Abstract
Xenotropic murine leukemia virus-related virus (XMRV) is a gammaretrovirus found in association with human prostate cancer and chronic fatigue syndrome, although these associations are controversial. XMRV shows at most 94% identity to known mouse retroviruses. Here we used XMRV-specific PCR to search for a more closely related source of XMRV in mice. While we could not find a complete copy, we did find a 3,600-bp region of XMRV in an endogenous retrovirus present in NIH/3T3 cells. These results show that XMRV has clear ancestors in mice and highlight another possible source of contamination in PCR assays for XMRV.
TEXT
Xenotropic murine leukemia virus-related virus (XMRV) was first detected in 2006 in human prostate cancer tissue (10). XMRV was later found to be produced by the prostate cancer cell line 22Rv1 (3), which was derived from prostate cancer cells obtained in 1991 (6, 8), suggesting that XMRV has been in humans since at least that time. More recently, XMRV was found in a high proportion of humans suffering from chronic fatigue syndrome (CFS) (4), further supporting a role for XMRV in human disease. However, the presence of XMRV in humans is controversial, and many recent reports fail to confirm its presence in those with prostate cancer or CFS (1, 7).
The similarity of XMRV to murine leukemia viruses (MLV) indicates that XMRV may have originated from a mouse retrovirus. However, current XMRV isolates that have been fully sequenced do not match any known retroviral sequences in the GenBank databases. At best, XMRV exhibits 94% overall sequence identity to known mouse retroviral sequences, while the existing XMRV sequences are ≥99.8% identical.
It is important to know the origin of XMRV regardless of XMRV's potential role in human disease. Here we have tested for the presence of XMRV-like viruses in nude mice to determine whether the XMRV in 22Rv1 cells may have originated from the mice in which these cells and their ancestors have been propagated. The exact strain of nude mice in which the tumor cells had been grown is uncertain, but we chose NU/J mice (stock no. 2019) from the Jackson Laboratory, Bar Harbor, ME, as representative. We also tested for XMRV in the genomes of a few common murine cell lines and mouse strains, including NIH/3T3 cells from Swiss mice, Mus dunni tail fibroblasts (MDTF), and tissue from the inbred mouse strains BALB/c and C57BL/6. XMRV-specific primers overlapping the characteristic XMRV glycosylated gag gene deletion (gap) between the left long terminal repeat (LTR) and the Gag coding region of XMRV (10) (GapF, 5′-CGAGTCGGACTTTTTGGAGTGG-3′) and to the R (repeat) region of the right LTR (RR, 5′-TTGCAAACAGCAAAAGGCTTTATTGG-3′) were used to amplify an ∼7.7-kb fragment from genomic DNA from these sources. PCR products of the correct size were identified for NIH/3T3, C57BL/6, and nude mice, but only sequences within amplified products from NIH/3T3 cells closely matched XMRV. Note that the NIH/3T3 cell DNA was from a frozen sample extracted in 1993 and was therefore unlikely to be contaminated by XMRV, which has been present in our lab only since 2009.
We next determined the complete sequence of the XMRV-like mouse endogenous virus in the NIH/3T3 cells. To clone the far left end of the provirus, we used a primer to the R region of the left LTR (RF, 5′-CCAATAAAGCCTTTTGCTGTTTG-3′) and a reverse-orientation primer that overlapped the glycosylated gag gene deletion (GapR, 5′-CCAACAAAGCCACTCCAAA-3′). The complete virus sequence (GenBank accession no. JF714652.1) contains open reading frames encoding all of the proteins typical of replication-competent gammaretroviruses. Bases 297 to 3922 in the left half of the virus are virtually identical (99.9%) to the same region of XMRV from 22Rv1 cells (GenBank accession no. FN692043.2) (Fig. 1); thus, we named the virus mERV-XL (mouse endogenous retrovirus, XMRV left half). Phylogenetic analysis of sequences similar to this region of mERV-XL identified by BLAST search of the GenBank databases (Fig. 2) shows the close relationship (≥99.5% identity) of this region of mERV-XL to all completely sequenced XMRV strains. In contrast, the next most similar GenBank sequence, an mERV on chromosome 9 (GenBank accession no. AC121813.3), is only 97.8% identical.
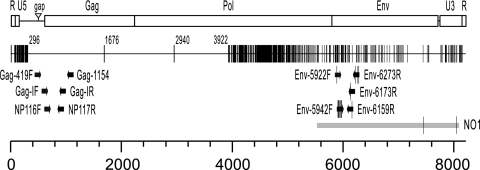
Fig. 1.
Structure of mERV-XL and comparison to XMRV and the polytropic retrovirus NO1. The genetic structure of the mERV-XL RNA genome is shown at top, with a scale in nucleotides at bottom. The deletion in the glycosylated gag coding region (10) characteristic of all XMRV isolates is present in mERV-XL (labeled “gap”). Under the mERV-XL structure is a depiction of mERV-XL, with nucleotide differences between mERV-XL (GenBank accession no. JF714652.1) and XMRV from 22Rv1 cells (GenBank accession no. FN692043.2) indicated by vertical lines. Numbers refer to the nucleotide position in mERV-XL of the nucleotide difference nearest to that number. The map was generated using Highlight software, available at http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter.html. The locations of PCR primers (arrows) used to detect XMRV in previous publications (3a, 4, 10) are shown, with sequence differences between the primers and mERV-XL indicated by vertical lines. The extent of mERV-XL identity with the NO1 polytropic mERV (gray bar) is shown, with sequence differences indicated by vertical lines. Abbreviations: R, retroviral repeat region at both ends of the retroviral RNA genome; U5, unique 5′ part of the retroviral long terminal repeat (LTR); U3, unique 3′ part of the retroviral LTR.
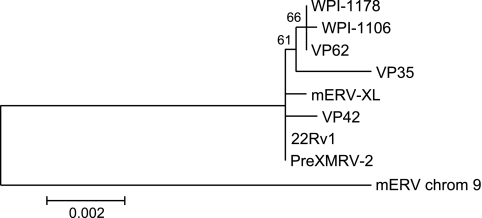
Fig. 2.
Phylogenetic tree based on XMRV sequences in the left half of mERV-XL. A BLAST search of the GenBank databases using bases 297 to 3932 of mERV-XL was conducted, and a maximum-likelihood phylogenetic tree was constructed from the resulting sequences using MEGA5 software (9). Bootstrap values are shown for 1,000 replicates, and the scale bar indicates evolutionary distance in base substitutions per site. chrom 9, chromosome 9.
A search for sequences similar to the right half of mERV-XL revealed that bases 5507 to 8093 are virtually identical (99.9%) to a partial sequence of the polytropic endogenous retrovirus NO1 from NFS/N (NIH Swiss) mice (GenBank accession no. AY219565.2) (2) (Fig. 1). Neither of the two nucleotide differences observed produces an amino acid change. The mERV-XL and NO1 proviruses may be the same because both are from Swiss mice. Bases 7075 to 7829 of mERV-XL also show high similarity (98.8%) to the same region of XMRV (Fig. 1).
We also attempted to PCR amplify sequences identical to regions of the right half of XMRV (bases 3988 to 4885, 5922 to 6880, and 7323 to 8185) from mouse DNA. Although stretches of up to 131 bases from nude mice were identical to XMRV, the regions sequenced were only 90 to 96% identical to XMRV overall and thus did not qualify as precursors of XMRV.
Highly sensitive PCR techniques to identify XMRV sequences in human samples have been performed using primers designed to amplify sections of the XMRV genome that were believed to be absent from the mouse genome. However, all primers used to amplify gag region sequences (3a, 4, 10) align perfectly with the mERV-XL sequence and amplify regions of mERV-XL that are 100% identical to XMRV (Fig. 1). Even some of the env primers align well with mERV-XL and could amplify products with the same size as those expected from XMRV. Because NIH/3T3 cells are in such widespread use, DNA from these cells is an obvious source of contamination that could lead to false-positive detection of XMRV.
We were unsuccessful in identifying a full-length copy or the right half of XMRV in the mouse strains we examined. A very recent publication also reports an inability to find an intact XMRV provirus in a wide range of mouse strains, but these authors were able to find two mERVs in nude mice that can generate XMRV following six recombination events (5). One of these proviruses, PreXMRV-2 (GenBank accession no. FR871850), is 99.9% identical to mERV-XL, and the other, PreXMRV-1 (GenBank accession no. FR871849), exhibits high similarity to the right half of XMRV. The phylogenetic tree in Fig. 2 shows that mERV-XL is slightly more distant from the 22Rv1 XMRV than is PreXMRV-2, as would be expected given the predicted source of 22Rv1 XMRV in nude mice and not from NIH/3T3 cells. In summary, it is clear that XMRV is not as distant from known mouse viruses as previously believed but has clear roots in endogenous retroviruses of mice.
Nucleotide sequence accession number.
The complete sequence of the XMRV-like mouse endogenous virus in NIH/3T3 cells was deposited in GenBank under accession no. JF714652.1.
Acknowledgments
This work was supported by training grant T32 CA080416 from the National Cancer Institute (R.M.), by pilot project funding from the Core Center of Excellence in Hematology, grant DK56465 (A.D.M.), and by funding from the Fred Hutchinson Cancer Research Center.
DNA sequencing was performed with help from shared resources at the FHCRC.
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Aloia A. L., et al. 2010. XMRV: a new virus in prostate cancer? Cancer Res. 70:10028–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Evans L. H., Lavignon M., Taylor M., Alamgir A. S. 2003. Antigenic subclasses of polytropic murine leukemia virus (MLV) isolates reflect three distinct groups of endogenous polytropic MLV-related sequences in NFS/N mice. J. Virol. 77:10327–10338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knouf E. C., et al. 2009. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J. Virol. 83:7353–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a. Lo S.-C., et al. 2010. Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors. Proc. Natl. Acad. Sci. U. S. A. 107:15874–15879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4. Lombardi V. C., et al. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585–589 [DOI] [PubMed] [Google Scholar]
- 5. Paprotka T., et al. 2011. Recombinant origin of the retrovirus XMRV. Science 333:97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pretlow T. G., et al. 1993. Xenografts of primary human prostatic carcinoma. J. Natl. Cancer Inst. 85:394–398 [DOI] [PubMed] [Google Scholar]
- 7. Shin C. H., et al. 2011. Absence of XMRV and other MLV-related viruses in patients with chronic fatigue syndrome. J. Virol. 85:7195–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sramkoski R. M., et al. 1999. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell. Dev. Biol. Anim. 35:403–409 [DOI] [PubMed] [Google Scholar]
- 9. Tamura K., et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. [Epub ahead of print.] doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Urisman A., et al. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]