Abstract
It is known that the human immune proteins APOBEC3G and -F (hA3G/F) can inhibit Vif-deficient HIV by G-to-A mutation; however, the roles of these enzymes in the evolution of HIV are debated. We argue that if evolutionary pressure from hA3G/F exists there should be evidence of their imprint on the HIV genome in the form of (i) underrepresentation of hA3G/F target motifs (e.g., TGGG [targeted position is underlined]) and overrepresentation of product motifs (e.g., TAGG) and/or (ii) an increase in the ratio of nonsynonymous to synonymous (NS/S) G-to-A changes among hA3G/F target motifs and a decrease of NS/S A-to-G changes among hA3G/F product motifs. To test the first hypothesis, we studied the representation of hA3G/F target and product motifs in 1,932 complete HIV-1 genomes using Markov models. We found that the highly targeted motifs are not underrepresented and their product motifs are not overrepresented. To test the second hypothesis, we determined the NS/S G↔A changes among the hA3G/F target and product motifs in 1,540 complete sets of nine HIV-1 genes. The NS/S changes did not show an increasing/decreasing trend within the target/product motifs, but the NS/S changes within the motif AG was exceptionally low. We observed the same pattern by analyzing 740 human genes. Given that hA3G/F do not act on the human genome, this suggests a small NS/S change within AG has arisen by other mechanisms. We therefore find no evidence of an evolutionary footprint of hA3G/F. We postulate several mechanisms to explain why the HIV-1 genome does not contain the hA3G/F footprint.
INTRODUCTION
The mutation of guanine (G) to adenine (A), the major error occurring during HIV reverse transcription, has been suggested to be responsible for the accumulation of A in the HIV genome (17). Observation of HIV sequences with extensive G-to-A mutations in HIV patients was later shown to be due to components of innate defense mechanism known as APOBEC3G and APOBEC3F (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G and 3F [hA3G/F]) (1). These are the host cytidine deaminase proteins that are incorporated into the Vif-deficient HIV virions and induce C-to-U (uracil) mutation in the minus strand of HIV. The plus strand is therefore enriched in A (30). It has been shown that the action of hA3G/F is highly dependent on the context of the sequence, i.e., on the type of nucleotides flanking a target C in the minus strand (G in the plus strand) (1, 14, 30). For example, hA3G preferentially mutates the underlined C in the context of CCCA (targeted position is underlined). This would result in a G-to-A mutation in the context of TGGG in the plus strand. Here, we refer to the hA3G/F target and product motifs in respect to the plus strand of the genome rather than the minus strand, where the actual cytosine deamination occurs.
While there is a consensus as to the mutagenicity of hA3G/F, the possible evolutionary pressure from these enzymes on the HIV genome is debated. The low frequency of the 4-mer CGGG, which is an hA3G target motif, in the HIV-1 genome has been attributed to the G-to-A mutation pressure from hA3G (30). Jern et al. (12) have hypothesized that those HIV-1 sequences within which the G-to-A mutations are the least deleterious (i.e., synonymous) can survive the selection pressure. Therefore, an evolutionary footprint of hA3G is expected to be observed in the form of a ratio of nonsynonymous (NS) to synonymous (S) G-to-A changes within the hA3G target motifs higher than that of a random G-to-A change. Using the results of a simulation study and in vitro passaging experiments, the authors have postulated a likely role for G-to-A mutation in the evolution of HIV-1. In a different study using several computational models of nucleotide misincorporation bias in HIV, no evidence was found to support an hA3G/F footprint (6). It was shown that a model in which misincorporation is explained only by a deoxynucleoside triphosphate (dNTP) imbalance describes 98% of the observed variation. Müller and Bonhoeffer (19) have argued that the preference for A over G is not related to the action of A3G but is a general characteristic of reverse transcription. They analyzed codon usage data to assess if there is a stronger G-to-A bias in viruses that infect cells that are known to express A3G than in those viruses that infect cells that do not express A3G. They also investigated if the strongest bias belongs to the viruses that lack Vif and infect A3G-expressing cells. They found no correlation between A3G and Vif with G-to-A mutation.
Here, we hypothesize that if the HIV genome has been changed due to evolutionary G-to-A pressure from hA3G/F, this should be indicated by an imprint of hA3G/F. The footprint would emerge in the form of underrepresentation of hA3G/F target motifs and overrepresentation of product motifs. This hypothesis is backed by two observations. The first is the strong motif dependency of mutation by hA3G/F (1, 14, 30), and the second is the evidence that implies hA3G/F have been in action against HIV for a long time (1). These two observations suggest that the nucleotide G must be depleted and A must be enriched in a motif-dependent manner in the HIV genome.
In order to test this hypothesis, the “observed” probabilities (pobs) of the hA3G/F target and product motifs need to be compared with the “expected” probabilities (pexp). For a given K-mer (i.e., motif, a short sequence consisting of K nucleotides), the pobs is defined as the ratio of the total count of the K-mer relative to the total count of all K-mers with the same length. The pexp of a K-mer could be defined in several different ways using the observed probabilities of the sub-K-mers. For example the pobs of 1-, 2-, and 3-mers can be used to define the pexp of a 3-mer. This is explained in detail below. It has been shown that the nucleotide sequences are described best by Markov chains of probabilities (15, 23). In this report, we develop Markov models for assessing the representation bias of the hA3G/F target and product motifs in a large number of complete HIV-1 sequences.
A second hypothesis to investigate the existence of an hA3G/F footprint is based on the type of G-to-A mutation. The deleterious (mainly NS) mutations, as opposed to nondeleterious (mainly S) mutations, are selected against and thus are less likely to be fixed in the virus population. Therefore, as recently proposed by Jern et al. (12), the hA3G/F target motifs, which are under high G-to-A mutation pressure, should contain a relative excess of NS G-to-A changes, since the motifs with an S G-to-A change are more easily removed.
HIV is under potential selective pressure arising from the G-to-A mutation by two major factors, namely, reverse transcriptase and hA3G/F. The former acts on all Gs within the HIV sequence in a random fashion, but the latter acts preferentially on those Gs within the context of hA3G/F target motifs (e.g., TGGG). The implication of the second hypothesis in relation to the evolutionary pressure of hA3G/F on the HIV genome is that a higher ratio of NS/S G-to-A changes would be expected for the Gs within the context of hA3G/F target motifs than for all Gs.
A G-to-A mutation within a “target” motif (e.g., TGGG) creates a “product motif” (e.g., TAGG); therefore, evolutionary pressure would result in an opposite NS/S pattern for hA3G/F product motifs. This means a lower ratio of NS/S A-to-G changes would be expected for As within the context of hA3G/F product motifs than for all As. It is noteworthy that “product” motifs are referred to as “putative ancestral target” motifs by Jern et al. (12). In order to test this hypothesis, we used all nine HIV-1 gene sequences and determined the ratio of NS/S G↔ A changes for all Gs and all As (we refer to them as random G and random A) and for the Gs and As within the context of hA3G/F target and product motifs, respectively. The same analysis was done on human genes to investigate whether an observed bias in the NS/S changes is specific in HIV-1 or if it is a common characteristic of both the virus and its host.
MATERIALS AND METHODS
Analysis of motif representation using Markov models.
A total of 1,932 complete HIV-1 sequences were obtained from the Los Alamos National Laboratory database in October 2009. A summary of the sequence compositions is given in Table 1. We calculated the pobs of each of the hA3G/F target and product motifs (K-mers are shown in Table 2) in each sequence.
Table 1.
Summary of HIV-1 sequencesa
| Parameter | Value |
||||
|---|---|---|---|---|---|
| A | G | C | T | Nonnucleotide | |
| Maximum count | 3,662 | 2,451 | 1,888 | 2,374 | 250 |
| Minimum count | 2,913 | 1,856 | 1,362 | 1,783 | 0 |
| Avg count | 3,256 | 2,152 | 1,585 | 1,994 | 4 |
| Maximum frequency (%) | 37.6 | 24.8 | 19.2 | 23.7 | |
| Minimum frequency (%) | 34.6 | 22.4 | 16.7 | 21.6 | |
| Avg frequency (%) | 36.3 | 23.9 | 17.6 | 22.2 | |
Sequence length (bp) was as follows: maximum, 10,280; minimum, 8,023; average, 8,991.
Table 2.
Target and product K-mer motifs of hA3G/Fa
| Target K-mer | Product K-mer |
|---|---|
| GG, GA, GT, GC | AG, AA, AT, AC |
| TGG, CGG, AGG, GGG, GGT, GGC, GGA | TAG, CAG, AAG, GAG, GAT, GAC, GAA |
| TGGG, TGGA, CGGG, TGGT, AGGG, GGGG, CGGT, AGGA, | |
| AGGT, GGGA, GGGT, TGGC, AGGC, GGGC, CGGA, CGGC | TAGG, TAGA, CAGG, TAGT, AAGG, GAGG, CAGT, AAGA, AAGT, GAGA, GAGT, TAGC, AAGC, GAGC, CAGA, CAGC |
Targeted positions are shown by an underline. Within each K-mer set, the motifs are ordered from highly targeted to the least targeted. NGGC motifs are the least favored 4-mer motifs (N is A, G, C, or T).
To assess the representation of a K-mer, the pobs of the K-mer was compared with its pexp by taking a ratio (D = pobs/pexp) (15). The pobs(K-mer) is defined as the ratio of the number of times the K-mer appears in the sequence to the total number of all K-mers with the same length. The pexp(K-mer) can be defined in different ways, as shown in equation 1 using a typical 3-mer example. In total there are 43 (64) different 3-mers. Therefore, the pexp of each 3-mer (e.g., ACG) may be defined as 1/64 (0.0156). This figure is not correct, partly because the 1-mers A, C, G, and T appear with different frequencies in the HIV sequence. Thus, a better estimation of the pexp(ACG) can be achieved if the pobs(A), pobs(C), and pobs(G) are taken into account (equation 1, first line). In this definition, the frequency bias within the 1-mers is considered but the possibility of a frequency bias within the 2-mers is ignored. A well-known example of a 2-mer frequency bias is the suppression of CG (i.e., CpG) in the HIV genome. Consequently any motif containing CG is infrequent in the HIV genome. Therefore, the pobs of 2-mers (AC and CG in this case) needs to be considered in estimating the pexp(ACG) (equation 1, second and third lines). Similar arguments apply for higher K-mers.
| (1) |
It has been shown that none of these combinations provides a correct estimation of pexp(K-mer) (2, 23). Instead, Markov models are known to best describe the sequences of nucleotides (2, 15, 23). In an nth-order Markov model, the probability of a given nucleotide at a particular position is determined by the conditional probabilities of its n preceding nucleotides. Equation 2 is the chain rule of probability for a 4-mer. The 1st- and 2nd-order Markov models of this sequence are given in equations 3 and 4. As shown in the 1st- and 2nd-order models, the probabilities are defined conditional on 1 and 2 preceding nucleotides, respectively. Markov models of the order 1 to K − 2 can be used to calculate the pexp of a K-mer.
| (2) |
| (3) |
| (4) |
The replacement of conditional probabilities with nonconditional probabilities in equations 3 and 4 results in equations 5 and 6, respectively, which are used to calculate the pexp.
| (5) |
| (6) |
In contrast to the models shown in equation 1, the Markov models consider the overlap between the constituents of K-mers. For example, in equation 5, the 2-mers AC and CG overlap at the 1-mer C. CG also overlaps with GT at the letter G. In the case of a 2nd-order model, shown in equation 6, the overlap between the 3-mers ACG and CGT is at the 2-mer CG. The use of (K − 1)- and (K − 2)-mer components to model a K-mer and also the consideration of overlap among them renders a model with a high prediction ability and minimum error. In this work, the ratio (D) of the pobs of the 2- to 4-mer hA3G/F target and product motifs to the pexp, obtained using the 0-, 1st-, and 2nd-order Markov models, respectively, were calculated. Over- and underrepresented K-mers were identified by D values of ≫1 and ≪1, respectively.
Analysis of NS/S G↔ A changes.
A total of 1,540 complete sets of nine HIV-1 genes (gag, pol, vif, vpr, tat, rev, vpu, env, and nef) were obtained from GenBank in August 2010. For each gene, we determined whether a G-to-A change within the hA3G/F (done separately for hA3G and hA3G) motifs is synonymous or nonsynonymous. We then summed, over all nine genes of each HIV-1 sequence, the total number of hA3G/F target motifs within which a G-to-A change is synonymous. The same summations were done for nonsynonymous G-to-A changes. For each of 1,540 HIV-1 sequences, a ratio of NS/S G-to-A changes within hA3G/F target motifs was calculated. We also calculated, for each HIV-1 sequence, a ratio of NS/S G-to-A changes for all Gs (i.e., random G as opposed to those within a particular target motif) within the HIV-1 genes. Thus, we could compare the NS/S G-to-A changes of Gs within the context of hA3G/F target motifs against that of Gs at random positions.
The same method was used to obtain a ratio of NS/S A-to-G changes for As within the context of the hA3G/F product motifs. We then obtained the ratio of NS/S A-to-G changes for all As to compare the NS/S changes of the hA3G/F product motifs to that of random A. We performed the same calculations for hA3F.
Figure 1 shows the method described above using a hypothetical HIV-1 gene. In the figure, only one of the hA3G target motifs, TGGG, and its product motif TAGG are described for brevity. Figure 1A shows how the NS/S G-to-A changes within TGGG and TAGG are calculated. Figure 1B shows the same calculations for random G and A.
Fig. 1.
Calculation of the ratio of nonsynonymous to synonymous G↔ A changes in a hypothetical HIV-1 gene. (a) NS/S G-to-A changes for Gs within the hA3G target motif TGGG and NS/S A-to-G changes for As within the hA3G product motif TAGG. (b) NS/S G-to-A changes for all Gs (random G) and NS/S A-to-G changes for all As (random A). The NS/S calculations of target and product motifs are shown below and above the hypothetical sequence in panel a, respectively. The NS/S calculations of all Gs and all As are shown below and above the hypothetical sequence in panel b, respectively. The codons are indicated by brackets.
The same method was used to calculate the ratio of NS/S G↔A changes for 740 human genes from the families of antigen processing and presentation, CD (cluster designation antigen), cellular immunity, chemokines and receptors, complement system, humoral immunity, inflammation, phagocytosis, and transcription factors. A summary of the compositions of these genes is given in Table 3.
Table 3.
Summary of human gene coding sequencesa
| Parameter | Value |
|||
|---|---|---|---|---|
| A | C | G | T | |
| Maximum frequency (%) | 0.41 | 0.4 | 0.39 | 0.35 |
| Minimum frequency (%) | 0.12 | 0.16 | 0.16 | 0.13 |
| Avg frequency (%) | 0.25 | 0.27 | 0.26 | 0.22 |
Sequence length (bp) was as follows: maximum, 9,172; minimum, 106; average, 1,332.
RESULTS
Representation of hA3G/F target and product motifs.
The mutation of G nucleotides by hA3G/F is sequence context dependent (1, 14, 30). It is known that hA3G preferentially mutates a G nucleotide that is flanked by a T at position −1 and Gs at positions +1 and +2 (compared to the target G at position 0), that is, the motifs GG, TGG, and TGGG. The motifs preferred by hA3F are GA, TGA, and TGAA. In both cases, NGNC (where N is A, C, G, or T) motifs are disfavored (1, 30). There are several independent studies in which the nucleotides flanking the target G are ranked based on their effects on the extent of the G-to-A mutation (1, 14, 30). Although there are minor differences among the reported ranks for the moderately to least mutated motifs, in almost all the studies, the favored and disfavored motifs are the ones described above. In addition, a strong preference for the most mutated motifs over the least mutated motifs has been demonstrated. The preference patterns of different 2- to 4-mer target motifs of hA3G and hA3F (Table 2) (1) are used here to investigate the hA3G and hA3F (hA3G/F) evolutionary footprint.
Given the observed hypermutation caused by hA3G/F and the data that suggest the long existence of hA3G/F (1), it is reasonable to expect the footprint of these restriction factors over the HIV genome. However, as indicated by the strong motif-dependent G-to-A mutations, the hA3G/F footprints should not be simply a higher frequency of A over C, G, and T across the entire HIV genome. Highly motif-dependent enzymes are expected to leave a highly motif-dependent footprint. This means the evolutionary pressure is expected to be evidenced by a negative correlation between the motif preference (by hA3G/F) and the representation, in the HIV sequence, of the motif. Alternatively an evolutionary footprint may be indicated by a positive correlation between the preference of the hA3G/F product motifs and their representations in the HIV genome.
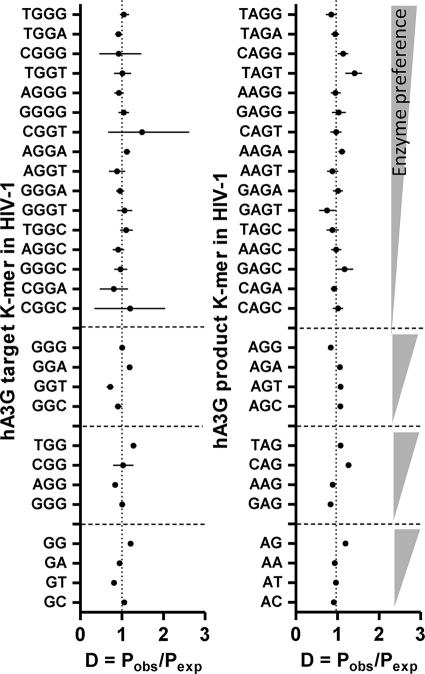
The results of the analysis of the hA3G footprint are shown in the text and those of hA3F in the supplemental material. The representations (D) of the hA3G and hA3F target K-mers are indicated on the left in Fig. 2 and in Fig. S1 in the supplemental material, respectively. The K-mers are shown on the vertical axis in order of enzyme preference, that is, within each K-mer group, the most and least preferred K-mers appear at the top and bottom, respectively. The horizontal axis displays the representation (D) of the K-mers. If the HIV-1 genome contains the footprint of hA3G/F, the most preferred K-mer motifs should be the most underrepresented (i.e., exhibit the lowest D) and the least preferred motifs are expected to be normal or overrepresented (D ≥ 1). This means that within each K-mer group, an increasing trend in the representation values of the K-mers from the top to the bottom would be expected.
Fig. 2.
Representation (D, the ratio of observed to expected probabilities) of hA3G target and product K-mers. K-mers of the HIV-1 genome are sorted on the vertical axis, within their own groups, from the most to the least preferred targets (left) and products (right). For example the first group on the left shows the 2-mers with a target G flanked from its 3′ end by G, A, T, or C. Within this group, GG and GC are targeted most and least by hA3G. Each data point is the average of the D values calculated from 1,932 complete HIV-1 sequences. The horizontal bars represent the 2.5 and 97.5 percentiles of HIV-1 sequences.
It is known that GG is strongly preferred by hA3G and also, to a lesser extent, by hA3F. The frequency of this motif is therefore expected to have declined during the evolution of HIV-1 so that the virus could evade the hA3G/F mutation pressure. Interestingly, GG is not notably underrepresented in the HIV-1 genome but actually appears to be overrepresented. The graph also does not show a continuous declining trend from GG to GC. The evolutionary footprint of hA3G is not evident from the data on 3- and 4-mers, either. No clear increasing trend from the most preferred 3- and 4-mer motifs (TGG/GGG and TGGG) to the least preferred motifs (GGG/GGC and CGGC) is observed. Also, the highly favored motifs TGG and TGGG are not underrepresented.
The representations of the hA3G and hA3F product motifs are shown on the right in Fig. 2 and in Fig. S1 in the supplemental material, respectively. In this case, an evolutionary footprint would be observed in the form of overrepresentation of the product of the motifs preferred by hA3G/F and underrepresentation of the product of those motifs that are disfavored by hA3G/F. This would result in a decrease in the representation values within each K-mer set. As shown, no such patterns are observed. Thus, analysis of the representation of target and product motifs provides no support for an hA3G/F footprint.
We also performed the same analysis described above on different HIV-1 subtypes and found results similar to those shown in Fig. 2 and Fig. S1 in the supplemental material. The plots of representation data for subtypes B and C are shown in Fig. S2 to S5 in the supplemental material.
Ratio of NS/S G↔A changes.
The ratio of nonsynonymous to synonymous (NS/S) G-to-A changes is influenced by at least three factors, namely, codon position, reverse transcriptase, and hA3G/F. According to the genetic codes, the G-to-A mutations in the first and second positions of a codon are nonsynonymous, while at the third position it is synonymous, except for two cases, TGG → TGA and ATG → ATA. In 14 out of the total 48 codons with at least one G, a G-to-A mutation is synonymous. Therefore, in a sequence with randomly distributed Gs and in the absence of any other mechanism, one would expect the NS/S G-to-A changes to be about 2.4 (34/14). This ratio can change due to the action of HIV reverse transcriptase. It is known that the most common error arising from the infidelity of the HIV reverse transcriptase is G-to-A mutation. If it is assumed that NS mutations are more likely to be deleterious than S mutations, then it is reasonable to expect an increase (>2.4) in the ratio of NS/S G-to-A changes in the HIV genome. By the same token, the G-to-A mutation by hA3G/F can further increase the ratio of NS/S G-to-A changes. However, because these enzymes preferentially mutate Gs within specific targets (e.g., TGGG), it is reasonable to expect a higher ratio of NS/S G-to-A changes for the Gs within hA3G/F target motifs than for all Gs (random Gs). We argue that if an evolutionary pressure from hA3G/F exists, there should be evidence of the imprint of hA3G/F on the HIV genome in the form of a higher ratio of NS/S G-to-A changes for the Gs within the context of highly targeted motifs than for random Gs. Also, there should be a decreasing trend in the ratio of NS/S G-to-A changes from the most targeted motifs (e.g., TGGG) to the least targeted motifs (e.g., CGGC). As shown on the left in Fig. 3 (for hA3G) and in Fig. S6 in the supplemental material (for hA3F), no such patterns are seen. The ratio of NS/S G-to-A changes for the underlined G in TGGG and TGAA, which are highly targeted by hA3G and hA3F, respectively, is not greater than that of a random G. Also no decreasing trend from TGGG and TGAA to CGGC and CGAC is observed.
Fig. 3.
Ratios of NS/S changes of hA3G target and product K-mers. K-mers of the HIV-1 genome are sorted on the vertical axis, within their own groups, from the most to the least preferred targets (left) and products (right). Each data point is the average of the NS/S values calculated from 1,540 complete sets of nine HIV-1 genes (gag, pol, vif, vpr, tat, rev, vpu, env, and nef). The horizontal bars represent the 2.5 and 97.5 percentiles of HIV-1 sequences.
An alternative way to look for an evolutionary footprint is to investigate the ratio of NS/S changes within the hA3G/F product motifs. Any G-to-A mutation within a target motif (e.g., TGGG) results in a product motif (e.g., TAGG). Therefore, in the absence of other mechanisms, an increase in the ratio of NS/S G-to-A changes within a target motif is proportional to a decrease in the ratio of NS/S A-to-G changes within its product motif. This means that in the presence of evolutionary pressure from hA3G/F, the products of the most targeted motifs are expected to have the lowest ratio of NS/S changes; hence, there should be evidence of the imprint of hA3G/F on the HIV genome in the form of a lower ratio of NS/S A-to-G changes for the As within the context of the product of highly targeted (i.e., highly produced) motifs than for random As. Also, there should be an increasing trend in the ratio of NS/S A-to-G changes from the most produced motifs (e.g., TAGG) to the least produced motifs (e.g., CGGC). As shown on the right in Fig. 3 and in Fig. S6 in the supplemental material, no increasing trend is observed for either hA3G or hA3F.
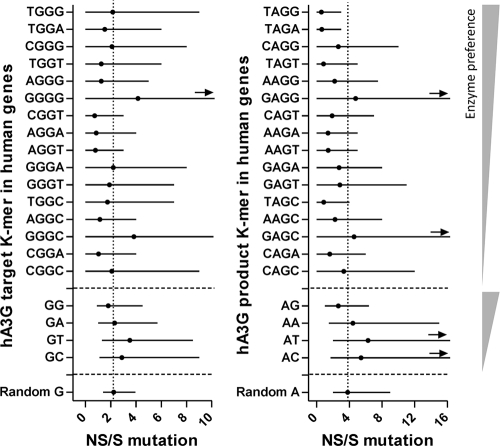
A notable trend in the analysis of hA3G product motifs is that the average ratio of NS/S A-to-G changes for almost all hA3G product 4-mers (an exception is GAGG) is less than that of random A. This appears to suggest a footprint; however, the fact that these different K-mer product motifs show more or less the same ratio of NS/S A-to-G changes could mean that a common feature among them is responsible for this pattern. To investigate if the low ratio of NS/S A-to-G changes is due to the 2-mer AG that exists in the middle of all these 4-mers, we calculated the ratio of NS/S A-to-G changes for the product 2-mers. As shown on the right in Fig. 3, among the four hA3G product 2-mers, only AG has a ratio of NS/S changes significantly lower than that of a random A. From the observations so far, one might argue that although the NS/S data of the 4-mer target and product motifs did not show an evolutionary footprint of hA3G, the NS/S data of the 2-mer AG may be evidence of such a footprint, as suggested by Jern et al. (12).
While the low ratio of NS/S changes in AG may appear to argue for a footprint effect, a number of features suggest this is not the case. First, there is no consistent increasing trend in the ratio of NS/S A-to-G changes within the 2-mer series from the most produced 2-mer, AG, to the least produced 2-mer, AC. The second piece of evidence derives from studying NS/S G↔ A changes in the human genome. If a low ratio of NS/S changes in AG is due to hA3G, this must be seen only in HIV genes and not in human genes. However, comparison of NS/S data from the human and HIV genomes shows the same pattern.
The NS/S results for human genes are shown in Fig. 4 and Fig. S7 in the supplemental material for hA3G and hA3F. In Fig. 4, each point is an average of 740 different human genes with diverse lengths (106 to 9,172 nucleotides [Table 3]). Despite such high diversity, the pattern of NS/S changes within the hA3G/F target and product motifs in human genes is very similar to those of HIV-1 shown in Fig. 3 and Fig. S6 in the supplemental material. Similar to the results for HIV-1, the ratio of NS/S A-to-G changes in Fig. 4 is less than that of a random A in the majority of hA3G 4-mer product motifs but only in the 2-mer AG. Therefore, the low ratio of NS/S changes of AG, and consequently the 4-mers containing AG, cannot be considered a footprint of hA3G unless it is assumed that the human genes have also evolved under the pressure of hA3G. Although a recent study has suggested a role for hA3A in editing the human genome, the study did not find editing by hA3G/F (25). Also there is no evidence to suggest that any member of the hA3 family has had an evolutionary impact on the human genome.
Fig. 4.
Ratios of NS/S changes of hA3G target and product K-mers. K-mers of human genes are sorted on the vertical axis, within their own groups, from the most to the least preferred targets (left) and products (right). Each data point is the average of the NS/S values calculated from 740 human genes. The horizontal bars represent the 2.5 and 97.5 percentiles of human gene sequences. The arrows indicate that the NS/S range of the K-mer extends beyond the upper limit of the horizontal axis (NS/S mutation axis).
DISCUSSION
The host immune proteins hA3G and hA3F have been shown to mutate G to A in a motif-dependent manner. This allows investigation of the hypothesis that the HIV-1 genome contains a footprint of these enzymes. A number of studies have claimed that hA3G has left its G-to-A footprint on the HIV-1 genome. Yu et al. (30) have reported that the hA3G target motif CGGG is underrepresented in the HIV-1 genome; however, they argue that the target TGGG is abundant because TGG is the only codon for tryptophan and therefore cannot be selected against without disturbing the protein sequence. They conclude that the evolutionary pressure from hA3G has shaped the HIV genome. We believe that the use of frequency instead of representation has led to this conclusion. These are two different concepts which could be misleading if used interchangeably. The frequency (i.e., abundance) of a K-mer is simply the observed probability of the K-mer. This is compared to the expected probability, on the assumption that all K-mers are equally probable (for example, since there are 256 possible 4-mers, the expected frequency of each is 1/256).
The representation of a K-mer, on the other hand, is defined in relation to the observed probabilities of its constituent sub-K-mers, as shown in equations 5 and 6. As an example, the 4-mer AAAA has a high frequency because the nucleotide A is frequent in the HIV genome; however, this motif is not necessarily overrepresented if the high frequency of monomer A is taken into account. The 4-mer CGGG is infrequent in the HIV-1 genome. However, this 4-mer motif is not extensively underrepresented, as evidenced by a representation value close to 1 (Fig. 2). The reason for the low abundance of the 4-mer CGGG is most likely the low frequency of the 2-mer CG, which resides within this 4-mer. It is well known that CG (also referred to as CpG) is extensively suppressed in the HIV genome (13), rendering any K-mer embracing CG infrequent. Thus, in working from 1-mers to 4-mers, we observe that C is less frequent than expected in the HIV genome, so we expect all K-mers with C to have a low frequency. The 2-mer CG is much more underrepresented than would be expected from the frequencies of C and G, suggesting some form of targeting. However, once we look at the various 4-mers made from CG, we find that CGGG has the representation expected. In other words the decreased representation was of the 2-mer CG, not of the 4-mer CGGG. Therefore, the low frequency of CGGG cannot be considered evidence for the evolutionary pressure of hA3G but is instead attributable to whatever mechanism has reduced the representation of CG. The use of frequency instead of representation to infer an evolutionary impact is also seen in the work of Kijak et al. (14). It is important to note that an assessment of the evolutionary footprints of enzymes requires an estimate of under- and overrepresentation, such as those given in equation 7, and cannot be achieved by simply using the frequency data.
Why have hA3G/F not left a footprint on the HIV-1 genome?
The lack of an evolutionary footprint left by hA3G/F on the HIV-1 genome may seem somewhat unexpected. The hypermutation caused by hA3G/F has been confirmed in several studies (1, 14, 30). It is believed that HIV Vif has evolved to counteract the deleterious effect of hA3G (12). Therefore, one might argue that even if there is an impact from hA3G/F, the actual effect is not realized due to the action of Vif, resulting in a complete blockage of hA3G/F from encapsidation in the virions. However, the recovery of hypermutated proviral sequences from HIV patients (1) suggests that despite the strong inhibition by Vif, hA3G/F can find their way into the newly infected cells, where they can perform their destructive hypermutation on the viral genome. They are therefore expected to have left their footprint on the HIV genome. The fact that a footprint is not observed may suggest that the major mechanism by which A3G/F inhibits HIV-1 is deaminase independent. An evolutionary G-to-A footprint may not be observed if G-to-A mutation by A3G/F is not the primary mechanism of viral control. There are several studies that support a deaminase-independent mechanism (10). Inhibition of hepatitis B virus (27) and mouse mammary tumor virus (MMTV) (22) by APOBEC3G with no or minimum G-to-A mutations has been reported. No correlation between the mutation levels in HIV-1 and the degree of viral inhibition has been observed (3). It has also been shown that catalytically inactive APOBEC3 mutants are able to inhibit HIV-1 (9). The deaminase-independent mechanisms proposed for APOBEC3 proteins include the reduction of tRNA priming (8), inhibition of the elongation of DNA (11) and reverse transcript (4), and several others. It is worth noting that there is no consensus as to which mechanism of HIV-1 inhibition by hA3G/F, deaminase dependent or independent, is dominant. For example, Browne et al. (5) have shown that active-site APOBEC3G mutants lack antiviral activity. They then concluded that the HIV inhibition by APOBEC3G is deaminase dependent. It is, however, reasonable to assume that if the major action of A3G/F against HIV-1 occurs by deaminase-independent mechanisms, the apparent lack of a G-to-A mutation footprint would not be unexpected.
An alternative hypothesis that could explain why hA3G/F have not left a footprint on the HIV-1 genome is that mutations by hA3G/F are highly deleterious and lead to the quick inactivation of the mutated sequences and their removal from the viral population. The number of A3G molecules per Vif-deficient HIV-1 virion has been variably reported to be 4 to 9 (21), 3 to 11 (29), and 17 to 22 (5). It has been shown that this number becomes 0.3 to 0.8 A3G molecules per virion in wild-type HIV (21). Browne et al. (5) have studied virions that contained 1 or 2 A3G molecules. They reported an average 2.3 mutations per kilobase for clones with an average 1.4 A3G molecules per virion. This equates to an average of 21 G-to-A mutations per full HIV sequence with an average of 9,000 bp. This may suggest that a single A3G/F molecule edits multiple sites per genome.
Compared to the hundreds of G-to-A replacements observed in “hypermutated” sequences (28) (most likely caused by multiple A3G/F), the small number of mutations (∼20 per full genome) caused by a single A3G/F might not seem significant; however, this number of mutations may be far beyond the tolerance of HIV, and therefore, even “lightly” mutated sequences will be selected against and thus are less likely to make a significant contribution to the evolution of HIV. There are several lines of evidence to support this argument. The first is the optimum mutation rate of HIV-1, which is believed to be about 0.3 mutations per genome per cycle (17); however, most of these mutations are deleterious (17, 20). The second line of evidence lies in the nature of the highly preferred A3G/F target motif TGG, which if edited creates a stop codon, TAG. It is reasonable to believe that even lightly mutated sequences with premature stop codons cannot survive. The third line of evidence is a series of reports that demonstrate the existence of mechanisms acting against mutated sequences. Browne et al. (5) have shown that an average of 2.3 mutations (caused by A3G) per kilobase reduces the infectivity of HIV-1 by about 40%. They suggest that the great reduction in infectivity is not merely due to the changes in the coding capacity of HIV-1 but rather is a direct effect of the reverse transcript, which could be caused by its degradation or a reduction in the elongation of the viral cDNA. It is also believed that most of the uracil-containing viral cDNA, produced as a result of cytosine deamination by A3G/F, is degraded by cellular enzymes before integration (7).
Given that on average each wild-type HIV-1 virion contains 0.6 A3G monomer (21), i.e., 0.3 A3G dimer, a Poisson distribution predicts that 74% of the wild-type virions do not have any A3G, 22% have a single A3G dimer, and 3% have two A3G dimers. This implies that the majority of the virions in a population do not experience any detrimental mutation caused by A3G/F. However, those virions that are affected experience high numbers of mutations. An extremely high number of mutations is not an optimal mechanism for selection, since most beneficial mutations are also accompanied by deleterious mutations and do not persist. Thus, APOBEC may exert too much pressure for optimal selection of mutations. However, we note that a recent study by Sadler et al. (24) suggests that hA3G can induce sublethal mutation.
A number of other minor hypotheses could also be invoked to explain the absence of a footprint, including the possible existence of reversionary (balancing) mechanisms or interactors/regulators that can reduce the catalytic activity of the enzymes. The former case would require that there be a G-to-A mutation pressure from hA3G/F but that the actions of other factors perturb the footprint on the HIV-1 genome. Such actions would need to reverse the effect of hA3G/F to totally eliminate the footprint. These could be in two forms. (i) In the first case, conversion of product motifs to target motifs by A-to-G mutation, the G-to-A mutations within the hA3G/F target motifs (e.g., TGGG and TGAA) need to have been reversed by a mechanism that specifically mutates A to G within the context of hA3G/F product motifs (e.g., TAGG and TAAA). (ii) The second possibility is independent removal of the product motifs and production of target motifs via nucleotide mutations of any kind, for example, by C-to-G mutation of TGCG, which results in TGGG, and independently by T-to-G mutation of TAGG, which results in GAGG. To the best of our knowledge, no such mechanisms with a motif preference order that is opposite to that of hA3G/F have been identified. An alternative possibility could be the existence of interactors/regulators that block or reduce the catalytic activity of hA3G/F (16). Thielen et al. (26) have observed a significantly lower deaminase activity for endogenous A3G than for exogenous A3G and have attributed this to an unknown factor that exists in T cells but not in epithelial-cell-derived cell lines.
Regardless of which mechanism is dominant, neither the representation of HIV-1 motifs nor the NS/S changes within the HIV-1 genes provide any evidence to support the idea that the HIV-1 genome contains the footprint of hA3G/F. Thus, although the footprint of major histocompatibility complex (MHC) recognition of the viral genome is discernible (18), the relationship between hA3G/F and the HIV genome appears more complex.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Australian National Health and Medical Research Council (NHMRC).
We thank Brynn Hibbert, Johnson Mak, Mark Tanaka, and Redmond Smyth for their instructive comments.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Armitage A. E., et al. 2008. Conserved footprints of APOBEC3G on hypermutated human immunodeficiency virus type 1 and human endogenous retrovirus HERV-K(HML2) sequences. J. Virol. 82:8743–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnold J., Cuticchia A. J., Newsome D. A., Jennings W. W., III, Ivarie R. 1988. Mono- through hexanucleotide composition of the sense strand of yeast DNA: a Markov chain analysis. Nucleic Acids Res. 16:7145–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bishop K. N., Holmes R. K., Malim M. H. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450–8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bishop K. N., Verma M., Kim E. Y., Wolinsky S. M., Malim M. H. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4:e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Browne E. P., Allers C., Landau N. R. 2009. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology 387:313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deforche K., et al. 2007. Estimating the relative contribution of dNTP pool imbalance and APOBEC3G/3F editing to HIV evolution in vivo. J. Comput. Biol. 14:1105–1114 [DOI] [PubMed] [Google Scholar]
- 7. Esnault C., et al. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430–433 [DOI] [PubMed] [Google Scholar]
- 8. Guo F., Cen S., Niu M., Saadatmand J., Kleiman L. 2006. Inhibition of tRNA3Lys-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710–11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holmes R. K., Koning F. A., Bishop K. N., Malim M. H. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation: comparisons with APOBEC3G. J. Biol. Chem. 282:2587–2595 [DOI] [PubMed] [Google Scholar]
- 10. Holmes R. K., Malim M. H., Bishop K. N. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32:118–128 [DOI] [PubMed] [Google Scholar]
- 11. Iwatani Y., et al. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35:7096–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jern P., Russell R. A., Pathak V. K., Coffin J. M. 2009. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. Plos Pathog. 5:e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karlin S., Doerfler W., Cardon L. R. 1994. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 68:2889–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kijak G. H., et al. 2008. Variable contexts and levels of hypermutation in HIV-1 proviral genomes recovered from primary peripheral blood mononuclear cells. Virology 376:101–111 [DOI] [PubMed] [Google Scholar]
- 15. Leung M. Y., Marsh G. M., Speed T. P. 1996. Over- and underrepresentation of short DNA words in herpesvirus genomes. J. Comput. Biol. 3:345–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malim M. H., Emerman M. 2008. HIV-1 accessory proteins: ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398 [DOI] [PubMed] [Google Scholar]
- 17. Mansky L. M., Temin H. M. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moore C. B., et al. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443 [DOI] [PubMed] [Google Scholar]
- 19. Müller V., Bonhoeffer S. 2005. Guanine-adenine bias: a general property of retroid viruses that is unrelated to host-induced hypermutation. Trends Genet. 21:264–268 [DOI] [PubMed] [Google Scholar]
- 20. Nowak M. 1990. HIV mutation rate. Nature 347:522. [DOI] [PubMed] [Google Scholar]
- 21. Nowarski R., Britan-Rosich E., Shiloach T., Kotler M. 2008. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat. Struct. Mol. Biol. 15:1059–1066 [DOI] [PubMed] [Google Scholar]
- 22. Okeoma C. M., Lovsin N., Peterlin B. M., Ross S. R. 2007. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445:927–930 [DOI] [PubMed] [Google Scholar]
- 23. Phillips G. J., Arnold J., Ivarie R. 1987. Mono- through hexanucleotide composition of the Escherichia coli genome: a Markov chain analysis. Nucleic Acids Res. 15:2611–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sadler H. A., Stenglein M. D., Harris R. S., Mansky L. M. 2010. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J. Virol. 84:7396–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suspènea R., et al. 2011. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc. Natl. Acad. Sci. U. S. A. 108:4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thielen B. K., et al. 2007. T Cells contain an RNase-insensitive inhibitor of APOBEC3G deaminase activity. PLoS Pathog. 21:1320–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Turelli P., Mangeat B., Jost S., Vianin S., Trono D. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 28. Vartanian J. P., Henry M., Wain-Hobson S. 2002. Sustained G→A hypermutation during reverse transcription of an entire human immunodeficiency virus type 1 strain Vau group O genome. J. Gen. Virol. 83:801–805 [DOI] [PubMed] [Google Scholar]
- 29. Xu H., et al. 2007. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology 360:247–256 [DOI] [PubMed] [Google Scholar]
- 30. Yu Q., et al. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435–442 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.