Abstract
In this paper we undertake an analysis of the antigenicity of influenza A virus hemagglutinin. We developed a novel computational approach to the identification of antigenically active regions and showed that the amino acid substitutions between successive predominant seasonal strains form clusters that are consistent, in terms of both their location and their size, with the properties of B-cell epitopes in general and with those epitopes that have been identified experimentally in influenza A virus hemagglutinin to date. Such an interpretation provides a biologically plausible framework for an understanding of the location of antigenically important substitutions that is more specific than the canonical “antigenic site” model and provides an effective basis for deriving models that predict antigenic escape in the H3N2 subtype. Our results support recent indications that antibodies binding to the “stalk” region of hemagglutinin are found in the human population and exert evolutionary pressure on the virus. Our computational approach provides a possible method for identifying antigenic escape through evolution in this region, which in some cases will not be identified by the hemagglutinin inhibition assay.
INTRODUCTION
Seasonal influenza virus epidemics have a significant impact on global health, with between 200,000 and 500,000 related deaths reported each year (38). This stems from the ability of influenza virus to escape host immunological memory and hence, over time, reinfect its hosts. This is accomplished through the mutation of those regions of the virion to which antibodies bind, a mechanism known as antigenic drift (37).
In influenza A virus, the hemagglutinin (HA) surface glycoprotein is the primary target of infection-neutralizing antibodies (33). Structurally, in the intact virion, HA is a homotrimer in which each monomer consists of two protein chains linked by a disulfide bond. These chains form the membrane-proximal HA2 domain and the membrane-distal HA1 domain. The host cell receptor binding site is near the membrane-distal tip of HA1 (35). Antibodies binding directly in the region of the receptor binding site (RBS), and also those binding to regions closer to the HA1/HA2 interface, have been shown to inhibit viral attachment to host cells (17). Antibodies binding to hemagglutinin can also neutralize the virus by inhibiting a structural transition required for membrane fusion (3, 6).
Knowledge, with a fair degree of precision, of the locations and characteristics of epitopes, that is to say, the identification of the specific residues participating in antibody binding, is of general relevance to vaccine design and diagnostics (11, 15).
Characteristics of antibody binding in influenza A virus hemagglutinin.
A recent structural analysis of a nonredundant set of 53 antibody-antigen complexes in the Protein Data Bank (PDB) (4) found that 75% of the epitopes consisted of between 15 and 25 amino acids and covered a contact surface area of between 600 and 1,000 Å2 (26). Previous mutation studies have demonstrated that a small number of the epitopic residues can contribute a majority of the binding energy, with the mutation of just a single key residue being sufficient in some cases to inhibit binding (1).
Our own analysis, confined to influenza A virus HA-antibody complexes in the PDB, is broadly in agreement with the above-described structural analysis, although in some cases, a larger buried surface area on the HA protein was observed: the number of identified epitopic residues ranges from 13 to 18, and the reported buried surface areas range from 640 to approximately 1,500 Å2 (see Table 3 below for a summary of structures considered). The epitopes are of irregular shape, with the longest distance between residues within a single epitope ranging from approximately 35 to 40 Å.
Table 3.
Comparison of entire HA/Fab fragment complexes from the Protein Data Bank
| PDB accession no. | Subtype | Binding region | Hemagglutination inhibiting | Description | Reference |
|---|---|---|---|---|---|
| 2VIR | H3 | RBS | Yes | Binds in regions A and B | 9 |
| 1KEN | H3 | RBS | Not known | Binds across 2 HA monomers. Fusion inhibiting | 3 |
| 1QFU | H3 | Midregion | Yes | 7 | |
| 1EO8 | H3 | Midregion | Yes | Epitope substantially overlaps with 1QFU | 8 |
| 3GBN | H1 | Midregion | No | Fusion inhibiting | 6 |
| Binds across HA1 and HA2 | |||||
| 3GBM | H5 | Midregion | No | Fusion inhibiting | 6 |
| Binds across HA1 and HA2 | |||||
| 3FKU | H5 | Midregion | No | Fusion inhibiting | 31 |
| Binds across HA1 and HA2 |
The ability for hemagglutinin to escape the binding action of an antibody by means of a small number of substitutions was demonstrated both experimentally and computationally (16, 41).
Experimental evidence for the location of influenza A virus hemagglutinin epitopes.
A small number of influenza A virus HA epitopes where particular antibodies are known to bind have been identified in crystallographic studies (see Table 3 in this report and Fig. 2A in reference 6 for a useful visual summary of binding locations). Other experimental evidence allows us to infer, with various degrees of precision, the location of HA epitopes. A series of experiments in the 1980s identified antigenically important regions in H1N1 and H3N2 HA1 by observing viral evolution in the presence of a binding monoclonal antibody, supplemented by observations of wild-type evolution (35–37). Given their wide coverage in the literature, we shall refer to these regions as the “canonical” antigenic regions. More recently, the ability to isolate and clone anti-HA antibodies from human volunteers prompted a number of studies (18, 24, 25, 39).
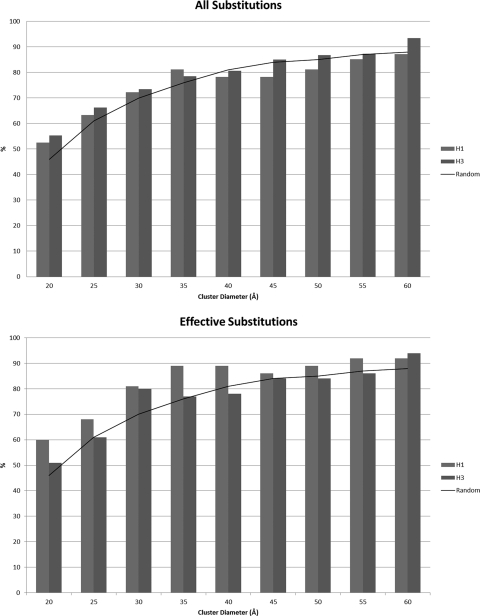
Fig. 2.
Proportion of substitutions lying within calculated clusters of various sizes compared to the proportion expected when substitutions are distributed at random on the H3 monomer. Effective substitutions follow the definition described previously by Shih et al. (28).
In the latter studies, the isolation of membrane-fusion-inhibiting antibodies is of particular interest. These antibodies bind to the “stalk” region near the HA1/HA2 interface and do not in all cases inhibit receptor binding. Although these methods allow the isolation and cloning of wild-type human antibodies from only a small number of subjects, the induction of stalk binding antibodies was noted previously in other studies (13, 34).
Computational analyses of antigenically effective substitutions at “immunodominant” locations.
The effectiveness of a selected influenza virus vaccine strain against a particular circulating strain is conventionally assessed by means of the hemagglutination inhibition (HI) assay. From this, the concept of antigenic distance between two strains has developed (19, 29). The computational prediction of the antigenic distance has the potential to assist with vaccine design by providing a rapid assessment of novel strains and by helping to understand the likelihood of the emergence of an epidemic strain. A number of computational models have been proposed; among these are models based on simple amino acid differences along the entire extent of HA1 (20), differences grouped according to amino acid type (22), and the identification of immunocritical residues (14). Our earlier work extended the number of residues that might reasonably be included in the canonical H3N2 binding regions and called into question the significance of these regions in terms of predictive performance (21).
Aim of this research.
In this study, we attempt to explain the pattern of successive substitutions observed between predominant circulating strains in a way that does not rely on canonical binding regions (i.e., is not constrained in terms of location) and that is consistent with our knowledge of epitope binding. By doing so, we aim to develop an understanding of the underlying drivers of HA evolution, in particular the binding of B-cell antibodies, and to develop improved computational models of antigenic distance.
MATERIALS AND METHODS
HA amino acid sequences of all available wild-type human H1N1 and H3N2 strains were downloaded from the Influenza Research Database (30) and the NCBI Influenza Virus Resource (2). Where multiple sequences for a given strain were available, a consensus sequence was derived. This resulted in a database containing sequences for 6,127 H1N1 strains and 7,337 H3N2 strains.
The predominant circulating strains of influenza A H1N1 and H3N2 viruses in each year between 1972 and 2009 were identified from the annual and semiannual influenza virus activity reports in the Weekly Epidemiological Record (http://www.who.int/wer/en/). The substitutions in HA1 between successive strains were deduced from amino acid sequences, and their relative positions in the protein structure were inferred from the X-ray structures of A/Aichi/2/68 (PDB accession number 1HGD) (27) for H3N2 strains and A/Puerto Rico/8/34 (PDB accession number 1RU7) (10) for H1N1 strains.
Effective substitutions (i.e., substitutions which are dominant in viral samples for a year or more and are therefore indicative of positive selection) were deduced by using methods previously described (28), with the sequence data set described above.
Substitutions between successive strains were examined for clusters as follows. First, a distance (here referred to as the “cluster distance”) was chosen. Next, the largest possible set of substitutions was found such that the Cα atoms of all substitutions in the set all lie within this distance of each other, and the set was identified as a cluster provided that it contained at least three substitutions. The process was repeated with the remaining substitutions (i.e., discounting those that had already been assigned to a cluster).
Where multiple clusters were identified in substitutions between adjacent circulating strains, and where sequences of intervening strains were available, phylogenetic trees derived from sequence data using PhyML (12) were used in conjunction with HI assay results compiled from data from journals and other sources to determine antigenic intermediates between the two epidemic strains. This allowed the evolutions of some multiple clusters to be separated.
Predictive models based on identified regions.
We examined the performances of predictive models of antigenic distance based on our identified antigenic regions in H3 strains. In these predictive models, we considered amino acid differences between two strains at the locations identified as participating in antigenic clusters. To provide a representation of the spatial distribution of the amino acid differences, we superimposed a three-dimensional grid onto the HA1 molecule, using the reference coordinates from the X-ray structure of A/Aichi/2/68 (PDB accession number 1HGD) (27). Each amino acid was assigned to the cell of the grid in which its Cα atom is found. Substitutions and other changes were accumulated to provide a score for each cell; for example, if, between two strains, there were three amino acid substitutions within a cell, the “difference” score for that cell would be 3. Antigenic distance was calculated as described above.
The scores obtained as described above are used as explanatory variables in a linear model. Coefficients for each variable were obtained by fitting against a “training” data set, and the predictive power of the model was then tested on a “validation” data set. In order to provide a benchmark comparison of performance, we have used the training and testing pairwise comparisons of strains previously reported by Liao et al. (22). We present results based both on the HI titers used in that study and on an extended set. Comparisons are given against the predictive results described previously by Liao et al.: in our own calculations, we did not observe a significant change in those previously reported results when the extended set of HI titers was adopted. Sequences and HI titers used in these models are provided in the supplemental material.
Being conscious that the selection of a particular grid cell size could lead to overfitting, we reviewed the impact of cell size on predictive properties. We present results for cubical grids with the length of each side varying between 2 Å and 22 Å. At 8 Å, 47 cells of the grid were occupied, giving an average of 1.6 residues per cell. At 22 Å, 10 cells were occupied, giving an average of 7.6 residues per cell. The predictive quality of the models was assessed by calculating the Matthews correlation coefficient (MCC) (23). Figure S4a in the supplemental material shows the predictive performance of the model at various cell sizes.
We were interested to know the extent to which substitutions in the midregion contributed to predictive performance. We therefore constructed a second model, in which only the 61 identified residues in the binding-site region were considered. Results for this model are presented in Fig. S4b in the supplemental material.
Both models provided relatively consistent performance across a broad range of cell sizes in which the residue density varied considerably. We feel that this indicates robustness in the underlying approach. The falloff in the predictive power in the validation set at cell sizes below 12 Å in the 76-location model and 8 Å in the RBS-only 61-residue model suggests that, at these small cell sizes, the model is overfitting to the training set by overweighting those locations that are significant in that set. The RBS-only model provides significantly improved results in the cell size range of 6 Å to 10 Å without significant degradation at a larger cell size. While this could be caused by overfitting in the 76-location model, the result is interesting in view of the discovery by Okada et al. (25) of wild-type antibodies binding in the midregion which are not hemagglutinin inhibiting.
Liao et al. (22) considered several predictive methods, reporting the best performance with a multiple-regression approach in which substitutions at roughly 20 locations were selected by the model during the analysis of the training set. To reduce noise induced by insignificant substitutions, amino acids were categorized into groups using one of six different approaches, with only substitutions that caused a switch of groups being considered significant. In the case of our models, we found that grouping approaches similar to those reported by Liao et al., or the introduction of additional explanatory variables based on properties such as charge or hydrophobicity, did not improve performance significantly, and we believe that this reflects the relatively low level of noise in difference scores at the locations that we have selected.
RESULTS
Mapping clusters to established regions.
Figures S1 and S2 in the supplemental material show the antigenic regions determined between the selected H1N1 and H3N2 intermediates with a cluster distance of 35 Å; this cluster distance was determined to be optimal in our analysis (see below) and is also in good agreement with crystallographically determined HA epitope dimensions. Interestingly, the predicted epitopes lie within quite closely defined regions of the monomer. All but 3 of the 21 H3N2 clusters and all but 4 of the 15 H1N1 clusters lie close to the sialic acid receptor binding site, with centroids positioned at 25 Å or less from the membrane-distal end of the monomer. The exceptions are grouped in a region closer to the viral membrane, with centroids positioned between 40 and 55 Å from the extreme membrane-distal end. We shall refer to these regions as the “receptor-binding-site (RBS) region” and the “midregion” in the remainder of this article.
According to this classification, of the 76 amino acid locations participating in clustered substitutions between the H3N2 strains considered (both the series shown in Fig. 1 and the series shown in Fig. 2), 61 occurred within RBS clusters, and 18 occurred within midregion clusters, with 3 locations occurring in both clusters (Table 1).
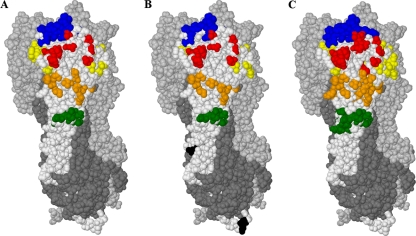
Fig. 1.
(A) Clustered substitutions observed in H3N2 strains considered in this study, superimposed onto one HA1 monomer (otherwise in white). (B) Locations undergoing effective frequency switches (28). (C) Canonical antigenic regions (5). Locations are colored according to the antigenic region: A (red), B (blue), C (green), D (yellow), E (orange), or unclassified (black). The remaining two HA1 monomers in the complex are shown in light gray, and the three HA2 monomers are shown in dark gray.
Table 1.
Clustered substitutions observed for H3N2 strains considered in this study, classified by the regions in which the clusters occur
| Region | Amino acid identifiers |
|---|---|
| RBS | 62, 75, 78, 80, 82, 83, 94, 96, 121, 122, 124, 126, 131, 133, 135, 137, 138, 139, 142, 143, 144, 145, 146, 155, 156, 157, 158, 159, 160, 163, 164, 182, 185, 186, 188, 189, 190, 192, 193, 194, 196, 197, 199, 201, 202, 207, 208, 213, 214, 216, 217, 219, 222, 223, 225, 226, 227, 233, 242, 244, 248 |
| Midregion | 47, 50, 53, 54, 57, 62, 63, 82, 83, 172, 174, 260, 262, 275, 276, 278, 299, 308 |
| Both | 62, 82, 83 |
Between successive H3N2 strains, we observed a number of substitutions in the midregion that did not meet the criteria for clusters developed above. We postulated that antigenicity could develop at a lower rate in this region and created a coarser-grained view in which we examined substitutions between the antigenic clusters identified previously by Smith et al. in their work on antigenic mapping (29), using the vaccine candidate identified in that work as being representative of each cluster. The results are shown in Fig. S3 in the supplemental material and identified midregion clusters in 6 out of 10 cases. A possible seventh candidate can be seen in the transition from Wuhan/359/1995 to Sydney/5/1997, where the substitution at position 121 could potentially be a member of either predicted epitope but has been assigned to the binding-site region by our algorithm.
Comparison with H3 canonical antigenic regions.
Table 2 lists the amino acid identifiers at which clustered substitutions were observed in the H3N2 strains considered, classified in terms of the canonical H3N2 binding regions. In previous work (21), we identified additional locations that should be considered additional members of these antigenic regions, in light of sequences not available at the time when the original list (5) was drawn up. Five locations (Table 2) are allocated to antigenic regions on the basis of this additional classification.
Table 2.
Clustered substitutions observed for H3N2 strains considered in this study, classified by canonical antigenic regiona
| Antigenic region | Amino acid identifiers |
|---|---|
| A | 122*, 124*, 126, 131*, 133*, 135, 137*, 138, 142, 143*, 144*, 145*, 146* (13/19) |
| B | 155*, 156*, 157, 158*, 159, 160*, 163, 164*, 186, 188*, 189*, 190*, 192, 193*, 194, 196*, 197*, 199 (18/22) |
| C | 47, 50*, 53*, 54*, 275, 276*, 278*, 299, 308 (9/27) |
| D | 96, 121, 172*, 174*, 182, 201*, 207*, 208, 213*, 214, 216, 217*, 219, 222*, 223, 225*, 226, 227, 233, 242, 244*, 248 (22/41) |
| E | 57, 62*, 63, 75*, 78, 80, 82*, 83*, 94, 260*, 262* (11/22) |
| Unclassified | 139, 185, 202* (3/0) |
See reference 5. Locations in italics are additional locations not classified in that work (see the text). Locations classified previously by Shih et al. (28) as carrying effective frequency switches are in boldface type. Locations identified previously by Smith et al. (29) as being cluster differentiators are indicated by asterisks. Numbers in parentheses show the numbers of substitutions observed in the region in this study and the total number of locations in the canonical H3 region identified previously by Bush et al. (5).
Of the 63 locations identified previously by Shih et al. (28) as undergoing frequency switches, 57 are represented in our list of 76 substitutions. Twenty-three of the 25 locations identified previously by Yang (40) as being under selective pressure (95% level, all models with listed locations) are included. Of the 45 locations identified previously by Smith et al. (29) as being cluster differentiating, 41 are included, as are all 6 locations forming the decision tree constructed previously by Huang et al. (14). These results give confidence that the regions identified by our technique are indeed those that are key to antigenic differentiation, indicated both by evidence of selective pressure and by their importance in antigenic cluster differentiation. Our list is roughly half the size of the 131 locations in canonical regions A to E. While our approach may not identify all antigenically sensitive regions (for example, in some cases escape may have occurred with substitutions of fewer than 3 residues), this significant reduction in the number and the broad agreement with residues identified by other techniques suggest that antigenic activity in wild-type strains takes place in a more constrained region than was observed for experiments with laboratory-grown strains in the 1980s (Fig. 1).
Optimal cluster distance versus epitope size.
To obtain an understanding of the sensitivity of the approach to the cluster distance, we examined the proportions of total and effective substitutions lying within the calculated clusters for cluster distances of between 20 Å and 60 Å. The results are presented in Fig. 2. With a cluster distance of 35 Å, approximately 80% of the substitutions between the strain transitions that we considered were contained within clusters. An increase of the cluster distance from 35 Å to 60 Å does not increase the percentage significantly, indicating that the remaining substitutions are sufficiently distant from clusters that they are unlikely to form part of a common region. We therefore consider a distance of 35 Å to be an optimal tradeoff between inclusion and specificity. Interestingly, the same pattern could be seen both for all substitutions and for effective substitutions.
The H3 epitopes derived from the crystallographic structures reported under PDB accession numbers 3GBN and 1EO8 described above require cluster distances of 32 Å and 40 Å, respectively, to accommodate all epitopic residues. The latter contains an outlier at residue 143P; if this is omitted, the required cluster distance is 31 Å. The optimal cluster distance suggested above is in good agreement with these experimentally deduced epitope sizes.
To understand the significance of these results, we compared them with simulated results obtained by distributing substitutions at random on the H3 monomer. Our H3 series contained 19 strain transitions with between 3 and 16 substitutions in each transition. We used bootstrap tests to examine the significance of the clusters of substitutions obtained compared to those expected from a random distribution of substitutions across the HA1 monomer. In these tests, 1,000 batches of 19 simulated strain transitions were created, with the number of substitutions reflecting the distribution of the H3 series and with the substitutions positioned randomly across the monomer.
With this approach, we observed a significant increase in the number of substitutions in a cluster at all diameters between 20 Å and 60 Å (P < 0.01 at 35 Å) compared to a random distribution of substitutions. Even if the randomly selected substitutions were chosen just from the 131 “canonical” residues in the 5 regions, this level of significance was found for H3 cluster distances of between 25 Å and 45 Å (the mean number of substitutions in an H3 cluster at 35 Å was 6.8 in our results, compared to 5.3 in the random simulation with substitutions selected from the 131 locations).
Comparison of cluster location with locations of known epitopes.
Okada et al. (25) previously isolated and cloned 98 antibodies to wild-type H3 HA from a single volunteer born in 1960. These antibodies were found to divide into three sets: one set that bound to strains isolated between 1968 and 1973, a second set that bound to strains isolated between 1977 and 1993, and a third set that bound to strains isolated between 1993 and 2004. This experimental study used a chimeric approach that was able to identify small subsets of residues containing partial epitopes of 95 of these antibodies within viral strains isolated within the three periods listed above.
We compared the residue locations obtained by Okada et al. with 35-Å clusters calculated from a predominant strain transition at or just after the end of each of the three identified periods of antibody binding, reasoning that substitutions in this transition would have led to escape.
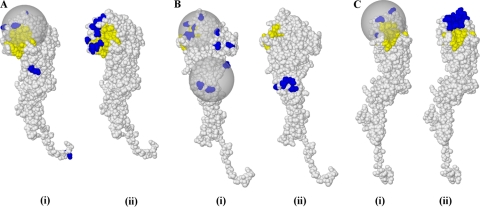
Okada et al. isolated 11 antibodies binding to viral strains isolated between 1968 and 1973; we compared their binding locations with clusters calculated for the transition between A/England/42/1972 and A/Port Chalmers/1/1973. Nine of the antibodies bound across regions B and D in a location that is consistent with that identified in our analysis. The remaining two bound in the RBS region, inside the cluster identified by our results. Figure 3A compares the results.
Fig. 3.
Comparison of clusters obtained in our results with data from an experimental study of antibodies isolated from a single individual (24). In each case, panel i shows clusters that we obtained from a transition in the predominant wild-type strain compared to those residues identified in an experimentally defined chimeric approach (ii), some of which are known to disrupt the binding of an antibody that became ineffective at that point in time. (A) Transition of A/England/42/1972 to A/Port Chalmers/1/1973; (B) transition of A/Beijing/32/1992 to A/Wuhan/359/1995 (Bii shows the midregion only); (C) transition of A/Fujian/411/2002 to A/Wellington/1/2004. Substitutions are in blue, and the receptor binding site is in yellow.
In the period of 1977 to 1993, most of the isolated antibodies bound in a region close to a “midcluster” that we calculated for the transition between A/Beijing/32/1992 and A/Wuhan/359/1995. The remainder bound in region E and region B. The set of locations identified in region E lie within the calculated midcluster, while those identified in region B lie within the calculated RBS cluster. The transition between our two selected strains shows a number of additional substitutions in the RBS area; this suggests that antibodies with additional epitopes with those identified in the study were active in the population in this period. Figure 3B provides a comparison.
In the period of 1993 to 2004, most antibodies isolated in the study showed binding activity in region B. Two of the substitutions in the cluster that we calculated for the transition from A/Fujian/411/2002 to A/Wellington/1/2004 are included in the set of residues from the study. The remaining antibodies from the study bound in the midregion; no substitutions in this region were observed for this strain transition. The experimental study also identified some binding activity in region A: the identified locations lie within the calculated cluster. The calculated cluster is compared with the study data in Fig. 3C.
A number of crystal structures of the entire HA complexed with antibody Fab fragments can be found in the Protein Data Bank, and these are summarized in Table 3. The number of crystal structures is clearly quite small, making it difficult to draw general conclusions from the sample. It is worth noting that the four H3 structures are based on the pandemic A/Hong Kong/1/1968 strain. The binding characteristics of this strain may not be representative of later strains that have undergone significant directed evolution to escape antibody binding. Finally, the reported structures represent structures of particular interest and therefore may not be representative of the general distribution of antibodies and epitopes. Nevertheless, some interesting comparisons can be drawn between these structures, our results, and the results reported by Okada et al. (25).
In two structures, the Fab fragment binds in the RBS region. The structure reported under PDB accession number 2VIR binds in antigenic regions A and B, and in terms of location, it is typical of many of the clusters that we have identified in H1 and H3 strain transitions and the RBS binding antibodies identified previously by Okada et al. The structure reported under PDB accession number 1KEN binds across antigenic regions A, B, and D in one HA monomer and regions A and B in another. The binding location is again typical, but our technique and that reported previously by Okada et al. do not specifically address cross-monomer binding.
The remaining structures are of midregion binding in H1 and H5 subtypes. These structures can be divided into two categories: one category (PDB accession numbers 1QFU and 1EO8) are hemagglutination inhibiting, while the other structures (accession numbers 3GBN, 3GBM, and 3KFU) inhibit the HA conformational change required for membrane fusion and are not hemagglutination inhibiting. Structures in both categories bind in similar locations (region B in H3 terms), but the latter structures bind slightly less than the former, with the epitope incorporating some locations in HA2.
Structurally, it is not possible to make a direct comparison between these antibodies and the midregion binding antibodies from the study reported previously by Okada et al., as the latter antibodies are H3 rather than H1/H5 antibodies. The antibodies reported by Okada et al. were not tested for binding to HA2. Their neutralization mechanism has not been explicitly determined, but they are not hemagglutination inhibiting. The existence of such wild-type antibodies has important implications for vaccine selection and epidemic forecasting, as their effect was not determined by the HI assay.
In summary, there is good agreement between the epitopes identified through crystal studies, the binding regions identified previously by Okada et al., and the locations of clusters identified in our study.
To obtain a wider comparison between the antigenically active regions identified in our study and those obtained by other researchers, we conducted a search of the Immune Epitope Database (http://www.immuneepitope.org/) (32) for conformational epitopes on human H1 and H3 HA1. Antibodies raised against synthetic peptides were excluded. We obtained references for a total of 16 additional studies (see Table S1 in the supplemental material). Epitopes in the region of the RBS were found in 15 of those studies, and the locations identified were in good agreement with the locations identified in this work. Midregion epitopes were found in three of those studies. Two of these midregion epitopes lie across HA1 and HA2; the third is confined to HA1 in the same region as that identified by our analysis and by Okada et al. and shown in Fig. 3C. One study identified an epitope that is distinct from those identified in our cluster analysis; interestingly, this epitope is from an antibody isolated from a human volunteer, which was found to bind to a relatively conserved region of H3 HA at positions 173 to 181 (18).
Predictive models.
Predictive models of antigenic distance based on the antigenically important regions identified in this study were able to meet or exceed the performances of previously reported models that are dependent on the selection of a much narrower set of critical locations. The best results were obtained with models that focused on the RBS region, which may reflect the derivation of antigenic distances from HI binding titers (Table 4 and see Table S1 in the supplemental material). For model details, please refer to Materials and Methods.
Table 4.
Comparison of the sensitivity and specificity of our predicted models when tested against the extended HI titer data set compared to those of the best multiple-regression models reported previouslya
| Model | Sensitivity (%) | Specificity (%) |
|---|---|---|
| Complete 76 locations, avg scores | 87.5 | 79.9 |
| Complete 76 locations, best result | 90.0 | 89.5 |
| RBS only, avg scores | 86.5 | 80.6 |
| RBS only, best result | 90.0 | 92.1 |
| Multiple regression, GM4b | 84.2 | 93.5 |
For our models we show both the average figures (averaged across 8 cell sizes from 8 Å to 22 Å) and the best result obtained (at 12 Å for the complete 76-location model and at 8 Å for the RBS-only model).
Previously published results (22).
DISCUSSION
Using a computational technique, and making use of the large number of previously reported hemagglutinin amino acid sequences and antigenic properties, we have demonstrated that approximately 80% of amino acid substitutions between successive seasonal strains of H1N1 and H3N2 subtypes lie within clusters whose sizes are consistent with those of conformational antibody epitopes and whose locations are consistent with experimental results from a number of sources. The approach is similar to the comparative sequence analysis experiments used in the laboratory in the 1980s to identify the canonical antigenic regions in H1 and H3, and our results are in broad agreement with those experiments, although we find that wild-type regions are more restricted than those determined in the laboratory. As a confirmation of our approach, we have demonstrated the development of simple predictive computational models that approach and in some cases exceed the performance of a “gold standard” selective model while retaining a much greater degree of generality.
Our study suggests that there are two predominant regions of antigenic importance in human wild-type strains of HA1: the binding-site region and the midregion. Viral strains, particularly H3 strains, appear to mutate rapidly in the binding-site region, and mutations in this region are generally responsible for the drift between successive seasonal strains. The development of clusters in the midregion occurs more slowly, at a rate that is comparable to that of the development of the cluster transitions identified by antigenic mapping. The cluster behavior observed with the H3N2 antigenic map may be accentuated by the interplay between activities in these two regions.
Our determination that antigenic distance is best predicted by considerations of substitutions in the RBS region alone rather than by considerations of changes in both the RBS and midregions lends weight to the idea that neutralizing but nonhemagglutinating antibodies of the type identified in the study by Okada et al. (25) are found at representative levels in the population as a whole, with potential implications for the use of the HI assay as the key determinant of antigenic distance. It would be useful to establish whether the current seasonal vaccines are capable of eliciting such antibodies, as the results of Okada el al. suggest that they can stay active over many seasons.
We followed other researchers in confining our analysis to the HA1 subunit, given its generally accepted dominant role in antigenic activity. However, given the frequent presence of antigenically active regions in the midregion, where epitopes can extend into HA2, we believe that antigenic analyses should be extended into this domain as well, and this will be a focus of future work.
The predictive approach which we have outlined can facilitate the development of more accurate predictive models of antigenic escape by directing the model more clearly to amino acid substitutions in antibody binding regions and by separating those responsible for escape in the two typical binding regions which we have identified. Because the approach can highlight regions of evolutionary change, it may prove useful in circumstances where antigenic escape is not detectable by means of the HI assay.
Supplementary Material
ACKNOWLEDGMENT
We thank Richard Goldstein of the United Kingdom National Institute for Medical Research for helpful discussions and suggestions.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Air G. M., Laver W. G., Webster R. G. 1990. Mechanism of antigenic variation in an individual epitope on influenza virus N9 neuraminidase. J. Virol. 64:5797–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bao Y., et al. 2008. The Influenza Virus Resource at the National Center for Biotechnology Information. J. Virol. 82:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbey-Martin C., et al. 2002. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology 294:70–74 [DOI] [PubMed] [Google Scholar]
- 4. Berman H. M., et al. 2002. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 58:899–907 [DOI] [PubMed] [Google Scholar]
- 5. Bush R. M., Fitch W. M., Bender C. A., Cox N. J. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 16:1457–1465 [DOI] [PubMed] [Google Scholar]
- 6. Ekiert D. C., et al. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fleury D., et al. 1999. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat. Struct. Biol. 6:530–534 [DOI] [PubMed] [Google Scholar]
- 8. Fleury D., Daniels R. S., Skehel J. J., Knossow M., Bizebard T. 2000. Structural evidence for recognition of a single epitope by two distinct antibodies. Proteins 40:572–578 [PubMed] [Google Scholar]
- 9. Fleury D., Wharton S. A., Skehel J. J., Knossow M., Bizebard T. 1998. Antigen distortion allows influenza virus to escape neutralization. Nat. Struct. Biol. 5:119–123 [DOI] [PubMed] [Google Scholar]
- 10. Gamblin S. J., et al. 2004. The structure and receptor-binding properties of the 1918 influenza hemagglutinin. Science 303:1838–1842 [DOI] [PubMed] [Google Scholar]
- 11. Gershoni J. M., Roitburd-Berman A., Siman-Tov D. D., Freund N. T., Weiss Y. 2007. Epitope mapping: the first step in developing epitope-based vaccines. BioDrugs 21:145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 13. Hardelid P., et al. 2010. Assessment of baseline age-specific antibody prevalence and incidence of infection to novel influenza AH1N1 2009. Health Technol. Assess. 14:115–192 [DOI] [PubMed] [Google Scholar]
- 14. Huang J., King C., Yang J. 2009. Co-evolution positions and rules for antigenic variants of human influenza A/H3N2 viruses. BMC Bioinformatics 10(Suppl. 1):S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Irving M. B., Pan O., Scott J. K. 2001. Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr. Opin. Chem. Biol. 5:314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin H., et al. 2005. Two residues in the hemagglutinin of A/Fujian/411/02-like influenza viruses are responsible for antigenic drift from A/Panama/2007/99. Virology 336:113–119 [DOI] [PubMed] [Google Scholar]
- 17. Knossow M., et al. 2002. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology 302:294–298 [DOI] [PubMed] [Google Scholar]
- 18. Kubota-Koketsu R., et al. 2009. Broad neutralizing human monoclonal antibodies against influenza virus from vaccinated healthy donors. Biochem. Biophys. Res. Commun. 387:180–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lapedes A., Farber R. 2001. The geometry of shape space: application to influenza. J. Theor. Biol. 212:57–69 [DOI] [PubMed] [Google Scholar]
- 20. Lee M., Chen J. S. 2004. Predicting antigenic variants of influenza A/H3N2 viruses. Emerg. Infect. Dis. 10:1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lees W. D., Moss D. S., Shepherd A. J. 2010. A computational analysis of the antigenic properties of haemagglutinin in influenza A H3N2. Bioinformatics 26:1403–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liao Y., Lee M., Ko C., Hsiung C. A. 2008. Bioinformatics models for predicting antigenic variants of influenza A/H3N2 virus. Bioinformatics 24:505–512 [DOI] [PubMed] [Google Scholar]
- 23. Matthews B. W. 1975. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta 405:442–451 [DOI] [PubMed] [Google Scholar]
- 24. Okada J., et al. 2010. Monoclonal antibodies in man that neutralized H3N2 influenza viruses were classified into three groups with distinct strain specificity: 1968–1973, 1977–1993 and 1997–2003. Virology 397:322–330 [DOI] [PubMed] [Google Scholar]
- 25. Okada J., et al. 2011. Localization of epitopes recognized by monoclonal antibodies that neutralized the H3N2 influenza viruses in man. J. Gen. Virol. 92(Pt. 2):326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rubinstein N. D., et al. 2008. Computational characterization of B-cell epitopes. Mol. Immunol. 45:3477–3489 [DOI] [PubMed] [Google Scholar]
- 27. Sauter N. K., et al. 1992. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography. Biochemistry 31:9609–9621 [DOI] [PubMed] [Google Scholar]
- 28. Shih A. C., Hsiao T., Ho M., Li W. 2007. Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc. Natl. Acad. Sci. U. S. A. 104:6283–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith D. J., et al. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376 [DOI] [PubMed] [Google Scholar]
- 30. Squires B., et al. 2008. BioHealthBase: informatics support in the elucidation of influenza virus host pathogen interactions and virulence. Nucleic Acids Res. 36:D497–D503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sui J., et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vita R., et al. 2010. The immune epitope database 2.0. Nucleic Acids Res. 38:D854–D862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang T. T., et al. 2010. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 6:e1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wei C., et al. 2010. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329:1060–1064 [DOI] [PubMed] [Google Scholar]
- 35. Wiley D. C., Skehel J. J. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365–394 [DOI] [PubMed] [Google Scholar]
- 36. Wiley D. C., Wilson I. A., Skehel J. J. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378 [DOI] [PubMed] [Google Scholar]
- 37. Wilson I. A., Cox N. J. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 8:737–771 [DOI] [PubMed] [Google Scholar]
- 38. World Health Organization 2009. Influenza WHO fact sheet no. 211. World Health Organization, Geneva, Switzerland [Google Scholar]
- 39. Wrammert J., et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208:181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang Z. 2000. Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus A. J. Mol. Evol. 51:423–432 [DOI] [PubMed] [Google Scholar]
- 41. Zhou R., Das P., Royyuru A. 2008. Single mutation induced H3N2 hemagglutinin antibody neutralization: a free energy perturbation study. J. Phys. Chem. B 112:15813–15820 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.