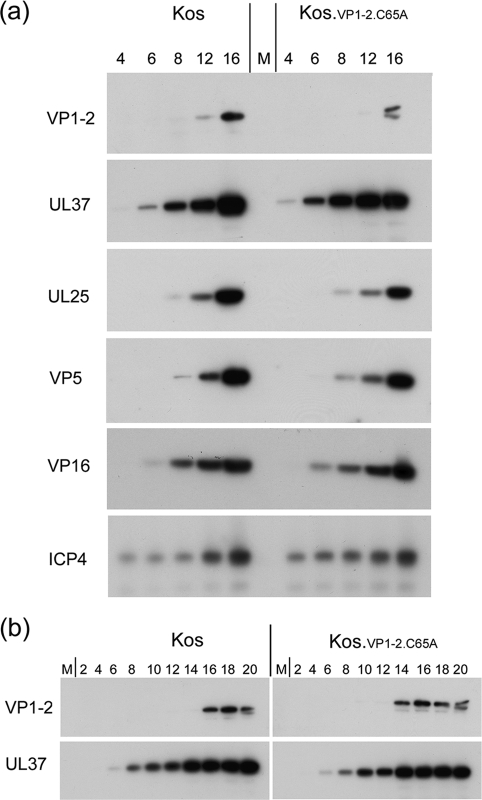
To examine the effect of the C65A mutation more definitively, we constructed viruses based on an HSV KOS bacmid expressing wt VP1-2 or the C65A mutation as described in Materials and Methods. Two strains termed KOS.VP1-2.wt and KOS.VP1-2.C65A were isolated and grown on normal noncomplementing cell lines. The mutant virus grew and formed plaques with no discernible difference in morphology or size (Fig. 8A). We further examined replication kinetics of the wt and C65A mutant in single-step growth assays with results which indicated no significant difference in the kinetics of virus production or the absolute virus yields (Fig. 8B). Finally, we examined the kinetics and levels of expression of VP1-2 and a number of candidate proteins which have been reported to interact with VP1-2 or are representative of the progression of infection. Neither the levels nor the kinetics of accumulation of VP1-2 were significantly altered after infection with KOS.VP1-2.C65A compared to the wt (Fig. 9a and b). Neither could we detect a significant difference in these parameters for the additional candidate proteins, UL37, UL25, VP16, VP5, or the immediate-early protein ICP4. With extended analysis every 2 h for 20 h, comparison of the kinetics of accumulation of VP1-2 and interacting partner UL37 is shown in Fig. 9b, again with no significant difference, although we note the relatively late accumulation of VP1-2 compared to UL37. This was not a quantitative difference, and while not the subject of the current investigation, it may reflect a qualitative difference in regulation of accumulation of the two proteins, with VP1-2 behaving more as a true late protein.
Fig. 8.
Growth characteristics of a recombinant virus expressing VP1-2.C65A. (A) KOS.wt and KOS.VP1-2.C65A viruses were constructed as described in Materials and Methods and grown in RSC cells. Typical examples of plaque formation after 72 h are shown. No significant difference in average plaque size was observed counting approximately 50 plaques from each strain (right panel). (B) RSC cells were infected with KOS.wt or KOS.VP1-2.C65A at an MOI of 5 and incubated for 2 h, after which time inoculum virus was inactivated by a low-pH wash and the cells were incubated and harvested every 2 h at the times indicated. Cultures were harvested by pooling medium and cell-associated virus, and the pooled lysate was subjected to 3 freeze-thaw cycles. Debris was then pelleted, and production of infectious virus was titrated in RSC cells.
Fig. 9.
Accumulation of virus proteins after infection with KOS.wt and KOS.VP1-2.C65A. (a) RSC cells were infected as in Fig. 8 and at the times indicated (h) lysed in SDS lysis buffer; accumulation of candidate proteins was then analyzed by gel electrophoresis and Western blotting. (b) More extended comparison showing accumulation of VP1-2 and UL37 at 2-h intervals at the times indicated.
DISCUSSION
VP1-2, encoded by the UL36 gene, is conserved across the herpesvirus family and essential in those viruses where it has been examined (4, 11, 14, 25, 26, 28). It is a very large multifunctional protein with important roles in many aspects of the virus life cycle including entry (1, 25, 34), assembly (10, 11, 14, 26), and transport (28, 33, 37, 43). VP1-2 also contains a novel ubiquitin-specific protease located at its N terminus, a domain which is present in all herpesviruses examined to date (22, 35). The core catalytic residues are completely conserved. Where examined, the isolated USP domains of several herpesviruses have been shown to exhibit specificity in ubiquitin binding, with little binding of ubiquitin-like proteins, and to be able to cleave polyubiquitin chains. Despite the striking conservation, variants in PRV, HCMV, MDV, and murid herpesvirus 68 (MHV-68) encoding catalytically inactive VP1-2 species remain viable, though growth defects are observed to different extents in tissue culture.
These observations notwithstanding, to date there have been few clues as to the role of the USP activity, and while it can have a dramatic effect in certain circumstances (21, 27), no rationale for the high conservation of the USP across viruses with diverse pathways of infection and pathogenesis. Furthermore, apart from cleavage of polyubiquitin chains in vitro, there has been little evidence that the USP can deubiquitinate a substrate and influence its degradation. In this study, we set out to examine the HSV-1 VP1-2 USP activity. We found that the core USP domain NT1(1-287) could cleave both K48- and K63-linked polyubiquitin chains in vitro with additional data indicating a modest preference for K48-linked chains. A similar bacterially expressed HSV USP (residues 1 to 533) was previously demonstrated to cleave K48-linked chains, although with little cleavage of K63-linked chains, while subsequent analysis of other herpesvirus VP1-2 USP domains by the same authors, consistent with our current results, indicated cleavage of both linkages with preference for K48 (21, 35, 36). As suggested (22) it could be that the difference in results stems from use of the core domain on our part, versus the extended region from 1 to 533 in previous analysis, with the extended region altering polyubiquitin cleavage properties. Direct comparison of the purified species will be required to examine any such effect on specificity of cleavage in vitro.
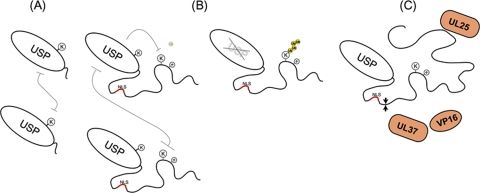
The main purpose of the work reported here was to characterize the VP1-2 USP activity in vivo and investigate potential cellular targets. In pursing these questions, we obtained unexpected results which lead us to focus on the activity of the USP on itself. A schematic summary of our observations is illustrated in Fig. 10.
Fig. 10.
Summary illustration of results, as described in the text, indicating the following. (A) The USP domain is ubiquitinated with relatively low efficiency and is able to deubiquitinate itself. (B) Determinants within the N-terminal region of NT3 confer efficient ubiquitin targeting, and protein accumulation is now dependent upon an active USP, removing ubiquitin in trans and potentially in cis. The inactive form of NT3 accumulates ubiquitin (yellow circles) and is targeted for degradation. (C) In virus-infected cells, alteration in processes or the presence of additional factors, e.g., known partners such as UL25, UL37, or VP16, antagonize ubiquitination, and as a consequence the protein is stabilized.
From both transient expression and in cell lines inducibly expressing the core VP1-2 USP (NT1), we did not observe loss of high-molecular-weight ubiquitin-conjugated species, or increases in total protein, though the resolution in our assays is limited and the USP could certainly have effects on cellular proteins. However, for the USP mutant NT1.C65A, we observed a novel species which the combined evidence indicates is due to ubiquitination of the USP domain itself. Proteosomal inhibition resulted in a further increase in this species and demonstrated the presence of at least monoubiquitinated NT1.C65A. Although we could not observe larger species of ubiquitinated NT1.C65A, with its relatively low abundance, we could not rule out formation of additional species indicative of polyubiquitination. In either case, absence of USP activity due to C65A did not have a major consequence for overall protein levels. The most straightforward interpretation of this result is that the USP domain in isolation, or with further extension, e.g., NT2(1-498), is relatively stable, subject to at least some degree of ubiquitination within the domain, and readily deubiquitinated by its own activity (Fig. 10A). It is possible that ubiquitination of the USP influences activity in some manner. Certain USPs have been reported to be themselves subject to mono- or polyubiquitination, and in some cases this is reversed by auto-deubiquitination, with this balance regulating enzyme activity (13, 29, 39). It could be that modification of the VP1-2 USP influences its activity, but this remains speculative. Future work will attempt to address this, though this will require isolation of ubiquitinated versus nonubiquitinated forms of the active USP for direct comparison on a suitable substrate.
Expression of extended versions, e.g., NT3(1-705), resulted in a dramatic difference and demonstrated ubiquitination which would target for degradation but which was actively counteracted by deubiquitination involving the functional USP itself. In the absence of USP activity (i.e., C65A mutation), the protein was efficiently ubiquitinated and degraded but could be stabilized by proteosomal inhibition (Fig. 10B). Although this instability was also observed for further extended versions of the protein and indeed for the full-length protein (though somewhat less so in this case), virus infection resulted in a relative stabilization of the USP-inactive proteins (Fig. 10C). An explanation consistent with these observations proposes the following: first, that determinants within the N-terminal region of the protein, in the absence of additional interacting partners, are either folded improperly or otherwise efficiently targeted for ubiquitination but can be efficiently deubiquitinated by the intrinsic USP; second, that individual protein partners, candidates for which are indicated (Fig. 10C), interact with VP1-2 resulting in a change in folding or direct masking of ubiquitination sites; and third, that the consequence of this is a reduction in the reliance of the USP for overall stability. Future work will address the proposal that virus infection stabilizes the unstable USP-inactive mutants and the requirements for such an effect, whether it is a consequence of infection generally on the processing pathways or whether it may require or be recapitulated, e.g., by coexpression of specific interacting partners. It could be that the instability of NT3.C65A is a nonspecific consequence of the lack of proper intra- or interprotein folding, and the result is efficient targeting for degradation (by nonspecific, we mean one which is not in a determinant or region involved in specific folding under the correct circumstances). Since USP dependence was not observed for NT1 or NT2, it could be that such a determinant within the NT2 and NT3 boundary folds improperly and is targeted for ubiquitination, resulting in dependence upon the USP. It is difficult to formally exclude this explanation proposing malfolding in a nonspecific manner. However, such an explanation would have to allow that such malfolded determinants did not affect folding and function of the USP domain itself within the context of the N-terminal proteins. Furthermore, similar N-terminal variants (containing the active USP) have been used to explore functional interactions of VP1-2, and isolated polypeptides almost identical to regions and subregions of the variants used here have been previously shown to have relevant and specific interaction with virus partners, including UL37 and VP16, that depends on specific amino acids (14, 24, 30, 40). These results all indicate that N-terminal variants of the sort used in those studies fold and interact with protein partners in a physiologically relevant way. We believe that, rather than exhibiting a random, nonspecific lack of folding, NT3 is folded normally but is targeted for ubiquitination (or malfolded) due to either a specific lack of determinants within the protein or a lack of interacting partners. Even if the explanation for the requirement for the USP reflects a nonspecific effect of malfolding in the N-terminal region, this may still be relevant for VP1-2 expression. For example, given its size of approximately 3,000 amino acids, VP1-2 may have unique issues associated with expression and folding. Although actual protein synthesis rates are unknown in virus-infected cells and will vary between mRNAs, translation of VP1-2 could require 10 min for completion even at the high end of estimates of translation rates and could be between 25 and 50 min at 1 to 2 amino acids/s (18). One especially conserved region of VP1-2 with important function, including possible folding determinants, is located at the extreme C terminus, which is obviously not translated until well after the N-terminal regions conferring ubiquitination. It could be that in VP1-2 intra- or interchain folding or interaction with partners is sensitive and more susceptible to recognition and ubiquitination. In infected cells, synthesis of appropriate partners or alterations to processing pathways may counteract such intrinsic sensitivity. In this scenario, the presence of the USP at the N terminus of VP1-2, the first domain to emerge from the ribosome, might act as an additional mechanism to counteract issues with VP1-2 synthesis and folding. Recent results suggest that the translation of at least certain late virus mRNAs is sensitive to conditions of limited access to translation factors and that virus-encoded factors (e.g., vhs) might prevent the limiting conditions or somehow facilitate translation initiation under these conditions (9).
Nevertheless, the absence of any obvious effect of the HSV VP1-2 C65A substitution on growth parameters in culture is perhaps surprising. Previous analyses of similar single mutants in other herpesviruses have revealed some defect, although a range of results has been reported: significant reduction in one-step growth curves of 20- to 30-fold in PRV (6), a 10-fold reduction for MHV-68 (17), a 4-fold reduction in MDV (21), little effect in HCMV (23), or a decrease and delay in virion production (41). We are currently examining the effect of the VP1-2 USP mutation on growth kinetics in other cell types, particularly in cells under stressed conditions, including limitations in nutrient provision or prior treatment with immune stimulators.
Our proposal that the USP may be involved in auto-deubiquitination of VP1-2 but that it may be obviated by additional mechanisms operating in virus-infected cells, while providing a unifying rationale for the conservation of the USP domain in all VP1-2s, remains speculative. Clearly the USP could function in trans on other cellular and indeed viral targets. In conclusion, our results now provide the first demonstration of the involvement of the VP1-2 USP in actively deubiquitinating a target substrate with a consequential reduction in ubiquitination and proteosomal degradation and also provide a framework for future mechanistic studies of the USP domain activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prashant Desai for HSV-1 KΔUL36, antibody, and the complementing cell line HS30; Val Preston and Tony Minson for antibodies; and Fraser Rixon for ΔARUL36 and the complementing line RSC-HAUL36. We are grateful to David Leib for provision of the KOS bacmid and to Nick Osterrieder and Greg Smith for bacmid reagents.
C.M.C. was supported by a University Research Fellowship from the Royal Society. F.A. was supported by a fellowship from the Spanish Ministerio de Educacion y Ciencia. This work was funded by Marie Curie Cancer Care.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Abaitua F., Daikoku T., Crump C. M., Bolstad M., O'Hare P. 2011. A single mutation responsible for temperature-sensitive entry and assembly defects in the VP1-2 protein of HSV. J. Virol. 85:2024–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abaitua F., O'Hare P. 2008. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J. Virol. 82:5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey D., Barreca C., O'Hare P. 2007. Trafficking of the bZIP transmembrane transcription factor CREB-H into alternate pathways of ERAD and stress-regulated intramembrane proteolysis. Traffic 8:1796–1814 [DOI] [PubMed] [Google Scholar]
- 4. Batterson W., Roizman B. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonifacino J. S., Weissman A. M. 1998. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 14:19–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bottcher S., et al. 2008. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J. Virol. 82:6009–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coller K. E., Lee J. I., Ueda A., Smith G. A. 2007. The capsid and tegument of the alpha herpesviruses are linked by an interaction between the UL25 and VP1-2 proteins. J. Virol. 81:11790–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins C. A., Brown E. J. 2010. Cytosol as battleground: ubiquitin as a weapon for both host and pathogen. Trends Cell Biol. 20:205–213 [DOI] [PubMed] [Google Scholar]
- 9. Dauber B., Pelletier J., Smiley J. R. 2011. The HSV-1 vhs protein enhances translation of viral true late mRNAs and virus production in a cell-type-dependent manner. J. Virol. 85:5363–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Desai P., Sexton G. L., Huang E., Person S. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J. Virol. 82:11354–11361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Desai P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608–116018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edelmann M. J., Kessler B. M. 2008. Ubiquitin and ubiquitin-like specific proteases targeted by infectious pathogens: emerging patterns and molecular principles. Biochim. Biophys. Acta 1782:809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fernandez-Montalvan A., et al. 2007. Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. FEBS J. 274:4256–4270 [DOI] [PubMed] [Google Scholar]
- 14. Fuchs W., Klupp B. G., Granzow H., Mettenleiter T. C. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gierasch W. W., et al. 2006. Construction and characterisation of bacterial artifical chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197–206 [DOI] [PubMed] [Google Scholar]
- 16. Gonzalez C. M., Wang L., Damania B. 2009. Kaposi's sarcoma-associated herpesvirus encodes a viral deubiquitinase. J. Virol. 83:10224–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gredmark-Russ S., et al. 2009. A gammaherpesvirus ubiquitin-specific protease is involved in the establishment of murine gammaherpesvirus 68 infection. J. Virol. 83:10644–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hershey J. W. 1991. Translational control in mammalian cells. Annu. Rev. Biochem. 60:717–755 [DOI] [PubMed] [Google Scholar]
- 19. Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 [DOI] [PubMed] [Google Scholar]
- 20. Isaacson M. K., Ploegh H. L. 2009. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5:559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jarosinski K., Kattenhorn L., Kaufer B., Ploegh H., Osterrieder N. 2007. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. U. S. A. 104:20025–20030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kattenhorn L. M., Korbel G. A., Kessler B. M., Spooner E., Ploegh H. L. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547–557 [DOI] [PubMed] [Google Scholar]
- 23. Kim E. T., Oh S. E., Lee Y. O., Gibson W., Ahn J. H. 2009. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J. Virol. 83:12046–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klupp B. G., Fuchs W., Granzow H., Nixdorf R., Mettenleiter T. C. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knipe D. M., Batterson W., Nosal C., Roizman B., Buchan A. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38:539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee J. I., Luxton G. W., Smith G. A. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086–12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee J. I., et al. 2009. A herpesvirus encoded deubiquitinase is a novel neuroinvasive determinant. PLoS Pathog. 5:e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luxton G. W., Lee J. I., Haverlock-Moyns S., Schober J. M., Smith G. A. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meray R. K., Lansbury P. T., Jr 2007. Reversible monoubiquitination regulates the Parkinson disease-associated ubiquitin hydrolase UCH-L1. J. Biol. Chem. 282:10567–10575 [DOI] [PubMed] [Google Scholar]
- 30. Mijatov B., Cunningham A. L., Diefenbach R. J. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26–31 [DOI] [PubMed] [Google Scholar]
- 31. Pasdeloup D., Blondel D., Isidro A. L., Rixon F. J. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pickart C. M., Eddins M. J. 2004. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695:55–72 [DOI] [PubMed] [Google Scholar]
- 33. Radtke K., et al. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roberts A. P., et al. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1 (HSV-1). J. Virol. 83:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schlieker C., Korbel G. A., Kattenhorn L. M., Ploegh H. L. 2005. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 79:15582–15585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schlieker C., et al. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shanda S. K., Wilson D. W. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J. Virol. 82:7388–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tischer B. K., von Einem J., Kaufer B., Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 39. Todi S. V., et al. 2009. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 28:372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vittone V., et al. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang J., Loveland A. N., Kattenhorn L. M., Ploegh H. L., Gibson W. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Whitehurst C. B., et al. 2009. The Epstein-Barr virus (EBV) deubiquitinating enzyme BPLF1 reduces EBV ribonucleotide reductase activity. J. Virol. 83:4345–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolfstein A., et al. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.