Abstract
Human APOBEC3B (A3B) has been described as a potent inhibitor of retroviral infection and retrotransposition. However, we found that the predominantly nuclear A3B only weakly restricted infection by HIV-1, HIV-1Δvif, and human T-cell leukemia virus type 1 (HTLV-1), while significantly inhibiting LINE-1 retrotransposition. The chimeric construct A3G/B, in which the first 60 amino acids of A3B were replaced with those of A3G, restricted HIV-1, HIV-1Δvif, and HTLV-1 infection, as well as LINE-1 retrotransposition. In contrast to the exclusively cytoplasmic A3G, which is inactive against LINE-1 retrotransposition, the A3G/B protein, while localized mainly to the cytoplasm, was also present in the nucleus. Further mutational analysis revealed that residues 18, 19, 22, and 24 in A3B were the major determinants for nuclear versus cytoplasmic localization and antiretroviral activity. HIV-1Δvif packages A3G, A3B, and A3G/B into particles with close-to-equal efficiencies. Mutation E68Q or E255Q in the active centers of A3G/B resulted in loss of the inhibitory activity against HIV-1Δvif, while not affecting activity against LINE-1 retrotransposition. The low inhibition of HIV-1Δvif by A3B correlated with a low rate of G-to-A hypermutation. In contrast, viruses that had been exposed to A3G/B showed a high number of G-to-A transitions. The mutation pattern was similar to that previously reported for A3B, with a preference for the GA context. In summary, these observations suggest that changing 4 residues in the amino terminus of A3B not only retargets the protein from the nucleus to the cytoplasm but also enhances its ability to restrict HIV while retaining inhibition of retrotransposition.
INTRODUCTION
The APOBEC3 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3) family of proteins is a major component of the innate immunity system, acting against a variety of viruses, including retroviruses, as well as other DNA pathogens such as LINE-1 and other retrotransposons (17, 20, 45). APOBEC3 (A3) proteins are cytidine deaminases acting on single-stranded DNA and inhibit retroviral infection by causing C-to-T transitions in the nascent minus strand during reverse transcription (RT) (10); however, other modes of action have also been described and may predominate in some APOBEC3/retrovirus combinations (9, 32, 33, 44).
The targets for the individual APOBEC3 family members vary and are a function of expression patterns and cellular localization. The best-characterized member of the APOBEC3 family is human APOBEC3G (A3G), which accumulates in the cytoplasm and has strong antiretroviral activity (1, 19, 25, 32, 33). The Vif protein of human immunodeficiency virus type 1 (HIV-1) has evolved to counteract A3G by causing its polyubiquitination and degradation (24, 40, 41). In contrast, human APOBEC3B (A3B) is localized predominantly in the nucleus and is known to inhibit LINE-1 retrotransposition potently (6, 29, 45). Although A3B has been reported to restrict HIV-1 infection, it is not targeted by Vif. The level of restriction of HIV-1 by A3B reported in the literature varies from about 2- to 40-fold (4, 14, 48). The reason for these discrepancies is not clear at this point. Expression of A3B in hematopoietic cells is low relative to that of most of the other A3 proteins (27, 37, 48) or is even absent (4, 14, 15). This has led to the hypothesis that HIV-1 normally is not exposed to A3B and hence has not evolved a mechanism to counteract A3B restriction.
APOBEC3 proteins consist of either one or two paralogous zinc-coordinating cytidine deaminase (CDA) domains (11); these so-called Z domains can be classified into Z1, Z2, or Z3 based on evolutionary relationship (28), and different arrangements of these CDAs make up the A3 proteins in different species. Humans have 3 genes encoding a single CDA and 4 genes encoding two CDAs called APOBEC3A (A3A) through APOBEC3H (A3H). A3A, APOBEC3C (A3C), and A3H are Z1, Z2, and Z3 single-domain proteins, respectively, while A3B and A3G share the Z2Z1 configuration (28), and APOBEC3D/E (A3D/E) and APOBEC3F (A3F) each consist of two Z2 CDAs. However, the first 58 amino acids of A3B are identical to those of A3D/E, while A3G and A3F differ from each other by only one residue within this region. A3B is almost exclusively located in the nucleus (7) based on immunofluorescence microscopy, although it has been demonstrated to shuttle between the cytoplasm and the nucleus. A3A and A3C are found dispersed throughout the cell, while the other A3 proteins largely partition to the cytoplasm (45). Stenglein et al. (46) recently showed that the first 60 amino acids control the cytoplasmic localization of A3G. They further delineated four of the 24 residues which differ between A3B and A3G within that stretch to be of particular importance. In spite of its nuclear localization, A3B is efficiently incorporated into HIV-1 particles (5, 14, 48). Packaging is not dependent on the presence of Vif, which has been reported to cause the translocation of A3B from the nucleus to the cytoplasm (31), as wild-type (wt) HIV-1 and HIV-1Δvif particles incorporate A3B to the same level (14, 48).
For this report we studied the effects of the amino terminus of A3B, which controls cellular localization, on the infectivity of HIV-1 and human T-cell leukemia virus type 1 (HTLV-1) virus-like particles (VLPs). While A3B itself showed only minimal restriction of infection in our single-cycle infectivity assay, an A3G/A3B hybrid (A3G/B) in which the first 60 amino acids of A3B were replaced with the corresponding sequences of A3G was highly inhibitory to HIV-1 and HTLV-1 infection. A3G/B caused higher levels of cytidine deamination of HIV-1 provirus than A3G with the consensus target sequence previously reported for A3B. A3B and A3G/B inhibited LINE-1 retrotransposition equally, and packaging levels of the two A3 proteins were comparable. These findings indicate that incorporation of A3 proteins, even with two active CDAs, does not necessarily result in inhibition of infection.
MATERIALS AND METHODS
Plasmid construction.
The hemagglutinin (HA)-tagged human A3B was kindly provided by Bryan R. Cullen (Duke University Department of Molecular Genetics and Microbiology), and the HA-tagged human A3G construct was a kind gift from Reuben S. Harris (University of Minnesota, Department of Biochemistry, Molecular Biology, and Biophysics). The following HA-tagged A3B/G and A3G/B chimeric constructs were generated by overlapping PCR: A3B1-60 A3G61-384 (5′ CCT TGA GTC GAC ATG AAT CCA CAG ATC AGA AAT CCG A, 5′TTC AAG CCT CAG TAC CAC GCG GAA TAC ACC TGG CCT CG, 5′ ACT CGA GAG CGT AAT CTG GAA CAT C) and A3G1-60 A3B61-382 (5′ GTC GAC ATG AAG CCT CAC TTC AGA AAC ACA, 5′ TTC AAG CCT CAG TAC CAC GCG GAA TAC ACC TGG CCT CG, 5′ ACT CGA GGA ATT GGT TTC CCT GAT TCT GGA G). Amino acid substitution mutants were created using site-directed mutagenesis (QuikChange; Stratagene). APOBEC3 genes were cloned into the cytomegalovirus (CMV) promoter-driven vector pcDNA3.1 (Invitrogen, Carlsbad) or pCMVpA, a mammalian expression vector with the immediate-early CMV promoter and the simian virus 40 (SV40) polyadenylation site. Packaging constructs pCMVHIV-1, pCMVHIV-1Δvif, and pCMVHT-1 MΔEnv and reporter constructs pUCHR-GFPLuc, pCMVRU5-GFPLuc, and pCMV-VSV-G for pseudotyping have been described previously (13).
For LINE-1 retrotransposition assays, we used the pLRE-3inLuc vector, which contains a 5′ untranslated region (5′UTR), ORF1, ORF2, CMV-driven luciferase in the antisense orientation which is disrupted by an intron, and a 3′UTR similar to those described by Moran et al. (34). For successful luciferase expression, LINE-1 transcription, splicing, reverse transcription, and integration are necessary.
Cells, transfections, and infections.
Human embryonic kidney 293T cells and HeLaP4 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. For transfection, 293T cells were seeded in 6-well plates at 24 h prior to transfection and reached 90% to 95% confluence at the time of transfection. For HIV-1 production, 5 × 105 293T cells were transfected with 0.5 μg packaging plasmid (pCMVHIV-1 or pCMVHIV-1Δvif), 1.5 μg reporter plasmid pUCHR-GFPLuc, and 0.125 μg pCMV-VSV-G. For HTLV-1 production, 0.5 μg packaging plasmid (pCMVHT-1MΔEnv), 1.5 μg reporter plasmid pCMVRU5-GFPLuc, and 0.125 μg pCMV-VSV-G were used. All transfections were performed using GenJet transfection reagent version II (SignaGen Laboratories), according to the manufacturer's protocol. Supernatants were collected at 48 h after transfection, filtered, and standardized by enzyme-linked immunosorbent assay (ELISA) (Zeptometrix, Buffalo NY). HeLaP4 cells were used as target cells for infection assays. Luciferase activity was measured at 72 h after infection using Promega luciferase reagent and a Lumat LB9501 luminometer (Berthold). For the LINE-1 assay, 293T cells were seeded in duplicates in 12-well plates and transfected with 0.75 μg of pLRE-3inLuc with or without different amounts of APOBEC3 constructs. After 72 h, cells were lysed and luciferase activity was measured using the Bright-Glo luciferase assay system kit (Promega) according to the manufacturer's protocols.
Immunoblotting.
Viruses from transfected 293T cells were collected by ultracentrifugation through a 20% glycerol cushion for 90 min at 100,000 × g or pelleted in a microcentrifuge at 20,000 × g for 90 min for smaller cultures. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer with protease inhibitors (Roche) for large cultures (10-cm dishes) or pelleted in a microcentrifuge at 20,000 × g for 90 min for smaller cultures. Cell and virus lysates were fractionated by SDS-PAGE under reducing conditions and transferred to Immobilon membranes (Millipore). HA-tagged APOBEC3 proteins were detected with mouse anti-HA monoclonal antibodies (MMS-101P; Covance). HIV-1 p24-CA (Zeptometrix or AIDS Research and Reference Reagent Program), HIV-1 p17-MA, and HTLV-1 p19-MA were detected with mouse monoclonal antibodies (Zeptometrix). Horseradish peroxidase-conjugated anti-mouse immunoglobulin G (Cell Signaling) was used as a secondary antibody. Cell lysates were also analyzed with antitubulin antibodies (Sigma).
Determination of APOBEC3 packaging into the virus core.
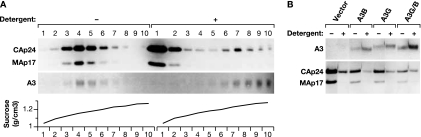
Pelleted viruses were dissolved in phosphate-buffered saline (PBS). Equal aliquots were left untreated or adjusted to 0.2% Triton X-100 for 5 min. Samples were layered on 20 to 70% sucrose gradients and pelleted for 1.5 h at 210,000 × g in a TLA55 rotor. Ten fractions were collected, and the refractive index in each fraction was determined. The entire gradient was analyzed by immunoblotting using anti-HA, anti-MAp17, and anti-CAp24 antibodies. Fractions with refractive indices of 1.39 and 1.43 of the gradients with untreated or detergent-treated samples, respectively, were further compared by immunoblotting as described above to demonstrate incorporation of A3 proteins into the core.
Infection and analysis of mutation frequency.
HeLaP4 target cells were infected with equal amounts of virus based on p24 CA levels determined by ELISA (Zeptometrix) and treated with recombinant DNase I (Roche). Cells were harvested 48 h after infection, and total cellular DNA was extracted using a QIAamp DNA Blood Minikit (Qiagen). Total isolated DNA was digested with DpnI enzyme, and luciferase gene sequences were PCR amplified. Primer sequences are available on request. PCR products were cloned into pCR2.1-TOPO (Invitrogen). Plasmid DNAs were isolated from single bacterial colonies and sequenced using the M13 sequencing primers (Invitrogen).
Immunofluorescence.
HeLaP4 cells were seeded on 4-well LabTec chambered cover glasses (Nunc). In the case of HA-tagged protein immunostaining, cells were fixed with 2% paraformaldehyde (PFA), permeabilized with 0.1% Triton X-100, and blocked in 3% bovine serum albumin (BSA)–PBS for 60 min, followed by incubation with primary antibody (anti-HA antibody) appropriately diluted in 3% BSA–PBS. Cells were rinsed 3 times and incubated for 30 min with secondary antibody. After 3 washes in PBS, cells were mounted using Prolong Gold antifade reagent (Invitrogen). Images were acquired using a DeltaVision RT deconvolution microscope.
Quantification of viral DNA synthesis in infected cells by real-time PCR.
A total of 3 × 106 293T cells were seeded in 10-cm dishes. After 24 h, cells were transfected with 2 μg of A3, 2 μg of pCMVHIV-1Δvif, 6 μg of pUCHR-GFPLuc, and 0.5 μg of pCMV-VSV-G. After 24 h the medium was changed, and after 48 h virus supernatants were collected, filtered through 0.45-μm polyvinylidene difluoride (PVDF) filters (GS-Tek), and pelleted through a 20% glycerol cushion to minimize DNA contamination. Pelleted virus was dissolved in PBS and treated with DNase I (Roche) for 2 h. HelaP4 cells were seeded at a density of 106 cells per 60-mm-diameter dish overnight and were infected with equal amounts of HIV-1Δvif p24 CA-normalized virus as previously described. Supernatant from transfected 293T cells with only pUCHR-GFPLuc was added to HelaP4 cells to determine the level of contamination with transfected DNA. Cells were harvested at 24 h after infection, and total cellular DNA was extracted using a QIAamp DNA Blood Minikit (Qiagen). Extracted DNA was digested with DpnI (NEB), which digests only methylated plasmid DNA and not cellular DNA. The amounts of unintegrated proviral DNA were quantified using an Applied Biosystems 7500 real-time PCR system. The primer-probe set for detection of late DNA synthesis products was designed as described previously (32) and purchased from IDT.
RESULTS
Targeting A3B to the cytoplasm enhances its antiviral activity.
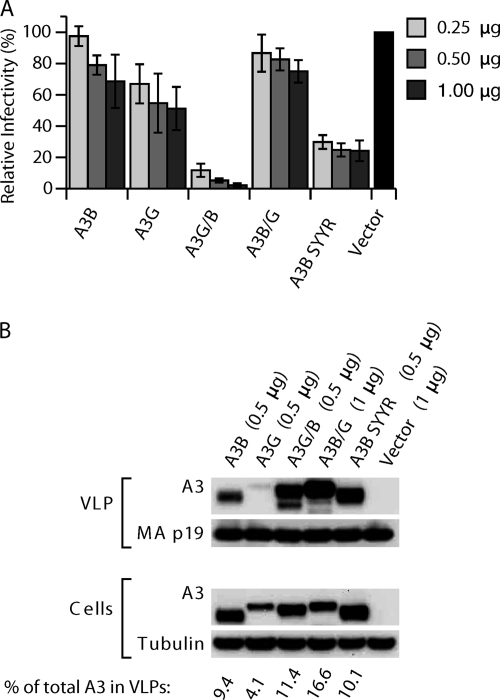
A3B and A3G (Fig. 1A) have the same subunit organization consisting of an amino-terminal Z2 domain and a carboxy-terminal Z1 domain (28), while A3F and A3D/E share the Z2Z2 configuration. A3B and A3D/E are identical to each other for the first 58 amino acids, while A3F and A3G differ from each other by only one residue in that region. A3B is a nuclear protein, while A3D/E, A3F, and A3G are in the cytoplasm (45). To investigate whether nuclear/cytoplasmic localization correlates with APOBEC3 antiviral activity, we constructed expression plasmids for chimeric proteins with the first 60 amino acids switched between A3G and A3B, similar to those described by Stenglein et al. (46) (Fig. 1A), including a carboxy-terminal hemagglutinin (HA) tag. As expected, A3G and A3B were located in the cytoplasm or nucleus, respectively, in HelaP4 cells (Fig. 1B) and in 293T cells (see supplemental Fig. A1B at http://home.ncifcrf.gov/hivdrp/Pak_et_al_SuppFig1_JVITMP-01576-11_1.pdf). A3G/B, which contains the first 60 amino acids of A3G on the A3B backbone, was located mainly in the cytoplasm both in HelaP4 cells (Fig. 1B) and 293T cells (see supplemental Fig. A1B at http://home.ncifcrf.gov/hivdrp/Pak_et_al_SuppFig1_JVITMP-01576-11_1.pdf), while the reciprocal construct A3B/G was predominantly nuclear, although the cellular distribution was not as exclusive as for the parental A3B and A3G proteins. We next tested the ability of these chimeras to restrict the infectivity of HIV-1Δvif VLPs produced in 293T cells. Empty vector plasmid (pcDNA3.1) or increasing amounts of plasmids (0.25, 0.5, and 1 μg) encoding the different HA-tagged APOBEC3s were cotransfected with HIV packaging construct pCMVHIV-1Δvif, HIV transfer vector pUCHR-GFPLuc (which expresses a green fluorescent protein [GFP]-luciferase fusion), and pCMV-VSV-G into 293T cells. After 48 h viral stocks were collected, normalized for antigen content by p24 ELISA, and used to infect HeLaP4 cells. The infected cells were lysed and analyzed for luciferase activity after 72 h. We observed that in this assay the restriction of HIV-1Δvif by A3B was not statistically different from that in control infections with empty vector even at the highest level of A3B input (Fig. 1C). In contrast, A3G/B restricted the virus 6- to 50-fold in the presence of increasing amounts of construct. As expected, A3G inhibited HIV-1Δvif 20- to 100-fold depending on the concentration of A3G (Fig. 1C). A3B/G showed no antiviral activity even though expression levels were high and packaging into VLPs was efficient. We tested several independently generated constructs, which all gave the same result. The results were similar when we used 293T cells as target cells for the infection.
Fig. 1.
The A3G/B chimera is located in the cytoplasm and active in HIV restriction. (A) Structural organization of and comparison between different APOBEC3 (A3) genes and chimeric constructs. The introns (thin lines), which vary in length in different genes, and exons (rectangles) of the double-CDA genes are indicated. Sequence homologies between different proteins (and genes) are represented by color. *, active CDA domain. (B) Cellular localization of A3G, A3B, and chimeras. HeLaP4 cells were transiently transfected with the expression vector for A3B-HA, A3G-HA, A3G/B-HA, and A3B/G-HA plasmids. Cells were fixed, permeabilized, and stained with mouse anti-HA antibodies and Alexa-Fluor-488 goat anti-mouse IgG. (C) Antiviral activities of A3G/B and A3B/G chimeras. Single-cycle replication assays were performed with VLPs produced in 293T cells cotransfected with varied amounts of plasmids encoding HA-tagged A3B, A3G, A3G/B, or A3B/G and pCMVHIV-1Δvif, pUCHR-GFPLuc, and pCMV-VSV-G. HeLaP4 cells were infected with ELISA-standardized amounts of VLPs, and productive infection was measured as luciferase activity. Values are presented as percent infectivity relative to virus produced in the absence of APOBEC3. Data are the means from five independent experiments, and error bars represent the standard deviations. (D) Western blots demonstrating the expression in cells and packaging of A3 proteins and chimeras into the VLPs. Cell and VLP extracts were immunoblotted and probed with anti-HA antibody. Tubulin and HIV-1 CA proteins were also detected to verify equal cell and virion loading.
To assess whether the difference in restriction between the mainly nuclear A3B and A3B/G constructs versus the cytoplasmic A3G and A3G/B was due to differential incorporation of the A3 proteins into VLPs, we analyzed equal amounts of VLP by immunoblot analysis. Figure 1D shows that irrespective of their cellular location or restriction activity, all four A3 proteins were packaged with about equal efficiency into VLPs. Parallel experiments performed with wt (Vif+) HIV packaging constructs also showed little restriction by A3B but significant restriction by A3G/B, while both were efficiently incorporated into VLPs. As expected, A3G was depleted from the cells in the presence of Vif and hence was incorporated substantially less efficiently (see supplemental Fig. A1C at http://home.ncifcrf.gov/hivdrp/Pak_et_al_SuppFig1_JVITMP-01576-11_1.pdf, and see below).
A3B and A3G as well as the chimera are in the viral core.
The antiretroviral activity of APOBEC3 proteins requires packaging into the HIV-1 virus core through the direct interaction with the Gag polyprotein and RNA. This interaction is mediated by the C-terminal Gag nucleocapsid (NC) region (2, 8). To explore the possibility that the lack of restriction activity of A3B was due to exclusion from the viral core, preparations of VLPs isolated from 293T cells cotransfected with pCMV-HIVΔvif and A3B-, A3G-, or A3G/B-expressing plasmids or empty vector were fractionated by centrifugation through 20 to 70% sucrose gradients with or without prior lysis by 0.2% Triton X-100. The detergent removes the lipid bilayer membrane, matrix protein, and surface protein from the virus while preserving the virus core, which contains about 40% of the CA present in the VLP, as described previously (26, 39, 47). We analyzed the gradients by immunoblotting and measurements of the refractive indices to identify the fractions containing VLPs and cores in the gradients of untreated or detergent-treated samples, respectively. Figure 2 A shows the analysis of two of the gradients with the A3B sample, showing that most of the A3B protein is associated with the core fraction after detergent treatment, while it is contained in the VLP without lysis. Figure 2B compares the fractions containing the cores from the gradients with detergent-treated samples (density, 1.25 g/cm3) to gradient fractions with the untreated VLPs (density, 1.16 g/cm3). All core fractions contained capsid but very little matrix protein, and A3B and A3G/B were packaged into the viral cores with the same or greater efficiency than A3G. There was no APOBEC3 protein detectable in the mock-treated viral pellet. Thus, this experiment suggested no difference in the incorporation between A3B, A3G, and the A3G/B chimera within the VLPs.
Fig. 2.
A3B and A3G/B are located in the core. VLPs were produced in 293T cells transfected with pCMVHIV-1Δvif with or without plasmids encoding the indicated A3 proteins. Equal aliquots were centrifuged through 20 to 70% sucrose in PBS gradients after treatment with or without detergent (0.2% Triton X-100). The presence of A3 proteins in VLPs or core pellets was analyzed by immunoblotting with anti-HA antibodies. Blots were reprobed with anti-MA p17 and anti-CA p24 HIV-1 antibodies to demonstrate the disruption of the viral envelope by the detergent. (A) Profile of the entire gradients of the A3B sample. The densities of the fractions were determined by refractive index and are given below the immunoblots. (B) Fractions 4 and 7 of the gradients loaded with untreated and detergent-treated samples, respectively, were analyzed by immunoblotting.
Residues 18, 19, 22, and 24 are responsible for localization of A3G/B and its increased antiviral effect on HIV-1Δvif.
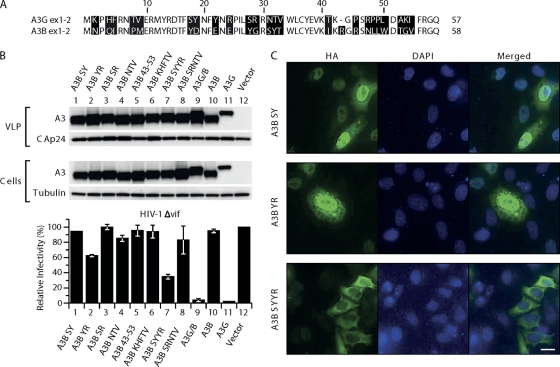
We next wanted to narrow down further which of the 60 amino acids within the A3G residues were responsible for its accumulation in the cytoplasm. There are 24 differences in the alignment of this stretch of the human A3B peptide sequence to the human A3G sequence (Fig. 3A). We probed the importance of the differences by mutagenesis, changing residues in A3B to those found at the corresponding positions in A3G. The initial set of five mutants had the following changes: A3B SY (Y18S, D19Y), A3B YR (E22Y, E24R), A3B SR (Y28S, G29R), A3B NTV (SYT31-33NTV), and A3B 43-53 (IKRGRSNLLWDTGV43-53TKGPSRPRLDAKI). None of the five A3B mutants were able to affect HIV-1Δvif infectivity significantly (Fig. 3B) or showed a significant change in cellular localization (see A3B SY and YR as examples in Fig. 3C). Next we combined some of these mutations by substituting four or five amino acids at a time, resulting in the following constructs: A3B KHFTV (N2K, Q4H, I5F, P8T, M9V), A3B SYYR (Y18S, D19Y, E22Y, E24R), and A3B SRNTV (Y28S, G29R, S31N, Y32T, T33V). Only the A3B SYYR protein was located mainly in the cytoplasm (Fig. 3C), and it was able to inhibit HIV-1Δvif infectivity about 3-fold (Fig. 3B). All constructs expressed comparable levels of protein in 293T cells and were equally well incorporated into VLPs when cotransfected with HIV-1Δvif viral vectors (Fig. 3B). These findings are in agreement with the results of Stenglein et al. (46), who found that changing the equivalent residues singly or in small patches in A3G to those present in A3B caused the resulting protein to be more nuclear in localization than A3G.
Fig. 3.
Mutational analysis of the first 60 amino acids in A3B. (A) Amino acids in A3B were replaced with residues present in A3G at corresponding positions. 293T cells were cotransfected with the A3B mutants with pCMVHIV-1Δvif, pUCHR-GFPLuc, and pCMV-VSV-G; HeLaP4 cells were infected with the resultant VLPs, and luciferase activity was measured at 48 h postinfection. The infectivity of virus produced in the presence of the different mutants relative to that in the absence of A3 proteins, set at 100%, is shown. Data are the means from 3 independent experiments, and error bars represent the standard deviations. (B) Expression of A3 mutants in cells and packaging into the VLPs. The presence of A3 proteins in virus and cell extracts was analyzed by immunoblotting with anti-HA antibody probing. Tubulin and HIV-1 CA proteins were also detected to verify equal cell and virion loading. (C) Localization of A3B mutants. HeLaP4 cells were transiently transfected with plasmids encoding the mutants A3B SY (Y18D, S19Y) A3B YR (E22Y, Y24R), and A3B SYYR (Y18D, S19Y, E22Y, Y24R). Cells were fixed, permeabilized, and stained with mouse anti-HA antibodies and Alexa Fluor 488 goat anti-mouse IgG. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
A3G/B has two active CDA domains.
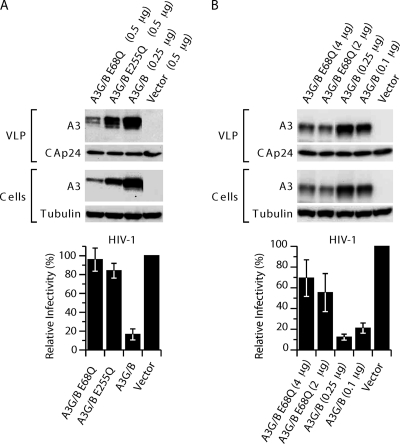
We next asked whether the catalytic activities of both domains of A3G/B were necessary for inhibition of HIV-1. In contrast to A3G, where only the C-terminal Z2 domain is catalytically active, both CDA domains in A3B have been shown to be functional and to contribute to the restriction activity of A3B (5). We generated mutations E68Q and E255Q in the active centers of the CDAs of A3G/B, as well as the double mutant. The glutamic acid-to-glutamine substitution in the coordination points of the zinc finger motif (H-X-E-X25-28-P-C-X2-4-C) has been shown to be inactivating (36). Cotransfection of these mutants with pCMVHIV-1, pUCHR-GFPLuc, and pCMV-VSV-G (Fig. 4A) showed that inactivation of either CDA resulted in a reduction of the inhibitory activity (Fig. 4A), as was previously described by others for A3B (5, 18). Similar results were obtained when pCMVHIV-1Δvif was used as the packaging construct in the experiment (data not shown). The results obtained with the E68Q mutants are complicated by the instability of this protein. Consistent with previous work, the CDA-inactive proteins, especially A3G/BE68Q, accumulated to lower steady-state levels in the cells and VLPs (5). Only after increasing the amount of transfected plasmid DNA to 4 μg did the level of the A3G/BE68Q protein become comparable to that obtained with 0.1 μg of A3G/B plasmid (Fig. 4B). However, even with these qualifications, it is clear that the overall restrictive capacity of CDA mutants is compromised, although not completely abolished, and that both CDAs contribute to HIV-1 restriction.
Fig. 4.
A3G/B inhibition of HIV-1 infection depends on catalytically active sites. (A) Single-cycle replication assays were performed with VLPs produced in 293T cells cotransfected with the A3G/B chimera and catalytically active site mutants, pCMVHIV-1, pUCHR-GFPLuc, and pCMV-VSV-G. HeLaP4 cells were infected with the resultant VLPs, and productive infection was measured by the luciferase activity. Values are presented as percent infectivity relative to virus produced in the absence of A3G/B and mutants. The data are the means from three independent experiments, and error bars represent the standard deviations. (B) Expression and virus packaging of A3 proteins and chimeras with catalytically active site mutations. The presence of A3 proteins in virus and cell extracts was analyzed by immunoblotting with anti-HA antibody probing. Tubulin and HIV-1 CA levels were detected to verify equal cell and virion extract loading.
Hypermutation of viral DNA by A3B and A3G/B proteins.
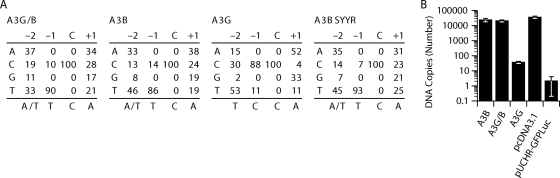
To determine whether A3G/B and A3B-SYYR caused cytidine deamination during reverse transcription, we infected HeLaP4 cells with virus preparations produced in 293T cells by cotransfection of pCMVHIV-1Δvif, pUCHR-GFPLuc, and pCMV-VSV-G with plasmids encoding these proteins. A fragment of the luciferase gene present in the transfer vector was amplified by PCR from the DNA of the infected HeLaP4 cells. For comparison we also performed the experiment with viruses produced in the presence of A3B and A3G or without A3 protein. The amplicons were cloned, and 20 independent clones were sequenced in each set. The results are shown in Table 1. A3B induced G-to-A mutations in 25% of HIV reverse transcripts, averaging 3.15 changes per clone. However, as only 25% of all proviruses experienced hypermutation, for those that were affected the rate was 12.5 changes/clone. In contrast, the A3G/B chimera caused hypermutations in 95% of the proviruses, with 18.7 G-to-A transitions per clone. As expected based on the lower level of HIVΔvif restriction by A3B-SYYR, a smaller proportion of the clones (61%) were mutated, showing 5.9 changes on average. The number of mutations induced by A3G averaged 5.5 G-to-A transitions/clone, and 90% of the proviruses had changes. Consistent with previous reports (4), with A3B 86% of the G-to-A transitions occurred in a GA context, while A3G preferred the GG context in 88% of the sequenced events. As expected, the sequence preference of A3G/B and A3B-SYYR was also GA, as 86 and 93% of the mutations occurred in that context, respectively (Fig. 5A).
Table 1.
G-to-A mutations induced by A3B, A3G/B, A3BSYYR, and A3Ga
| Parameter | Value for sample |
||||
|---|---|---|---|---|---|
| Vector | A3B | A3G/B | A3BSYYR | A3G | |
| Infectivity (%) | 100 | 80 | 2 | 13 | 1 |
| Total no. of bp sequenced | 23,084 | 21,734 | 22,958 | 23,391 | 22,398 |
| % Clones mutated | 5 | 25 | 95 | 61 | 90 |
| No. of G-to-A mutations | |||||
| Total | 1 | 63 | 374 | 82 | 110 |
| Per clone | 0.05 | 3.15 | 18.7 | 5.9 | 5.5 |
293T cells were transfected with pCMVHIV-1Δvif, pUCHR-GFPLuc, pCMV-VSV-G, and the indicated A3 construct or empty vector. HeLaP4 cells were infected with the resultant virus, and genomic DNA was isolated at 48 h postinfection. Proviral DNA was amplified and cloned, and 20 independent clones were sequenced. The numbers of G-to-A mutations were counted, and mutation rates were calculated.
Fig. 5.
Comparison of the sequence contexts preferred by A3B, A3G, and chimera A3G/B. (A) The matrices show the frequency (in percent) of the four bases at positions −2, −1, and + 1 relative to the dC residue, which was deaminated. The consensus sequences are shown on the bottom of each matrix. (B) Amounts of late RT products quantified by the U5Ψ primer-probe set. Viral DNA levels were determined at 24 h after infection with HIV-1Δvif virus produced in the presence of the indicated A3 proteins or pcDNA3.1. To determine DNA contamination, pUCHR-GFPLuc vector was used. Error bars in the graph represent the standard deviations from the means from three independent experiments.
A3B and A3G/B do not inhibit viral DNA synthesis in infected cells.
Although the high degree of hypermutation alone could explain the loss of infectivity caused by A3G/B, we tested whether A3G/B also interferes with reverse transcription as has been reported for A3G (32, 33). For virus production, 293T cells were transiently transfected with packaging construct pCMVHIV-1Δvif, transfer vector pUCHR-GFPLuc, and pCMV-VSV-G envelope without or with the different A3 constructs. The pcDNA3.1 vector was used as an empty vector control, and the pUCHR-GFPLuc reporter vector was transfected alone as a control for plasmid DNA contamination. The resulting virus stocks were normalized for p24 CA and used to infect HelaP4 cells. DNA was extracted from infected cells at 24 h postinfection, and the number of reverse transcription products was quantified by real-time PCR using the standard late RT product primer-probe set U5Ψ (32). The effects of A3B and A3G/B on HIV-1Δvif virus replication were compared with that of A3G. As shown on Fig. 5B, neither A3B nor A3G/B inhibited viral DNA synthesis in infected cells; only A3G drastically reduced late viral reverse transcripts, by about 1,000-fold. The effect of A3G was higher than previously reported; however, the study by Mbisa et al. (32) was performed under conditions where A3G was present at physiological levels in the cell. In contrast, we transfected large amounts of the A3-expressing construct to ensure that any effects would be apparent.
A3G/B inhibits LINE-1 retrotransposition.
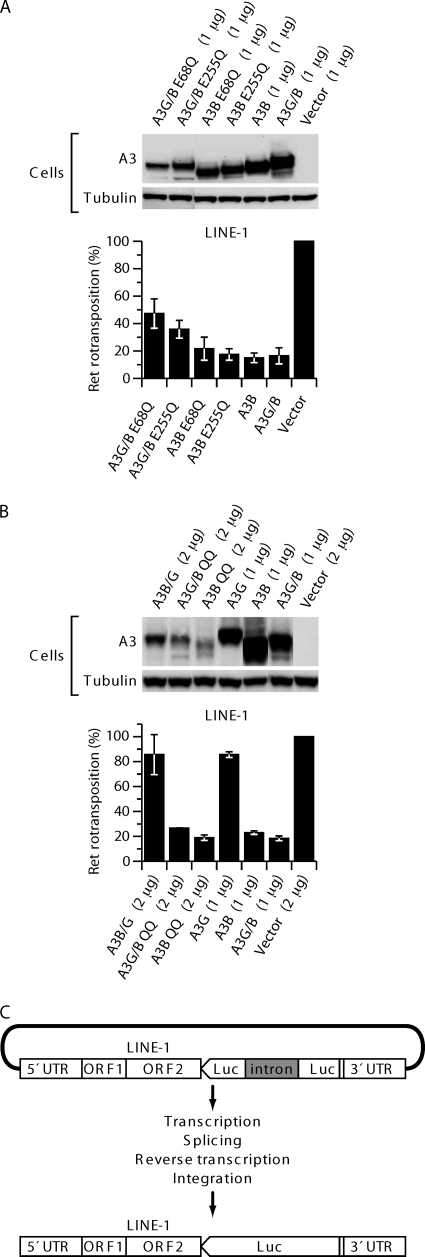
A3B has been shown to inhibit LINE-1 retrotransposition efficiently, while A3G has little effect on LINE-1 activity (6, 29, 45). The A3B inhibition is not dependent on functional CDA, and LINE-1 elements that have undergone retrotransposition in the presence of A3B show almost no traces of cytidine deamination (6, 45). To compare the activities of the A3 constructs in 293T cells, we used pLRE-3inLuc to measure retrotransposition. The pLRE-3inLuc construct (Fig. 6C) is similar to previously described vectors used to assess LINE-1 retrotransposition based on GFP or NeoR gene expression (21, 34), except that at the end of the entire LINE-1 element it contains a firefly luciferase expression cassette transcribed in the opposite orientation to that of the LINE-1 element. The luciferase gene is inactivated by an insert, which can be spliced from the LINE-1 transcript but not from the luciferase gene mRNA in the transfected cells. Luciferase activity in the culture is dependent on complete retrotransposition events, including transcription, splicing, and translation of LINE-1 sequences as well as integration-coupled reverse transcription. Figure 6A shows that A3B, A3B E68Q, and A3B E255Q inhibited retrotransposition about 6-fold. A3G/B was as inhibitory as A3B, while A3B/G and A3G did not inhibit retrotransposition (Fig. 6B). Mutations E68Q and E255Q in the active centers of A3G/B impaired the restriction activity only slightly, and again the effects were comparable to those seen with A3B with the corresponding mutations. Similar results were obtained with A3B and A3G/B double mutants (A3B QQ and A3G/B QQ) with inactivating mutations in both CDA active centers. Even though the steady-state levels of the double mutant proteins were even lower than those of the single mutants, restriction was as active as that observed with the wild-type proteins (Fig. 6B). These results show that the activities of A3G/B and A3B are similar with respect to retrotransposition and are in agreement with what others have reported for A3B (7, 45).
Fig. 6.
A3B and the A3G/B chimera inhibit LINE-1 retrotransposition in a catalytically active site-independent manner. (A) 293T cells were transfected in the presence or absence of A3B and catalytically active site mutants or chimera A3G/B and catalytically active site mutants of A3G/B with LINE-1 indicator plasmid pLR-3inLuc. Luciferase activity was measured as an indicator of productive retrotransposition events. The data represent the means from five independent experiments, and error bars represent the standard deviations. (B) Expression of A3 proteins in 293T cells cotransfected with pLR-3inLuc. Tubulin was also detected to confirm equal cell extract loading. (C) Schematic of the replication-dependent retrotransposition assay. pLRE-3inLuc contains an intron-interrupted firefly luciferase (FF luc) expression cassette transcribed in opposite orientation to the LINE-1 element. The splice donor and acceptor signals are in sense orientation on the LINE-1 transcript. Thus, the LINE-1 transcription, splicing, reverse transcription, and integration are required to generate an uninterrupted FF luc gene.
Effects of A3G, A3G/B, and A3B/G proteins on HTLV-1.
We also investigated whether A3B, A3G, A3G/B, A3B/G, and A3BSYYR can restrict human T-cell leukemia virus type 1 (HTLV-1) infection. Like HIV, HTLV-1 is T-cell tropic in vivo and causes adult T-cell leukemia and HTLV-associated myelopathy/tropical spastic paraparesis (HAM/TSP). We and others have previously shown that HTLV-1 resists A3G antiretroviral effects by preventing packaging of A3G and other A3 proteins (13, 35, 42). The exclusion is dependent on a stretch of amino acids in the C-terminal region of the nucleocapsid protein (NC). Analysis of proviral sequences from patients with HAM/TSP showed that there is no evidence for human A3 induced G-to-A hypermutations in vivo (30). In agreement with our previous report, A3G did not affect HTLV-1 infectivity in this experiment (Fig. 7A) (13), and it is clearly packaged less efficiently than the other A3 proteins (Fig. 7B). Interestingly, the activities of the other A3 proteins against HTLV-1 mirrored those against HIV-1. A3B and the chimeric protein A3B/G also did not inhibit HTLV-1 transmission, but the A3G/B chimera decreased HTLV-1 infectivity from 6- to 50-fold in a dose-dependent manner. Mutant A3B SYYR protein also inhibited infectivity of HTLV-1 about 3-fold. Except for A3G, all A3 proteins were efficiently packaged into the HTLV-1 virions (Fig. 7B). Given that HIV-1 and HTLV-1 have evolved very different mechanisms to counteract the effects of A3G, this result indicates that the A3G/B chimera is not recognized by either viral component.
Fig. 7.
Antiviral activities of A3B, A3G, and chimeric proteins against HTLV-1. (A) 293T cells were cotransfected in the presence or absence of the indicated A3 plasmids with pCMVHT-1Δenv, pCRU5-GFPLuc, and pCMV-VSV-G. HeLaP4 cells were infected with ELISA-standardized amounts of VLPs, and productive infection was measured by luciferase activity. Values are presented as percent infectivity relative to virus produced in the absence of A3 protein. The data are the means from five independent experiments, and error bars represent the standard deviations. (B) Expression in cells and packaging of A3 proteins and chimeras into the VLPs. Cell and virus extracts were immunoblotted with anti-HA antibodies. Tubulin and HTLV-1 MA proteins were also detected to verify equal cell and virion loading.
DISCUSSION
The role of A3B in restricting HIV-1 in vivo is under debate; on one hand there is evidence that individuals who are homozygous deficient in A3B due to a naturally occurring deletion of the entire gene are more susceptible to HIV-1 infection and generally have a faster disease progression (3), while others find that the deletion has no consequences for HIV-1-induced disease (23). Similarly, the effect of A3B on HIV-1 infection in experimental settings varies significantly between different groups, from the 15% inhibition we observed to 50-fold restriction (5, 14). Most studies show only 2- to 3-fold restriction with A3B (4, 12, 38, 48), clearly less than with A3G. The clear differences in the antiviral effect of A3B and A3G in our system facilitated the analysis of hybrid constructs. In this study we focused on the effect of reciprocal changes in the amino-terminal end of the proteins, highlighting the effect of that region on cellular localization and antiviral activity.
The fact that all A3 constructs were packaged nearly equally well into HIV-1 VLPs, and even into cores, could indicate that there are subtle differences in the RNA-Gag-A3 complexes formed by the different proteins, which affect whether the A3 protein can exert antiviral activity during reverse transcription. We showed recently how differences in the assembly can affect the activity of the complex: incorporation of an A3G construct with a mutation in the CDA1 RNA-binding site can be rescued by recruiting it to RNA by the addition of a different RNA-binding motif; this construct was efficiently incorporated into VLPs but has no HIV-1 inhibitory activity (16), showing that the precise formation of the complex is necessary for function.
A3G/B and A3B SYYR differ from A3B by only 24 and 4 residues in the amino terminus, respectively, and are no longer mainly localized to the nucleus. The changes in the amino-terminal end also confer restriction activity to the proteins when incorporated into VLPs produced by our HIV vector system. It is unclear why the translocation of the protein should have this result, as the nuclear localization of A3B does not interfere with its incorporation into VLPs, indicating that A3B can be recruited to the RNA-A3B-Gag complex efficiently while A3B shuttles between the nucleus and the cytoplasm (6). However, it is possible that variation in the complex, for instance, the degree of multimerization, is caused by differences in local concentrations. The change from a mainly nuclear to a cytoplasmic localization may result in high levels of incorporation into a few particles versus lower, but sufficient, incorporation into many particles. This difference would not be apparent at the level of immunoblotting. Alternatively, the cellular localization may be not the cause but the consequence of altered interactions with cellular components or of a varying degree of homomultimerization. A recent paper by Huthoff et al. (22) presented a model based on the APOBEC2 structure that suggested that the N-terminal arginine residues R24 and R30 in A3G are part of a positively charged pocket involved in RNA binding as well as a homodimer interface. While R30 is conserved in both proteins, A3B has a glutamic acid at position 24, which would be expected to weaken such a pocket. The SYYR mutation in the A3B N terminus not only makes the E24R change but also replaces D19 and E22 with tyrosines, thus removing additional negatively charged residues from the area. In addition to the change in charge, reconstituting the aromatic amino acids at positions 19 and 22 may enhance the cooperation with residues Y125/W127 in A3G, as discussed by Stenglein et al. (46). As the main residues in the RNA-binding site around Y125/W127 are conserved between A3B and A3G, these changes may be sufficient to create a more A3G-like amino-terminal interface, resulting in better complex formation and thus causing increased retention of the mutant A3B in the cytoplasm. It is noteworthy in this context that of the five mutations, only the E22Y, E24R replacement at the N terminus resulted in a slight increase (30%) of antiviral activity. The lack of activity of the A3B/G chimera would also fit this explanation; in this chimera the amino-terminal sequences cannot be recruited into complexes necessary for A3G activity or for retention in the cytoplasm. The finding that A3B/G was also inactive in preventing LINE-1 retrotransposition while A3G/B was still active suggests that it may not only be cellular localization that is important for this activity but that other factors not resident in the amino-terminal end are necessary for this function.
Although A3B and A3G share the Z2Z1 configuration, a more detailed sequence comparison reveals that the two genes are not direct paralogues. The N-terminal Z2 domain of A3B is, in fact, more closely related to the N-terminal domains of A3D/E and A3F. Ignoring the first 58 amino acids for now (see below), the rest of the A3B-Z2 domains shares 77 and 92% sequence identity with A3D/E and A3F, respectively, compared to only 46% when the same regions of A3B and A3G are compared. Alignments of the C-terminal domains reveal a 90% identity between A3B and A3A, while both are about equidistant from A3G, with about 65% sequence identity. Another feature that sets A3B and A3G apart is the fact that the Z2 domain of A3G is catalytically inactive, while A3B-Z2 still has deamination activity (5). Based on the homologies, one could envision that A3B arose by a relatively recent duplication of the first two exons of A3D/E, the next two exons of A3F, and the entire A3A.
The conservation between the first two exons of human A3G and human A3F is striking; sequence identity for the first 60 codons is 99.4% and then drops to 70% for the remainder of the 5′ half of the open reading frame (ORF) (98% versus 45% at the amino acid level). As detailed above, the amino-terminal halves of huA3B and huA3D/E are more closely related throughout; however, even here the homology drops from 99.4% for the first two exons to 85% for the remainder of the first CDA sequence (100% versus 78% at the amino acid level). Interestingly, the first 60 amino acids in other primate species do not show this pairwise sequence conservation but have the same extent of sequence divergence as the remainder of the genes (or proteins), indicating that positive selective pressure was active throughout the sequences (43). The differences, which were acquired during the evolution of the Old World monkeys and apes, were lost in the hominid lineage. Taken together these data suggest that two gene conversion events in the hominid lineage lead to the identity between the first two exons of huA3G and huA3F and between the first two exons of huA3B and huA3D/E.
In summary, we present data which show that a change of 4 amino acids results in the redirection of the normally nuclear A3B to the cytoplasm and in activation of the restrictive activity against HIV-1 and HTLV-1. The restriction can be explained by the high level of deamination that occurs on the minus strand during reverse transcription. Except for the gain of function against HIV-1, A3G/B showed the typical activity pattern for A3B, with high specific activity and the ability to restrict LINE-1 retrotransposition independent of the activity of the CDA domains.
ACKNOWLEDGMENTS
We dedicate this paper to the memory of David Derse.
We are grateful to Stephen Hughes and Reuben Harris for discussions, to Thomas Fanning for reading the manuscript, and to Richard Frederickson for help with the graphics. We thank B. Cullen for the A3B plasmid and Patricia Lloyd for help with plasmid constructions. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 p24 Gag Monoclonal (no. 24-4) from Michael H. Malim.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Albin J. S., Harris R. S. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med. 12:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alce T. M., Popik W. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083–34086 [DOI] [PubMed] [Google Scholar]
- 3. An P., et al. 2009. APOBEC3B deletion and risk of HIV-1 acquisition. J. Infect. Dis. 200:1054–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bishop K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396 [DOI] [PubMed] [Google Scholar]
- 5. Bogerd H. P., Wiegand H. L., Doehle B. P., Cullen B. R. 2007. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology 364:486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bogerd H. P., Wiegand H. L., Doehle B. P., Lueders K. K., Cullen B. R. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34:89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bogerd H. P., et al. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 103:8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burnett A., Spearman P. 2007. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 81:5000–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiu Y. L., Greene W. C. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317–353 [DOI] [PubMed] [Google Scholar]
- 10. Conticello S. G., Langlois M. A., Yang Z., Neuberger M. S. 2007. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv. Immunol. 94:37–73 [DOI] [PubMed] [Google Scholar]
- 11. Cullen B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dang Y., Wang X., Esselman W. J., Zheng Y. H. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522–10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derse D., Hill S. A., Princler G., Lloyd P., Heidecker G. 2007. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. U. S. A. 104:2915–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doehle B. P., Schafer A., Cullen B. R. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281–288 [DOI] [PubMed] [Google Scholar]
- 15. Doehle B. P., Schafer A., Wiegand H. L., Bogerd H. P., Cullen B. R. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 79:8201–8207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friew Y. N., Boyko V., Hu W. S., Pathak V. K. 2009. Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology 6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goila-Gaur R., Strebel K. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hakata Y., Landau N. R. 2006. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J. Biol. Chem. 281:36624–36631 [DOI] [PubMed] [Google Scholar]
- 19. Harris R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 20. Harris R. S., Liddament M. T. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868–877 [DOI] [PubMed] [Google Scholar]
- 21. Heidmann T., Heidmann O., Nicolas J. F. 1988. An indicator gene to demonstrate intracellular transposition of defective retroviruses. Proc. Natl. Acad. Sci. U. S. A. 85:2219–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huthoff H., Autore F., Gallois-Montbrun S., Fraternali F., Malim M. H. 2009. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 5:e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Itaya S., Nakajima T., Kaur G., Terunuma H., Ohtani H., Mehra N., Kimura A. 2010. No evidence of an association between the APOBEC3B deletion polymorphism and susceptibility to HIV infection and AIDS in Japanese and Indian populations. J. Infect. Dis. 202:815–816 (Author reply, 202:816–817.) [DOI] [PubMed] [Google Scholar]
- 24. Iwatani Y., et al. 2009. HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc. Natl. Acad. Sci. U. S. A. 106:19539–19544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jern P., Russell R. A., Pathak V. K., Coffin J. M. 2009. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 5:e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiernan R. E., Ono A., Freed E. O. 1999. Reversion of a human immunodeficiency virus type 1 matrix mutation affecting Gag membrane binding, endogenous reverse transcriptase activity, and virus infectivity. J. Virol. 73:4728–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koning F. A., et al. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. LaRue R. S., et al. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83:494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lovsin N., Peterlin B. M. 2009. APOBEC3 proteins inhibit LINE-1 retrotransposition in the absence of ORF1p binding. Ann. N. Y. Acad. Sci. 1178:268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mahieux R., et al. 2005. Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J. Gen. Virol. 86:2489–2494 [DOI] [PubMed] [Google Scholar]
- 31. Marin M., Golem S., Rose K. M., Kozak S. L., Kabat D. 2008. Human immunodeficiency virus type 1 Vif functionally interacts with diverse APOBEC3 cytidine deaminases and moves with them between cytoplasmic sites of mRNA metabolism. J. Virol. 82:987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mbisa J. L., et al. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mbisa J. L., Bu W., Pathak V. K. 2010. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 84:5250–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moran J. V., et al. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927 [DOI] [PubMed] [Google Scholar]
- 35. Navarro F., et al. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374–386 [DOI] [PubMed] [Google Scholar]
- 36. Newman E. N., et al. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170 [DOI] [PubMed] [Google Scholar]
- 37. Refsland E. W., et al. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38:4274–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rose K. M., Marin M., Kozak S. L., Kabat D. 2005. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res. Hum. Retroviruses 21:611–619 [DOI] [PubMed] [Google Scholar]
- 39. Rulli S. J., Jr., et al. 2008. Interactions of murine APOBEC3 and human APOBEC3G with murine leukemia viruses. J. Virol. 82:6566–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Russell R. A., Moore M. D., Hu W. S., Pathak V. K. 2009. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Russell R. A., Pathak V. K. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sasada A., et al. 2005. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology 2:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sawyer S. L., Emerman M., Malik H. S. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith J. L., Bu W., Burdick R. C., Pathak V. K. 2009. Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends Pharmacol. Sci. 30:638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stenglein M. D., Harris R. S. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837–16841 [DOI] [PubMed] [Google Scholar]
- 46. Stenglein M. D., Matsuo H., Harris R. S. 2008. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J. Virol. 82:9591–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang S., et al. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357–9366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu Q., et al. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379–53386 [DOI] [PubMed] [Google Scholar]