Abstract
As obligate intracellular parasites, viruses not only hijack cellular machinery, they also deregulate host stress responses for their infection. Human cytomegalovirus (HCMV) modulates the endoplasmic reticulum (ER) stress response, due at least in part to the viral protein pUL38, and one of the consequences is to maintain the viability of infected cells. Consequently, pUL38-deficient virus induces premature cell death during infection. In addition, pUL38 activates mammalian target of rapamycin complex 1 (mTORC1), which may also antagonize other detrimental cellular stresses (N. J. Moorman et al., Cell Host Microbe 3:253–262, 2008). It remains elusive how pUL38 inhibition of cell death is related to mTORC1 activation. In this study, we defined the interplay of the two pUL38 activities. We constructed a series of pUL38 truncation mutants based on the secondary structure prediction and evolutionary conservation of its sequence. We found that the N-terminal 239 residues of pUL38 were necessary and sufficient to block cell death induced by pUL38-deficient virus or by the ER stress inducer tunicamycin. However, this pUL38 domain was unable to activate mTORC1 when expressed alone. Importantly, small-molecule inhibitors of mTORC1, rapamycin or torin 1, did not compromise pUL38 activity to block cell death in isolation or in virus infection. Expression of a constitutively active variant of an mTORC1 activator, Rheb (Ras homolog enriched in brain), could not prevent cell death induced by pUL38-deficient virus. Collectively, we provide genetic and biochemical evidence that pUL38 prevents ER stress-induced cell death independent of its role in mTORC1 activation.
INTRODUCTION
To establish successful replication in the host, viruses have developed elegant strategies to modify the cellular environment and overcome the host defense systems. Human cytomegalovirus (HCMV) is a slow-growing betaherpesvirus that has a broad cellular tropism. It is the leading viral cause of congenital diseases in newborns and a common cause of severe infectious complications in immunocompromised individuals (reviewed in references 1 and 25). HCMV requires 48 to 72 h to complete a replication cycle. This protracted life cycle makes HCMV highly exposed to host defense responses. However, HCMV is one of the most prevalent viruses and establishes a lifelong latent and recurrent infection in the human host. It is not surprising that HCMV has evolved mechanisms to subvert multiple types of host antiviral responses, including cellular stress responses.
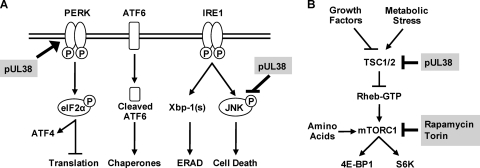
The endoplasmic reticulum (ER) is a key organelle involved in sensing and responding to stressful conditions, such as the accumulation of excessive glycoproteins or perturbations of Ca2+ homeostasis in the ER. HCMV produces an abundant amount of viral glycoproteins and promotes the release of Ca2+ from the ER to the cytosol during infection (36). Therefore, HCMV has the potential to induce multiple cellular stress responses and profoundly impacts the ER function during infection (16, 40). Once the ER detects the stress, three signaling pathways collectively termed the unfolded protein response (UPR) are activated to enhance the ER function to alleviate the stress or induce cell death if the stress cannot be resolved (Fig. 1A). The UPR is initiated by three ER lumen-located sensor molecules, the protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), the activating transcription factor 6 (ATF6), and inositol-requiring enzyme-1 (IRE1) (24, 34). HCMV modulates the UPR by targeting all three signaling pathways during infection (16). It induces PERK-dependent phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF-2α) and translation of the activating transcriptional factor 4 (ATF4) while also preventing the attenuation of global translation. HCMV blocks ATF6 proteolytic activation but induces ATF6-independent expression of a subset of downstream target genes, such as Bip and GRP94. In addition, HCMV activates IRE1-mediated splicing of the XBP-1 transcript but it subsequently blocks XBP-1-dependent gene expression. It is conceivable that HCMV uses this modulatory strategy to hijack the components of the UPR that are beneficial to its infection but blocks others that are detrimental to its infection (16).
Fig. 1.
(A) Schematic diagram of the ER stress response. Three sensors on the ER membrane, PERK, ATF6, and IRE1, are activated upon ER stress. Activated PERK phosphorylates eIF-2α to attenuate global protein translation but selectively enhance ATF4 translation. Activated ATF6 is cleaved and translocated to the nucleus, where it upregulates ER chaperone expression. Once activated, the endoribonuclease activity of IRE1 mediates splicing of Xbp-1, which encodes a transcriptional activator to induce transcription of ERAD components. The kinase activity of IRE1 phosphorylates JNK, which can ultimately lead to cell death. Viral protein pUL38 activates PERK-dependent ATF4 expression, suppresses persistent JNK phosphorylation, and inhibits ER stress-induced cell death. Modulation of other components of the ER stress response by HCMV is described in the text. (B) Schematic diagram of the mTOR pathway. Growth factors and metabolic stresses regulate mTORC1 activation by acting on the TSC1/2 complex. The TSC1/2 complex functions as a GTPase-activating protein for Rheb, which activates mTORC1 in its GTP-bound form. Once activated, mTORC1 phosphorylates 4E-BP1 and S6K, two well-characterized substrates regulating protein translation. Amino acids activate mTORC1 through a parallel pathway independent of TSC1/2 or Rheb. pUL38 binds to TSC2 and activates mTORC1. Rapamycin inhibits mTORC1 by binding to mTOR as a rapamycin-FKBP12 complex to interfere with its substrate binding capability. Torin 1 directly inactivates mTOR kinase activity by competing with ATP binding.
Several studies have shed light on the mechanism that HCMV uses to accomplish this modulation of the UPR during its infection. HCMV induces the accumulation of the ER chaperone protein Bip at both transcriptional and translational levels (5). Bip is involved in cytoplasmic virion assembly and egress during HCMV infection (4). It accomplishes this by maintaining the integrity of viral cytoplasmic assembly centers (3) and inducing nuclear lamina phosphorylation and rearrangement (2). In addition, ER chaperones, including Bip, also facilitate HCMV US2 and US11 protein-mediated degradation of the major histocompatibility complex (MHC) class I heavy chain to dismantle the antiviral immune response (14, 28, 39). Moreover, we have identified the HCMV protein pUL38 as another factor that helps the virus to modulate the UPR and inhibit ER stress-induced cell death (40) (Fig. 1A). pUL38-deficient HCMV fails to upregulate PERK and eIF-2α phosphorylation and thus results in a loss of robust ATF4 accumulation. pUL38 also allows HCMV to suppress persistent phosphorylation and activation of c-Jun N-terminal kinase (JNK), which can be induced by IRE1 and is a major cell death inducer during prolonged ER stress. Importantly, ATF4 overexpression and JNK inhibition markedly reduced premature cell death induced by infection of pUL38-deficient virus. Thus, pUL38 appears to be critical for HCMV to remodel the ER function and protect host cells from ER stress-induced cell death during virus infection.
Another major stress response elicited during infection results from virus-induced metabolic stress. Viruses hijack the host cell to produce large quantities of progeny viruses in a relatively short period of time, thus demanding high levels of energy production and biological macromolecule synthesis. Mammalian target of rapamycin complex 1 (mTORC1) is a central factor regulating cellular macromolecule synthesis (6, 35) (Fig. 1B). When energy and nutrients are ample, mTORC1 is activated to promote biosynthesis of macromolecules, such as proteins and lipids. Under metabolically stressful conditions, such as nutrient withdrawal or energy depletion, the activity of mTORC1 is inhibited, thus coupling cell growth with the metabolism status. Due to the high demand of energy production and macromolecule biosynthesis, lytic virus infection has the potential to induce metabolic stress, which would lead to inactivation of mTORC1 and consequently the inability of the host cell to sustain robust virus production. Therefore, viruses such as HCMV have evolved mechanisms to deal with metabolic stress and maintain mTORC1 activation regardless of the nutrient or energy status during infection. In particular, HCMV induces a rapamycin-resistant mTORC1 activity to phosphorylate and inactivate eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1), a translation suppressor that otherwise would bind to eukaryotic translation initiation factor eIF-4E and suppress cap-dependent protein translation. The importance of mTORC1 in HCMV infection is underscored by several studies showing that inhibition of mTORC1 by rapamycin, torin 1, or RNA interference (RNAi) knockdown of raptor (a subunit of mTORC1) inhibits HCMV replication in fibroblasts (10, 20–22, 27, 29).
In a proteomic study searching for pUL38-interacting proteins, tuberous sclerosis protein 2 (TSC2), a negative regulator of mTORC1, was identified as an interacting target of pUL38 (26) (Fig. 1B). Together with TSC1, TSC2 forms tuberous sclerosis protein complex (TSC1/2) that acts as a GTPase-activating protein for Rheb (Ras homolog enriched in brain), a small GTPase that activates mTORC1 in its GTP-bound form. Activation of Rheb GTP hydrolysis by TSC1/2 keeps Rheb in the GDP-bound form and therefore renders it unable to activate mTORC1. pUL38 binds to TSC2 and activates mTORC1, which has been proposed to allow HCMV to antagonize cellular stresses such as nutrition and energy depletion during lytic infection (26).
These studies suggest that pUL38 is important for HCMV to inhibit not only ER stress-induced cell death but also the negative effects of metabolic stress during infection. In the present study, we sought to determine whether these two activities of pUL38 are the result of a common mechanism. We have found that a pUL38 truncation mutant lacking the C-terminal 92 amino acid residues fails to activate mTORC1 but retains the ability to prevent cell death. Moreover, inhibition of mTORC1 activation does not affect pUL38 activity to block cell death, and activation of mTORC1 does not prevent cell death induced by infection of pUL38-deficient HCMV. Collectively, we present genetic and biochemical evidence that the cell death inhibition function of pUL38 is independent of its mTORC1 activation function, thus providing further insight into the role of this protein during HCMV infection.
MATERIALS AND METHODS
Plasmids and reagents.
The following retroviral overexpression vectors were derived from pRetro-EBNA (19). pYD-C163 carried the UL38 open reading frame (ORF) (37) and pYD-C596 carried the UL38 ORF that was tagged with hemagglutinin (HA) at its amino terminus. pYD-C548 (expressing a pUL38 segment from amino acid 57 to amino acid 331 [pUL3857–331]), pYD-C549 (pUL3886–331), pYD-C550 (pUL38120–331), pYD-C551 (pUL38150–331), pYD-C553 (pUL381–277), pYD-C554 (pUL381–257), pYD-C556 (pUL381–225), pYD-C557 (pUL381–184), pYD-C559 (pUL381–146), pYD-C561 (pUL381–239), and pYD-C562 (pUL381–248) carried individual HA-tagged UL38 truncation mutants. pYD-C331 carried the coding sequence of a myc-tagged constitutively active variant of Rheb (S16H Rheb) (41).
Plasmid pYD-C191 carried a kanamycin cassette bracketed by two FLP recombination target (FRT) sites and was used for the first step of linear recombination during bacterial artificial chromosome (BAC) recombineering (see below). Pharmaceutical agents used in the present study included the ER stress-inducing agent tunicamycin (Sigma) and mTORC1 inhibitors rapamycin (Sigma) and torin 1 (a generous gift from David Sabatini at Whitehead Institute for Biomedical Research). Primary antibodies used in the present study included anti-cleaved poly(ADP-ribose) polymerase (PARP) (19F4), anti-ribosomal S6 kinase (S6K), anti-phosphorylated S6K, anti-eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1), and anti-phosphorylated 4E-BP1 (all from Cell Signaling); anti-HA (Sigma); anti-myc (Santa Cruz); anti-actin (AC15; Abcam); and anti-pUL38, anti-pp28, and anti-IE1 (gifts from Thomas Shenk at Princeton University).
Cells and viruses.
Primary human foreskin fibroblasts (HFFs) were propagated in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum. To create cells expressing various forms of the UL38 protein or myc-(S16H Rheb), HFFs were transduced three times with retrovirus reconstituted from the aforementioned retroviral vectors. Control cells expressing an empty vector were made by transduction with retrovirus reconstituted from pRetro-EBNA. To compare cells expressing pUL38, pUL381–239, or pUL381–257, HFFs were either transduced with the empty retroviral vector five times (vector), with pUL381–239- or pUL381–257-expressing vector five times (pUL381–239 or pUL381–257), or with pUL38-expressing vector three times followed by the empty vector for two additional times (pUL38). This achieved equal expressions of pUL38, pUL381–239, and pUL381–257.
Three BAC-HCMV clones were used in the present study to reconstitute recombinant HCMV viruses. pAD-GFP carried the green fluorescent protein (GFP)-tagged genome of the HCMV AD169 strain and was used to produce wild-type virus ADwt. pADpmUL38 was used to reconstitute ADpmUL38, the mutant virus that carried multiple point mutations engineered to specifically disrupt pUL38 expression (40). pADtrunUL381–239 was used to reconstitute ADtrunUL381–239, the mutant virus in which the coding sequence for the C-terminal 92 amino acid residues of pUL38 was deleted. To create pADtrunUL381–239, a cassette that contained a stop codon followed by the FRT-bracketed kanamycin selection marker was amplified by PCR from pYD-C191 with a pair of 70-bp primers. The 50-bp sequences at the 5′ end of these primers were homologous to the viral DNA sequences immediately upstream or downstream of the amino acid residue 239 of the UL38 ORF: 5′-GGAAAATTCCTATGACCTTCGTGGATCGCGACTCGCTGCGAGCCAATTCGTGAACCACGTCGTGGAATGCCTTC-3′ and 5′-TGCGCCGGGGCGTGAGAAGGCTGAGCCCCGGTGGCCTGGATGTGGGCCAAAAGGACGACGACGACAAGTAA-3′. The cassette was recombined into the UL38 gene carried in pAD-GFP, and the kanamycin sequence was subsequently removed by Flp/FRT recombination as previously described (30). The final clone (pADtrunUL381–239) contained a stop codon along with a small FRT site inserted at the residue 240 of the UL38 ORF.
To reconstitute virus, 2 μg of the BAC-HCMV DNA and 1 μg of the pp71 expression plasmid were transfected into normal HFFs or pUL38-expressing HFFs that were made by retroviral transduction by electroporation as described previously (42). Culture medium was changed 24 h later, and virus stock was prepared by harvesting cell-free culture supernatant when the entire monolayer of cells was lysed. Alternatively, virus stocks were produced by collecting cell-free culture medium from infection at a multiplicity of infection (MOI) of 0.001. Virus titers were determined in duplicate by a plaque assay in pUL38-expressing HFFs unless otherwise noted.
Analysis of viral growth kinetics.
Normal HFFs were seeded in 12-well dishes overnight to produce a subconfluent monolayer. Cells were then inoculated with recombinant HCMV viruses for 1 h at an MOI of 0.01 for multistep growth analysis. The inoculum was removed, the infected monolayers were rinsed with phosphate-buffered saline (PBS), and fresh medium was replenished. At various times postinfection, cell-free virus was collected from cells by harvesting medium from infected cultures and was titered by 50% tissue culture infectious dose (TCID50) assay in pUL38-expressing HFFs.
Protein analysis.
Proteins were analyzed by immunoblotting as described previously (40). Briefly, cells were collected, washed, and lysed in the sodium dodecyl sulfate (SDS)-containing sample buffer. Proteins from equal cell numbers were resolved by electrophoresis on an SDS-containing polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, hybridized with primary antibodies, reacted with horseradish peroxidase-conjugated secondary antibodies, and visualized by using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
Assays of cell death/apoptosis.
Premature cell death was induced in HFFs either by infection of pUL38-deficient HCMV or by treatment of cells with the ER stress-inducing agent tunicamycin. For HCMV-induced cell death, confluent HFFs were infected at an MOI of 1 in DMEM containing 10% fetal calf serum. For drug-induced cell death, confluent HFFs were treated with 2 μg/ml tunicamycin at 36 h after seeding. At times indicated in figure legends, cells were analyzed for morphological and biochemical changes characteristic of induction of premature cell death as previously reported (37, 40). These assays included microscopic analysis for cell morphology, immunoblot analysis for elevated PARP [poly(ADP ribose) polymerase] cleavage, and flow cytometry analysis for phosphatidylserine translocation together with nuclear dye TO-PRO3 permeability. For microscopic analysis, HFFs were examined under a phase-contrast microscope at 72 h postinfection (hpi) for their cell morphology. Dying cells were detached from the culture dish and fragmented, while surviving cells remained attached and maintained a flat, fibroblast-like morphology. For induction of elevated PARP cleavage, cell lyate was analyzed by immunoblotting using the antibody specific to the cleaved forms of PARP. To quantify the percentages of cells in which phosphatidylserine was translocated from the inner to the outer surface of the cell membrane, cells were labeled with fluorescein-conjugated annexin V, a phospholipid binding protein with high affinity for phosphatidylserine, according to the manufacturer's instructions (Roche). In addition, cells were also costained with TO-PRO3 to identify dying cells with compromised membrane integrity. The percentages of annexin V-positive cells and TO-PRO3-positive cells were determined by flow cytometry.
Assays of mTORC1 activation.
Immunoblotting of S6K phosphorylation was used in most of the experiments in this study as the readout for mTORC1 activation. In some experiments, immunoblot analysis of 4E-BP1 phosphorylation was included as an additional readout for mTORC1 activation. To determine mTORC1 activation in expressing cells, HFFs overexpressing various UL38 constructs or myc-S16H Rheb were cultured in serum-free medium for 48 h or in PBS for two additional hours following 48 h of serum starvation. Cell lysate was then prepared for immunoblot analysis. To determine mTORC1 activation in infected cells, HFFs were starved for 48 h and infected with recombinant HCMV at an MOI of 1 in serum-free medium, and cell lysate was collected at indicated times for immunoblot analysis.
RESULTS
Construction and analysis of the UL38 serial truncation mutants.
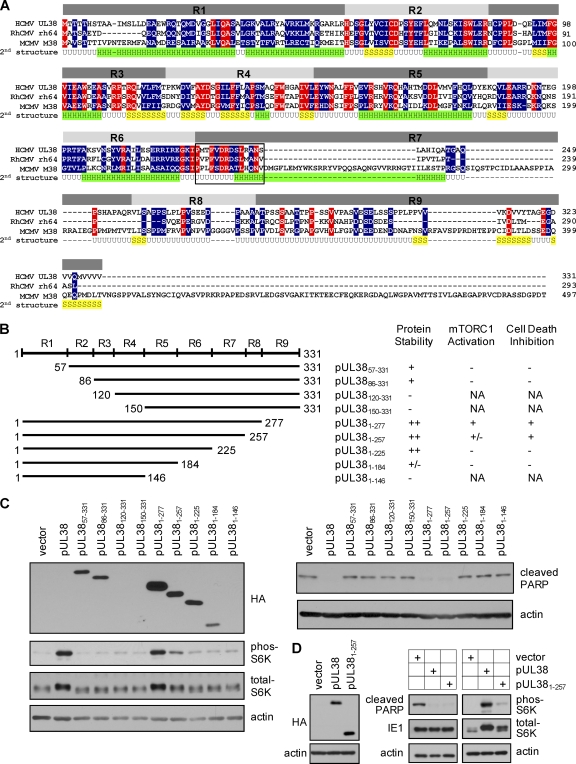
One powerful approach to dissect a multifunctional protein is to employ mutagenic analysis and identify functional domains or sequences responsible for each function. Based on the predicted protein secondary structure and sequence alignment of pUL38 with its homologs in rhesus CMV (RhCMV) and murine CMV (MCMV), we divided the 331-amino-acid UL38 coding sequence into nine regions (R1 to R9) and targeted them for truncational mutagenesis (Fig. 2A). We constructed four amino (N)-terminal truncations in which the regions R1 to R4 were progressively deleted (Fig. 2B). In addition, we also created five carboxyl (C)-terminal truncations in which the regions R9 to R5 were progressively deleted (Fig. 2B). All of the truncation variants of pUL38 were also tagged with HA at their N terminus to facilitate protein detection.
Fig. 2.
Construction and analysis of pUL38 serial truncation mutants. (A) Sequence alignment of the UL38 coding sequence and its homologs in rhesus CMV (rh64) and murine CMV (M38). Amino acid residues conserved in all three species are indicated in red, and residues conserved between the UL38 and rh64 or M38 coding sequences only are indicated in blue. Also shown is the putative second structure of pUL38 predicted using the JUFO program (http://www.meilerlab.org/index.php/servers/show). H, α-helix; S, β-sheet; U, unstructured random coil. Based on the sequence conservation among pUL38 homologs and the predicted secondary structure, the UL38 coding sequence was divided into nine regions (R1 to R9) that were targeted for mutagenic analysis. Conserved 14 amino acids between residues 226 and 239 within R7 are bracketed by a box. (B) Construction and analysis of UL38 serial truncation mutants. The left panel shows the scheme of constructed UL38 mutants. The start and terminal amino acid residues for each mutant are indicated. Each UL38 mutant was also tagged with HA at the N terminus. The right panel summarizes the protein stability of each UL38 mutant and its ability to activate mTORC1 or inhibit cell death when expressed in primary human foreskin fibroblasts (HFFs). NA, Not applicable due to protein instability. (C) Analysis of mTORC1 activation and cell death inhibition in HFFs expressing various UL38 mutants. HFFs expressing various forms of the UL38 protein were generated by retroviral transduction. To determine mTORC1 activation, HFFs expressing indicated UL38 proteins were cultured in serum-free medium for 48 h before being collected for immunoblot analysis for S6K phosphorylation. To analyze cell death, HFFs expressing UL38 mutants were infected with pUL38-deficient HCMV (ADpmUL38) at a multiplicity of infection (MOI) of 1, and cell lysate was collected at 72 h postinfection (hpi) for immunoblot analysis for PARP cleavage. The protein level of each UL38 mutant was analyzed using the anti-HA antibody. (D) pUL381–257 inhibits cell death induced by pUL38-deficient virus infection but only partially activates mTORC1. HFFs were transduced five times with an empty retroviral vector (vector) or pUL381–257-expressing vector (pUL381–257), or three times with pUL38-expressing vector followed by two additional times with the empty vector (pUL38). This allowed equal expression levels of pUL38 and pUL381–257 as determined by immunoblot analysis for HA (left panel). The ability of transduced cells to inhibit cell death induced by UL38-deficient virus infection or activate mTORC1 under the serum starvation condition was determined by PARP cleavage or S6K phosphorylation (right panel). IE1 and actin were used as the infection and loading controls, respectively.
Human foreskin fibroblasts (HFFs) expressing these mutant variants of pUL38 were created by retroviral transduction (40). These cells were tested for their ability to activate mTORC1 under the serum starvation condition or to inhibit premature cell death induced by pUL38-deficient virus (ADpmUL38) (40). HFFs expressing the untagged full-length pUL38 or an empty vector were also made by retroviral transduction and included as the positive and negative controls, respectively. Some mutant UL38 proteins, particularly the N-terminal truncation mutants (e.g., pUL38120–331 and pUL38150–331), were unstable, as we failed to detect their accumulation using the HA antibody (Fig. 2B and C). On the other hand, most of the C-terminal truncation mutants were relatively stable even though their levels varied. Such protein stability appears to be consistent with the secondary structure prediction that the N-terminal portion of pUL38 predominantly consists of structured α-helices and β-sheets and may therefore represent its core structure, whereas the C terminus of the protein is largely disordered (Fig. 2A).
To test the ability of mutant UL38 proteins to prevent cell death, transduced cells were infected with pUL38-deficient HCMV (ADpmUL38). Cell death was evaluated by examining induction of elevated PARP [poly(ADP ribose) polymerase] cleavage, a biochemical marker of apoptosis (Fig. 2C). The mutant UL38 protein containing only the N-terminal 257 residues (pUL381–257) or 277 residues (pUL381–277) retained the ability to prevent elevated PARP cleavage at a level comparable to that of full-length pUL38. However, all other UL38 truncation mutants failed to prevent elevated PARP cleavage (Fig. 2C) and displayed morphology characteristic of cell death (37) (data not shown).
To test the ability of mutant UL38 proteins to activate mTORC1, serum-starved, transduced cells were examined for S6K (ribosomal S6 kinase) phosphoryation, a biomarker of mTORC1 activation (Fig. 2C). Among all the mutants tested, only pUL381–277 retained the full ability to induce S6K phosphorylation. Deletion of the C-terminal 106 residues (i.e., pUL381–225) completely knocked out the ability of pUL38 to induce S6K phosphorylation. Interestingly, pUL381–257 induced only partial S6K phosphorylation even though it was able to fully prevent PARP cleavage (Fig. 2C). Consistently, total S6K in pUL381–257-expressing cells appeared to migrate faster than that in pUL38- or pUL381–277-expressing cells, albeit this effect was subtle. In this experiment, we noted that the level of pUL381–257 was somewhat reduced compared to that of pUL38 proteins capable of fully activating mTORC1 (e.g., pUL381–277). To rule out the possibility that the reduced accumulation of pUL381–257 was the reason for its inability to activate mTORC1, we transduced HFFs five times with pUL381–257-expressing retrovirus. As a control, we transduced cells three times with retrovirus expressing the HA-tagged full-length pUL38 followed by transduction with the empty vector for two additional times. Under this transduction condition, almost all transduced cells expressed pUL38 or pUL381–257 as determined by flow cytometry (data not shown), and importantly, they expressed pUL381–257 at a level no less than that of pUL38 (Fig. 2D). As anticipated, pUL381–257 continued to efficiently block pUL38-deficient virus-induced PARP cleavage. Importantly, the ability of pUL381–257 to induce S6K phosphorylation remained markedly compromised compared to that of pUL38. Together, our results suggested that the ability of mutant UL38 proteins to block cell death does not correlate entirely with its ability to activate mTORC1.
The N-terminal 239 residues are sufficient for pUL38 to block premature cell death during HCMV infection.
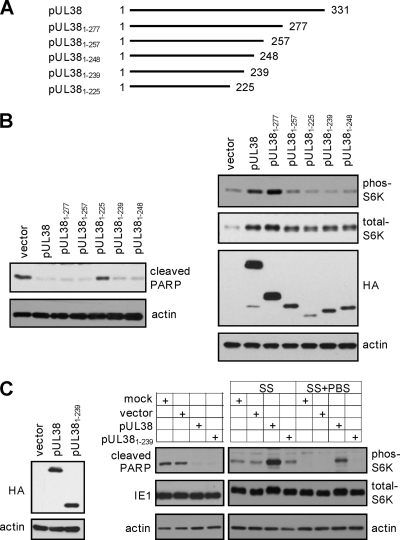
As pUL381–257 showed a differential ability to block cell death and activate mTORC1 but pUL381–225 lost both functions (Fig. 2), we hypothesized that the sequence between residues 226 and 257 was critical to uncouple these two activities of pUL38. We noticed that the first 14 residues of this region (i.e., residues 226 to 239) were highly conserved among pUL38 homologs, while the remainder (i.e., residues 240 to 256) varied between pUL38 and its homologs (Fig. 2A). We constructed two additional truncation mutants, pUL381–239 and pUL381–248, and both were also tagged with HA at their N termini (Fig. 3A). pUL381–239 carried the N-terminal 239 amino acids of pUL38, therefore containing these conserved residues, while pUL381–248 contained 10 additional nonconserved residues at its C terminus. An N-terminally HA-tagged full-length pUL38 was used as a control. We found that both pUL381–239 and pUL381–248 were unable to induce S6K phosphorylation as compared to full-length pUL38, indicating that they lost the ability to activate mTORC1 (Fig. 3B). However, both pUL381–248 and particularly pUL381–239 were fully capable of preventing elevated PARP cleavage, suggesting that these 239 amino acids of pUL38 were sufficient for its ability to block virus-induced cell death.
Fig. 3.
The 92 amino acid residues at the C terminus of pUL38 are dispensable for cell death inhibition but are essential for mTORC1 activation when expressed in isolation. (A) The scheme of the C-terminal serial truncation mutants of pUL38. Indicated are start and terminal amino acid residues of each mutant. (B) mTORC1 activation and cell death inhibition in HFFs expressing C-terminal serial truncation mutants of pUL38 were analyzed as described in the legend to Fig. 2. (C) pUL381–239 fails to activate mTORC1 but inhibits cell death induced by pUL38-deficient virus infection. HFFs were either mock transduced, transduced five times with the empty retroviral vector (vector) or with pUL381–239-expressing vector (pUL381–239), or transduced three times with pUL38-expressing vector followed by two additional times with the empty vector (pUL38) to achieve equal expression levels of pUL38 and pUL381–239. The protein level of UL38 proteins was validated (left panel) and the ability of transduced cells to inhibit cell death induced by pUL38-deficient virus infection was determined by immunoblot analysis (right panel). To determine mTORC1 activation, these HFFs were either treated with serum-free medium for 48 h (SS) or incubated with PBS for two additional hours after serum starvation (SS+PBS). Cells were then collected for immunoblot analysis for S6K phosphorylation.
In this experiment, we noted that the level of pUL381–239 was also reduced relative to that of full-length pUL38 (Fig. 3B). In an additional experiment, we transduced HFFs with pUL381–239-expressing retrovirus for five times, or with retrovirus expressing full-length pUL38 three times followed by the empty vector for two additional times. Under these transduction conditions, almost all transduced cells expressed pUL38 or pUL381–239 as determined by flow cytometry (data not shown), and importantly, they expressed equal levels of pUL38 or pUL381–239 (Fig. 3C). As anticipated, pUL381–239-expressing cells blocked pUL38-deficient virus-induced PARP cleavage as efficiently. However, compared to what was the case for control cells expressing the empty vector, no elevated S6K phosphorylation was observed in pUL381–239-expressing cells under two different serum starvation conditions. Therefore, the N-terminal 239 amino acids of pUL38 were sufficient to block cell death but were insufficient to activate mTORC1. We hypothesized that the ability of pUL38 to block cell death was independent of its ability to activate mTORC1 under stress conditions.
pUL38 blocks ER stress-induced cell death even when mTORC1 activity is inhibited.
pUL38 blocks premature cell death, particularly ER stress-induced cell death, both in the context of virus infection and when expressed alone (40). Even though the cell death inhibition activity of pUL381–239 was evident during infection of pUL38-deficient virus, it was formally possible that other virally encoded factors potentiated its ability to inhibit cell death during infection. Therefore, we wanted to test if pUL381–239 preserved the ability to block ER stress-induced cell death in the absence of any other viral factors. We treated HFFs that expressed pUL381–239, pUL38, or the empty vector with the ER stress inducer tunicamycin and analyzed induction of cell death either by scoring cells that were positive for annexin V and/or TO-PRO3 staining via flow cytometry or by determining PARP cleavage via immunoblot analysis (Fig. 4A). As previously reported (40), PARP cleavage and the percentage of cells positive for annexin V and/or TO-PRO3 were markedly reduced in both pUL38- and pUL381–239-expressing cells relative to those in vector-expressing control cells upon tunicamycin treatment. Therefore, pUL381–239 was sufficient to inhibit ER stress-induced cell death in the absence of any other viral factors despite its inability to activate mTORC1.
Fig. 4.
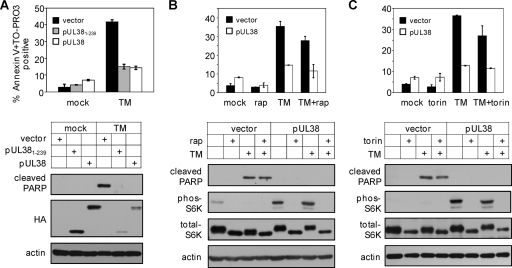
Inhibition of mTORC1 does not compromise the ability of pUL38 to block ER stress-induced cell death. (A) pUL381–239 inhibits tunicamycin (TM)-induced cell death. Confluent HFFs expressing the empty vector, pUL38, or pUL381–239 were treated with 2 μg/ml tunicamycin at 36 h after seeding and stained with fluorescein-conjugated annexin V and TO-PRO3 at 72 h after treatment. The percentage of annexin V- and/or TO-PRO3-positive cells was determined by flow cytometry. PARP cleavage and S6K phosphorylation were analyzed by immunoblotting. (B) Inhibition of mTORC1 by rapamycin (rap) does not compromise the ability of pUL38 to inhibit tunicamycin-induced cell death. HFFs expressing the empty vector or pUL38 were treated with rapamycin (10 nM) only or tunicamycin only (2 μg/ml) or pretreated with rapamycin for 6 h and subsequently treated with rapamycin and tunicamycin. At 72 h after treatment, the percentage of annexin V- and/or TO-PRO3-positive cells, PARP cleavage, and S6K phosphorylation were determined as described for panel A. (C) Inhibition of mTORC1 by torin 1 does not compromise the ability of pUL38 to inhibit tunicamycin-induced cell death. HFFs expressing the empty vector or pUL38 were treated with torin 1 (250 nM) only, tunicamycin only (2 μg/ml), or torin 1 and tunicamycin. At 72 h after treatment, the percentage of annexin V- and/or TO-PRO3-positive cells, PARP cleavage, and S6K phosphorylation were determined as described for panel A.
To provide additional evidence that the inhibitory activity of pUL38 in ER stress-induced cell death is independent of its activation of mTORC1, we treated pUL38-expressing HFFs with tunicamycin in the presence of rapamycin or torin 1, two inhibitors of mTORC1. Rapamycin or torin 1 treatment alone did not induce premature cell death in cells expressing the empty vector or pUL38 (Fig. 4B and C). On the other hand, rapamycin and torin 1 effectively prevented S6K phosphorylation even in the presence of pUL38, indicating that mTORC1 activation was inhibited by these two inhibitors. However, the ability of pUL38 to inhibit elevated PARP cleavage and reduce the percentage of annexin V- and/or TO-PRO3-positive cells upon tunicamycin treatment was not compromised in the presence of rapamycin or torin 1. Collectively, our data indicate that pUL38 expression alone is able to prevent ER stress-induced cell death even when its ability to activate mTORC1 is inhibited.
Mutant HCMV virus expressing only the N-terminal 239 residues of pUL38 is able to block premature cell death and activate mTORC1 during infection.
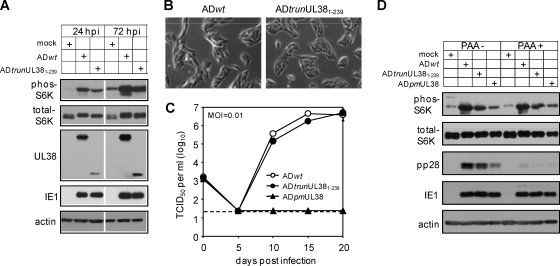
As pUL381–239 blocked cell death induced by pUL38-deficient virus but failed to activate mTORC1 when expressed alone (Fig. 3C), we went on to determine if this phenotype could be recapitulated in the context of virus infection. We constructed a recombinant HCMV (ADtrunUL381–239) that expressed pUL381–239 in place of the full-length pUL38 by BAC recombineering. As anticipated, the mutant virus showed no evidence for elevated premature cell death during infection (Fig. 5B). However, to our surprise, while expression of pUL381–239 alone failed to activate mTORC1 in serum-starved cells (Fig. 3C), ADtrunUL381–239 induced mTORC1 activation as indicated by S6K phosphorylation, albeit to a lesser extent than did wild-type HCMV (Fig. 5A). Multistep growth analysis showed that the mutant virus replicated at wild-type levels (Fig. 5C), suggesting that mTORC1 activity induced by ADtrunUL381–239 was sufficient to maintain virus growth.
Fig. 5.
Recombinant HCMV lacking the C-terminal 92 amino acid residues of pUL38 (ADtrunUL381–239) retains an ability to activate mTORC1 during infection. (A) HFFs were cultured in serum-free medium for 48 h and then infected with wild-type HCMV (ADwt) or UL38 recombinant HCMV (ADtrunUL381–239) in serum-free medium at an MOI of 1. Infected cells were collected at indicated times to determine S6K phosphorylation. (B) The cell morphology of HFFs infected with ADwt or ADtrunUL381–239 at 72 hpi was analyzed with a phase-contrast microscope. (C) Multistep growth analysis of ADtrunUL381–239. HFFs were infected with indicated recombinant HCMV viruses at an MOI of 0.01. Supernatants of infected cells were collected at indicated times and analyzed for the presence of virus by the TCID50 assay. The detection limit of the TCID50 assay is indicated by the dashed line. (D) The ability of ADtrunUL381–239 to activate mTORC1 is independent of late viral gene expression. HFFs were cultured in serum-free medium for 48 h before being infected with indicated recombinant HCMV viruses at an MOI of 1 in the presence or absence of the viral DNA synthesis inhibitor phosphonoacetic acid (PAA) (100 μg/ml). Infected cells were collected at 72 hpi for immunoblot analysis.
As the UL381–239 mutant protein in isolation failed to activate mTORC1 but the recombinant HCMV expressing this pUL38 variant retained this ability, other viral activities were likely to be involved in mTORC1 activation during HCMV infection. To determine if an early or late viral protein was required for mTORC1 induction, we infected cells with wild-type virus (ADwt), virus expressing pUL381–239 (ADtrunUL381–239), or virus deficient in UL38 (ADpmUL38), treated cells with the viral DNA synthesis inhibitor phosphonoacetic acid (PAA), and analyzed S6K phosphorylation at 72 hpi (Fig. 5D). Inhibition of viral DNA synthesis did not compromise the ability of ADtrunUL381–239 or ADwt to activate mTORC1, suggesting that mTORC1 induction was not dependent on late viral proteins. Interestingly, compared to mock infection, pUL38-deficient virus appeared also to induce a low-level but reproducible increase of S6K phosphorylation during infection. Therefore, an early viral activity, which may act synergistically with or independently of pUL38, induces activation of mTORC1 during infection.
mTORC1 activation is not required for HCMV to prevent premature cell death during infection.
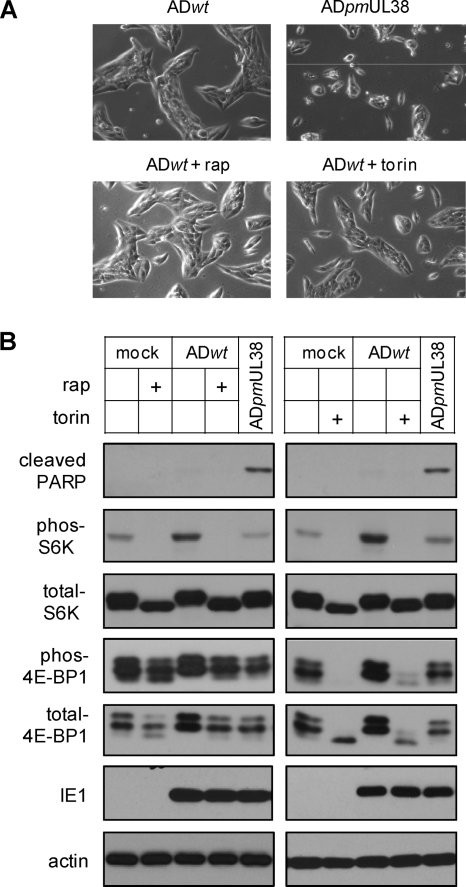
ADtrunUL381–239 retained the ability to block premature cell death and induce mTORC1 activation and therefore could not allow us to uncouple the two activities of pUL38 in the context of HCMV infection. To test if mTORC1 activity was involved in pUL38's ability to prevent premature cell death during HCMV infection, we treated infected HFFs with mTORC1 inhibitors (rapamycin or torin 1) and determined the levels of cell death in these cells. We reasoned that if mTORC1 activity was necessary for pUL38 to prevent cell death during infection, we should observe induction of cell death upon rapamycin or torin 1 treatment. Both rapamycin and torin 1 effectively blocked S6K phosphorylation in uninfected control cells as well as HCMV-infected cells (Fig. 6B). In addition, consistent with previous reports (8, 9, 12, 38), rapamycin treatment reduced but did not completely block 4E-BP1 phosphorylation (another marker of mTORC1 activation) in uninfected cells. It had an even lesser effect on 4E-BP1 phosphorylation in infected cells (22). Importantly, torin 1 effectively prevented 4E-BP1 phosphorylation in both uninfected and infected cells as previously reported (10, 27, 38). However, we observed no appreciable change of morphology characteristic of elevated death in infected cells treated with rapamycin or torin 1 (Fig. 6A). Consistently, PARP cleavage was not elevated in wild-type HCMV-infected cells regardless of whether they were treated with rapamycin or torin 1 (Fig. 6B).
Fig. 6.
mTORC1 inhibition does not induce premature cell death during HCMV infection. (A) HFFs were infected with ADwt at an MOI of 1 in the presence or absence of rapamycin (10 nM) or torin 1 (250 nM). The morphology of infected cells was analyzed with a phase-contrast microscope at 72 hpi. (B) HFFs were either pretreated with rapamycin for 6 h and then infected with ADwt in the presence of rapamycin, or infected with ADwt in the presence of torin 1. At 72 hpi, infected cells were collected and analyzed for cell death (PARP cleavage) and mTORC1 activation (phosphorylation of S6K and 4E-BP1). HFFs infected with ADpmUL38 were included as the control for mutant virus-induced cell death.
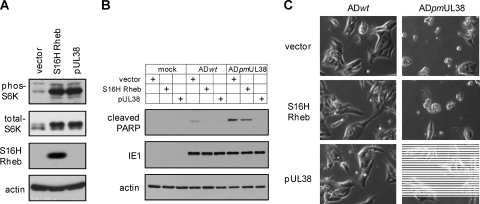
Finally, we wanted to determine if activation of mTORC1 would prevent premature cell death induced by pUL38-deficient virus infection. In this experiment, we overexpressed a myc-tagged, constitutive active variant of Rheb (S16H Rheb) (41), an activator of mTORC1. Indeed, overexpression of S16H Rheb-induced S6K phosphorylation to a level comparable to that by pUL38, indicating induction of mTORC1 activation (Fig. 7A). However, during infection of pUL38-deficient virus, robust PARP cleavage (Fig. 7B) and a markedly elevated level of cells with morphology characteristic of cell death were readily observed in S16H Rheb-expressing HFFs (Fig. 7C), comparable to those seen in vector-expressing control HFFs. Therefore, activation of mTORC1 was not sufficient to inhibit premature cell death induced by infection with pUL38-deficient virus.
Fig. 7.
mTORC1 activation does not prevent premature cell death induced by pUL38-deficient virus. HFFs expressing empty vector (vector), pUL38, or myc-tagged, constitutively active variant of Rheb (S16H Rheb) were made by transduction with expression retroviral vectors. (A) To confirm mTORC1 activation by S16H Rheb overexpression, transduced HFFs were cultured in serum-free medium for 48 h, and cell lysate was analyzed for myc-Rheb expression and S6K phosphorylation. (B and C) To determine the effect of mTORC1 activation on pUL38-deficient virus-induced premature cell death, transduced cells were either mock infected, or infected with ADwt or ADpmUL38 virus at an MOI of 1. Cells were collected at 72 hpi and analyzed for PARP cleavage by immunoblotting (B) or for the morphology of infected cells by a phase-contrast microscope (C).
Taken together, our results indicate that the multifunctional HCMV protein pUL38 prevents cell death independent of its induction of mTORC1 activation, both in isolation and during virus infection. Its cell death inhibitory activity is genetically and biochemically separable from its mTORC1 induction activity.
DISCUSSION
The hypothesis of “one gene, one enzyme” has been conventional wisdom in the molecular biology field for nearly 70 years. However, an overwhelming body of evidence indicates that this is an overly simplified view of a complex reality. Genes and proteins, even those in organisms with a large genome size, are regulated at multiple levels to perform different functions under various circumstances. This is particularly true for viruses, which are obligate intracellular parasites with a limited coding capacity compared to other living organisms. In order to replicate in an otherwise hostile host environment, the limited number of viral proteins need to be multifunctional in nature, thereby allowing viruses to subvert host machinery and resist host antiviral defense responses. In this postgenomic era, understanding the complexity of protein functions is becoming an important quest in almost every area of molecular biology.
pUL38 is one such viral protein that plays multiple critical roles during HCMV infection. It modulates the unfolded protein response and prevents ER stress-induced cell death (37, 40). It also activates mTORC1 to promote anabolic metabolism which is required for virus production (26). To test our hypothesis that the different domains of a multifunctional protein are responsible for its individual activities, we dissected the UL38 coding sequence into nine regions based on its predicted secondary structure and conservation with its homologs (Fig. 2). Analysis of serial truncation mutants, where each region was progressively deleted, has generated three major findings defining the sequence and domain requirements for pUL38 function. (i) Regions R3, R4, and R5 are critical for protein stability as the mutant UL38 proteins lacking these regions appear to be highly unstable (Fig. 2). These three regions are well conserved among pUL38 and its homologs in other CMV species and thus likely represent the core structure of the protein. (ii) The domain composed of the most conserved N-terminal 239 residues of pUL38 is necessary and sufficient to prevent cell death induced by pUL38-deficient virus infection or by the ER stress inducer tunicamycin (Fig. 3 and 4). However, this domain is not sufficient to activate mTORC1 in the absence of other viral factors (Fig. 3). (iii) The less conserved 38 amino acids at the C-terminal portion of pUL38 between residues 240 to 277 are necessary for full mTORC1 activation in isolation (Fig. 2 and 3). Thus, the ability to activate mTORC1 may be a new function that pUL38 has acquired during evolution of HCMV with its host.
Our study also provides several lines of evidence supporting the hypothesis that pUL38 prevents cell death independent of its function to activate mTORC1. When expressed alone, the truncation mutant protein pUL381–239 completely lost the ability to activate mTORC1 but it blocked ER stress-induced cell death as effectively as the wild type (Fig. 3 and 4). Moreover, suppression of mTORC1 activity by inhibitors did not compromise the ability of pUL38 to suppress ER stress-induced cell death in overexpressing cells (Fig. 4) or to prevent premature cell death during infection (Fig. 6). Activation of mTORC1 by a constitutively active Rheb does not prevent premature cell death induced during pUL38-deficient virus infection (Fig. 7). Therefore, our results provide strong evidence that mTORC1 activation is not required for pUL38 to prevent cell death either in isolation or in the context of virus infection.
A surprising finding of our study was that a recombinant HCMV expressing the mutant protein pUL381–239, which failed to activate mTORC1 in isolation, retained the ability to activate mTORC1 during infection (Fig. 5). In addition, pUL38-deficient virus also appeared to induce some mTORC1 activity, although at levels reduced from those seen for wild-type virus or virus expressing pUL381–239. These observations led to us to speculate that HCMV may have multiple ways, in addition to the pUL38-dependent mechanism, to activate mTORC1. Other viral factors or activities may also be involved in mTORC1 activation during infection. The identity of these factors and their means of mTORC1 activation remain elusive, but our data suggest that these are early events of HCMV infection (Fig. 5). At least two parallel signaling pathways responding to growth factors or nutrients, respectively, control mTORC1 activity. HCMV is known to activate the PI3K-Akt pathway during infection, which could allow the virus to exploit Akt-mediated TSC2 inhibition and activate mTORC1 (11, 15, 18, 32). HCMV also increases intracellular calcium concentration during infection (36), which could potentiate mTORC1 via a mechanism similar to that by amino acid stimulation (13). Precedents exist for HCMV to use multiple mechanisms or factors to modulate a single cellular pathway. It is sensible that HCMV may have built in a functional redundancy to maintain mTORC1 activity for a successful infection under various conditions.
How the N-terminal 239-residue region of pUL38 is involved in mTORC1 activation during HCMV infection remains unknown. The inability of this pUL38 region to activate mTORC1 in isolation but its ability to do so during infection signifies the involvement of other viral factor or activities. This pUL38 region may potentiate the activity or regulate the expression of other viral factors to activate mTORC1. Alternatively, it could enhance mTORC1 activity by inhibiting cell death or facilitating viral growth. Further studies are required to elucidate how exactly pUL38 and other viral factors interplay to induce mTORC1 activation and how they contribute to HCMV replication.
The ER and mTORC1 are two important hubs for maintaining cellular homeostasis. In a broader sense, both the ER and mTORC1 are involved in the regulation of macromolecular biosynthesis pathways that are critical to efficient virus production. mTORC1 enhances protein translation, mainly at the initiation steps, while the ER facilitates protein glycosylation and folding, the very last stages of protein production. The ER is the organelle for lipid biosynthesis and regulation, and the role of mTORC1 in lipogenesis has also recently been reported (7, 23, 31). The ER is the major intracellular calcium store and maintains calcium homeostasis within the cell, while calcium is an important signaling molecule that activates mTORC1. Both the ER and mTORC1 are sensitive to stress and respond to environmental cues to maintain a healthy intracellular environment. Importantly, an increasing body of evidence suggests the presence of cross talk between the ER and mTORC1. Protein translation induced by mTORC1 may result in the overloading of newly synthesized polypeptides into the ER, thus inducing the ER stress response. Conversely, the ER stress response induces eIF-2α phosphorylation to attenuate protein translation and can negatively regulate the mTORC1 pathway (17, 33). It appears that the ER and mTORC1 regulate and keep each other in check in order to maintain cellular homeostasis. HCMV uses pUL38 to modulate both the ER and mTORC1 functions by rewiring the circuit in a way that now stimulates a high mTORC1 activity and also induces a modified ER stress response. This will remodel ER function and enhance its capacity without eliciting the detrimental effect of the stress response, such as cell death, thus making an infected cell a remarkably efficient factory for virus production. In this study, we have identified the important domains of the UL38 protein and defined the relationship between its two known functions by using combined genetic and biochemical approaches. This sets the stage for further elucidating how pUL38 modulates these two very important cellular functions to benefit HCMV infection.
ACKNOWLEDGMENTS
We thank Herbert Virgin and the members of his laboratory for helpful discussions and invaluable advice, David Sabatini for torin 1, Thomas Shenk for the antibodies, Richard Lamb for the S16H Rheb plasmid, the High Speed Cell Sorter Core at the Siteman Cancer Center for excellent technical assistance, and members of the Yu lab for critical reading of the manuscript.
This study was supported by a Public Health Service grant (R01CA120768) and in part by a grant from the American Heart Association (09GRNT2290199) and an Interdisciplinary Research Initiative grant from Children's Discovery Institute at Washington University. D.Y. holds an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund. B.X. is a Berg-Morse Fellow of the Department of Molecular Microbiology, Washington University School of Medicine.
Footnotes
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Britt W. J., Alford C. A. (ed.). 1996. Cytomegalovirus, 3rd ed Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 2. Buchkovich N. J., Maguire T. G., Alwine J. C. 2010. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J. Virol. 84:7005–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buchkovich N. J., Maguire T. G., Paton A. W., Paton J. C., Alwine J. C. 2009. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. J. Virol. 83:11421–11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buchkovich N. J., et al. 2008. Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J. Virol. 82:31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchkovich N. J., Yu Y., Pierciey F. J., Jr., Alwine J. C. 2010. Human cytomegalovirus induces the endoplasmic reticulum chaperone BiP through increased transcription and activation of translation by using the BiP internal ribosome entry site. J. Virol. 84:11479–11486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchkovich N. J., Yu Y., Zampieri C. A., Alwine J. C. 2008. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 6:266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chakrabarti P., English T., Shi J., Smas C. M., Kandror K. V. 2010. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes 59:775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choo A. Y., Yoon S. O., Kim S. G., Roux P. P., Blenis J. 2008. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl. Acad. Sci. U. S. A. 105:17414–17419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chresta C. M., et al. 2010. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 70:288–298 [DOI] [PubMed] [Google Scholar]
- 10. Clippinger A. J., Maguire T. G., Alwine J. C. 2011. The changing role of mTOR kinase in the maintenance of protein synthesis during human cytomegalovirus infection. J. Virol. 85:3930–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dan H. C., et al. 2002. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J. Biol. Chem. 277:35364–35370 [DOI] [PubMed] [Google Scholar]
- 12. Feldman M. E., et al. 2009. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gulati P., et al. 2008. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 7:456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hegde N. R., et al. 2006. The role of BiP in endoplasmic reticulum-associated degradation of major histocompatibility complex class I heavy chain induced by cytomegalovirus proteins. J. Biol. Chem. 281:20910–20919 [DOI] [PubMed] [Google Scholar]
- 15. Inoki K., Li Y., Zhu T., Wu J., Guan K. L. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4:648–657 [DOI] [PubMed] [Google Scholar]
- 16. Isler J. A., Skalet A. H., Alwine J. C. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 79:6890–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin H. O., et al. 2009. Activating transcription factor 4 and CCAAT/enhancer-binding protein-beta negatively regulate the mammalian target of rapamycin via Redd1 expression in response to oxidative and endoplasmic reticulum stress. Free Radic. Biol. Med. 46:1158–1167 [DOI] [PubMed] [Google Scholar]
- 18. Johnson R. A., Wang X., Ma X. L., Huong S. M., Huang E. S. 2001. Human cytomegalovirus up-regulates the phosphatidylinositol 3-kinase (PI3-K) pathway: inhibition of PI3-K activity inhibits viral replication and virus-induced signaling. J. Virol. 75:6022–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kinsella T. M., Nolan G. P. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405–1413 [DOI] [PubMed] [Google Scholar]
- 20. Kudchodkar S. B., Del Prete G. Q., Maguire T. G., Alwine J. C. 2007. AMPK-mediated inhibition of mTOR kinase is circumvented during immediate-early times of human cytomegalovirus infection. J. Virol. 81:3649–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kudchodkar S. B., Yu Y., Maguire T. G., Alwine J. C. 2006. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc. Natl. Acad. Sci. U. S. A. 103:14182–14187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kudchodkar S. B., Yu Y., Maguire T. G., Alwine J. C. 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J. Virol. 78:11030–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laplante M., Sabatini D. M. 2010. mTORC1 activates SREBP-1c and uncouples lipogenesis from gluconeogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:3281–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marciniak S. J., Ron D. 2006. Endoplasmic reticulum stress signaling in disease. Physiol. Rev. 86:1133–1149 [DOI] [PubMed] [Google Scholar]
- 25. Mocarski E. S., Shenk T., Pass R. F. (ed.). 2007. Cytomegaloviruses, 5th ed., vol. 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 26. Moorman N. J., et al. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moorman N. J., Shenk T. 2010. Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J. Virol. 84:5260–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oresic K., Tortorella D. 2008. Endoplasmic reticulum chaperones participate in human cytomegalovirus US2-mediated degradation of class I major histocompatibility complex molecules. J. Gen. Virol. 89:1122–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perez C., McKinney C., Chulunbaatar U., Mohr I. 2011. Translational control of the abundance of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. J. Virol. 85:156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perng Y., Qian Z., Fehr A. R., Xuan B., Yu D. 2011. Human cytomegalovirus gene UL79 is required for the accumulation of late viral transcripts. J. Virol. 85:4841–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Porstmann T., et al. 2008. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8:224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Potter C. J., Pedraza L. G., Xu T. 2002. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4:658–665 [DOI] [PubMed] [Google Scholar]
- 33. Qin L., Wang Z., Tao L., Wang Y. 2010. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 6:239–247 [DOI] [PubMed] [Google Scholar]
- 34. Schroder M., Kaufman R. J. 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74:739–789 [DOI] [PubMed] [Google Scholar]
- 35. Sengupta S., Peterson T. R., Sabatini D. M. 2010. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40:310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sharon-Friling R., Goodhouse J., Colberg-Poley A. M., Shenk T. 2006. Human cytomegalovirus pUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc. Natl. Acad. Sci. U. S. A. 103:19117–19122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Terhune S., et al. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81:3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thoreen C. C., et al. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284:8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tirosh B., et al. 2005. Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J. Virol. 79:2768–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xuan B., Qian Z., Torigoi E., Yu D. 2009. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J. Virol. 83:3463–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yan L., et al. 2006. Hyperactivation of mammalian target of rapamycin (mTOR) signaling by a gain-of-function mutant of the Rheb GTPase. J. Biol. Chem. 281:19793–19797 [DOI] [PubMed] [Google Scholar]
- 42. Yu D., Smith G. A., Enquist L. W., Shenk T. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]