Abstract
The structure of VRC01 in complex with the HIV-1 gp120 core reveals that this broadly neutralizing CD4 binding site (CD4bs) antibody partially mimics the interaction of the primary virus receptor, CD4, with gp120. Here, we extended the investigation of the VRC01-gp120 core interaction to the biologically relevant viral spike to better understand the mechanism of VRC01-mediated neutralization and to define viral elements associated with neutralization resistance. In contrast to the interaction of CD4 or the CD4bs monoclonal antibody (MAb) b12 with the HIV-1 envelope glycoprotein (Env), occlusion of the VRC01 epitope by quaternary constraints was not a major factor limiting neutralization. Mutagenesis studies indicated that VRC01 contacts within the gp120 loop D, the CD4 binding loop, and the V5 region were necessary for optimal VRC01 neutralization, as suggested by the crystal structure. In contrast to interactions with the soluble gp120 monomer, VRC01 interaction with the native viral spike did not occur in a CD4-like manner; VRC01 did not induce gp120 shedding from the Env spike or enhance gp41 membrane proximal external region (MPER)-directed antibody binding to the Env spike. Finally, VRC01 did not display significant reactivity with human antigens, boding well for potential in vivo applications. The data indicate that VRC01 interacts with gp120 in the context of the functional spike in a manner distinct from that of CD4. It achieves potent neutralization by precisely targeting the CD4bs without requiring alterations of Env spike configuration and by avoiding steric constraints imposed by the quaternary structure of the functional Env spike.
INTRODUCTION
The HIV-1 envelope glycoprotein (Env) consists of the exterior gp120 and the transmembrane Env gp41, which comprise the functional trimer (10, 20, 21, 40, 54, 67, 68). The gp120 subunit interacts initially with the primary receptor, CD4, present on the surface of target cells (12, 30, 42). Receptor interaction results in substantial conformational changes in gp120, exposing the coreceptor binding site and permitting high-affinity binding to one of the coreceptors, either CCR5 or CXCR4 (1, 11, 13, 17, 19, 22, 63, 69). This receptor-and-coreceptor interaction triggers extensive rearrangements within gp41 (23, 29, 33, 60), which then mediates virus-to-target-cell membrane fusion and the entry of viral genomic information into susceptible target cells. During the course of natural infection, the HIV-1 Env elicits both type-specific and broadly cross-reactive neutralizing antibodies (24, 41, 50, 55, 66, 72). The latter response occurs in about 15% to 25% of HIV-1-infected individuals and can mediate neutralization of diverse viral isolates (14, 18, 37, 57, 61, 62). It has proven difficult to elicit such cross-reactive neutralizing antibody responses via Env immunization, in part due to the immune-dominance of gp120 variable regions and the apparent limited immune recognition of conserved regions of the viral Env (50, 72). To gain better insights that might apply to immunization strategies, investigators have focused on those HIV-1-infected individuals that mount neutralizing antibody responses to conserved regions of the viral Env (3, 14, 25, 37, 38, 57). Therefore, isolation of novel broadly neutralizing monoclonal antibodies (MAbs) and characterization of interactions with their cognate epitopes have received renewed interest.
Several broadly neutralizing monoclonal antibodies isolated from HIV-infected individuals define conserved epitopes on the HIV Env. These include the membrane proximal external region (MPER) of gp41, which is targeted by the MAbs 4E10, 2F5, and Z13 (5, 52, 75, 76), the carbohydrate-specific outer domain epitope which is targeted by 2G12 (5, 8, 56, 58, 64), a V2-V3-associated epitope which is targeted by PG9/PG16 (65), and the CD4 binding site (CD4bs) (7) targeted by several antibodies. The CD4bs overlaps with the conserved region on gp120, which is involved with engagement of CD4. The prototypical CD4bs-directed MAb b12 can neutralize ∼40% of circulating primary isolates, and its structure in complex with the core of gp120 is defined (74). Interestingly, viral resistance to b12 only partially correlates with residue variation within the structurally defined epitope (71). Several b12-resistant viruses display an intact b12 epitope on their respective gp120 subunits (71), suggesting that quaternary packing of Env also confers resistance to b12. These observations emphasize that in addition to the structural information provided by X-ray crystallography of the antibody-Env interaction, phenotypic studies may provide additional insights into mechanisms of antibody-mediated neutralization and viral resistance.
We recently isolated three CD4bs-directed MAbs, VRC01, -02, and -03 (70), from a donor whose serum neutralization activity maps predominantly to the CD4bs (37). VRC01 and VRC02 are a closely related pair of somatic variants that neutralize over 90% of diverse HIV-1 primary isolates. The structure of VRC01 in complex with the gp120 core reveals that the VRC01 heavy chain binds to the gp120 CD4bs in a manner similar to that of the primary receptor, CD4 (73). However, the gp120 core lacks the major variable regions, as well as the N and C termini, and may only partially reflect the VRC01 interaction with full-length gp120, or with gp120 in the context of the native viral spike. In this study, we sought to further characterize VRC01 neutralization in the context of full-length gp120, its impact on the architecture of the viral Env functional spike upon binding, and viral factors associated with the relatively few cases of HIV-1 neutralization resistance.
MATERIALS AND METHODS
Generation of Env pseudovirus and neutralization assays.
A panel of 119 JRCSF gp160 plasmid DNAs containing single point mutations in gp120 (51) or VRC01-resistant virus gp160 plasmid DNA was used in combination with a luciferase reporter plasmid containing the essential HIV structural genes to produce mutant Env pseudoviruses as described previously (35). In the case of making viruses treated with kifunensin, kifunensin (Cayman Chemical, Ann Arbor, MI) at 25 μM was added into the cell culture media 2 h before the transfection. The harvested cell culture supernatant containing the pseudovirus was stored at −80°C prior to use to perform either virus neutralization assays or enzyme-linked immunosorbent assay (ELISA)-based binding analysis. HIV-1 Env pseudoviruses were also used for neutralization assessment in a single round of an entry assay using TZM-bl target cells as previously described (35).
Env protein and antibody expression and purification.
HIV-1 gp120 was expressed in Drosophila S2 cells and purified with 17b-coupled protein A affinity columns as previously described (38). Antibody 2G12 was obtained from Polymun Scientific, Inc. (Vienna, Austria). Four-domain soluble CD4 (D1 to D4) of CD4 was purchased from Progenics (Tarrytown, NY). The plasmid encoding CD4-Ig contains the D1D2 domain fused with the human Fc fragment was provided by Joseph Sodroski (Dana Farber Cancer Institute) and used to transfected 293 FreeStyle cells as previously described to express CD4-Ig secreted into cell culture supernatant (38), followed by purification with a protein A affinity column (GE Healthcare). IgG b12 has been described previously (7). V3 region-specific 447D was provided by Susan Zolla-Pazner (New York University CFAR), and the CD4i antibodies 17b, 48D, 2.1c, and A32, along with the C1/C5 conformational antibody C11, were provided by James Robinson (Tulane University).
gp120 and CD4bs ligand-binding ELISA.
MAb and CD4 binding to monomeric gp120 dissociated from Env pseudovirus proteins was measured using antibody capture and evaluated by sandwich ELISA as previously described (43), with minor modifications (71). Relative affinities (avidities) were calculated as the antibody concentration at half-maximal binding (50% effective concentration [EC50]) derived by curve fitting with software GraphPad Prism, version 5.0 (GraphPad Software Inc., La Jolla, CA) using the sigmoidal dose-response model with a variable slope. 2G12 binding to virus monomeric gp120 was used to normalize the amount of virus gp120 in each virus lysate preparation. In cases of poor 2G12 recognition of gp120 due to 2G12 epitope mutations, such as N332A, HIV-Ig, or MAb 447, was used for normalization. The effect of a given mutation on antibody binding was represented by apparent affinity (avidity) relative to those for wild-type (WT) gp120, calculated with the formula ([(EC50_WT/EC50_mutant)/(EC50_WT for 2G12/EC50_mutant for 2G12)] × 100). Competition ELISAs of biotin-labeled antibodies were carried out with recombinant YU2 gp120 captured in ELISA wells coated with the sheep D7324 polyclonal antibody, which is specific for the gp120 C5 region as described previously (70).
ITC.
Isothermal titration calorimetry (ITC) measurements were carried out by using the ITC200 microcalorimeter system (MicroCal, Inc.). All reactions were performed at 37°C as previously described (70). The concentration of gp120 in the sample cell was approximately 5 μM, and that of CD4-Ig, VRC01, or 17b in the syringe was approximately 50 μM. The molar concentrations of the proteins were calculated using the following molar extinction coefficients: gp120, 1.52; VRC01, 1.53; CD4-Ig, 1.20; and 17b, 1.36. For the second step of a two-step reaction, the gp120 concentration was recalculated per the final volume at the end of the first reaction, which is equal to the sample cell volume plus the total volume of the first injectate. Before the second titration, the sample volume equivalent to the volume of the first injectate was removed from the sample cell. Data were analyzed with Microcal ORIGIN software using a single-site binding model.
HIV-1 Env gp120 shedding.
293T cells were transfected with the full-length JRFL or JRCSF Env-expressing plasmid pSVIII as described previously (49). Forty-eight hours posttransfection, approximately 2 × 107 cells were collected, washed with PBS, and concentrated to 1 × 107cells/ml. Two-hundred-microliter aliquots of the cell suspension were deposited into microcentrifuge tubes for each ligand at a given concentration. The tubes containing the cells were subjected to a 5-min centrifugation at 335 × g at room temperature to pellet the cells. A total of 160 μl of the supernatant was removed and discarded. A total of 20 μl of the ligand solution prepared in a serial dilution was added to the cell pellets, and following mixing, the cells and ligand were incubated at 37°C for 1 h with gentle mixing every 20 min. The cells were then subjected to centrifugation, and the supernatants were transferred to fresh tubes. A total of 30 μl of each supernatant was loaded onto SDS-PAGE gels and run for 1.5 h at 200V. The proteins were transferred to nitrocellulose membranes for Western blotting. To identify gp120, the V3-specific anti-gp120 human monoclonal 39F was used as a primary antibody (1 μg/ml), followed by a horseradish peroxidase (HRP)-conjugated anti-human secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) at a 1:10,000 dilution, and development with substrate followed.
Antibody self-antigen recognition assays.
Two assays were used for evaluation of the MAb reactivity to self-antigens as previously described (27). Briefly, the luminex AtheNA Multi-Lyte ANA test (Wampole Laboratories, Princeton, NJ) was used to test for MAb reactivity to self antigens: systemic lupus erythematosus autoantigens SSA and SSB, sphingomyelin (Sm), ribonucleoprotein (RNP), sclerosis autoantigen (Scl-70), histidine-tRNA ligase (Jo-1), double-stranded DNA (dsDNA), and centromere B (CentB). Reactivity to HIV-1-negative human epithelial HEp-2 cells was determined by indirect immunofluorescence on HEp-2 slides using Evans Blue as a counterstain and fluorescein isothiocyanate (FITC)-conjugated goat anti-human IgG (Zeus Scientific, Raritan NJ). Antibody reactivity with phospholipid was assessed with activated partial thromboplastin time (aPTT) automated reagent (Diagnostica Stago, Parsippany, NJ), and time to clot formation was measured as an indicator of phospholipid reactivity. Anti-respiratory syncytial virus (RSV) monoclonal antibody, Synagis (palivizumab; MedImmune, MD) and plasma from normal donors were used as negative controls.
RESULTS
Characterization of VRC01 binding and neutralization specificity using a panel of Env alanine mutants.
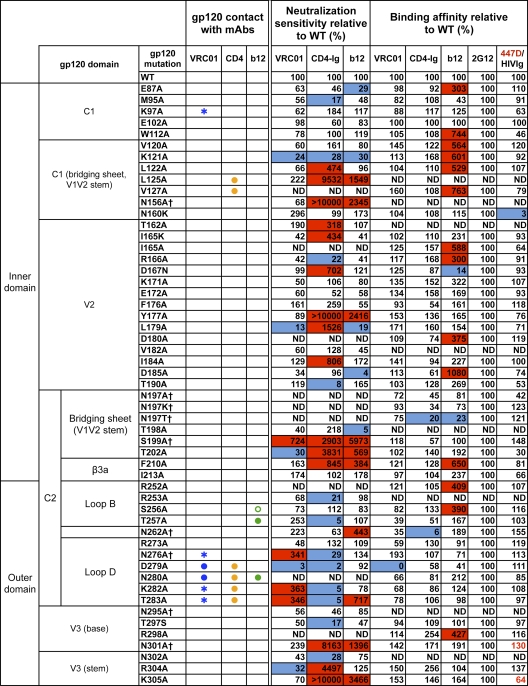
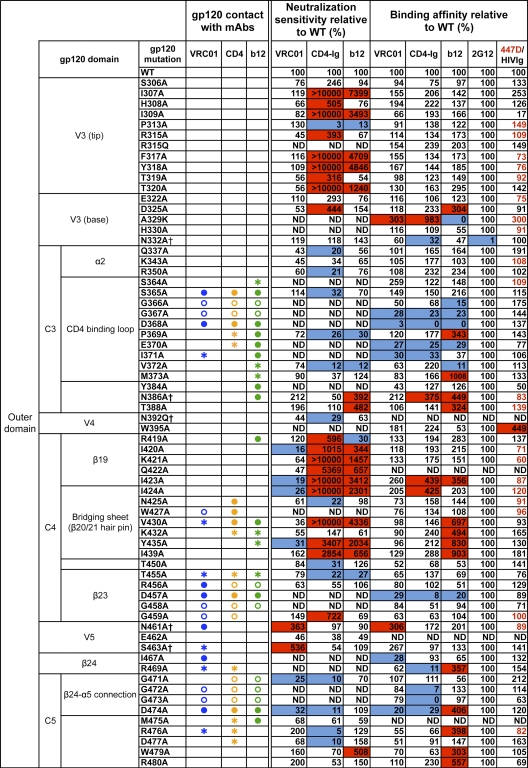
The previously published crystal structure of VRC01 in complex with a monomeric gp120 core revealed the antibody contact sites on the trimmed protein lacking the major variable regions V1, V2, and V3 and portions of the N and C termini (73). To study the VRC01 interaction with the functional viral spike, we used a panel of JRCSF alanine (Ala) scanning Env pseudovirus mutants (51) to assess the neutralization potency of VRC01 in comparison to that of CD4-Ig and the prototypic CD4bs MAb b12 (Fig. 1). Since some of the JRCSF Env mutants were not entry competent, we also studied VRC01 binding to Ala scanning mutants in the context of full-length monomeric JRCSF gp120. Consistent with the structural analysis, the mutational analyses confirmed that mutations at residues in loop D (N terminal to the V3 region), the CD4 binding loop, and the V5-β24-α5 region diminished VRC01-mediated binding or neutralization (Fig. 1). In some cases, single mutations, such as D279A (loop D) and D474A (β24-α5), diminished both VRC01-mediated neutralization and binding to the corresponding gp120. For other contact sites, such as those in the CD4 binding loop, neutralization could not be tested since these Envs did not support entry, but Ala mutations at G367, D368, and I371 resulted in diminished binding to gp120. However, for many of the predicted contact sites, individual Ala mutations did not result in a measurable decrease in either VRC01 binding or neutralization. This is perhaps not surprising, as single residue mutations may be insufficient to disrupt high-affinity antibody-ligand binding if the specific contact does not contribute substantial binding energy to the ligand-ligand interaction. In addition, the energetics of binding contributed by individual residues is not always fully evident from the structurally defined protein-protein interface alone. Of note, gp120 loop D and V5 regions contained substitutions uniquely affecting VRC01 binding, but not b12 or CD4-Ig binding (Fig. 2A).
Fig. 1.
VRC01 neutralization and binding on a panel of primary isolate JRCSF functional Env and gp120 mutants. Amino acid numbering of mutants is based on the HIV-1 HXBc2 sequence. The gp120 structurally defined contact residues for the VRC01, CD4, and b12 complexes are indicated in blue, orange, and green, respectively, with open circles (○) designating gp120 main-chain-only contacts, asterisk (*) designating gp120 side chain-only contacts, and filled circles (●) designating both main-chain and side-chain contacts. Mutations that knock out putative N-glycosylation sites (NXS/T motifs) of gp120 are labeled with a dagger symbol (†). Relative ELISA binding affinities to captured gp120s were calculated based on the antibody concentration at half-maximal binding (EC50). The effect of each mutation on antibody binding was normalized using 2G12 to control for the amount of captured gp120. Mutations that resulted in decreased gp120 binding (<33% relative to that of wild type [WT]) are highlighted in blue, and those that resulted in increased ELISA binding (>300% relative to that for the wild type) are highlighted in red. HIVIg or the anti-V3 MAb 447D (values shown in red) were used as the controls for gp120s with poor 2G12 binding. Neutralization sensitivity of each mutant to MAb VRC01, CD4-Ig, or MAb b12 was assessed and compared to neutralization of the wild-type Env pseudovirus. The color scheme is the same as that used for the gp120 binding data.
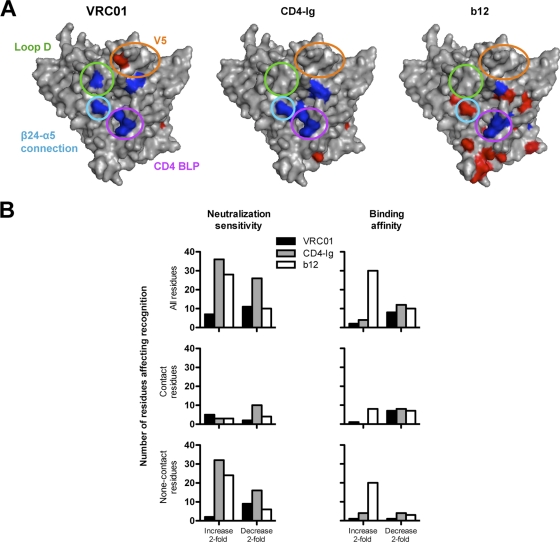
Fig. 2.
(A) Surface model of the JRCSF gp120 core showing mutants resulting in decreased binding (blue) or increased binding (red). The gp120 domains that contain mutations most affecting affinities for VRC01 are denoted in circles of the following colors: green, loop D; gold, V5/β24; purple, CD4 BLP (CD4 binding loop); and blue, β24-α5 connection. (B) Histograms showing the number of mutant residues affecting virus neutralization or gp120 binding for VRC01, b12, and CD4-Ig.
We also observed that Ala mutations beyond the structurally defined VRC01, CD4, and b12 contact sites and scattered within V2, V3, the bridging sheet, and β19 regions significantly affected neutralization sensitivity to these CD4bs ligands (Fig. 1). In most cases, these mutations had more pronounced effects on viral neutralization than on monomeric gp120 binding affinity, suggesting effects on functional spike quaternary interactions or “packing” (9). In a few cases, Ala mutations at such “noncontact” residues diminished neutralization by b12, CD4-Ig, or VRC01 (e.g., K121A and L179A). However, in most cases, these mutations resulted in a marked enhancement of viral neutralization sensitivity, especially for ligands CD4-Ig and b12. Specifically, mutations located in V2 (L179A) and V3 (R304A) and clustered in the bridging sheet (S199A, T202A, I424A, and Y435A) and β19 (Ι420Α and Ι423Α) facilitated increases in CD4-Ig or/and b12 neutralization potency while imbuing VRC01 potency with less quantitative increases (Fig. 1). A few mutations that eliminated N-glycosylation sites in the V1V2 stem (S199A), loop D (N276A), or V5 (N461A or S463A) resulted in enhanced VRC01 potency (Fig. 1). Perhaps these Ala substitutions better expose the VRC01 epitope in the spike context as most of these mutations had a minor effect on binding affinity to the gp120 monomer. Overall, the Ala scanning mutagenesis data revealed that there were many more noncontact mutations leading to increased viral neutralization sensitivity for CD4-Ig and b12 than for VRC01, especially in the major variable regions V1, V2, and V3 (Fig. 2B).
Steric hindrance is not a major factor limiting VRC01 neutralization.
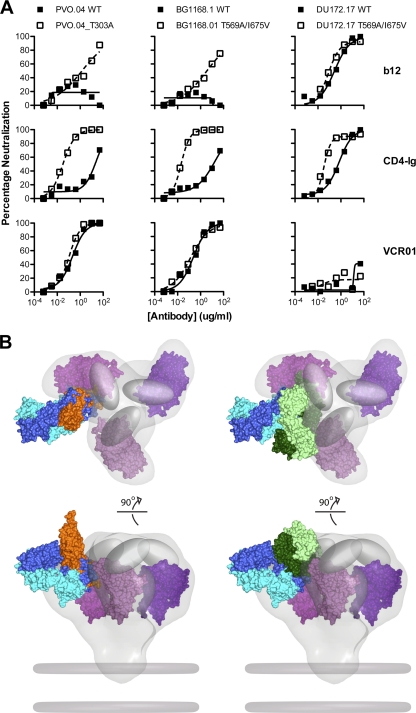
It has previously been reported that limitations to antibody access to the CD4bs may restrict neutralization by CD4bs antibodies, including the neutralizing MAb b12 (4, 31, 32, 36). This steric hindrance is thought to be a result of the three-dimensional structure of the functional viral spike, including packing of the variable loops and N-linked glycan shielding. Thus, viral resistance to b12 can often occur due to limitations of the antibody access to the CD4bs, even when the MAb cognate contact residues are present on a given gp120 (71). Since VRC01 can neutralize approximately 90% of virus isolates by precisely targeting the CD4bs (70), we hypothesized that access to the CD4bs may not be a major restriction to its neutralization breadth. Stated in another way, neutralization by VRC01 should correlate closely with gp120 binding affinity, and few mutations on the Env glycoprotein should result in increased binding or neutralization by VRC01. This hypothesis is consistent with our neutralization and binding results conducted with the JRCSF mutant virus panel shown in Fig. 1. Here, to extend the analysis with more circulating virus strains and various Env regions, we also evaluated the impact of previously described mutations that alter the Env structure or neutralization sensitivity and which also result in an increased neutralization potency by either MAb b12 or by CD4-Ig. Removal of a V3 loop N-linked glycan at position 301 (via a T303A mutation) (31) or mutations in gp41 (T569A/I675V) (4) resulted in marked enhancement in the potency of neutralization by b12 and CD4-Ig, but these mutations did not significantly affect VRC01-mediated neutralization (Fig. 3A).
Fig. 3.
Impact of conformational constraints on neutralization by b12, CD4-Ig, and VRC01. (A) Env pseudoviruses PVO.04 (left) and BG1168.1 (middle) are relatively resistant to b12 and CD4-Ig. In the setting of the gp120 T303A mutation or the gp41 T569A/I675V mutations that result in a more open Env conformation, these viruses show enhanced neutralization sensitivity to b12 and CD4-Ig. In contrast, both the wild-type and mutant versions of these viruses were equally sensitive to VRC01-mediated neutralization. Virus DU172.17 (right) was resistant to VRC01, and this resistance was not affected by the Env mutations that alter conformation. (B) Model of VRC01, CD4, and b12 binding to the HIV-1 functional spike. Top (top) and front (bottom) views of the density map surface of the HIV-1 spike, presenting three gp120 core protomers in magenta (39) (Electron Microscopy Database [EMDB]: EMD-5019; Protein Data Bank [PDB]: 3DNN) and viral membrane in dark gray. To analyze the relative binding orientations of b12, CD4, and VRC01, the X-ray coordinates for gp120, in the b12, VRC01, and CD4/17b/bound conformations (PDB: 2NY7, 3NGB, and 2NY3), respectively, were superimposed on the same gp120 core protomer (PDB: 3DNN). The molecular surface of the b12-Fab fragment is green; the VRC01 Fab is blue. Within each color-coded Fab, the heavy chains are shaded darker than the light chains. The molecular surface of sCD4 is orange, and the variable loops are represented as gray ovals. Note that in this model, b12 and CD4 are more loop proximal than is VRC01.
Taken together, these results described immediately above, along with the JRCSF Ala virus mutant panel data, suggest that VRC01 can approach its cognate epitope on the functional spike with less steric hindrance than does b12 and, surprisingly, with less hindrance than the soluble form of the virus receptor CD4 itself (Fig. 3B). These differences might be related to the quite different angle of approach to the CD4bs employed by VRC01, in contrast to the more loop-proximal approach employed by CD4 and b12 (see the trimer model for these ligands shown in Fig. 3B).
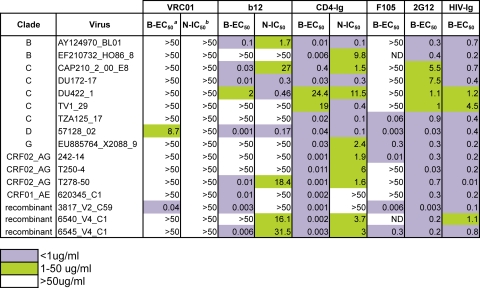
Further analysis of 16 VRC01-resistant viral isolates (from a panel of 190) (70) showed that VRC01 bound poorly to gp120 C5-specific antibody-captured gp120 derived from 15/16 primary isolate viruses (Table 1). VRC01 demonstrated relatively strong binding to the captured gp120 of only one neutralization-resistant virus, 3817_V2_C59. This is in contrast to our previous finding for b12, in which b12 bound with high relative affinity to eight of the 14 b12-resistant viral gp120s (71). Taken together, these data demonstrate that the relatively infrequent cases of VRC01 natural resistance arise largely from antigenic variation at the direct gp120 contact site itself, rather than by steric restrictions on access to the CD4bs that is more commonly observed for naturally occurring b12 resistance. These data are consistent with the Env functional spike model shown in Fig. 3B, in which V loop packing impacts b12 access to its epitope to a greater degree than it does VRC01 epitope access.
Table 1.
VRC01-resistant virus binding/neutralization profile
aB-EC50 (μg/ml), half-maximal effective concentration of ELISA binding assay.
bN-IC50 (μg/ml), 50% inhibitory concentration of neutralization assay.
Antigenic and sequence variation of gp120 displayed by VRC01-resistant viruses.
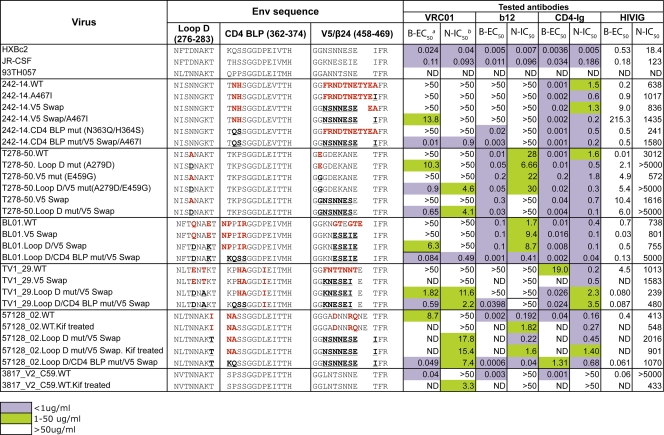
Our prior analysis of the sequence of 17 VRC01-resistant isolates, based upon the structurally defined contact sites of VRC01 on the gp120 core, demonstrated that the most resistant isolates contained alterations in residues in one of two regions of gp120: loop D or the V5 region (73) (see Fig. S1 in the supplemental material). In addition to these two sites, we also observed primary amino acid sequence variation within the CD4 binding loop region (see Fig. S1 in the supplemental material). In contrast, the gp120 of most VRC01-sensitive viruses contained intact VRC01 contact sites. Specifically, sequence variation was located within a 5-amino acid motif of 278T(D/N)NAK283 of loop D, within residues 362 to 365 of the CD4 binding loop and flanking potential N-linked glycosylation, and within residues 458 to 467 of the V5/β24 region. One interpretation of these observations is that the motif variations are linked, suggesting that multiple alterations in more than one of the gp120 hotspots might be required to generate full VRC01 neutralization resistance. For example, the resistant virus 242-14 contains variations in the CD4 binding loop and in V5/β24; and the resistant isolate, T278_50, possessed variations in both loop D and V5.
To test the hypothesis that multiple changes in the described motifs contribute to full VRC01 resistance, we altered specific residues and made chimeric “swaps” to restore VRC01-sensitive viral sequences (e.g., that of the reference virus Env from HXBc2) to these resistant viruses (Table 2). We selected five viruses with the typical features of variation described above and tested this hypothesis, as described below.
Table 2.
VRC01-resistant virus reversion mutant mutagenesis and binding/neutralization profilec
aB-EC50 (μg/ml), half-maximal effective concentration of ELISA binding assay.
bN-IC50 (μg/ml), 50% inhibitory concentration of neutralization assay.
cThree VRC01-sensitive virus gp120 sequences are listed at top as references. VRC01-resistant virus gp120 variations from the reference sequences are listed in bold red type. The mutated residues are marked in bold black type and underlined.
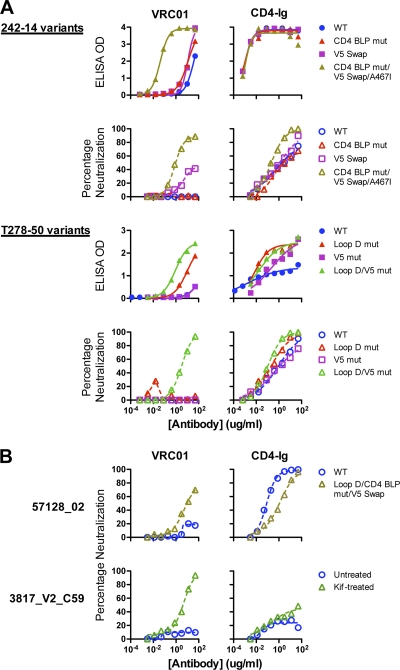
For resistant virus 242-14, full sensitivity to VRC01 was restored by a set of mutations that included a V5 swap, a V5/β24 Α467Ι mutation, and replacement of residues flanking the CD4 binding loop with wild-type HXBc2 sequences. Individual mutations were insufficient to produce a sensitive virus, whereas the full set of mutations resulted in both high binding to the captured gp120 and potent neutralization (Fig. 4A; Table 2). These data suggest that each single alteration restores VRC01 recognition to a degree, but all are required for full sensitivity.
Fig. 4.
Binding and neutralization of reversion mutants and chimeras of VRC01-resistant viruses. (A) VRC01-resistant viruses 242-14 and T278-50 and their reversion mutants. Curves show binding and neutralization with VRC01 or CD4-Ig. The specific reversion mutants and chimeras are described in Table 2. (B) Analysis of two Env pseudoviruses that were resistant to VRC01 despite recognition of the viral gp120. gp120 of virus 57128_02 bound to VRC01 with an EC50 of 8.7 μg/ml (Table 1) yet was resistant to VRC01 neutralization. With additional reversion mutations in loop D, CD4 binding loop, and V5, the mutant virus was moderately neutralized by VRC01 (top). Virus 3817_V2_C59 was resistant to VRC01 neutralization (bottom) despite strong recognition of its gp120 by VRC01 (EC50 of 0.04 μg/ml, Table 1). When the virus was produced in the presence of the glycan modification inhibitor kifunensin, it became sensitive to VRC01-mediated neutralization (bottom).
Similarly, the viral isolate T278-50 contained variations from gp120s derived from VRC01-sensitive viruses in both the loop D region (residue A279) and the V5 region (residue E459). Altering both sites to the wild-type sequence was required to restore VRC01 binding to gp120 and to recover full sensitivity to VRC01-mediated neutralization (Fig. 4A; Table 2). In addition, the viral isolates BL01 and TV1_29 both contained variations in the structurally defined VRC01 contact sites in loop D, in the CD4 binding loop, in the V5 region, and in the β24-α5 connection (see Fig. S1 in the supplemental material). Restoring the loop D residues, CD4 binding loop residues, and the V5 region residues to VRC01-sensitive viral sequences restored both VRC01 binding to gp120 and sensitivity to VRC01-mediated neutralization (Table 2), strongly suggesting that the aforementioned resistance motifs within gp120 contribute to the VRC01-resistant phenotype of these isolates. We also performed mutagenesis on isolate 57128_02, which displays variability at residue 283 (I283T) in loop D, in residues 362 to 365 of the CD4 binding loop, and in the V5-β24 region (Table 2). Reversion to putative VRC01 contact sequences in these three regions resulted in a gp120 with restored VRC01 binding recognition and a virus displaying moderate VRC01-mediated neutralization sensitivity (Fig. 4B; Table 2).
It is of note that, with the exception of the TV1-29 isolate, all the VRC01 neutralization reversion mutants are more resistant to HIV-Ig neutralization (Table 2). This might be explained by the likely possibility that the majority of the neutralization specificity in HIVIG, a pool of IgGs derived from HIV-1-infected individuals, is distinct from that of the monospecific VRC01 MAb.
As noted above, VRC01 bound with high relative affinity (avidity) to the monomeric gp120 of only one resistant isolate, 3817_V2_C59. Sequence analysis indicated that the structurally defined VRC01 epitope on this gp120 was intact and, as opposed to the 15 other cases, that resistance might be due to indirect conformational influences resulting in occlusion of the VRC01 epitope in the context of the viral functional spike. To test this possibility, we generated a 3817_V2_C59 Env pseudovirus in the presence of kifunensin. Kifunensin treatment of the viral producer cells inhibits formation of complex N-glycans on the viral Env (2, 16, 59). Compared to the virus generated from untreated cells, the kifunensin-treated virus was more sensitive to VRC01-mediated neutralization (Fig. 4B; Table 2). Thus, in this one case, viral resistance to VRC01-mediated neutralization was likely due to a glycan-dependent limitation of VRC01 accessibility to its epitope in the context of the functional Env spike.
VRC01 and CD4 interaction with gp120 displays differential effects on the coreceptor binding site.
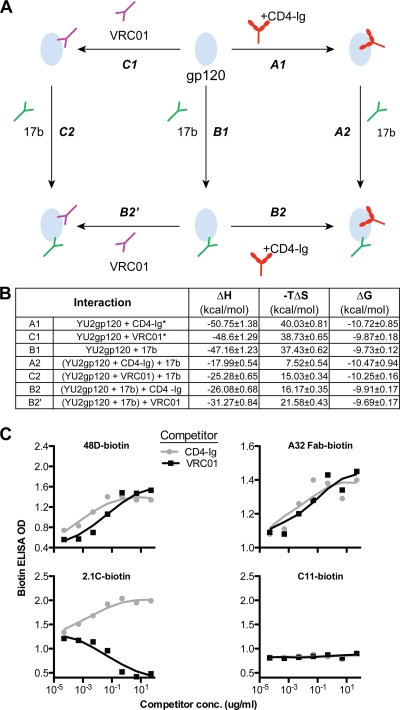
Previously, we demonstrated that VRC01 induced substantial conformation changes upon binding to monomeric gp120, as assessed by isothermal titration calorimetry (ITC) (70), in a manner very similar to that displayed by sCD4 interacting with gp120 (47). Here, we more fully characterized the effects of VRC01 binding to gp120, compared to those of the viral receptor CD4 and the coreceptor mimetic MAb 17b. We did this comparison by performing a complete thermodynamic cycle analysis using VRC01 and CD4-Ig in parallel, along with 17b (Fig. 5A). The change in enthalpy (ΔH) and entropy (−TΔS) of gp120 upon binding to the CD4bs or coreceptor binding site ligands was determined. These thermodynamic values are thought to reflect conformational changes imposed by the binding of each of these gp120 ligands. As reported previously, following gp120 interaction with CD4-Ig, VRC01, or 17b, individually, gp120 undergoes a large conformational change as assessed by the observed large change in enthalpy (∼50 kcal/mol for both CD4 and VRC01) (Fig. 5B). However, when CD4-Ig or VRC01 was mixed with gp120 to form stable complexes prior to the addition of 17b, the change in enthalpy of 17b binding to gp120 was reduced more than 50% compared to the 17b-gp120 ΔH value, without the prior addition of either CD4-Ig or VRC01 (Fig. 5B). These data are consistent with the previous observation that both CD4 and VRC01 can stabilize monomeric gp120 in a 17b-preferred binding conformation. Although VRC01 induction of the 17b epitope was slightly less than that of CD4, the overall signatures comprised by the full thermodynamic cycles derived from these ligands were quite similar (Fig. 5B).
Fig. 5.
VRC01 effects on monomeric gp120, compared with CD4-Ig. (A) Scheme of thermodynamic cycles showing enhancement of binding to gp120 between CD4bs ligands CD4-Ig and VRC01 and the CD4i antibody 17b. (B) Thermodynamic values of CD4-Ig, VRC01, and 17b interactions with gp120 measured by ITC at 37°C, including the change in enthalpy (ΔH) and entropy (−TΔS) upon binding of gp120 to the coreceptor mimetic antibody 17b, antibody VRC01, and CD4-Ig, a surrogate for the primary receptor, CD4. *, data extracted from reference 70. (C) Effect on other CD4i antibodies binding to gp120, captured by sheep anti-gp120 C5 antibody D7324 on ELISA plate. The binding of biotin-labeled CD4i antibodies, 48D, A32 Fab, and 2.1C to gp120 was tested in the presence of CD4-Ig or VRC01, with signals detected by the streptavidin-HRP conjugate. The MAb C11, which recognizes a C1-to-C5 conformational epitope on gp120, was used as negative control.
To further characterize the effects of the VRC01-gp120 interaction on coreceptor-directed antibodies as a class, we analyzed the impact of gp120 recognition by CD4i antibodies not previously evaluated. Recognition of these antibodies to gp120 was assessed in the presence or absence of the positive control, CD4-Ig, or the VRC01 MAb. The CD4i MAb 48D displayed enhanced binding to gp120 in the presence of both CD4-Ig and VRC01 (Fig. 5C). Similarly the CD4i MAb A32, which binds to the gp120 C1-to-C4 region (46), rather than the coreceptor binding region, also displayed enhanced recognition of gp120 in the presence of VRC01 (Fig. 5C). In contrast, 2.1C, a CD4i antibody whose epitope overlaps with coreceptor binding site, displayed enhanced binding in the presence of CD4-Ig; however, its binding to gp120 was decreased in the presence of VRC01 (Fig. 5C). This is consistent with the fact that both VRC01 and 2.1C appear to interact with residue V430 on gp120 (15, 73) and likely have overlapping epitopes. Despite this single exception, the interaction of VRC01 with monomeric gp120 in the antibody induction context appears to be similar to that of CD4.
VRC01 and CD4 have markedly different effects on the functional Env spike.
While VRC01 and CD4-Ig have similar effects on monomeric gp120, the Ala scanning of gp120 in the context of the functional viral spike revealed distinct differences between VRC01 and CD4-Ig interactions. Therefore, we sought to analyze additional interactions of these ligands with gp120 in the context of the functional spike. It was shown previously that in vitro, upon addition of sCD4 to Env-expressing cells or to free virus, the gp120 subunit dissociates from gp41 and is “shed” from the spike into the surrounding tissue culture media (6, 26, 44, 45, 48) (so-called “CD4-induced shedding”). To investigate if VRC01 can similarly induce shedding of gp120 from the functional trimer in vitro, we transfected 293T cells with plasmids encoding the JRCSF or JRFL Env to generate HIV-1 trimers on the surface of these cells. Following transient expression, sCD4 or VRC01 was added to the trimer-displaying cells, and immune-precipitation using the anti-V3 antibody 39F was performed to detect any gp120 that was shed into the supernatant. The shed, monomeric gp120 was resolved by SDS-PAGE gels and quantified by Western blotting (Fig. 6A). As expected, sCD4 induced significant gp120 shedding from both the cell surface JRFL and JRCSF Env trimers, in a dose-dependent manner. However, there was no measurable shedding of gp120 induced by the VRC01 MAb. Interestingly, although MAb b12 induces little conformational change to gp120 as assessed by thermodynamic analysis (34, 74), it induced gp120 shedding from JRFL cell surface Env trimers (Fig. 6A). The lack of VRC01-induced gp120 shedding, even in the context of the labile JRFL cell surface Env spike, implies that VRC01 interacts with the functional virus spike in a manner that causes less alteration of the configuration of the viral Env spike than does either CD4 or the MAb b12.
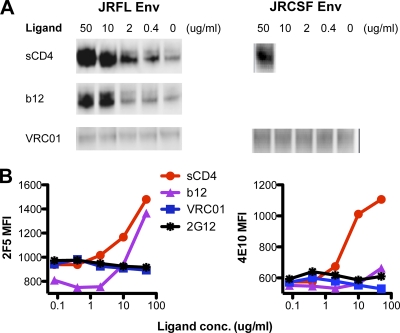
Fig. 6.
VRC01 effects on HIV envelope glycoprotein functional trimer. (A) Western blot analysis of Env from supernatants of 293T cells transfected with cleavage competent gp160 JRFL or JRCSF Env plasmid DNA. HIV-1 gp120 shedding upon binding of ligands, including sCD4, b12, and VRC01, from the Env trimer on the surface of 293T cells was evaluated by probing the blot with the anti-gp120 human monoclonal antibody 39F (V3 specific). (B) Effect of VRC01, b12, and sCD4 on gp41 MPER-directed antibodies 2F5 and 4E10 binding to trimeric Env. 293T cells transfected with JRFL-ΔCT Env expression plasmid were stained with biotin-labeled 2F5 or 4E10. Binding of 2F5 or 4E10 to JRFL Env in the presence of the CD4bs ligands VRC01, b12, and sCD4 was detected with streptavidin-APC conjugate. MFI, mean fluorescence intensity.
To further investigate this apparent difference between MAbs VRC01 and sCD4 in their respective interaction with gp120 in the context of the trimeric spike, we assessed the effects on the gp41 subunit following incubation of these two CD4 binding site ligands. The rationale for this experiment is as follows. As a key functional component of the functional spike, gp41 undergoes significant rearrangement after gp120 engages CD4 (and perhaps the coreceptor CCR5) to mediate viral entry. The membrane proximal external region (MPER) is a highly conserved and important functional region of gp41 and is the target of the broadly neutralizing antibodies 2F5 and 4E10. The MPER of primary isolates has been demonstrated to be occluded on the envelope trimer prior to receptor engagement (9, 76) and becomes more accessible to 2F5 or 4E10 after CD4 binding (9, 53), presumably by substantial conformational changes induced in the overall Env spike architecture by CD4 interaction with the gp120 subunit, which is intimately associated with the gp41 subunit. To investigate the effect of VRC01 on accessibility of the gp41 MPER to antibody, we assessed 2F5 or 4E10 binding to JRFL Env-expressing cells in the presence, or absence, of the CD4bs ligand subset of VRC01, b12, and sCD4. Consistent with our prior findings and the shedding data presented here, both sCD4 and b12 enhanced 2F5 binding to the Env functional trimer (9) (Fig. 6B). In contrast, VRC01 had no effect on 2F5 binding to the functional spike (Fig. 6B), similar to the negligible effects on MPER recognition induced by the glycan-dependent negative-control antibody 2G12. Similarly, preincubation of sCD4 with the JRFL Env-expressing cells enhanced recognition of cell surface Env by the other MPER-directed neutralizing MAb, 4E10 (Fig. 6B). Preincubation of the cell surface Env with b12 slightly enhanced 4E10 binding, but VRC01 had no effect. These data again suggest that unlike CD4, binding by VRC01 to gp120 does not induce conformational changes in the context of the functional spike. Thus, although the conformational signatures of VRC01 and CD4 are very similar on the soluble gp120 monomer, they have distinctly different signatures on the architecture of the viral functional spike.
Lack of self-reactivity or polyreactivity.
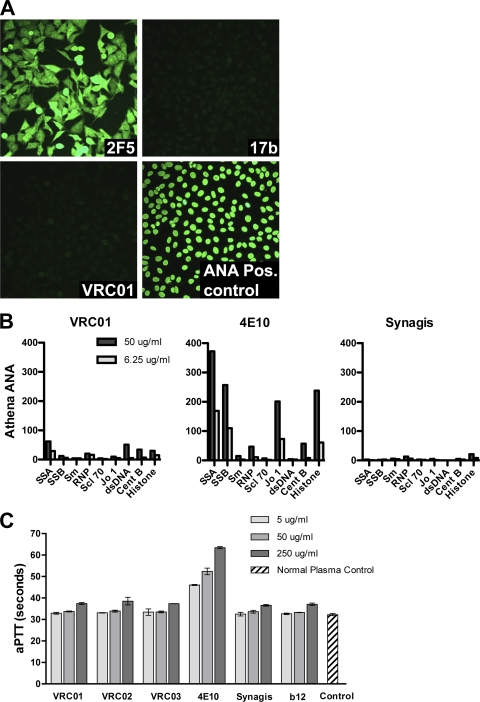
VRC01 was also tested for evidence of self- or polyreactivity. We observed no evidence of VRC01 binding to human HEp-2 epithelial cells, which are often used for the detection and identification of antinuclear antibodies (27) (Fig. 7A). VCR01 also displayed little or no reactivity by the Luminex assay to a diverse panel of autoantigens (Fig. 7B) and no reactivity by ELISA to cardiolipin (not shown). Lastly, VRC01 was tested for antiphospholipid activity using the clinical activated partial thromboplastin test (aPTT). No prolongation of the clotting time was observed for VRC01 (Fig. 7C), indicating that VRC01 does not have significant antiphospholipid binding properties.
Fig. 7.
Lack of VRC01 self-reactivity. (A) Reactivity of MAbs 2F5, 17b, and VRC01 with human HEp-2 epithelial cells. The MAb 2F5 served as a positive control and reacted in a diffuse cytoplasmic and nuclear pattern. (B) Luminex AtheNA Multi-Lyte ANA test for a panel of nuclear antigens: systemic lupus erythematosus autoantigens SSA and SSB, sphingomyelin (Sm), ribonucleoprotein (RNP), sclerosis autoantigen (Scl-70), histidine-tRNA ligase (Jo-1), double-stranded DNA (dsDNA), and centromere B (CentB). (C) The clinical assay, activated partial thromboplastin time (aPTT), was used to assess potential antiplatelet, antiphospholipid activity. The MAbs were added to normal sera, and the time to clot formation was measured. The clotting times for VRC01, VRC02, VRC03, b12, and the anti-RSV monoclonal antibody Synagis (palivizumab) were all in the normal range. In contrast, the known polyreactive MAb 4E10 caused a prolongation of the aPTT.
DISCUSSION
In this study, we assessed the interaction of the broadly neutralizing MAb VRC01 with the HIV-1 native viral spike, and where informative, to full-length monomeric gp120. We measured the effect of gp120 Ala scanning point mutations on virus neutralization and recognition of gp120 by VRC01 and the prototypic CD4bs ligands b12 and CD4-Ig. We also analyzed the contribution of amino acid variation within specific regions of Env to viral resistance to VRC01. In addition, we compared the thermodynamic properties of VRC01 and CD4-Ig (or sCD4) and assessed their impact on the conformation of gp120 and the native viral spike. Finally, we determined that there was little detectable interaction of the human MAb VRC01 with human antigens commonly recognized by self-reactive antibodies that may have escaped the normal regulatory mechanisms of B-cell self-tolerance.
There are several important findings highlighted by the results presented in our current study. We confirmed that mutations of structurally defined contact residues in loop D (N terminal to the V3 region), the CD4 binding loop, and the V5-β24-α5 region diminished VRC01-mediated binding or neutralization. With regard to natural resistance, in most cases, combinations of mutations at these sites were required to restore full VRC01 neutralization sensitivity. Interestingly, many Ala substitutions at noncontact residues increased the potency of CD4- or b12-mediated neutralization, but few of these substitutions enhanced VRC01-mediated neutralization. This suggests that VRC01 targets its epitope on the functional spike in a highly precise manner, overcoming the steric constraints that restrict the binding of many Env ligands. The observation that VRC01 targeting appears to be superior to the primary virus receptor itself is somewhat paradoxical, as one might expect that HIV-1 would maintain optimal recognition of its primary receptor, CD4. We speculate that selective pressure imposed by CD4 binding site antibodies may select for viruses that occlude optimal access to the CD4 binding region, even for CD4 itself. Obviously, the favorable energetics of the CD4-gp120 interaction allow the viral spike to engage host cell CD4, triggering the conformational changes in Env necessary for viral entry. In contrast, the relatively uncommon CD4bs ligand, VRC01, appears to target the functional spike in a manner that requires limited Env spike rearrangement to accomplish access and neutralization. The lack of VRC01-induced gp120 shedding and the lack of conformational changes of the native virus spike observed after VRC01 binding are consistent with this interpretation. Somewhat paradoxically, VRC01-induced conformational changes on the free gp120 monomer generated a signature that was very similar to that of CD4, implying that the conformation of free gp120 is considerably different than its conformation on the functional spike. Alternatively, VRC01 may indeed induce conformational changes on the functional trimer that are not detected by the assays used in this study but that serve to constrain gp120 in a (low-energy) state not compatible with coreceptor interaction or liberation of the gp41 fusion peptide required for entry.
Our studies of the mechanism of neutralization resistance to VRC01 reveal that a minority of viruses do contain alterations in key contact sites that diminish VRC01 binding and neutralization. However, we found only one example of a viral isolate that appears to resist VRC01 neutralization by restricting the antibodies' access to the CD4bs. These observations are consistent with the model in which VRC01 has achieved nearly optimal access to the CD4bs on the functional spike (70, 73). We further defined specific aspects of neutralization resistance among a panel of 16 VRC01-resistant Env pseudoviruses. We demonstrated that full VRC01 neutralization sensitivity could be restored to several viruses possessing natural escape variations in their respective Envs. However, restoration of VRC01 sensitivity was accomplished only after multiple changes in three gp120 hotspots, suggesting that viral resistance might require multiple, and perhaps sequential, alterations. Ongoing studies will evaluate the effect of these VRC01 resistance mutations on the efficiency of CD4-mediated cell entry and on viral replicative fitness.
In our previous study, we reported that VRC01 could enhance CD4i antibody 17b and coreceptor CCR5 binding to monomeric gp120 (70). In the current study, we extended such analysis and observed similarly that the binding of other CD4i antibodies to gp120 was enhanced by prior VRC01 interaction, a property similar to that of CD4. However, when we compared the effects of VRC01 to sCD4 interaction in the context of the functional trimer by other means of analysis, again there were striking differences from the monomeric context. We found that sCD4 binding to the functional trimer can trigger gp120 shedding from the Env trimer and enhancement of gp41 MPER exposure, confirming previous work that CD4 can induce substantial rearrangements of the functional spike, likely necessary to permit entry. In contrast, we did not see the induction of gp120 shedding or enhancement of gp41 MPER exposure after VRC01 bound to the Env functional trimer. These data support a model in which VRC01 locks the Env functional trimer into a low-energy conformation (or trough) from which it cannot “escape” to undergo the further rearrangements that eventually coalesce in virus entry. Finally, the observation that VRC01 does not display substantial self-reactive or polyreactive properties will facilitate the use of passive protection studies of nonhuman primates and humans. This outcome is perhaps not so surprising since VRC01, as noted previously, does not possess an exceptionally long or hydrophobic HCDR3 loop (28), both of which have correlated with some level of self-reactivity previously.
In summary, the data presented here support the concept that VRC01 interacts with the functional spike in a manner distinct from that of CD4. VRC01 achieves potent neutralization by precisely targeting a highly conserved region of the CD4bs without requiring the alterations of the Env functional spike configuration that occur upon CD4 ligation. This helps to explain how VRC01 can access the CD4bs on the large majority of virus isolates and why VRC01 resistance based on the quaternary structure of the HIV-1 Env is uncommon. These studies highlight the unique features of this broadly neutralizing CD4bs-directed antibody and point to further efforts regarding HIV immunogen design based on its gp120 epitope.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Christian Poulsen for help with molecular modeling, Christina Corbaci for help with manuscript preparation, James Robinson (Tulane University) for providing antibodies, including C11, 17b, 48D, 2.1c, and A32, Susan Zolla-Pazner (New York University) for providing antibody 447D, and Joseph Sodroski (Dana Farber Cancer Institute) for providing the plasmid encoding CD4-Ig.
This study was supported by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and the International AIDS Vaccine Initiative.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Alkhatib G., et al. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958 [DOI] [PubMed] [Google Scholar]
- 2. Binley J. M., et al. 2010. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J. Virol. 84:5637–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Binley J. M., et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blish C. A., Nguyen M. A., Overbaugh J. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 5:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchacher A., et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10:359–369 [DOI] [PubMed] [Google Scholar]
- 6. Bugelski P. J., Ellens H., Hart T. K., Kirsh R. L. 1991. Soluble CD4 and dextran sulfate mediate release of gp120 from HIV-1: implications for clinical trials. J. Acquir. Immune Defic. Syndr. 4:923–924 [PubMed] [Google Scholar]
- 7. Burton D. R., et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 8. Calarese D. A., et al. 2005. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. U. S. A. 102:13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakrabarti B. K., et al. 2011. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res. Hum. Retroviruses [Epub ahead of print.] doi:10.1089/aid.2010.0281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan D. C., Fass D., Berger J. M., Kim P. S. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273 [DOI] [PubMed] [Google Scholar]
- 11. Choe H., et al. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148 [DOI] [PubMed] [Google Scholar]
- 12. Dalgleish A. G., et al. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767 [DOI] [PubMed] [Google Scholar]
- 13. Deng H., et al. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666 [DOI] [PubMed] [Google Scholar]
- 14. Dhillon A. K., et al. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 81:6548–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diskin R., Marcovecchio P. M., Bjorkman P. J. 2010. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat. Struct. Mol. Biol. 17:608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doores K. J., Burton D. R. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84:10510–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doranz B. J., et al. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158 [DOI] [PubMed] [Google Scholar]
- 18. Doria-Rose N. A., et al. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dragic T., et al. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673 [DOI] [PubMed] [Google Scholar]
- 20. Earl P. L., et al. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Earl P. L., Doms R. W., Moss B. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 87:648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feng Y., Broder C. C., Kennedy P. E., Berger E. A. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877 [DOI] [PubMed] [Google Scholar]
- 23. Furuta R. A., Wild C. T., Weng Y., Weiss C. D. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276–279 [DOI] [PubMed] [Google Scholar]
- 24. Gray E. S., et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gray E. S., et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hart T. K., et al. 1991. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U. S. A. 88:2189–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haynes B. F., et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906–1908 [DOI] [PubMed] [Google Scholar]
- 28. Haynes B. F., Moody M. A., Verkoczy L., Kelsoe G., Alam S. M. 2005. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum. Antibodies 14:59–67 [PMC free article] [PubMed] [Google Scholar]
- 29. He Y., et al. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 77:1666–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klatzmann D., et al. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768 [DOI] [PubMed] [Google Scholar]
- 31. Koch M., et al. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387–400 [DOI] [PubMed] [Google Scholar]
- 32. Kolchinsky P., Kiprilov E., Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 75:2041–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koshiba T., Chan D. C. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J. Biol. Chem. 278:7573–7579 [DOI] [PubMed] [Google Scholar]
- 34. Kwong P. D., et al. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 35. Li M., et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y., et al. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., et al. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Y., et al. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu M., Blacklow S. C., Kim P. S. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075–1082 [DOI] [PubMed] [Google Scholar]
- 41. Mascola J. R., Montefiori D. C. 2010. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 28:413–444 [DOI] [PubMed] [Google Scholar]
- 42. McDougal J. S., et al. 1986. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science 231:382–385 [DOI] [PubMed] [Google Scholar]
- 43. Moog C., Fleury H. J., Pellegrin I., Kirn A., Aubertin A. M. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J. Virol. 71:3734–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moore J. P., McKeating J. A., Norton W. A., Sattentau Q. J. 1991. Direct measurement of soluble CD4 binding to human immunodeficiency virus type 1 virions: gp120 dissociation and its implications for virus-cell binding and fusion reactions and their neutralization by soluble CD4. J. Virol. 65:1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moore J. P., McKeating J. A., Weiss R. A., Sattentau Q. J. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142 [DOI] [PubMed] [Google Scholar]
- 46. Moore J. P., Sodroski J. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Myszka D. G., et al. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. U. S. A. 97:9026–9031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Orloff S. L., Kennedy M. S., Belperron A. A., Maddon P. J., McDougal J. S. 1993. Two mechanisms of soluble CD4 (sCD4)-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) infectivity and their relation to primary HIV-1 isolates with reduced sensitivity to sCD4. J. Virol. 67:1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pancera M., Wyatt R. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145–156 [DOI] [PubMed] [Google Scholar]
- 50. Pantophlet R., Burton D. R. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24:739–769 [DOI] [PubMed] [Google Scholar]
- 51. Pantophlet R., et al. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Parker C. E., et al. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75:10906–10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Peachman K. K., Wieczorek L., Polonis V. R., Alving C. R., Rao M. 2010. The effect of sCD4 on the binding and accessibility of HIV-1 gp41 MPER epitopes to human monoclonal antibodies. Virology 408:213–223 [DOI] [PubMed] [Google Scholar]
- 54. Pinter A., et al. 1989. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J. Virol. 63:2674–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Richman D. D., Wrin T., Little S. J., Petropoulos C. J. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sanders R. W., et al. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sather D. N., et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scanlan C. N., et al. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scanlan C. N., et al. 2007. Inhibition of mammalian glycan biosynthesis produces non-self antigens for a broadly neutralising, HIV-1 specific antibody. J. Mol. Biol. 372:16–22 [DOI] [PubMed] [Google Scholar]
- 60. Si Z., et al. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 101:5036–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Simek M. D., et al. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stamatatos L., Morris L., Burton D. R., Mascola J. R. 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15:866–870 [DOI] [PubMed] [Google Scholar]
- 63. Trkola A., et al. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co- receptor CCR-5. Nature 384:184–187 [DOI] [PubMed] [Google Scholar]
- 64. Trkola A., et al. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Walker L. M., et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wei X., et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 67. Weiss C. D., Levy J. A., White J. M. 1990. Oligomeric organization of gp120 on infectious human immunodeficiency virus type 1 particles. J. Virol. 64:5674–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430 [DOI] [PubMed] [Google Scholar]
- 69. Wu L., et al. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183 [DOI] [PubMed] [Google Scholar]
- 70. Wu X., et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu X., et al. 2009. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J. Virol. 83:10892–10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wyatt R., Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888 [DOI] [PubMed] [Google Scholar]
- 73. Zhou T., et al. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhou T., et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zwick M. B., et al. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 79:1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zwick M. B., et al. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892–10905 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.