Abstract
In Epstein-Barr virus (EBV) latency III (LTIII) infection, BHRF1 encodes three microRNAs (miRNAs). Herein we report that Drosha cleavage of LTIII BHRF1 RNA and cis-acting splicing effects inhibit splicing and inhibit BHRF1 RNA and protein expression. Evidence shown here supports the view that Drosha cleavage to generate mature miRNAs and cis-acting sequences that prevent mRNA maturation are independent processes that prevent LTIII BHRF1 expression in lymphoblastoid cell lines.
INTRODUCTION
Eukaryotic RNA polymerase II (RNAP II) synthesizes pre-messenger RNAs (pre-mRNAs), which usually contain multiple exons and introns. Pre-mRNAs undergo processing steps, including 5′ capping, 3′ polyadenylation, and splicing to remove introns in the nucleus prior to transport to the cytoplasm for translation. These RNA-processing steps are coordinated and coupled to RNAP II transcription (37, 40, 45).
MicroRNAs (miRNAs) are a class of approximately 22-nucleotide (nt) small noncoding RNAs which regulate protein expression in eukaryotes (5, 9, 20, 30). Mature miRNA is processed largely from RNAP II 5′-capped and 3′-polyadenylated primary RNA (pri-miRNA) (6, 36). The pri-miRNA is cleaved in the nucleus by an RNase III enzyme, Drosha, and other interacting proteins, such as DGCR8 (23, 35), to release an approximately 70-nt hairpin pre-miRNA, which is exported into the cytoplasm and processed by the RNase III enzyme Dicer complex (11, 25) to produce an approximately 22-nt duplex (16). In addition to canonical pre-mRNA processing in the nucleus, Drosha cleavage occurs on pre-mRNAs that encode miRNA as well as protein. This noncanonical processing occurs on transcripts encoding miRNAs in introns or on the independently transcribed miRNA-coding transcripts (4, 19, 31, 42). However, for RNA transcripts that encode miRNAs in exons, the correlation of Drosha cleavage with canonical pre-mRNA processing steps, mRNA accumulation, and encoded protein expression is not clear.
Epstein-Barr virus (EBV), a human gammaherpesvirus, establishes latency III (LTIII) infection in B lymphocytes (29), causing infected cell proliferation into lymphoblastoid cell lines (LCLs) in vitro or lymphoproliferative diseases in vivo (29, 50). In LTIII infection, EBV expresses LTIII BHRF1 RNA that encodes three BHRF1 miRNAs (miR-BHRF1-1, miR-BHRF1-2, and miR-BHRF1-3) (7, 47). LTIII BHRF1 RNA is distinct from the EBV replication-associated 1.4-kb BHRF1 mRNA that is fully spliced and initiated by the virus replication-activated BHRF1 promoter (15). LTIII BHRF1 RNA has a long 5′-untranslated region (UTR) and is not spliced (3, 44, 48). Transcripts from the Cp or Wp promoter initiate alternatively spliced EBNA transcripts in LTIII infection and can be polyadenylated after the BHRF1 3′-UTR to produce LTIII BHRF1 RNAs (29). miR-BHRF1-1 is in the 5′-UTR of the LTIII BHRF1 mRNA and overlaps the TATA box of the EBV replication-activated BHRF1 promoter (15), whereas miR-BHRF1-2 and miR-BHRF1-3 are located in the 3′-UTR. While the BHRF1 miRNAs are produced in LTIII infection (57), BHRF1 protein has not been detected in LTIII-infected LCLs using effective BHRF1 antibodies (1, 29).
BHRF1 miRNAs inhibit apoptosis, favor cell cycle progression in early-phase infection of primary B cells (52), and strongly enhance B-cell transformation (21). In addition, miR-BHRF1-3 suppresses expression of the interferon-inducible T-cell-attracting chemokine CXCL-11/I-TAC in primary lymphoma, suggesting that the BHRF1 miRNA also participates in immunomodulatory processes (56).
BHRF1 is a 17-kDa homologue of human Bcl-2 (14, 44), is expressed early in EBV replication (13, 44), and exerts antiapoptotic effects by binding to Bim (18), VRK2 (human vaccinia virus B1R kinase-related kinase 2) (38), hPRA1 (human prenylated rab acceptor 1) (39), Bid, Puma, and Bak (33).
The purpose of this report was to investigate the mechanisms that prevent LTIII BHRF1 mRNA and protein expression.
MATERIALS AND METHODS
Cell culture and antibodies.
HEK293 cells (22) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% Fetal Plex animal serum complex (FPASC; Gemini). B95-8 cells and EBV-infected and uninfected BJAB cells were maintained in RPMI 1640 medium (Gibco-BRL) supplemented with 10% Fetal Plex animal serum complex (Gemini). Monoclonal antibody (MAb) 3E8 (13) or JL8 (Clontech) is specific for EBV BHRF1 protein or EGFP (enhanced green fluorescent protein), respectively. MAb against human β-actin or Drosha was purchased from Sigma.
Plasmids.
The EBV BHRF1 gene was amplified by PCR by using primers PmiBH-1f and PmiBH-1r (Table 1) from virus replication-induced Akata cells (54), cloned into pENTR/D-TOPO (Invitrogen), and transferred into p47F by LR homologous recombination (Invitrogen) to generate construct aLTIII. In the same way, the construct b or c was generated by using primer pair BHRF1-1/PmiBH-2r or PmiBH-1f/PmiBH-2r. Mutations were created using a QuickChange kit (Stratagene) with primers PmiBHmu1, Loop1, and Loop2 for miR-BHRF1-1, PmiBHmu2 for miR-BHRF1-2, and PmiBHmu3 for miR-BHRF1-3. The deletions were created by PCR in the 5′-UTR of LTIII BHRF1 mRNA. Briefly, the PCR products obtained using primer pairs Not F1/NheR1, Not F2/NheR1, Not F3/NheR1, Not F4/NheR1, and Not F5/NheR1 were digested by NotI and NheI and replaced a 0.814-kb NotI/NheI fragment of the construct aLTIII to generate constructs a7, a8, a9 a10, and a11, respectively. For the luciferase reporter assay, the DNA fragments containing the cytomegalovirus (CMV) promoter and the BHRF1 5′-UTR sequences in the constructs aLTIII, a7, a8, a9, and a11 (see Fig. 6) were released by MluI/BglII double digestion and then inserted into MluI/BglII doubly digested pGL3-basic vector (Promega) to produce the constructs rLTIII, r1, r2, r3, and r4, respectively. The CMV promoter alone was cloned into MluI/HindIII doubly digested pGL3-basic vector as a positive control (p). The pGL3-basic vector without eukaryotic promoter was used as a negative control (nc). All of the constructs were confirmed by sequencing.
Table 1.
Primers used in PCR
| Primer | Sequences |
|---|---|
| PmiBH-1f | 5′-CACCTTAGGAAGCACCACGTCC-3′ |
| PmiBH-2f | 5′-CACCGGACGGCTCCTTATTAAC-3′ |
| PmiBH-1r | 5′-AAACCTACTACCCCATTG-3′ |
| PmiBH-2r | 5′-TTAGTGTCTTCCTCTGGA-3′ |
| PmiBH-3r | 5′-TCAGGTTAATAAGGAGC-3′ |
| BHRF1-1 | 5′-CACCATGGCCTATTCAACAAGG-3′ |
| Loop1 | 5′-CAGCCCCGGAGTTGCCTTTCATCACTAACCC-3′ |
| Loop2 | 5′-GGGTTAGTGATGAAAGGCAACTCCGGGGCTG-3′ |
| PmiBHmu1 | 5′-CCTGTTTCATCACTAACCTTAATTAAGAAGAGGTTGACAAGAAG-3′ |
| PmiBHmu2 | 5′-GATAGCTGATACCCAATGTCTCGAGAATTGGCAGAAATTGAAAGTG-3′ |
| PmiBHmu3 | 5′-CACGTAATTTGCAAGCGGGATATCTGTTTCTTCGTTAAAATAACAC-3′ |
| NheR1 | 5′-CTTGCTGCTAGCTCCAACA-3′ |
| Not F1 | 5′-CGCTGCGGCCGCTATTAACCTGATCAGCC-3′ |
| Not F2 | 5′-CGCTGCGGCCGCGGTCAAGGTTTCGTCTG-3′ |
| Not F3 | 5′-CGCTGCGGCCGCCGAGATGTCACGTTGTG-3′ |
| Not F4 | 5′-CGCTGCGGCCGCTCCCACCCTCAATTTTC-3′ |
| Not F5 | 5′-CGCTGCGGCCGCAGATCTTGTAGAGCAAGA-3′ |
| pmiR1 | 5′-CTTAACCTGATCAGCCCCGGAGTAAA-3′ |
| pmiR2 | 5′-CGCTATCTTTTGCGGCAGAAATTGAAAGTG-3′ |
| pmiR3 | 5′-GCTAACGGGAAGTGTGTAAGCACACAA-3′ |
| pmiR16 | 5′-CGCTAGCAGCACGTAAATATTGGCGAA-3′ |
| pactbF | 5′-CAGATCATGTTTGAGACCTT-3′ |
| pactbR | 5′-GGTGACCCCGTCACCGGAGT-3′ |
| pF1F | 5′-GTCCCGGGGACGGAAGGGGAC-3′ |
| pF1R | 5′-CTTATTCTTGCTCATATTTCC-3′ |
| pF2F | 5′-GTCTCCGGGTGGGCAGGCAG-3′ |
| pF2R | 5′-GACACAGACCTGAAACACAAC-3′ |
| pF3F | 5′-GATATTTCACCGTGGAGACCCA-3′ |
| pF3R | 5′-TGACAGGTCCACAACATAGTAA-3′ |
| pF4F | 5′-GGACAAGATACTAAAGAAATAA-3′ |
| pF4R | 5′-TCCCCCCGAGTCTGGGCTGCAG-3′ |
| pF5F | 5′-GGAAATATGAGCAAGAATAAG-3′ |
| pF5R | 5′-AGACGAAACCTTGACCCTTC-3′ |
| pF6F | 5′-ACAAGAATAACATGCCAATGA-3′ |
| pF6R | 5′-TATCCCACCTAGGACACCCAA-3′ |
| pF7F | 5′-AAGGGCAGGGGCTGTTGGGTG-3′ |
| pF7R | 5′-CCGGAGACCTGCATCTGCACA-3′ |
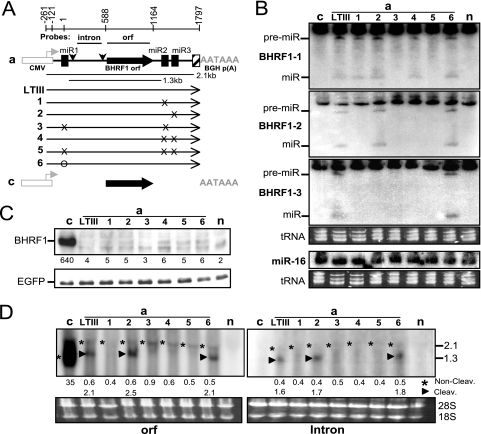
Fig. 6.
Splicing is key to BHRF1 protein expression. Transfection of the 293 cells and analysis of the protein, miRNAs, and messenger RNAs were done as described in the Fig. 3 legend. (A) Diagram of the constructs. The numbers at the deleted BHRF1 region (dotted lines) correspond to the first nucleotide position on either side of the undeleted regions. The constructs are named on the left. (B) Northern analysis of the pre-miRNA (pre-miR) and mature miRNA (miR). n, mock transfection. (C) Western analysis of protein expression. Numbers on the bottom indicate the relative intensity of specific BHRF1 protein bands. n, mock transfection. (D) Northern analysis of the BHRF1 RNA transcripts by using 32P-labeled ORF probe. The positions of 2.1-, 1.3-, and 0.9-kb RNA bands are indicated on the left in kilobases. The sizes of Millennium RNA size makers (Ambion) are shown on the right in kilobases. The numbers on the bottom indicate the relative intensities of noncleavage (*) or Drosha cleavage (▶) BHRF1 RNA bands. 18S, 18S rRNA; 28S, 28S rRNA; n, mock transfection. (E) 5′-RACE analysis of BHRF1 RNA isolated from transfected 293 cells. Reverse transcription was primed with Racer1 primer (57). The resulting cDNAs were poly(dC) tailed and amplified by PCR. PCR products were separated in 1.5% agarose gels. PCR products were purified, cloned, and sequenced. Sequencing-confirmed EBV-specific PCR products are labeled with asterisks on the left of the bands. M, 1-kb Plus DNA ladder (Invitrogen). The sizes of the 1-kb Plus DNA ladder are shown on the left in base pairs. The lower panel shows the partial sequences of the PCR products. (F) Subcellular localization of the noncleavage (*) or Drosha cleavage (▶) BHRF1 transcripts. Cytoplasmic (Cyto) and nuclear (Nuc) RNAs were separately isolated from 293 cells at 24 h posttransfection. The subcellular localization of BHRF1 transcripts in transfected cells was determined by Northern hybridization using 32P-labeled BHRF1 ORF probe. The BHRF1 transcript Fig. 3A from construct c was used as cytoplasmic RNA control. The sizes of Millennium RNA markers (Ambion) are shown on the right in kilobases. n, mock transfection; 18S, 18S rRNA; 28S, 28S rRNA.
Transfection.
The plasmids were transfected into HEK293 cells or EBV-infected and uninfected B cells by using Effectene (Qiagen) or Gene Pulser (Bio-Rad). All of the experiments were repeated independently at least three times.
Western blot analysis.
The cells were harvested 24 h posttransfection and lysed in 2× SDS-PAGE loading buffer. Western blot analysis was done as described previously (57). The specific signals in the Western blots were quantitated using Kodak IM Network software (Kodak).
Northern blot analysis.
The cytoplasmic, nuclear, and total RNA were prepared as described previously (57) by using Trizol reagent (Invitrogen). Northern analysis of the total RNAs was performed as described previously (57). Cellular miR-16 (34) was included as a loading control. The specific signals in the Northern blots were quantitated using PhosphorImager Imagequant software (Amersham Biosciences).
5′-RACE.
5′-RACE (rapid amplification of cDNA 5′ ends) analysis was done with a 5′-RACE kit (Invitrogen) as described previously (57).
siRNA.
Control small interfering RNA (siRNA) was purchased from Ambion. An siRNA targeting human Drosha was designed as reported (35), purchased from Invitrogen, and transfected into 293 cells. The plasmid DNA then was transfected into siRNA-treated cells 2 days later. At 24 h posttransfection of plasmid DNA, the total RNA was isolated from the whole-cell lysate by using Trizol reagent, and the poly(A)-tailed mRNA was purified from cytoplasmic RNA by using oligo(dT) cellulose (Ambion). RNA was treated by Turbo DNase (Ambion) prior to reverse transcription reactions. The random primers plus the 18mer oligo(dT) primed the reverse transcription of total RNA or the cytoplasmic mRNA.
Real-time RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed in triplicate on the Applied Biosystems 7500 fast real-time PCR system. For mature miRNA, real-time RT-PCR was done using an NCodeVILO miRNA cDNA synthesis kit and Express SYBR green ER miRNA quantitative RT-PCR kits (Invitrogen). The primers pmiR1, pmiR12, pmiR13, and pmiR16 (Table 1) are specific to miR-BHRF1-1, miR-BHRF1-2, miR-BHRF1-3, and cellular miR-16, respectively. For non-miRNA RNAs, real-time RT-PCT was done using a QuantiTect SYBR green PCR kit according to the manufacturer's instructions (Qiagen). RT-PCR products F1 to F7 were amplified using primer pairs pF1F/pF1R, pF2F/pF2R, pF3F/pF3R, pF4F/pF4R, pF5F/pF5R, pF6F/pF6R, and pF7F/pF7R, respectively (Table 1). The threshold cycle (CT) values were normalized to that of the human β-actin mRNA in each corresponding sample.
Dual luciferase reporter assay.
Ten micrograms of each luciferase reporter construct plus 4 μg of plasmid expressing β-galactosidase under the control of the simian virus 40 (SV40) promoter were electroporated into B95-8 cells or EBV-infected or uninfected BJAB cells. Twenty-four hours posttransfection, the cells were harvested to determine the luciferase activity (Promega) and the β-galactosidase activity (Galacton Plus; Tropix) by using luminometry.
Statistical analysis.
Statistical analysis was performed by Student's t test. P < 0.05 was considered significant and was indicated with asterisks.
RESULTS
Abolishing Drosha cleavage of the exon does not change either mRNA level or encoded protein expression.
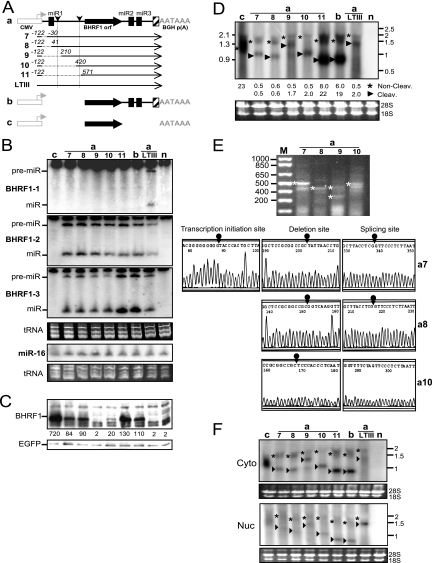
Since three BHRF1 miRNAs are located in the LTIII BHRF1 RNA and flank the BHRF1 open reading frame (ORF) (3, 47, 48), we first determined if Drosha cleavage at the miRNA sites restricts protein expression by destabilizing the mRNA. The BHRF1 gene of 1,918 bp (nt −121 to 1797) was isolated from the EBV Akata strain and then placed under the control of the CMV promoter (Fig. 1). The miRNA loci then were mutated (Fig. 2). The BHRF1 ORF also was cloned into the same vector (Fig. 3, lanes c) as a control. Resulting constructs were individually cotransfected into HEK 293 cells with an EGFP-expressing plasmid as an internal control. At 24 h posttransfection, the levels of BHRF1 protein, miRNA, and messenger RNA were examined.
Fig. 1.
Diagram of the partial sequences of the construct aLTIII. The 1,918-bp EBV sequence was placed under the control of the CMV promoter (open box) and BGH polyadenylation signal, and the nucleotides are numbered between positions −121 and 1797. A small arrowhead shows the start site and orientation of transcription initiated by the CMV promoter. The CMV promoter initiated RNA transcript starts at nt −261 and has 139-nt non-EBV leader sequences. The dotted lines represent sequences not shown. The sequences of pre-miR-BHRF1-1, pre-miR-BHRF1-2, and pre-miR-BHRF1-3 are underlined and indicated as miR1, miR2, and miR3. The TATA box (TATTA) and transcription start site (nucleotide position 1) of the EBV lytic replication-activated BHRF1 promoter and polyadenylation signal (AATAAA) are in boldface. The splice donor, acceptor, and 439-nt intron in brackets are indicated. The partial BHRF1 amino acid sequences are shown under corresponding codons.
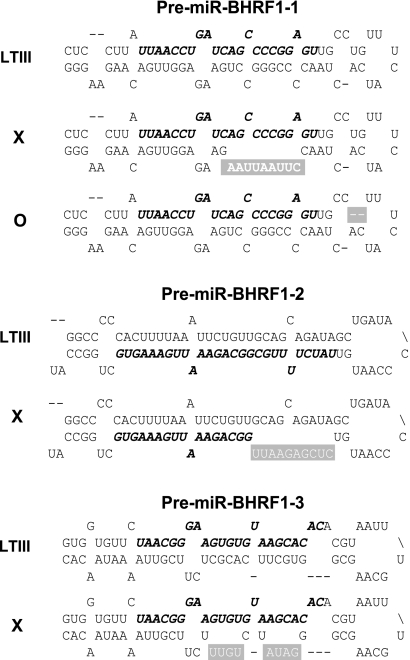
Fig. 2.
Mutations disrupt the hairpin structures of pre-miR-BHRF1-1, pre-miR-BHRF1-2, and pre-miR-BHRF1-3. Shown are the mature miRNA sequences (italics and boldface), mutated sequences (white letters), and deleted sequences (white dashes). Mutated pre-miRNAs are represented by the symbols on the left.
Fig. 3.
Blocking Drosha cleavage of the exon does not affect the mRNA level and encoded protein expression. The 293 cells were cotransfected by an EGFP-expressing plasmid and the wild-type or mutant BHRF1 construct. At 24 h posttransfection, the levels of BHRF1 protein or miRNA and mRNA were determined by Western or Northern analysis. The expression of EGFP was examined as an internal control. (A) Diagram of BHRF1 constructs. Shown are the CMV promoter (open box) and its transcription orientation (horizontal arrowhead), BGH polyadenylation signal (AATAAA), authentic BHRF1 polyadenylation signal (hatched box), ORF and intron probes, BHRF1 ORF (filled arrow), pre-miR-BHRF1-1 (miR1), pre-miR-BHRF1-2 (miR2), pre-miR-BHRF1-3 (miR3), splice donor and acceptor (vertical arrowheads), 2.1- and 1.3-kb BHRF1 RNA transcripts, and mutations (× or O). The BHRF1 constructs are named on the left. (B) Northern analysis of the pre-miRNA (pre-miR) and mature miRNA (miR). n, mock transfection. (C) Western analysis of protein expression. Numbers on the bottom indicate the relative intensity of specific BHRF1 protein bands. n, mock transfection. (D) Northern analysis of the BHRF1 RNA transcripts by using 32P-labeled ORF or intron probe. The positions of 2.1- and 1.3-kb RNA bands are indicated on the right in kilobases. The numbers on the bottom indicate the relative intensities of noncleavage (*) or Drosha cleavage (▶) BHRF1 RNA bands. 18S, 18S rRNA; 28S, 28S rRNA; n, mock transfection.
In cells transfected by construct aLTIII, BHRF1 miRNAs (Fig. 3B) but not the BHRF1 protein (Fig. 3C) were detectable. This duplicates EBV LTIII infection in that BHRF1 miRNA rather than the protein is produced (57). BHRF1 ORF and intron probes detected a 2.1- and a 1.3-kb RNA in Northern analysis (Fig. 3D). The 2.1-kb RNA band is weak and consistent in size with the noncleavage full-length transcript (Fig. 3A and D). The 1.3-kb RNA is consistent in size and Northern hybridization characteristics with the BHRF1 transcript cleaved by Drosha at the miR-BHRF1-1 and miR-BHRF1-2 sites and is at least 3-fold more abundant than the 2.1-kb RNA from which the 1.3-kb RNA was derived. In cells transfected with mutant construct a1, a3, a4, or a5, neither the corresponding miRNA (Fig. 3B) nor the 1.3-kb Drosha cleavage RNA (dcRNA) (Fig. 3D) was detectable, indicating that the mutations abolished Drosha cleavage. However, both BHRF1 protein (Fig. 3C) and noncleavage 2.1-kb RNA (Fig. 3D) remain at the same low level as that of aLTIII, demonstrating that Drosha cleavage of the RNA does not alone affect mRNA or encoded protein expression. Since both miRNAs (Fig. 3B) and Drosha cleavage 1.3-kb BHRF1 RNA (Fig. 3D) were detectable in the cells transfected by construct a6, the deletion in this construct (Fig. 2 and 3A, lane a6) did not alone affect Drosha cleavage. The mutation of the pre-miR-BHRF1-2 sequences resulted reproducibly in undetectable miR-BHRF1-3 (Fig. 3A and B, lane a1) and Drosha cleavage 1.3-kb RNA (Fig. 3D, lane a1). Since miR-BHRF1-3 is separated by only 72 nt from miR-BHRF1-2 in the primary transcript, the mutation in miR-BHRF1-2 may block Drosha cleavage at the miR-BHRF1-3 site by altering the structure of the primary RNA.
Depletion of Drosha does not affect the LTIII BHRF1 mRNA level and encoded protein expression.
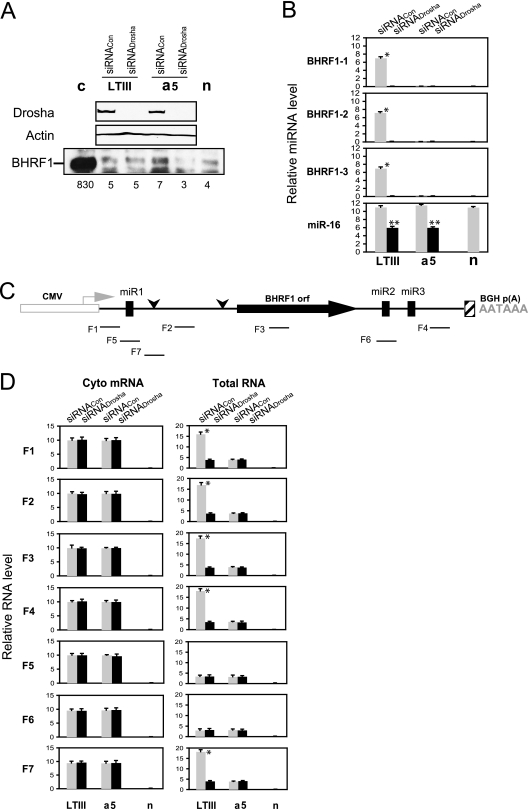
To further examine the effect of Drosha cleavage on mRNA accumulation and protein expression, we depleted cellular Drosha by transfecting Drosha siRNA (siRNADrosha) into 293 cells (Fig. 4). The siRNADrosha resulted in undetectable Drosha in the cells (Fig. 4A). Quantitative real-time RT-PCR detected three BHRF1 miRNAs only in the control siRNA (siRNACon) and aLTIII-transfected cells (Fig. 4B), confirming that depletion of Drosha abolished the cleavage of the LTIII BHRF1 RNA. However, BHRF1 protein was not increased in cells transfected by either wild-type aLTIII or the miRNA locus-mutated a5 (Fig. 4A). The cellular miR-16 was decreased but still readily detectable in Drosha-depleted cells (Fig. 4B), likely due to the longer half-life of the mature miRNA (26).
Fig. 4.
Depletion of Drosha does not alone affect the LTIII BHRF1 cytoplasmic or total RNA level or encoded protein expression. The 293 cells were transfected by siRNADrosha or negative control siRNACon and 2 days later were transfected by the indicated expression plasmid described in Fig. 3Aa, LTIII, c, or a5. “n” is vector only. At 24 h posttransfection of plasmid DNA, the protein lysates, RNA, and cDNA were prepared. The levels of miRNA and RNA were measured by real-time RT-PCR. Cellular miR-16 or β-actin mRNA was measured and used as a reference for normalization of the threshold cycle values (CT). (A) Western analysis of protein expression. Numbers on the bottom indicate the relative intensity of BHRF1 protein. (B) miRNA determined by real-time RT-PCR analysis of total RNA. (C) Diagram showing the relative positions of PCR products amplified by real-time RT-PCR. (D) Real-time RT-PCR analysis of BHRF1 transcripts in cytoplasmic poly(A) mRNAs (cyto mRNA) or in total RNAs. The averages of triplicates ± standard deviations were plotted. n, vector transfection. *, P < 0.001 for comparisons of siRNACon to siRNADrosha; **, P < 0.01 for comparisons of siRNACon to siRNADrosha.
The BHRF1 transcripts were analyzed by real-time RT-PCR using seven primer pairs (Fig. 4C). All primer pairs detected similar levels of RNA in cytoplasmic poly(A) RNA isolated from either control siRNA or Drosha siRNA-transfected cells (Fig. 4D), indicating that abolishing Drosha cleavage by either the knockout of Drosha or the mutation of miRNA loci, such as in construct a5, does not affect mRNA accumulation in the cytoplasm. The primer pairs for PCR products F1, F2, F3, F4, and F7 detected more than 4-fold increases of RNA in the total RNAs isolated from the control siRNA and aLTIII-transfected cells but not from the cells where Drosha cleavage has been abolished by the knockout of the cellular Drosha (Fig. 4D, lane aLTIII with Drosha siRNA) or by mutation of the miRNA loci (Fig. 4D, lanes a5). The RT-PCR using primer pairs for PCR products F5 and F6 that span miR-BHRF1-1 and miR-BHRF1-2, respectively (Fig. 4C), detected similarly low levels of RNA in total RNA isolated from the transfected cells (Fig. 4D), suggesting that the high level of accumulated Drosha-cleaved RNA contributes to the increased RNAs detected by the primer pairs for PCR products F1, F2, F3, F4, and F7 in the total RNA isolated from the aLTIII-transfected Drosha-competent cells (Fig. 4D). Taking these results together, the depletion of Drosha does not affect the LTIII BHRF1 mRNA level and encoded protein expression in this context.
Dynamic accumulation of Drosha-cleaved RNA and uncleaved LTIII BHRF1 mRNA.
Since PCR products F2 and F5 may reflect Drosha cleavage and noncleavage LTIII BHRF1 transcripts, respectively, in construct aLTIII-transfected Drosha-competent cells (Fig. 4C and D), we did real-time RT-PCR using the primer pairs for F2 and F5 to analyze the accumulation of Drosha-cleaved RNA and the uncleaved RNA. As a control, the cells were transfected by miRNA locus-mutated construct a5 as well. Total RNA was isolated at the indicated time points posttransfection. In the cells transfected by a5, both F2 and F5 were undetectable until 12 h posttransfection and accumulated to a low level even at 48 h posttransfection (Fig. 5, a5). In contrast, in the wild-type construct aLTIII-transfected cells, F2 was readily detectable as early as 4 h posttransfection and quickly accumulated to a high level, but F5 was undetectable until 12 h posttransfection and accumulated to a low level at 48 h posttransfection (Fig. 5, aLTIII). This observation further supports the view that Drosha cleavage is not correlated with mRNA accumulation.
Fig. 5.
Accumulation of the Drosha-cleaved and uncleaved BHRF1 transcripts. The 293 cells were transfected by construct aLTIII or a5 (Fig. 3). Total RNA was isolated at the indicated time points posttransfection and analyzed by real-time RT-PCR using primer pairs for PCR products F2 and F5 (Fig. 4D).
Splicing controls protein expression of the LTIII BHRF1 RNA.
We examined the effect of the 5′-UTR of LTIII BHRF1 RNA on protein expression by analyzing a series of deletion mutations (Fig. 6A). Transcripts initiated by the CMV promoter all start at the same site and have a common 139-nt leader sequence (Fig. 1 and 6A), thus excluding effects of transcription initiation. miR-BHRF1-1 was detected only in the aLTIII-transfected cells, whereas miR-BHRF1-2 and miR-BHRF1-3 were expressed from all mutated constructs (a7 to a11) (Fig. 6B). The deletion between nt −122 and −30 in construct a7 left all pre-miR-BHRF1-1 sequences but still blocked miR-BHRF1-1 expression. This is consistent with the previous report that sequences immediately adjacent to pre-miRNA sequences are necessary for Drosha cleavage (35). The deletion of the first 91 or 161 nt of BHRF1 sequence in construct a7 or a8 (Fig. 6A) resulted in an approximately 40-fold increase in BHRF1 protein (Fig. 6C), although the noncleavage RNA levels were almost the same as that of aLTIII (Fig. 6D). However, an approximately 1.5-kb, rather than the expected 2.0-kb, noncleavage RNA, or an approximately 1.1-kb, rather than the expected 1.5-kb, Drosha cleavage RNA was detected in Northern analysis of construct a7- or a8-transfected cell RNA (Fig. 6D). In 5′-RACE analysis, a BHRF1-specific RT-PCR product of about 0.5 or 0.45 kbp rather than the expected 0.9 kbp was amplified. The sequencing demonstrates that these PCR products lack the 439-nt intron (Fig. 6E), indicating that the BHRF1 RNAs produced from the constructs a7 and a8 have been spliced as the EBV replication-associated 1.4-kb BHRF1 mRNA is (57). These data reveal 5′ cis-acting effects on splicing as key to regulating protein expression from LTIII BHRF1 RNA.
The deletion of the first 330 or 540 nt of BHRF1 in construct a10 slightly increased the BHRF1 protein level (Fig. 6C), while the noncleavage RNA accumulated at a level similar to that of aLTIII (Fig. 6D). Both constructs lack the splice donor and contain partial intron sequences (Fig. 6A). In the construct a10-transfected cell RNA, a 1.6-kb noncleavage RNA and an approximately 1.0-kb dcRNA were detected by ORF probe in Northern analysis (Fig. 6D). Furthermore, a 0.5-kbp PCR product with the expected size and sequence content was amplified by 5′-RACE (Fig. 6E). In the construct a9-transfected cell RNA, in addition to the expected 1.8-kb noncleavage RNA and 1.2-kb dcRNA, a smaller RNA (about 1.0 kb in size) was detectable (Fig. 6D). In this RNA pool, the 5′-RACE identified two BHRF1-specific PCR products (0.32 and 0.45 kb) (Fig. 6E), both of which lack the 139-nt common leader and contain partial intron sequences, suggesting that the BHRF1 mRNAs in the construct a9-transfected cells underwent degradation at the 5′ end (Fig. 6C). Overall, the observations indicate that 5′ sequences affect LTIII BHRF1 RNA processing and translation.
The deletion of the first 690 nt out of the total 708 nt of the BHRF1 5′-UTR in construct a11 (Fig. 6A) completely removed the intron and increased BHRF1 protein (Fig. 6C) and mRNA (Fig. 6D) to levels as high as those of construct b, demonstrating that intron content can affect BHRF1 protein expression at the transcript level as well.
Both Drosha cleavage and noncleavage BHRF1 transcripts from the constructs a7, a8, a9, a10, a11, aLTIII, and b were detected mainly in the cytoplasm, except the 5′ and 3′ Drosha double-cleavage 1.3 kb RNA, which was detected mainly in the nucleus (Fig. 6F, lane aLTIII), indicating that the entire intron in LTIII BHRF1 RNA (aLTIII) inhibits export of RNA to the cytoplasm.
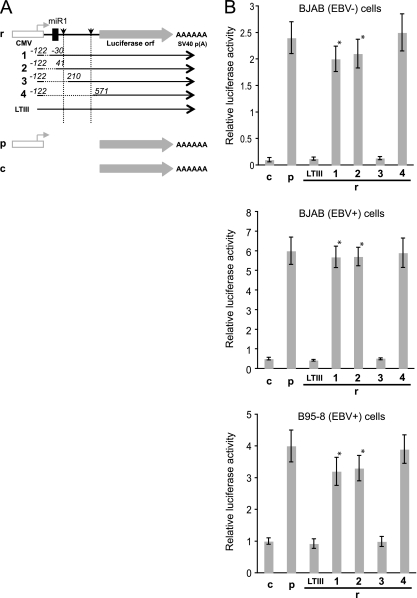
LTIII BHRF1 RNAs are produced in the EBV latency III-infected LCLs (3, 48). To test if the 5′-UTR of LTIII BHRF1 transcripts regulates protein expression in B cells in the same way as in HEK293 cells, the BHRF1 5′-UTR sequences with the deletions shown in Fig. 6A were placed between a CMV promoter and a firefly luciferase ORF (Fig. 7A). The resulting plasmids were transfected into EBV-infected or uninfected BJAB cells or EBV-transformed marmoset B lymphoblastoid cells (B95-8) (8). The dual luciferase reporter assay was done at 24 h posttransfection. The construct rLTIII, or r3, contains the entire intron content (0.4 kb) or a large part of the intron content and resulted in dramatically reduced expression of luciferase in either EBV-infected or uninfected B cells (Fig. 7B). The deletion of the first 91 nt (between BHRF1 nt −122 and −30) or 161 nt (between BHRF1 nt −122 and 41) resulted in spliced BHRF1 transcripts, dramatically increased BHRF1 protein expression in 293 cells (Fig. 6, a7 and a8), and significantly increased luciferase activity in construct r1- or r2-transfected EBV-infected or uninfected B cells (Fig. 7A and B). As expected, deletion of the entire intron in construct r4 (Fig. 7A) led to levels of luciferase activity in transfected B cells as high as those of constructs r1 and r2 (Fig. 7B). These observations confirm that the intron is an important regulator of BHRF1 protein expression in B cells.
Fig. 7.
Luciferase reporter assay in EBV-infected or uninfected B cells. The EBV-infected (EBV+) B95-8 and BJAB cells or uninfected (EBV−) BJAB cells were cotransfected by firefly luciferase reporter constructs and plasmid expressing β-galactosidase. Twenty-four hours posttransfection, the β-galactosidase activity and luciferase activity were measured. (A) Diagram of the firefly luciferase reporter constructs. Shown are the firefly luciferase ORF (gray arrow), SV40 polyadenylation signal (AAAAAA), start site and orientation (smaller horizontal arrow) of transcription initiated by CMV promoter (open box), positions of pre-miR-BHRF1-1 (miR1) and splice donor and acceptor (vertical arrowheads), and BHRF1 5′-UTR sequences (boldface line), as depicted in the Fig. 3 legend. The mutant constructs are named on the left. Nucleotide numbers at the deletion region (dotted line) correspond to the first nucleotide on either side of the undeleted BHRF1 regions. c, construct without promoter. (B) Firefly luciferase reporter assay. The luciferase activity was measured and normalized with β-galactosidase activity. The average of triplicate experiments ± the standard deviation was plotted. *, P < 0.001.
DISCUSSION
Drosha cleavage of the exon does not affect mRNA level and encoded protein expression.
We found that the expression of BHRF1 protein was silenced by a small intron (0.4 kb) immediately upstream of the BHRF1 ORF (Fig. 8). This observation is in agreement with a genomic analysis suggesting that the Drosha cleavage could not directly destabilize mRNAs that encode miRNA in the exon (53), except for DGCR8 mRNA (24). The finding that blocking Drosha cleavage, particularly in the case of EBV LTIII BHRF1 RNA, in which Drosha cleavage RNA was accumulated at at least 3-fold-higher levels than noncleavage RNA (Fig. 3 to 6, lanes aLTIII), did not increase noncleavage mRNA accumulation (Fig. 3 to 6) suggests that Drosha cleavage and mRNA synthesis derive from different RNA molecules. We propose that unstable transcripts may be the major substrates for Drosha processing. This proposal is reminiscent of the defective ribosomal protein products (DRiPs) or truly short-lived proteins (SLiPs), which result from mistakes in fidelity of translating mRNA into proteins (58). The imperfect, unstable DRiPs or SLiPs are the major source of antigenic peptides presented by major histocompatibility complex class I molecules in induction of cytotoxic T-cell responses (58).
Fig. 8.
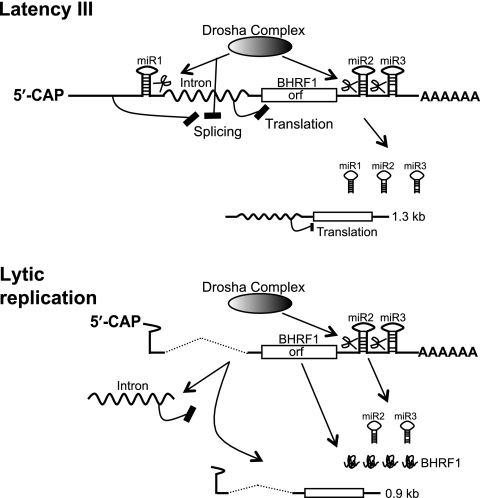
Splicing and Drosha RNA processing coordinately control the selective expression of miRNA from the LTIII BHRF1 RNA. In EBV latency III infection, the inhibition of RNA splicing in cis by the sequences upstream of the miR-BHRF1-1 site and in trans by the Drosha cleavage of the miR-BHRF1-1 site results in the presence of intron in the LTIII BHRF1 transcripts, thereby blocking the translation of the LTIII BHRF1 RNA while miR-BHRF1-1, miR-BHRF1-2, and miR-BHRF1-3 and the 1.3-kb dcRNA are produced (57). During EBV lytic replication, the EBV lytic replication-activated BHRF1 promoter initiates transcription from a new start site within the pre-miR-BHRF1-1 sequences. The transcripts have neither the splicing inhibition sequences nor the Drosha cleavage site upstream of the intron. Thus, the resultant splicing enables the EBV replication-associated 1.4-kb BHRF1 mRNA to be translated into BHRF1 protein. Additionally, the Drosha cleavage of the spliced RNA transcripts at the miR-BHRF1-2 and miR-BHRF1-3 sites produces miR-BHRF1-2 and miR-BHRF1-3 and a spliced 0.9-kb dcRNA that was identified in EBV-infected B cells during virus replication (57).
The observation that only about 10% of the miRNAs are encoded in the exon of the protein-coding and noncoding RNA transcripts and that more than 60% are encoded in the intron implies that the Drosha cleavage of the exon has regulatory effects on mRNA synthesis and encoded protein expression that are usually avoided (31, 41, 42). Using a conserved exon to encode miRNAs to target another conserved exon could impose an evolutionary obstacle. An intron may be evolutionarily advantageous as a host for miRNAs due to its flexibility in both sequence composition and fate (51).
Drosha cleavage RNA other than pre-miRNA (dcRNA) has an unusual stability character.
dcRNA is always regarded as the junk produced in the process of miRNA biogenesis. In this report, we found that the dcRNA could accumulate to a level severalfold greater than that of the corresponding mRNA (Fig. 3 to 6, lanes aLTIII, a11, and b), indicating that the dcRNA can be stable. The biological significance of the high-level accumulation of dcRNA remains to be investigated. This phenomenon seems to be in contrast to the postulation that Drosha cleavage facilitates degradation of introns by recruiting exonucleases to sites of Drosha cleavage (4, 42). The 3′ dcRNA (Fig. 6F, lanes a7 to 11) was readily detectable in the cytoplasm but barely in the nucleus, indicating that this RNA was efficiently exported from the nucleus into the cytoplasm, similarly to the noncleavage RNA. In contrast, the 3′ and 5′ 1.3-kb dcRNA was readily detectable in the nucleus but barely detected in the cytoplasm (Fig. 6F, aLTIII). These observations suggest that the 5′ end that is critical to mRNA export (12) is also critical for dcRNA export.
One of the widely studied viral non-protein-coding RNAs (ncRNAs) is the herpes simplex virus type 1 latency-associated transcript (HSV-1 LAT), which is the only viral gene expressed throughout latent infection (27). Like EBV LTIII BHRF1 RNA, HSV-1 LAT encodes several miRNAs (46). Moreover, while the primary 8.3-kb LAT is highly unstable, LAT fragments of 2.0, 1.5, and 1.4 kb are stable (27). This phenomenon is reminiscent of the aforementioned proposal that Drosha would cleave unstable RNAs to produce stable RNA and pre-miRNA. The role of Drosha cleavage in this stabilizing process is worthy of being investigated. For eukaryotes, dcRNA may represent one source of long noncoding RNAs that are pervasively transcribed (10). Long noncoding RNAs play important roles in nuclear architecture or in regulation of gene expression (10).
Drosha cleavage at the miR-BHRF1-1 site inhibits downstream RNA splicing.
Drosha cleavage and splicing are cotranscriptional processes (40, 42, 45). Here, we show evidence indicating that Drosha cleavage at the miR-BHRF1-1 site can inhibit downstream splicing. The deletion of the first 91 nt of the BHRF1 5′-UTR sequence resulted in RNA splicing (Fig. 6D), indicating that sequences upstream of the miR-BHRF1-1 site inhibit splicing in cis. However, the removal of these sequences by Drosha cleavage at the miR-BHRF1-1 site produced an intron-containing 1.3-kb dcRNA (Fig. 6D), suggesting that Drosha cleavage at this site concomitantly inhibits splicing in trans (Fig. 8), which prevents leaky BHRF1 protein expression from high-level, BHRF1 ORF-containing, Drosha cleavage 1.3-kb RNA (57). The inhibition of splicing by Drosha cleavage at the miR-BHRF1-1 site could be via physical interference in which Drosha complex binding at the miR-BHRF1-1 site makes the splice donor inaccessible to the spliceosome or interrupts the coupling of transcription to spliceosome assembly (17). Because an intact 5′ cap is required for efficient splicing as well as polyadenylation and transcription (43, 49), the removal of the 5′ cap by Drosha cleavage to produce miR-BHRF1-1 may contribute to inhibition of splicing as well.
Drosha cleavage may occur after splicing event.
The splice donor and acceptor are located far upstream of miR-BHRF1-2 and miR-BHRF1-3 in the LTIII BHRF1 RNA. Thus, cotranscriptional splicing may be done before the transcription goes through the miR-BHRF1-2 and miR-BHRF1-3 sites. The identification of both spliced and Drosha-cleaved BHRF1 RNAs in the cells transfected with the constructs a7 and a8 (Fig. 6D) indicates that Drosha cleavage does not necessarily occur only prior to (19, 42) or simultaneously with (31) RNA splicing, and may occur after a splicing event.
Intron inhibits translation of LTIII BHRF1 RNA.
The roles of the intron in the regulation of gene expression were confined mainly to the nucleus (37, 59). In this report, we found that the BHRF1 intron may act as a cis-acting regulator to downregulate BHRF1 protein expression (Fig. 6). The four extra translation start codons (ATG) in the intron would contribute to the downregulation of protein expression by interfering with translation initiation at the authentic start site. In addition, the intron may inhibit translation by forming structure or by recruiting other trans-acting factors (32).
In addition to its antiapoptotic role in EBV replication, BHRF1 protein may have a role in initial cell transformation (2) or in the pathogenesis of Wp-restricted Burkitt lymphomas (28, 55). The expression of BHRF1 protein has been confirmed in a subset of Burkitt lymphomas (28). Consistently with the finding in this report that the 0.4-kb intron immediately upstream of the BHRF1 ORF inhibits the translation of the LTIII BHRF1 RNA (Fig. 8), the BHRF1 protein in the Wp-restricted Burkitt lymphoma was expressed from a variant of latency BHRF1 transcripts in which the 0.4-kb intron immediately upstream of the BHRF1 ORF has been excised by an alternative splicing distinct from that of the EBV lytic replication-associated 1.4-kb BHRF1 mRNA (28).
Conclusion.
The TATA box and transcription start site of the virus replication-activated BHRF1 promoter (15) overlap with the pre-miR-BHRF1-1 sequences. EBV replication-associated BHRF1 RNAs initiated by this promoter have neither the upstream splicing inhibition sequences nor the intact pre-miR-BHRF1-1 sequences for Drosha cleavage, thus enabling the virus replication-associated BHRF1 mRNA to be spliced to remove the intron and express BHRF1 protein in early virus replication (57) (Fig. 8).
The evidence in this report reveals that EBV selectively expresses BHRF1 miRNAs from a potentially protein-coding RNA by Drosha cleavage of RNA during latency III infection (Fig. 8).
ACKNOWLEDGMENTS
We are grateful to Mei-Ying Liu and Jen-Yang Chen from the National Taiwan University for generously providing MAb 3E8.
The research was supported in part by grants CA47006, CA131354, and CA085180 from the National Cancer Institute of the U.S. Public Health Service to E.K.
The authors declare no competing financial interests.
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Alfieri C., Birkenbach M., Kieff E. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595–608 [DOI] [PubMed] [Google Scholar]
- 2. Altmann M., Hammerschmidt W. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 3:e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Austin P. J., Flemington E., Yandava C. N., Strominger J. L., Speck S. H. 1988. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 85:3678–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ballarino M., et al. 2009. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell. Biol. 29:5632–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cai X., Hagedorn C. H., Cullen B. R. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10:1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai X., et al. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callard R. E., et al. 1988. The marmoset B-lymphoblastoid cell line (B95-8) produces and responds to B-cell growth and differentiation factors: role of shed CD23 (sCD23). Immunology 65:379–384 [PMC free article] [PubMed] [Google Scholar]
- 9. Carthew R. W., Sontheimer E. J. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen L. L., Carmichael G. G. 2010. Decoding the function of nuclear long non-coding RNAs. Curr. Opin. Cell Biol. 22:357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chendrimada T. P., et al. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436:740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng H., et al. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127:1389–1400 [DOI] [PubMed] [Google Scholar]
- 13. Chou S. P., Tsai C. H., Li L. Y., Liu M. Y., Chen J. Y. 2004. Characterization of monoclonal antibody to the Epstein-Barr virus BHRF1 protein, a homologue of Bcl-2. Hybrid. Hybridomics 23:29–37 [DOI] [PubMed] [Google Scholar]
- 14. Cleary M. L., Smith S. D., Sklar J. 1986. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell 47:19–28 [DOI] [PubMed] [Google Scholar]
- 15. Cox M. A., Leahy J., Hardwick J. M. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cullen B. R. 2004. Transcription and processing of human microRNA precursors. Mol. Cell 16:861–865 [DOI] [PubMed] [Google Scholar]
- 17. Das R., et al. 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev. 20:1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Desbien A. L., Kappler J. W., Marrack P. 2009. The Epstein-Barr virus Bcl-2 homolog, BHRF1, blocks apoptosis by binding to a limited amount of Bim. Proc. Natl. Acad. Sci. U. S. A. 106:5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edwards R. H., Marquitz A. R., Raab-Traub N. 2008. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J. Virol. 82:9094–9106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eiring A. M., et al. 2010. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140:652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feederle R., et al. 2011. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 7:e1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Graham F. L., Smiley J., Russell W. C., Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74 [DOI] [PubMed] [Google Scholar]
- 23. Han J., et al. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125:887–901 [DOI] [PubMed] [Google Scholar]
- 24. Han J., et al. 2009. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136:75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hutvágner G., et al. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834–838 [DOI] [PubMed] [Google Scholar]
- 26. Hwang H. W., Wentzel E. A., Mendell J. T. 2007. A hexanucleotide element directs microRNA nuclear import. Science 315:97–100 [DOI] [PubMed] [Google Scholar]
- 27. Jones C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kelly G. L., et al. 2009. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog. 5:e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kieff E., Rickinson A. B. 2007. Epstein-Barr virus and its replication, p. 2603–2654 In Knipe D. M., Howley P. M. (ed.), Fields virology. Lippincott Williams & Wilkins Publishers, Philadelphia, PA [Google Scholar]
- 30. Kim V. N., Han J., Siomi M. C. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10:126–139 [DOI] [PubMed] [Google Scholar]
- 31. Kim Y. K., Kim V. N. 2007. Processing of intronic microRNAs. EMBO J. 26:775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kozak M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13–37 [DOI] [PubMed] [Google Scholar]
- 33. Kvansakul M., et al. 2010. Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1. PLoS Pathog. 6:e1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294:853–858 [DOI] [PubMed] [Google Scholar]
- 35. Lee Y., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419 [DOI] [PubMed] [Google Scholar]
- 36. Lee Y., et al. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Le Hir H., Nott A., Moore M. J. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28:215–220 [DOI] [PubMed] [Google Scholar]
- 38. Li L. Y., Liu M. Y., Shih H. M., Tsai C. H., Chen J. Y. 2006. Human cellular protein VRK2 interacts specifically with Epstein-Barr virus BHRF1, a homologue of Bcl-2, and enhances cell survival. J. Gen. Virol. 87:2869–2878 [DOI] [PubMed] [Google Scholar]
- 39. Li L. Y., Shih H. M., Liu M. Y., Chen J. Y. 2001. The cellular protein PRA1 modulates the anti-apoptotic activity of Epstein-Barr virus BHRF1, a homologue of Bcl-2, through direct interaction. J. Biol. Chem. 276:27354–27362 [DOI] [PubMed] [Google Scholar]
- 40. Moore M. J., Proudfoot N. J. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136:688–700 [DOI] [PubMed] [Google Scholar]
- 41. Morin R. D., et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18:610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morlando M., et al. 2008. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 15:902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Orphanides G., Reinberg D. 2002. A unified theory of gene expression. Cell 108:439–451 [DOI] [PubMed] [Google Scholar]
- 44. Pearson G. R., et al. 1987. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology 160:151–161 [DOI] [PubMed] [Google Scholar]
- 45. Perales R., Bentley D. 2009. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36:178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pfeffer S., et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276 [DOI] [PubMed] [Google Scholar]
- 47. Pfeffer S., et al. 2004. Identification of virus-encoded microRNAs. Science 304:734–736 [DOI] [PubMed] [Google Scholar]
- 48. Pfitzner A. J., Tsai E. C., Strominger J. L., Speck S. H. 1987. Isolation and characterization of cDNA clones corresponding to transcripts from the BamHI H and F regions of the Epstein-Barr virus genome. J. Virol. 61:2902–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Proudfoot N. J., Furger A., Dye M. J. 2002. Integrating mRNA processing with transcription. Cell 108:501–512 [DOI] [PubMed] [Google Scholar]
- 50. Rickinson A. B., Kieff E. 2007. Epstein-Barr virus, p. 2655–2700 In Knipe D. M., Howley P. M. (ed.), Fields virology. Lippincott Williams & Wilkins Publishers, Philadelphia, PA [Google Scholar]
- 51. Roy S. W., Gilbert W. 2006. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 7:211–221 [DOI] [PubMed] [Google Scholar]
- 52. Seto E., et al. 2010. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 6:e1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shenoy A., Blelloch R. 2009. Genomic analysis suggests that mRNA destabilization by the microprocessor is specialized for the auto-regulation of Dgcr8. PLoS One 4:e6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takada K., et al. 1991. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes 5:147–156 [DOI] [PubMed] [Google Scholar]
- 55. Watanabe A., et al. 2010. Epstein-Barr virus-encoded Bcl-2 homologue functions as a survival factor in Wp-restricted Burkitt lymphoma cell line P3HR-1. J. Virol. 84:2893–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xia T., et al. 2008. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 68:1436–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xing L., Kieff E. 2007. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J. Virol. 81:9967–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yewdell J. W., Nicchitta C. V. 2006. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 27:368–373 [DOI] [PubMed] [Google Scholar]
- 59. Zhu J., et al. 2010. A novel role for minimal introns: routing mRNAs to the cytosol. PLoS One 5:e10144. [DOI] [PMC free article] [PubMed] [Google Scholar]