Abstract
The current model of flavivirus membrane fusion is based on atomic structures of truncated forms of the viral fusion protein E in its dimeric prefusion and trimeric postfusion conformations. These structures lack the two transmembrane domains (TMDs) of E as well as the so-called stem, believed to be involved in an intra- and intermolecular zippering reaction within the E trimer during the fusion process. In order to gain experimental evidence for the functional role of the stem in flavivirus membrane fusion, we performed a mutagenesis study with recombinant subviral particles (RSPs) of tick-borne encephalitis virus, which have fusion properties similar to those of whole infectious virions and are an established model for viral fusion. Mutations were introduced into the stem as well as that part of E predicted to interact with the stem during zippering, and the effect of these mutations was analyzed with respect to fusion peptide interactions with target cells, E protein trimerization, trimer stability, and membrane fusion in an in vitro liposome fusion assay. Our data provide evidence for a molecular interaction between a conserved phenylalanine at the N-terminal end of the stem and a pocket in domain II of E, which appears to be essential for the positioning of the stem in an orientation that allows zippering and the formation of a structure in which the TMDs can interact as required for efficient fusion.
INTRODUCTION
Flaviviruses form a genus within the family Flaviviridae and include important human pathogens such as the dengue viruses, Japanese encephalitis virus, West Nile virus, yellow fever virus, and tick-borne encephalitis virus (TBEV) (41). These small enveloped viruses enter cells by receptor-mediated endocytosis and fuse their membrane with that of the endosome. Fusion is triggered by the acidic pH of this compartment and is mediated by the viral envelope protein E, a class II viral fusion protein (37). During virus assembly, heterodimers of E and the precursor membrane protein prM together with the nucleocapsid bud into the lumen of the endoplasmic reticulum (ER), thus leading to the formation of nonfusogenic immature particles (10, 24). In the course of their exocytosis, the prM protein is processed by the host protease furin in the trans-Golgi network, and only the membrane-anchored part of M remains associated with secreted mature virions (42, 43). The maturation process results in the formation of smooth-surfaced particles with a herringbone-like arrangement of metastable E homodimers on the virus surface (18, 28).
X-ray crystallography of carboxy-terminally truncated soluble E (sE) proteins revealed an organization into three structurally distinct domains (domain I [DI], DII, and DIII) linked by short flexible regions (Fig. 1A) (15, 25, 27, 30, 31, 46). DII contains the highly conserved fusion peptide (FP) loop at its tip. In the full-length protein, DIII is followed by the ∼50-amino-acid-long “stem” that connects the ectodomain to the membrane anchor and consists of two amphipathic alpha helices (H1 and H2) flanking a central conserved sequence (CS) element (Fig. 1A and F) (45). As shown by cryo-electron microscopy (EM) reconstructions of mature dengue virus particles, the stem lies essentially flat on the viral membrane underneath the ectodomain and is half buried in the outer lipid leaflet (45).
Fig. 1.
Representations of the flavivirus fusion model (A to E) and sequence alignment of the stem regions of the E fusion proteins of different flaviviruses (F) and the postfusion E trimer (G and H). Color coding for E is as follows: red, domain I (DI); yellow, domain II (DII); blue, domain III (DIII); orange, fusion peptide; cyan, stem helix (helix 1 [H1] and H2); green, transmembrane domain (TMD). Color coding for membranes is as follows: light gray, virus; dark gray, cell. (A) E dimer on the virion surface. (B) Low-pH-induced E dimer dissociation, lifting of the stem, and FP interaction with the target membrane. (C) DIII relocation to the side of the molecule, E protein trimerization, and initiation of stem zippering. (D) Hemifusion intermediate with fused outer leaflets. (E) Postfusion hairpin-like E trimer with FPs and TMDs at the same side of the molecule. (F) Amino acid sequence alignment of DIIs (amino acids 212 to 230) and stem regions (amino acids 402 to 449) of several flavivirus E proteins (TBEV numbering): TBEV (GenBank accession number U27495), Powassan virus (POWV) (GenBank accession number L06436), dengue virus (DENV) type 1 (GenBank accession number FJ687432), DENV type 2 (GenBank accession number NC_001474), DENV type 3 (GenBank accession number FJ850055), DENV type 4 (GenBank accession number AY618990), Japanese encephalitis virus (GenBank accession number EF571853), West Nile virus (WNV) (GenBank accession number DQ211652), and yellow fever virus (YFV) (GenBank accession number AY640589). Identical amino acids are highlighted in yellow. Residues which were mutated in this study are labeled with black stars. The predicted stem helices and the conserved sequence (CS) between the helices are indicated below the alignment. (G) Surface representation of truncated sE postfusion trimer of TBEV (side view) (Protein Data Bank [PDB] accession number 1URZ). Stem helices added schematically to the structure are shown as cylinders in a groove provided by neighboring DII domains. (H) Schematic representation of the full-length E trimer (side view), displaying the mutations introduced into the stem (right side) and DII (left side). Mutations that gave rise to mature particles are labeled in black, and mutations that abolished either particle secretion or maturation are labeled in gray.
During cell entry E functions as a receptor-binding and fusion protein (reviewed in reference 17). The acidic pH in the endosome induces structural changes in E that mediate fusion. Based on the pre- and postfusion sE crystal structures that lack the stem-anchor region (5, 15, 25–27, 29–31, 46) and biochemical analyses (22, 36, 40), a mechanistic model for flavivirus membrane fusion has been developed (Fig. 1A to E) (11). The process is initiated by the acidic-pH-induced dissociation of the dimers that leads to the exposure of the FP and its insertion into the target membrane (Fig. 1B). This step is presumably supported by the stem that lifts off the viral membrane, as suggested by cryo-EM studies using West Nile virus and monoclonal antibody (MAb) fragments (16) (Fig. 1B). The fusion model then proposes that a trimeric intermediate is formed in which DIII is folded back to the side of the molecule but with the stem not in its final position (Fig. 1C). The following zippering of the stem along DIIs of this intermediate, starting at the carboxy terminus of DIII and proceeding toward the FPs, together with DIII relocation, is believed to drive membrane fusion (Fig. 1C to E). Consistent with this hypothesis, DIII and stem-derived peptides were shown previously to inhibit fusion and/or infectivity (13, 19, 22, 33, 34). Specifically, helix 2 peptides were demonstrated to be able to bind to virions already at a neutral pH, presumably by interacting with the viral membrane, and to block fusion by interacting with an E intermediate generated in the course of acidic-pH-induced conformational changes (33, 34). The arrangement of the stem in the postfusion trimer is not known, but modeling of the stem helix 1 suggests that it follows a groove formed by neighboring DIIs (Fig. 1G) and that conserved stem residues are directed toward DII of the same polypeptide chain (5). This includes F403, which was supposed to interact with a conserved patch in DII that forms a pocket in the same protein subunit (5).
Since structural or experimental data for stem zippering (Fig. 1C and D) were not available so far, we targeted this reaction in a mutagenesis study with recombinant subviral particles (RSPs) of TBEV. These capsid-free noninfectious particles contain a lipid bilayer in which the two viral glycoproteins M and E are integrated. Like virions, they are assembled in the ER in an immature form and undergo the same maturation prM-to-M cleavage in the secretory pathway (32). Most importantly, they display fusion characteristics similar to those of virions and represent an established model for studying fusion and fusion-related processes of flaviviruses (2, 7–9). Using this system we introduced mutations into the stem as well as that part of DII predicted to interact with the stem in the trimer (Fig. 1H) and analyzed the effects of the mutations on different stages of the fusion process.
None of these modifications had an effect on the early stages of membrane fusion, including FP-mediated attachment to target membranes and E trimerization. However, our data suggest that the proper positioning of the stem for initiating the zippering reaction is critically dependent on the interaction of F403 at the beginning of the stem with a pocket in DII that is formed by W219 and L223.
MATERIALS AND METHODS
Mutagenesis of RSPs.
Mutations were introduced into the SV-PE WT plasmid by site-directed mutagenesis using the Gene Tailor kit (Invitrogen) according to the manufacturer's instructions. SV-PE WT contains the TBEV prM and E genes under the control of the simian virus 40 (SV40) early promoter (1). To confirm the presence of only the desired mutations, the whole prM/E sequence of each mutant plasmid was verified.
Production of RSPs.
The recombinant plasmids were electroporated into COS-1 cells for RSP production as described previously (32). At 48 h posttransfection, RSPs were harvested from cell culture supernatants and pelleted by ultracentrifugation. For coflotation assays, RSPs were further purified by sucrose gradient centrifugation (32). For membrane fusion assays, RSPs were metabolically labeled with 1-pyrene hexadecanoic acid (Invitrogen) as described previously (7).
Quality controls of RSPs.
RSPs secreted from transfected cells were quantified in a four-layer enzyme-linked immunosorbent assay (ELISA) after solubilization with 0.4% sodium dodecyl sulfate (SDS) at 65°C for 30 min (12), and their particulate nature was assessed by ultracentrifugation (32). The hemagglutination (HA) activity of RSPs was measured at pH 6.4 according to a method described previously by Clarke and Casals using goose erythrocytes (6). The maturation state (presence of prM) of RSPs was analyzed by Western blotting using prM-specific polyclonal sera (8, 9) and by ELISA using prM-specific monoclonal antibody (MAb) 8H1 (14). The structural integrity and proper folding of E were confirmed by epitope mapping with a panel of E-protein-specific MAbs (8, 9).
Liposomes.
Phosphatidylcholine, phosphatidylethanolamine (Avanti Polar Lipids), and cholesterol (Sigma-Aldrich) were mixed at a molar ratio of 1:1:2 in chloroform, dried to a thin film in a high vacuum, and hydrated in liposome buffer (10 mM triethanolamine [TEA], 140 mM NaCl [pH 8.0]) by 5 freeze-thaw cycles (38). Prior to use, liposomes were subjected to 21 extrusion cycles through polycarbonate membranes (200-nm pore size) using a LiposoFast-Basic extruder (Avestin).
Coflotation assay.
Purified RSPs were incubated with liposomes at a ratio of 1 μg E to 200 nM lipids (8, 9). The mixture was acidified to pH 5.4 by the addition of 300 mM morpholineethanesulfonic acid (MES) and incubated for 15 min at 37°C. The samples were back-neutralized with 150 mM TEA to pH 7.8 and mixed with sucrose to yield a concentration of 20% (wt/wt). This sample (0.6 ml) was put onto a 1-ml 50% sucrose cushion and further overlaid with 1.4 ml of 15% sucrose and 1 ml of 5% sucrose. All sucrose solutions were prepared in TAN buffer (50 mM TEA, 100 mM NaCl) (pH 8.0). The gradients were centrifuged for 2 h at 4°C at 50,000 rpm in an SW55 Beckman Coulter rotor. Fractions of 200 μl were collected by upward displacement with a Piston gradient fractionator (BioComp Instruments Inc.), and the E protein in each fraction was determined by a four-layer ELISA after treatment with 0.4% SDS at 65°C for 30 min (12).
Sedimentation analysis.
RSPs (3 μg E protein) in TAN buffer were incubated for 10 min at 37°C either at pH 8.0 or after acidification with MES at pH 5.4 (8, 9). The samples were then back-neutralized with 150 mM TEA and solubilized with Triton X-100 at a final concentration of 1%. The mixtures were incubated for 1 h at room temperature and applied onto 7 to 20% (wt/wt) continuous sucrose gradients containing 0.1% Triton X-100. The gradients were centrifuged for 20 h at 38,000 rpm at 15°C in an SW40 rotor (Beckman Coulter). Fractions of 600 μl were collected by upward displacement and analyzed by a quantitative four-layer ELISA (12).
To test the thermostability of trimer preparations, acidic-pH-treated and solubilized RSPs were incubated for 10 min at 70°C before sedimentation analysis (8, 9).
Fusion assay.
The fusion of pyrene-labeled RSPs with liposomes was measured by monitoring the decrease in pyrene excimer fluorescence caused by the dilution of the RSP membrane (inserted probe) into unlabeled liposome membranes as described previously (8, 9). Fluorescence was recorded continuously for 60 s at 480 nm with a Perkin-Elmer LS 50B fluorescence spectrophotometer at an excitation wavelength of 343 nm. RSPs were mixed with liposomes in a continuously stirred fluorimeter cuvette at 37°C and acidified to pH 5.4 by the addition of 300 mM MES. The initial excimer fluorescence after mixing was defined as 0% fusion. To determine the residual excimer fluorescence at an infinite dilution of the probe (defined as 100% fusion for calculating the fusion extents), the detergent n-octa(ethylene glycol)n-dodecyl monoether (Sigma-Aldrich) was added to a final concentration of 10 mM to disperse the viral and liposomal membranes.
RESULTS
Generation of stem and DII mutant RSPs.
To provide direct experimental data for the role of the stem in flavivirus membrane fusion, we used TBEV RSPs and engineered single and double mutations into the stem and that part of DII which was predicted to form the interface with the stem in the postfusion trimer (5). The targets for mutagenesis were conserved residues in both helices of the stem, in the linker between the two helices, and in DII (Fig. 1H and Table 1). The chosen substitutions were either predicted not to affect the overall folding of the protein (4) (W219Y, L223I, L223V, T406A, and G434N) or occurred naturally in a certain flavivirus strain (F403I in dengue virus 2 New Guinea C). In addition, we introduced mutations that might theoretically influence the local structure more strongly (W219A, W219I, L223A, L223Y, F403A, T406I, and W421I).
Table 1.
Quality controls of TBEV mutant RSPsa
| Mutation | Location(s) in E | Particle secretion | Maturation cleavage | Reactivity with MAbs | Oligomeric state of E | Hemagglutination activity |
|---|---|---|---|---|---|---|
| L223I | DII | Reduced | + | + | Dimer | + |
| F403I | Stem (H1) | + | + | + | Dimer | + |
| T406A | Stem (H1) | + | + | + | Dimer | + |
| W421I | Stem (CS) | + | + | + | Dimer | + |
| G434N | Stem (H2) | + | + | + | Dimer | + |
| L223I-F403I | DII + H1 | Reduced | + | + | Dimer | + |
| L223A | DII | No | NA | NA | NA | NA |
| L223V | DII | No | NA | NA | NA | NA |
| L223Y | DII | No | NA | NA | NA | NA |
| W219A | DII | No | NA | NA | NA | NA |
| W219V | DII | No | NA | NA | NA | NA |
| W219Y | DII | No | NA | NA | NA | NA |
| W219I | DII | No | NA | NA | NA | NA |
| F403A | Stem (H1) | No | NA | NA | NA | NA |
| T406I | Stem (H1) | + | Reduced | NA | NA | NA |
H1, helix 1; CS, conserved sequence element in the stem; H2, helix 2; +, WT-like activity; NA, not analyzed.
Since mutations can potentially also interfere with processes unrelated to fusion, such as assembly and/or maturation, we subjected all mutant RSPs to a set of quality control experiments to ensure that particle formation and prM cleavage as well as the proper folding of E were unaffected (see Materials and Methods). These controls included (i) the ultracentrifugation of transfected cell culture supernatants to verify that particles were formed, (ii) Western blots with prM-specific polyclonal sera and ELISAs with a prM-specific monoclonal antibody (MAb) to confirm wild-type (WT)-like prM cleavage, (iii) sedimentation analyses to control the dimeric state of E, and (iv) reactivity with a set of conformation-sensitive MAbs to assess the proper folding of E. RSPs that were indistinguishable from the WT according to the criteria listed above were obtained with mutations in DII at position 223 and in the stem at positions 403, 406, 421, and 434 (Table 1 and Fig. 1H). Nine further mutations either abolished particle formation or impaired prM cleavage and could therefore not be further analyzed with respect to the stem-zippering reaction (Table 1 and Fig. 1H).
No effect of the introduced mutations on early steps of membrane fusion.
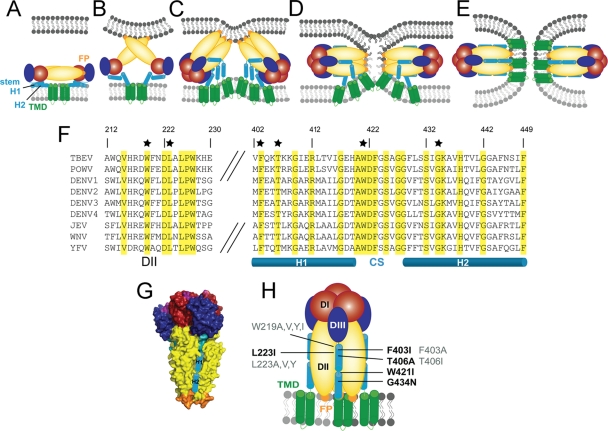
To allow conclusions regarding the effect of the mutations on stem zippering along the body of the trimer (Fig. 1C and D), it was first necessary to find out whether any of the mutations affected stages of fusion that occur before this reaction. According to the fusion model, these steps include E dimer dissociation, FP exposure, and FP-mediated membrane binding as well as trimer formation (Fig. 1B and C). FP exposure and the stable insertion of the FPs into target membranes were assessed by coflotation with liposomes, and the dimer-to-trimer transition was assessed by sedimentation analysis (Materials and Methods). Previous studies have shown that these methods allow the quantification of an impairment of early fusion functions (2, 9). The acidification of WT and mutant RSPs in the presence of liposomes led to a quantitative association with the target membranes in all cases (Fig. 2A). In addition, all mutants were able to undergo the dimer-to-trimer transition to the same extent as that of the WT, as revealed by sedimentation analysis of acidic-pH-treated and solubilized WT and mutant RSPs in sucrose gradients in the presence of Triton X-100 (Fig. 2B and see Materials and Methods).
Fig. 2.
Low-pH-induced coflotation (A) and E protein trimerization (B) of WT and mutant RSPs. (A) RSPs were exposed to acidic pH in the presence of liposomes, back-neutralized, and subjected to sucrose step gradient centrifugation. The gradients were fractionated, and the amount of E protein in each fraction was determined by a quantitative four-layer ELISA. Results are expressed as a percentage of E found in the top fractions relative to the WT (set to 100%). (B) RSPs were exposed to acidic pH, back-neutralized, solubilized, and subjected to sedimentation in sucrose gradients. The gradients were fractionated, and the amount of E protein in each fraction was determined by a four-layer ELISA. Results are expressed as a percentage of E present in the trimer peak fractions relative to the WT (set to 100%). The data display the means of data from two independent experiments, and the error bars represent the observed ranges.
Taken together, these experiments indicated that the introduced mutations did not affect those steps of fusion that precede the putative stem-zippering reaction.
Thermostability of mutant trimers.
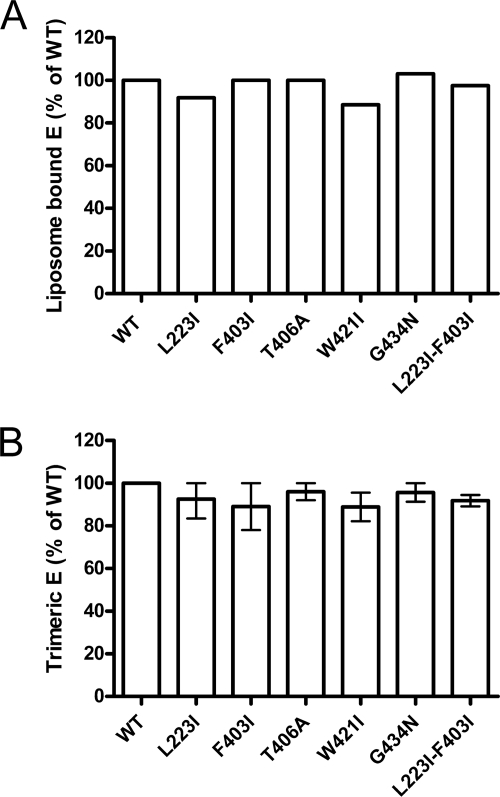
It was shown previously that full-length E trimers were more thermostable than truncated sE trimers that lack the whole stem-anchor region (39). In order to assess whether the introduced mutations had an effect on the stability of mutant trimers, we performed thermal denaturation experiments. For this purpose, WT and mutant RSPs were exposed to acidic pH, solubilized, heated for 10 min to 70°C, and then subjected to sedimentation analysis as described in Materials and Methods. Under these conditions, the sedimentation behavior of the mutant T406A trimer was unaffected up to 70°C like the WT, and the W421I as well as G434N trimers displayed only slightly reduced trimer stabilities (Fig. 3). In contrast, trimers containing the L223I (DII) or F403I (stem H1) single mutations were significantly denatured at this temperature, as indicated by a smaller residual trimer peak (Fig. 3) and aggregated material accumulated in the pellets of the gradients (data not shown).
Fig. 3.
Thermostability of WT and mutant E trimers. Trimers were exposed to 37°C or 70°C and subjected to rate-zonal sucrose gradient centrifugation. The extents of trimer stability of WT and mutant trimers are expressed as percentages of E found in the trimer peak fraction after incubation at 70°C relative to the amount of E present in the trimer peak fraction after incubation at 37°C (set to 100%). The data display the means of data from two independent experiments, and the error bars represent the observed ranges.
Together with the data presented in Fig. 2B, these results provide experimental evidence that the disturbance of specific contacts between DII and the stem can lead to a reduced trimer stability, although the same changes did not affect the acidic-pH-induced trimerization per se.
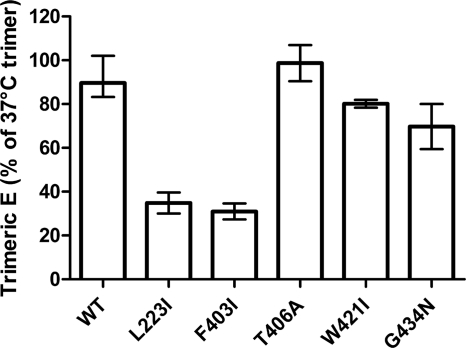
Fusion activity of mutant RSPs and partial compensation of fusion deficiency.
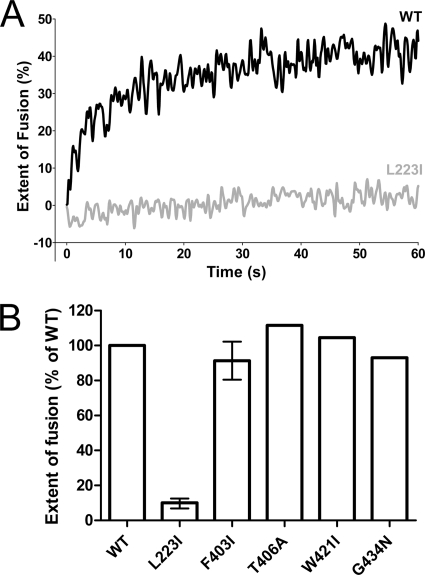
To assess the effect of mutations in the proposed stem-DII interface on the overall fusion activity, we used an in vitro fusion assay with pyrene-labeled RSPs and liposomes (see Materials and Methods). Only the replacement of L223 by isoleucine in DII resulted in a strong impairment of both the rate and the extent of fusion (Fig. 4A), whereas all of the stem mutants (F403I, T406A, W421I, and G434N) displayed an unimpaired fusion activity and behaved like the WT (Fig. 4B). Especially for the F403I mutant, this was unexpected, because its E trimer stability was as reduced as that of the L223I mutant (Fig. 3).
Fig. 4.
Fusion activity of pyrene-labeled WT and mutant RSPs with liposomes at acidic pH. (A) Kinetic fusion curves of WT RSP (black line) and the L223I mutant (gray line) at acidic pH. (B) Extent of fusion of mutant RSPs relative to that of WT RSP (set to 100%) at acidic pH after 60 s. Experiments with mutants that exhibited reduced trimer stability (L223I and F403I) were performed at least twice, and the error bars represent the observed ranges.
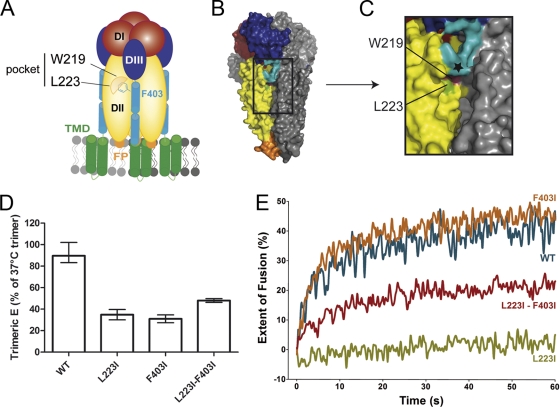
In the trimer, L223 forms part of a pocket (together with W219) that was previously suggested to accommodate F403 and thus might allow the initiation of zippering (Fig. 5A to C). We therefore speculated that the L223I mutation changed the geometry of the mutated pocket in such a way that it could no longer serve its function of correctly positioning H1 for zippering along DII through its interaction with F403. In order to corroborate this hypothesis, we created a double mutant combining the L223I substitution and the already characterized and fusion-active F403I mutation in one protein. We reasoned that isoleucine, due to its smaller size than that of phenylalanine, might compensate for the defects observed for the single L223I mutant by encountering less steric hindrance in the mutated W219/L223I pocket. As shown in Fig. 5, a partial compensatory effect was indeed observed with the double mutant, with respect to both trimer stability (Fig. 5D) and the overall fusion activity (Fig. 5E).
Fig. 5.
Schematic representation and structures of the predicted DII/stem interaction site (A to C), mutant trimer thermostability (D), and fusion activity (E) compared to those of the WT. (A) DII residues W219 and L223 form a pocket hypothesized to interact with the stem residue F403 in the postfusion trimer. (B and C) Surface view of the TBEV sE trimer (PDB accession number 1URZ) (B) and the details of the W219/L223 pocket in the groove formed by DIIs of neighboring subunits (C). Color coding is as follows: magenta, W219; green, L223 (PDB accession number 1URZ). The C terminus of the crystallized sE trimer is indicated by a black star. (D) Thermostability of the trimers was analyzed as described in the legend of Fig. 3. (E) Kinetic fusion curve of pyrene-labeled WT RSP (blue line) and L223I mutant (green line), F403I mutant (orange line), and L223I-F403I double-mutant RSPs (red line). The data are representative examples of data from two or more independent experiments.
These experiments are consistent with a molecular interaction in which F403 in H1 interdigitates with the pocket formed by DII residues W219 and L223 and thus places the stem in a position that allows further zippering along the grooves between DIIs.
DISCUSSION
In this study, we provide experimental evidence for a specific molecular interaction between the stem and DII of the TBEV fusion protein that appears to be essential for initiating the so-called zippering reaction in the late phase of the flavivirus fusion process. Specifically, we propose that F403, following the C-terminal end of DIII and located at the beginning of H1 of the stem, interdigitates with a pocket in DII (formed by W219 and L223) after the acidic-pH-induced trimerization and relocation of DIII, thus positioning the stem in such a way that it can zipper down completely along DII to form the final postfusion conformation of E (Fig. 5A). This model is based on the fact that the changing of the pocket by the replacement of L223 with isoleucine results in a complete loss of fusion activity and a lower stability of the full-length E trimer, although this mutation has an effect on neither FP exposure, interactions with target membranes, nor E trimerization. Since L and I have similar volumes and hydrophobicities, these effects are apparently due to the isomeric differences between the two amino acids that affect the interaction with F403. Consistent with the interpretation that the structurally modified pocket would no longer be able to accommodate the bulky F403 (with a volume of 190 Å3 [44]) for the proper positioning of the stem, we found that the defects observed for the L223I mutant could be at least partially compensated for by replacing F403 with isoleucine, having a volume of only 167 Å3 (44).
In previous studies, it was found that mutations affecting trimer stability also impaired fusion activity (8, 9). It was therefore surprising that the F403I mutant, despite its reduced trimer stability, had retained its wild-type-like fusion activity. One possible explanation for this seeming discrepancy could be that the most important function of the zippering reaction is not so much to provide additional energy but to position the stem in such a way that the transmembrane domains (TMDs), following the C terminus of the stem, can interact to form the final postfusion conformation. Recent work has indeed shown that interactions of the TMDs are essential for the final stabilization of the full-length postfusion trimer and efficient fusion (8). In the context of the findings with the F403I mutant, one might therefore speculate that most of the energy required for membrane fusion is already provided by the relocation of DIII and the dimer-trimer conversion of the E ectodomain. Previous thermostability analyses had shown that truncated E trimers lacking the stem and TMDs are already significantly more stable than full-length prefusion E dimers albeit less stable than full-length trimers (35, 39). Also, previous work with the structurally closely related class II fusion protein E1 of the alphavirus Semliki Forest virus (SFV) indicated that an E1 homotrimer in which DIII, but not the stem, has completed its packing was already as SDS and trypsin resistant as the final homotrimer (21). It is therefore possible that the F403I mutation in the stem, although somewhat decreasing trimer stability, still allowed the zippering reaction, which is a prerequisite for bringing the TMDs into proper contact. Indeed, experimental evidence suggests that both intra- and intermolecular TMD interactions contribute to the final stages of membrane fusion (8).
It was an intriguing finding of our study that none of the mutations introduced into the stem, including F403I, which displayed reduced trimer stability, resulted in an impairment of fusion. This is reminiscent of data obtained with the E1 fusion protein of SFV. The introduction of major changes into the stem of this protein had either no or only slight effects on fusion, but several of these mutations affected the budding of virus particles (20). Similarly, we identified stem mutations that interfered with either particle formation (F403A) or the prM maturation cleavage (T406A) of immature particles. These findings are consistent with previous work in which we demonstrated that the stem can contribute to the stabilization of the prM-E heterodimers formed in the ER as building blocks of immature particles (3) and a recent mutagenesis study in which the disruption of the helical structure of the stem was shown to affect not only the assembly of dengue virus-like particles but also that of whole infectious viruses (23). The fact that mutations in DII also had an effect on particle formation, through an as-yet-unknown mechanism, corroborates the importance of assembly/maturation controls of RSPs to be included in functional studies of membrane fusion.
It has to be kept in mind that the RSP liposome fusion assay used in this study measures lipid mixing and not fusion pore formation. It can therefore not be excluded that some of the stem mutations analyzed in our study, despite their WT-like fusion properties in the liposome fusion assay, had a defect in the final merger of both membranes. This question could be the subject of future investigations with similar mutations introduced into whole infectious virions that would allow the use of content mixing assays and thus would allow conclusions to be drawn about effects on fusion pore formation. The limiting factor may be the amount of virus required for such studies, given the fact that mutations affecting fusion and/or assembly are prone to reduce the replication capacity of the virus and, consequently, virus yields, as observed in recent studies with TMD mutants of TBEV (8).
ACKNOWLEDGMENT
This work was supported by the Austrian Science Fund (FWF) (P19843-B13).
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Allison S. L., Mandl C. W., Kunz C., Heinz F. X. 1994. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes 8:187–198 [DOI] [PubMed] [Google Scholar]
- 2. Allison S. L., Schalich J., Stiasny K., Mandl C. W., Heinz F. X. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allison S. L., Stiasny K., Stadler K., Mandl C. W., Heinz F. X. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bordo D., Argos P. 1991. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J. Mol. Biol. 217:721–729 [DOI] [PubMed] [Google Scholar]
- 5. Bressanelli S., et al. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clarke D. H., Casals J. 1958. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg. 7:561–573 [DOI] [PubMed] [Google Scholar]
- 7. Corver J., et al. 2000. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology 269:37–46 [DOI] [PubMed] [Google Scholar]
- 8. Fritz R., et al. 2011. The unique transmembrane hairpin of the flavivirus fusion protein E is essential for membrane fusion. J. Virol. 85:4377–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fritz R., Stiasny K., Heinz F. X. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 183:353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guirakhoo F., Heinz F. X., Mandl C. W., Holzmann H., Kunz C. 1991. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J. Gen. Virol. 72(Pt. 6):1323–1329 [DOI] [PubMed] [Google Scholar]
- 11. Harrison S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heinz F. X., et al. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109–117 [DOI] [PubMed] [Google Scholar]
- 13. Hrobowski Y. M., Garry R. F., Michael S. F. 2005. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iacono-Connors L. C., Smith J. F., Ksiazek T. G., Kelley C. L., Schmaljohn C. S. 1996. Characterization of Langat virus antigenic determinants defined by monoclonal antibodies to E, NS1 and preM and identification of a protective, non-neutralizing preM-specific monoclonal antibody. Virus Res. 43:125–136 [DOI] [PubMed] [Google Scholar]
- 15. Kanai R., et al. 2006. Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 80:11000–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaufmann B., et al. 2009. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog. 5:e1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaufmann B., Rossmann M. G. 2011. Molecular mechanisms involved in the early steps of flavivirus cell entry. Microbes Infect. 13:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuhn R. J., et al. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liao M., Kielian M. 2005. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. J. Cell Biol. 171:111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao M., Kielian M. 2006. Functions of the stem region of the Semliki Forest virus fusion protein during virus fusion and assembly. J. Virol. 80:11362–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liao M., Kielian M. 2006. Site-directed antibodies against the stem region reveal low pH-induced conformational changes of the Semliki Forest virus fusion protein. J. Virol. 80:9599–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liao M., Sanchez-San Martin C., Zheng A., Kielian M. 2010. In vitro reconstitution reveals key intermediate states of trimer formation by the dengue virus membrane fusion protein. J. Virol. 84:5730–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin S. R., et al. 2011. The helical domains of the stem region of dengue virus envelope protein are involved in both virus assembly and entry. J. Virol. 85:5159–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindenbach B. D., Thiel H. J., Rice C. M. 2007. Flaviviridae: the viruses and their replication, p. 1101–1152 In Knipe D. M., et al. (ed.), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25. Modis Y., Ogata S., Clements D., Harrison S. C. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 100:6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Modis Y., Ogata S., Clements D., Harrison S. C. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319 [DOI] [PubMed] [Google Scholar]
- 27. Modis Y., Ogata S., Clements D., Harrison S. C. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79:1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mukhopadhyay S., Kim B. S., Chipman P. R., Rossmann M. G., Kuhn R. J. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 29. Nayak V., et al. 2009. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J. Virol. 83:4338–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nybakken G. E., Nelson C. A., Chen B. R., Diamond M. S., Fremont D. H. 2006. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 80:11467–11474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rey F. A., Heinz F. X., Mandl C., Kunz C., Harrison S. C. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291–298 [DOI] [PubMed] [Google Scholar]
- 32. Schalich J., et al. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmidt A. G., Yang P. L., Harrison S. C. 2010. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog. 6:e1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmidt A. G., Yang P. L., Harrison S. C. 2010. Peptide inhibitors of flavivirus entry derived from the E protein stem. J. Virol. 84:12549–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stiasny K., Allison S. L., Mandl C. W., Heinz F. X. 2001. Role of metastability and acidic pH in membrane fusion by tick-borne encephalitis virus. J. Virol. 75:7392–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stiasny K., Allison S. L., Schalich J., Heinz F. X. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stiasny K., Heinz F. X. 2006. Flavivirus membrane fusion. J. Gen. Virol. 87:2755–2766 [DOI] [PubMed] [Google Scholar]
- 38. Stiasny K., Koessl C., Heinz F. X. 2003. Involvement of lipids in different steps of the flavivirus fusion mechanism. J. Virol. 77:7856–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stiasny K., Kossl C., Heinz F. X. 2005. Differences in the postfusion conformations of full-length and truncated class II fusion protein E of tick-borne encephalitis virus. J. Virol. 79:6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stiasny K., Kossl C., Lepault J., Rey F. A., Heinz F. X. 2007. Characterization of a structural intermediate of flavivirus membrane fusion. PLoS Pathog. 3:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thiel H. J., et al. 2005. Family Flaviviridae, p. 981–998 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA [Google Scholar]
- 42. Yu I. M., et al. 2009. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J. Virol. 83:12101–12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu I. M., et al. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837 [DOI] [PubMed] [Google Scholar]
- 44. Zamyatnin A. A. 1972. Protein volume in solution. Prog. Biophys. Mol. Biol. 24:107–123 [DOI] [PubMed] [Google Scholar]
- 45. Zhang W., et al. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., et al. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]