Abstract
The replication of plus-strand RNA viruses depends on many cellular factors. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is an abundant metabolic enzyme that is recruited to the replicase complex of Tomato bushy stunt virus (TBSV) and affects asymmetric viral RNA synthesis. To further our understanding on the role of GAPDH in TBSV replication, we used an in vitro TBSV replication assay based on recombinant p33 and p92pol viral replication proteins and cell-free yeast extract. We found that the addition of purified recombinant GAPDH to the cell extract prepared from GAPDH-depleted yeast results in increased plus-strand RNA synthesis and asymmetric production of viral RNAs. Our data also demonstrate that GAPDH interacts with p92pol viral replication protein, which may facilitate the recruitment of GAPDH into the viral replicase complex in the yeast model host. In addition, we have identified a dominant negative mutant of GAPDH, which inhibits RNA synthesis and RNA recruitment in vitro. Moreover, this mutant also exhibits strong suppression of tombusvirus accumulation in yeast and in virus-infected Nicotiana benthamiana. Overall, the obtained data support the model that the co-opted GAPDH plays a direct role in TBSV replication by stimulating plus-strand synthesis by the viral replicase.
INTRODUCTION
Plus-strand RNA [(+)RNA] viruses replicate in the cytosol of infected cells using membrane-bound replicase complexes. The viral replicase is a multisubunit complex consisting of viral-coded components, including RNA-dependent RNA polymerase (RdRp), one or more viral auxiliary replication proteins, the viral RNA, and co-opted host proteins and membranes (1, 19, 22, 34, 35, 52, 57). However, the functions of many host components of the viral replicase complex are currently unknown.
Among the co-opted host proteins for (+)RNA virus replication are the abundant cellular RNA-binding proteins, such as poly(C)- and poly(A)-binding proteins, nucleolin, elongation factor 1A (eEF1A), and the heterogeneous nuclear ribonucleoprotein (hnRNP) A1 (15, 22–24, 29, 50, 63). Additional subverted RNA-binding proteins include ribosomal proteins, translation factors, and RNA-modifying enzymes (1, 36, 39). Although most of these host RNA-binding proteins have been shown to associate with the viral positive-strand RNAs, there is a small number of host proteins that preferentially bind to the (−)RNA intermediate template in the replicase complex. For example, the T-cell intracellular antigen-1 (TIA-1) and the TIA-1-related protein (TIAR), which are stress granule proteins, that bind specifically to the 3′-stem-loop (SL) of West Nile virus (−)RNA (20). Mutations in the 3′-SL, which reduce TIA1/TIAR binding, can greatly decrease genomic RNA amplification, suggesting that TIA1/TIAR facilitate efficient (+)RNA synthesis (12). Similarly, the cellular hnRNP C, which binds specifically to the 3′ end of the poliovirus (−)RNA (6, 51), has also been proposed to affect positive-strand synthesis. The hnRNP C may function to maintain the single-stranded form of poliovirus (−)RNA via its RNA chaperone activity and recruit viral 3CD replication protein to form an initiation complex for (+)RNA synthesis (6).
Tomato bushy stunt virus (TBSV) is a small (+)RNA virus that infects a wide range of host plants. Its genome codes for five proteins, including the overlapping p33 and p92pol viral replication proteins (67). p92pol, the viral RdRp protein, shares a common N-terminal region with p33. TBSV has emerged as a model virus to study virus replication and recombination based on the development of yeast (Saccharomyces cerevisiae) as a model host (34, 38, 42, 67). Systems biology approaches in yeast have facilitated the identification of over 300 host genes affecting either TBSV replication or recombination (17, 21, 23, 27, 30, 32, 39, 55, 56). A proteomics analysis of the highly purified tombusvirus replicase complex has revealed the presence of p33 and p92pol viral replication proteins and 4 to 10 host proteins in the replicase complex. These host proteins included a protein chaperone (heat shock protein 70 [HSP70]), eEF1A, Cdc34p E2 ubiquitin-conjugating enzyme, ESCRT proteins, and metabolic enzymes (Tdh2/3p and Pdc1p) (2, 21, 23, 24, 46, 54, 64–66). The subverted proteins play various roles in TBSV replication, such as facilitating the recruitment of the viral RNA for replication, promoting the assembly of the replicase complex or affecting RNA synthesis.
Among the identified permanent resident host factors in the tombusvirus replicase complex is the glyceraldehyde-3-phosphate dehydrogenase (GAPDH; also known as Tdh2p and Tdh3p in yeast). GAPDH is a highly conserved, abundant, and ubiquitous protein (59, 62). Aside from being a key component of cytosolic energy production, GAPDH is also involved in many additional activities, including apoptosis, endocytosis, nuclear tRNA transport, vesicular secretary transport, nuclear membrane fusion, modulation of the cytoskeleton, DNA replication and repair, maintenance of telomere structure, and transcriptional control of histone gene expression (59, 60, 62). Furthermore, GAPDH exhibits distinct binding to AU-rich sequences at the 3′ terminus of mRNAs, which leads to their stabilization in cells (5). In yeast, GAPDH is coded by the highly similar, functionally redundant TDH2 and TDH3 genes, which can complement one another.
GAPDH has been shown to bind to AU-rich sequences present in various RNA viruses, including TBSV, hepatitis A virus (HAV), hepatitis C virus (HCV), Japanese encephalitis virus, transmissible gastroenteritis coronavirus, hepatitis delta virus RNA, and human parainfluenza virus type 3 (9, 10, 13, 25, 37, 44, 50, 53, 58, 64, 68). In addition, GAPDH also binds to the internal ribosome entry site (IRES) element in HAV, which could be involved in suppressing cap-independent translation of HAV RNA (69). GAPDH might also be involved in the posttranscriptional regulation of hepatitis B virus gene expression (11, 70).
Previous functional studies of GAPDH in tombusvirus replication have revealed a regulatory role in asymmetric viral RNA synthesis (64). It has been shown that depletion of GAPDH preferentially inhibits the accumulation of (+)RNA, resulting in a 1:1 ratio of (+)RNA and (−)RNA products in yeast and in Nicotiana benthamiana host. GAPDH binds selectively to an AU pentamer sequence close to the 3′ terminus of TBSV (−)RNA. Deletion of this AU pentamer in a TBSV replicon (rep)RNA led to the production of a 1:1 (+)RNA/(−)RNA ratio even in the presence of wild-type levels of GAPDH. Therefore, GAPDH was proposed as the host factor that binds to and retains TBSV (−)RNA for multiple rounds of (+)RNA synthesis inside the tombusviral replicase complex (64). On the other hand, the viral (+)RNA progeny, which are not bound by GAPDH, are released from the viral replicase complex into the cytosol for further cycles of replication or other viral processes.
To further advance our understanding of the roles of GAPDH in TBSV replication, we utilized the recently developed in vitro viral replication assay based on yeast cell-free extracts (CFE) that are prepared from yeast mutant strains depleted of GAPDH. Our data show that the in vitro-assembled TBSV replicase produces less positive-strand viral RNAs in the GAPDH-depleted CFE, resulting in symmetric replication of the two strands. Addition of purified recombinant GAPDH to the GAPDH-depleted CFE restored asymmetric RNA synthesis by producing 4- to 5-fold more positive than negative strands in vitro. Interaction of GAPDH with p92pol was needed for the recruitment of GAPDH into the viral replicase complex. Finally, the two RNA binding regions of GAPDH, when expressed in the absence of the C-terminal catalytic domain, acted in a dominant negative manner in vitro, in yeast, and in plants, too. Altogether, we highlight here the direct role of GAPDH during TBSV replication.
MATERIALS AND METHODS
Yeast strains and expression plasmids.
Saccharomyces cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was from Open Biosystems. Yeast strain (tdh2Δ TET::TDH3) was previously generated (64), while strain (tdh1-3Δ) was obtained from Hans Lehrach (Max Planck Institute, Germany) (49). While tdh-1 to -3 are deleted in this strain, it expresses E. coli GAPDH.
The plasmids pHisGBK-CUP-His33/GAL-DI72 and pGAD-CUP-Hisp92 used for in vivo viral replication experiments were created previously (16, 21). The plasmids used in the in vitro RdRp assay—pGAD-ADH-HFp92, pYC-GAL-DI72, and pHisGBK-ADH-HFp33—were cloned according to the method of Serva and Nagy (54).
To generate the Escherichia coli expression plasmids for GST-Tdh2 and its deletion derivatives, Tdh-ΔRBD1, Tdh-ΔRBD2, Tdh-ΔRBD1+2, Tdh-RBD1+2, and Tdh-ΔC, the restriction sites used for ligation into pGEX-2T vector were BamHI and EcoRI. PCR amplification of Tdh-ΔRBD1, Tdh-ΔRBD1+2, Tdh-RBD1+2, and Tdh-ΔC were performed with the primer sets 1979/4155, 1843/4155, 1839/4152, and 1839/4151 (Table 1), respectively. PCR products were digested with BamHI and EcoRI and ligated into pGEX-2T using the same enzymes. PCR fragment of Tdh-ΔRBD2 required two cloning steps. First, the primer sets 1839/4188 and 4189/4155 were used to amplify coding sequence without the RBD2 domain, and then the two separate PCR products were combined in a ligation mixture, followed by PCR amplification with the primers 1839 and 4155.
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| 1839 | CGCGGGATCCGTTAGAGTTGCTATTAAC |
| 1843 | CGCGGGATCCCCATCTTCCACCGCCCC |
| 1979 | CGCGGGATCCTTCAAGTACGACTCTACTCA |
| 3782 | CCGGCTCGAGTTAGCTAGCAGCCTTGGCAACGTGTTCAACCAAGTC |
| 3783 | GCCGGATCCGCTAGCGGTCATCATCATCATCATCACGACAAGCAGAAGAACGGC |
| 3784 | GGATCCGCTAGCGGTCATCATCATCATCATCACGTGAGCAAGGGCGAGGAG |
| 3785 | CGGCCTGCAGGCTCGAGTTAGGCGGTGATATAGACGTTG |
| 3786 | CGGCCTGCAGGCTCGAGTTACTTATACAGCTCGTCCATGCCGAG |
| 4151 | CGCGGAATTCGGAAGCGGTTCTACCACCTCTCCR |
| 4152 | CGCGGAATTCAGCAGTGATGACAACCTTCTTGGCACCAGCGTC |
| 4155 | CGCGGAATTCTTAAGCCTTGGCAACGTGTTCAACC |
| 4188 | GACGAACATTGGGGCGGTGGAAGATGGCATGTAAGCGGAGTAGTCGTTAGAGATGAAAGC |
| 4189 | CTTTCATCTCTAACGACTACTCCGCTTACATGCCATCTTCCACCGCCCCAATGTTCG |
| 4354 | CGCGCTCGAGAGCAGTGATGACAACCTTCTTGGCACCAGCGTC |
| 4355 | CGCGCTCGAGCCTTCTCGTTGACACCCACAACAAACATGG |
| 4364 | CGCGGGATCCATGGACTACAAAGACGATGACGACAAGGTTAGAGTTGCTATTAACGGTTT |
| 4365 | CGCGGGATCCATGGACTACAAAGACGATGACGACAAGAGATCGGAATCAATGGATTTGGA |
pESC-CUP-HFTdh2 and pESC-CUP-HFRBD1+2 used to express 6×His/FLAG-tagged Tdh2p and 6×His/FLAG-tagged RBD1+2 in yeast for the pulldown assays was generated by PCR using the primer pairs 4364/4354 and 1839/4155, respectively. The PCR products were digested with BamHI and EcoRI and ligated into pESC plasmid digested with the same set of restriction enzymes.
To express Tdh-RBD1+2 and N. benthamiana RBD1+2 truncated proteins in plants, PCR was performed using the primer set 4364/4354 for the amplification of Tdh-RBD1+2 fragment, which was then digested with BamHI and XhoI and ligated into pGD plasmid predigested with the same enzymes. The sequence of N. benthamiana-RBD1+2 was obtained by a BLAST search against the N. benthamiana PUTs database and using the primers 4365 and 4356 to PCR amplify the cloning fragment. The PCR product of N. benthamiana RBD1+2 was also digested with BamHI and XhoI for ligation as described above.
Analysis of protein-protein interaction using the split-ubiquitin assay.
The split-ubiquitin assay is based on the Dualmembrane Kit3 (Dualsystems) and performed as previously described (3, 27). The bait constructs, pGAD-BT2-N-His33 and pGAD-BT2-N-His92, expressing tombusvirus replication proteins p33 and p92, respectively, were as described previously (21). TDH2 was amplified by using the primers 1839 and 3782 and pGBK-Tdh2-YFP as a template. The TDH2 PCR product was digested with BamHI and NheI and ligated into the pPRN-N-RE and pPRN-C-RE (21) vectors digested with the same enzymes. Yeast strain NMY51 was cotransformed with pGAD-BT2-N-His33 and pPR-N-RE (NubG) or one of the prey constructs carrying TDH2 and plated onto Trp− Leu− (TL) synthetic minimal medium plates for plasmid selection. Yeast colonies were then resuspended in 50 μl of water and spotted onto Trp− Leu− His− Ade− (TLHA) plates to detect p33/p92-host protein interactions as described previously (23). Plasmid containing SSA1 sequence was as previously described (4, 27) and used as the positive control in this assay.
Confocal laser microscopy.
To visualize the cellular localization of Tdh2p in yeast in the presence of the viral proteins p33 and p92, yeast was transformed with pGBK-Tdh2-YFP, in combination with pYES-CFP-p33 or pGAD-CFP-p92 (37). As a positive control, yeast was transformed with pGAD-CFP-p92 and pESC-p33/DI72 (37), which allowed tombusvirus replication. Confocal laser microscopy was performed on an Olympus FV1000 (Olympus America, Inc., Melville, NY) as described earlier (18).
Bimolecular fluorescence complementation assay (BiFC).
PCR products of Venus N-terminal (VN) and C-terminal (VC) fragments were amplified from pYES-Venus-p33 plasmid (37) with the primer sets 3784/3785 and 3783/3786, respectively. Both products were digested with BamHI and XhoI and ligated into pESC and pYES vectors using the same enzymes (unpublished data). PCRs amplifying the VN and VC fragments for construction of pESC-VN-p33, pESC-VC-p33, pGAD-VN-p92, and pGAD-VC-p92 plasmids used the primer sets 1292/3762 and 3763/3764 and were digested with NcoI and BamHI for cloning as described previously (unpublished data). The PCR was used to fuse the TDH2 sequence with VN and VC with the primers 1839 and 3782, followed by NheI digestion to ligate the NheI-digested VN/VC fragment. PCR amplification of Tdh-VN and Tdh-VC were carried out with the primer sets 1839/3785 and 1839/3786, respectively, and digested with BamHI and XhoI and ligated into pESC and pYES vectors digested with the same enzymes. Live cell imaging with confocal microscopy was carried out as described above.
Analysis of protein-protein interaction in vitro.
The maltose-binding protein (MBP)-tagged p33/p33C, MBP-tagged p92/p92C, and MBP-tagged p88/p88C were purified from E. coli as described previously (47). Amylose columns were used to bind the MBP-tagged viral proteins or MBP (negative control). The columns were washed three times with cold column buffer prior to addition of the yeast lysate. For the pulldown assay, 100 mg of yeast pellets containing His6- and Flag-tagged Tdh2 were resuspended in 150 μl of chilled buffer I (20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 200 mM NaCl, 10% [vol/vol] glycerol, 0.1% [vol/vol] NP-40, 10 mM β-mercaptoethanol, 1% [vol/vol] yeast protease inhibitor cocktail [Ypic]) and 1 μl of RNase A (1 mg/ml). Cells were broken in a Genogrinder with a 250-μl volume of acid-washed glass beads for 2 min at 1,500 rpm, followed by the addition of 600 μl of buffer I. The yeast lysate was centrifuged at 100 × g at 4°C for 5 min, and the supernatant was transferred to a prechilled Eppendorf tube and centrifuged at 15,000 rpm at 4°C for 5 min before being loaded onto amylose columns and incubated for 3 h at 4°C. The columns were washed five times with cold column buffer, and the bound protein complexes were eluted with column buffer supplemented with 10 mM maltose. The presence of His6/Flag-Tdh2 protein in the eluate was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by Western blotting with an anti-His6x antibody (42). The amount of MBP-tagged viral proteins in the eluate was visualized by Coomassie blue staining of the SDS-PAGE gels.
Analysis of TBSV repRNA replication in yeast.
Replication assays in yeast were performed as previously described (40). Detection of DI-72 replicon RNA (repRNA) accumulation was visualized on a 1.5% ethidium bromide-stained agarose gel and quantified by Northern blot using RNA probes complementary to region III-IV of DI-72(+) (38).
Yeast strain (tdh2Δ TET::TDH3) was transformed with plasmids pGAD-CUP-Hisp92, pHisGBK-CUP-Hisp33/GAL-DI72, and pYC (low copy number)/YES (high-copy number)-GAL-Tdh-RBD1+2. Yeast transformants were first grown in synthetic dropout (SD) minimal medium (Ura− Leu− His− [ULH−]) containing 2% galactose, 100 μM bathocuproinedisulfonic acid (BCS), and 10 μg of doxycycline/ml at 23 and 29°C overnight, respectively. The cells were harvested the following day by centrifugation at 3,850 rpm for 4 min to collect yeast pellets and washed three times with ULH− medium containing 2% galactose to remove BCS and then grown in ULH− medium containing 2% galactose and 10 μg of doxycycline/ml for 24 h at 23 and 29°C, respectively.
In vitro replication assay using the whole-cell extract.
The yeast strain (tdh2Δ TET::TDH3) used for cell extract (CFE) were grown in yeast extract-peptone-dextrose (YPD) medium with or without 10 μg of doxycycline/ml at 23°C for 6 h to a final optical density of 1.5, followed by 30 min at 37°C before harvesting and prepared as described previously (46). Similarly, the tdh1-3Δ TEF::Sir2 yeast strain was grown in the same conditions without doxycycline in the medium. The CFE assay (25 μl), containing 3 μl of CFE, 0.5 μg of DI-72 (+)repRNA transcript, 700 ng of purified MBP-p33, 1.4 μg of purified MBP-p92pol (both recombinant proteins were purified from E. coli), was as described previously (46). In addition to the recombinant viral replication proteins, MBP-p33 and MBP-p92pol, 800 ng of GST-Tdh2 or its deletion derivatives was also included in the assay. Reactions containing only GST or Tdh2p without CFE served as negative controls. The reaction mixture was incubated at 25°C for 2 h (46). The newly synthesized 32P-labeled RNA products were separated by electrophoresis in a 5% polyacrylamide gel (PAGE) containing 0.5× Tris-borate-EDTA (TBE) buffer with 8 M urea. To detect the double-stranded RNA (dsRNA) in the cell-free replication assay, the 32P-labeled RNA samples were loaded onto the PAGE gel, without heat treatment, and run for 4.5 h at 150 V (23).
Protein purification from E. coli.
Expression and purification of the recombinant TBSV p33 and p92 and Turnip crinkle virus (TCV) p88/p88C replication proteins from E. coli were carried out as described earlier, with modifications (47, 48). Purification of glutathione S-transferase (GST)-tagged proteins was carried out using glutathione resin and eluted with 10 mM glutathione and 10 mM β-mercaptoethanol in the column buffer according to the same protocol as for the MBP proteins (47, 48). The E. coli lysates were treated with RNase one to remove RNA from the protein samples. The MBP-tagged proteins were eluted with a low-salt column buffer containing 0.18% (vol/wt) maltose and 6% (vol/vol) glycerol and divided into aliquots for storage at −80°C. Protein fractions used for the replication assays were at least 95% pure, as determined by SDS-PAGE. The purified protein preparations were RNase free since we did not observe repRNA degradation at the end of the replicase assays, as judged by ethidium-bromide stained RNA gels (not shown).
Replicase purification from yeast and in vitro RdRp assay.
Yeast strain (tdh2Δ TET::TDH3) was transformed with the plasmids pGAD-ADH-HFp92, pYC-GAL-DI72, and pHisGBK-ADH-HFp33 expressing 6×His- and Flag-tagged tombusvirus p33 and p92. Copurification was done according to a previously described procedure with the following modification (23). Briefly, 2 g of yeast cells was resuspended and homogenized in TG buffer (50 mM Tris–HCl [pH 7.5], 10% glycerol, 15 mM MgCl2, 10 mM KCl, 0.5 M NaCl, 0.5% Triton, and 1% [vol/vol] yeast protease inhibitor cocktail [Ypic]) by glass beads using a FastPrep homogenizer (MP Biomedicals). The yeast cell lysate was cleared by centrifugation at 500 × g for 5 min at 4°C to remove unbroken cells and debris. The membrane fraction containing the viral replicase complex was collected by centrifugation at 35,000 × g for 15 min at 4°C and then solubilized in 1 ml of TG buffer with a buffer containing 2% Triton and 1% (vol/vol) Ypic via gentle rotation for 3 h at 4°C. The solubilized membrane fraction was centrifuged at 35,000 × g for 15 min at 4°C, and the supernatant was incubated with 100 μl of anti-FLAG M2-agarose affinity resin (Sigma) pre-equilibrated with 1 ml of TG buffer. After 3 h of gentle rotation at 4°C, the column was washed five times with TG buffer containing 0.5% Triton. The resin-bound replicase complex was eluted in 700 μl of elution buffer (50 mM Tris-HCl [pH 7.5], 10% glycerol, 15 mM MgCl2, 10 mM KCl, 0.05 M NaCl, 0.5% Triton, 1% Ypic, and 0.15 mg of Flag peptide [Sigma]/ml) after overnight rotation at 4°C. An in vitro RdRp activity assay was performed using (−)DI-72 RNA template transcribed in vitro by T7 transcription (23).
In vitro TCV p88C RdRp assay.
The TCV RdRp reaction was carried out as previously described for 2 h at 25°C (48). The RdRp reactions were performed in a 20-μl mixture containing 500 ng of (−)DI72 RNA template and 250 ng of affinity-purified MBP-p88C. Reactions containing only 800 ng of GST-Tdh2 protein without p88C were used as negative controls in the assay. The 32P-labeled RNA products were analyzed by electrophoresis in a 5% polyacrylamide–8 M urea gel (31).
In vitro viral RNA recruitment assay.
The in vitro RNA recruitment reaction was performed according to (45, 46), except that 32P-labeled DI72 (+)repRNA was used and rCTP, rUTP, 32P-labeled UTP, and actinomycin D were omitted from the reaction. As a negative control, E. coli purified GST protein was used in the absence of yeast CFE. After a 2-h incubation at 25°C, 1 ml of reaction buffer was added to the in vitro reaction mixture and left on ice for 10 min before centrifugation at 35,000 × g for 45 min. The membrane-bound 32P-labeled repRNA was extracted from the pellet by adding 0.1 ml of stop buffer and 0.1 ml of phenol-chloroform, followed by brief vortexing at a high setting and centrifugation at 27,000 × g for 4 min. The RNA samples were analyzed by denaturing PAGE and phosphorimaging as described previously (45, 46).
Agroinfiltration and plant infection.
A culture of Agrobacterium tumefaciens strain C58C1 carrying pGD-Flag-Tdh-RBD1+2/pGD-Flag-N. benthamiana-RBD1+2 and pGD-p19 was prepared and infiltrated together into leaves of N. benthamiana as previously described (61). Agrobacterium cultures containing pGD-p19 and pGD plasmid were used as a negative control. For Cucumber necrosis virus (CNV) infection experiments, young N. benthamiana plants were infiltrated with Agrobacterium containing pGD-CNV at an optical density at 600 nm of 0.1 following 24 h of preinfiltration of host protein and p19. Infiltrated and systemic new leaves were collected at various days postinfiltration for viral repRNA analyses as previously described (2). Detection of viral repRNA accumulation was visualized on a 1.5% ethidium bromide-stained agarose gel and quantified by Northern blot using RNA probes complementary to region III-IV of the CNV viral genome. The CNV expression plasmids pGD-CNV and pGD-p19 were as described previously (7).
Northern and Western blot analyses.
Total RNA isolation and Northern blot analysis were performed as described previously (38). Protein analysis was performed as described previously using an anti-His6 antibody (42).
RESULTS
Yeast cell extract with depleted GAPDH supports symmetrical TBSV RNA synthesis in vitro.
To obtain yeast cell-free extract (CFE) with depleted GAPDH, we used TDH3/THC (TET::TDH3 tdh2Δ) strain, which lacks the TDH2 gene, and the expression of the TDH3 gene is under the control of a doxycycline-titratable promoter in the yeast genome (64). Therefore, the expression of the TDH3 gene can be downregulated by the addition of doxycycline to the yeast growth medium (28). This approach allowed us to culture yeast expressing either a high level of Tdh3p (when yeast was grown without added doxycycline) or a reduced level of Tdh3p (when yeast was grown in the presence of doxycycline), leading to the depletion of cellular GAPDH (Fig. 1B and C) (28).
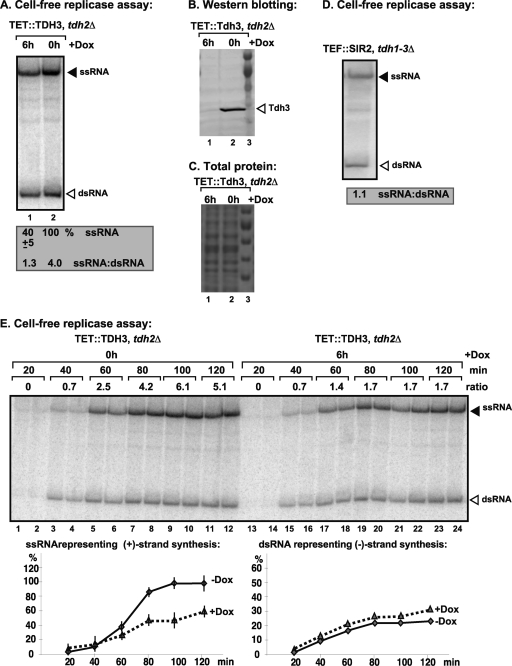
Fig. 1.
The cell-free TBSV replicase assay supports a role for GAPDH in plus-strand synthesis. (A) Purified recombinant p33 and p92pol replication proteins of TBSV in combination with TBSV DI-72 (+)repRNA were added to the whole-cell extract (CFE) prepared from GAPDH-depleted (lane 1, 6 h of treatment with 25 μg of doxycycline/ml to downregulate GAPDH expression) or yeast expressing GAPDH as shown (lane 2). A nondenaturing PAGE analysis of the 32P-labeled repRNA products obtained was performed. The amount of ssRNA and the ratio of ssRNA to dsRNA in the samples are shown. The value represents the percentage of ssRNA and dsRNA compared to the treatment in the absence of doxycycline, which is arbitrarily set at 100% for ssRNA. Note that the dsRNA product represents the annealed (−)RNA and the (+)RNA, while the ssRNA products represents the newly made (+)RNA products. (B) Western blot analysis of the whole-cell extracts for endogenous GAPDH (Tdh3p) based on a specific antibody. (C) A Coomassie-blue stained SDS-PAGE gel shows the comparable levels of total proteins in the whole-cell extracts prepared from the indicated yeast. (D) Nondenaturing PAGE analysis of the 32P-labeled repRNA products obtained in CFE assay as shown in panel A, except using the CFE from yeast expressing E. coli GAPDH (all three yeast TDH genes are deleted, and E. coli GAPDH is expressed). (E) Kinetic analysis of the 32P-labeled repRNA products produced in CFE assay as shown in panel A based on nondenaturing PAGE analysis. The ssRNA/dsRNA ratio in the samples is shown. Each experiment was repeated three times.
To study the role of GAPDH in TBSV replication, we obtained CFE with depleted GAPDH and control CFE with a high level of GAPDH and programmed these CFEs with purified recombinant p33 and p92pol viral replication proteins and DI-72(+) repRNA to induce viral replication (Fig. 1A). The CFE with depleted GAPDH (Fig. 1A, lane 1) supported 40% level of TBSV repRNA replication. Moreover, the ratio of positive- and negative-strand RNAs was close to 1 in CFE with depleted GAPDH compared to four times more positive- than negative-strand RNAs in the control CFE (lane 2) containing a high level of GAPDH (Fig. 1B, lane 2). Similarly, CFE prepared from the SIR2 (tdh1-3Δ) strain lacking any yeast GAPDH, but expressing the E. coli GAPDH, also supported symmetrical TBSV replication (Fig. 1D). Thus, these in vitro experiments established that GAPDH-depleted yeast CFEs support reduced TBSV replication and that the viral RNA synthesis is close to symmetrical, producing only slightly more positive strands than negative strands. The ratio of (+)RNA to (−)RNA was lower in the control CFE than was previously observed with CFEs from the wild-type (wt) BY4741 strain (20:1 ratio) (46); this was likely due to a different level of GAPDH expression in the TDH3/THC (TET::TDH3 tdh2Δ) strain than in the wt strain carrying both TDH2 and TDH3 genes under regulation by their native promoters.
To analyze the kinetics of RNA synthesis in CFEs, we performed time course experiments, as shown in Fig. 1E. The negative-strand RNA synthesis mostly occurred during the first 60 min in both types of CFE, producing only a slightly more negative strand in CFE with depleted GAPDH than in the control CFE containing GAPDH (Fig. 1E). In contrast, the majority of positive-strand RNA synthesis took place between the 40- and 80-min time points, leading to an ∼2-fold-higher amount of (+)RNA in the control CFE than in the CFE with depleted GAPDH (Fig. 1E). Thus, the kinetics data confirm that GAPDH mostly affects positive-strand RNA synthesis.
Purified GAPDH stimulates positive-strand RNA synthesis in the yeast cell extract.
To obtain evidence on the direct role of GAPDH in TBSV replication, we added purified recombinant GAPDH to CFEs, which were depleted of GAPDH. Interestingly, the recombinant GAPDH stimulated positive-strand RNA synthesis by ∼3-fold and 5-fold depending on the type of CFE used (Fig. 2B, lane 4, and Fig. 2C, lane 3). On the other hand, the (−)RNA synthesis was only slightly affected. Therefore, the recombinant GAPDH directly affected the positive-strand/negative-strand ratio during TBSV replication in CFE assays.
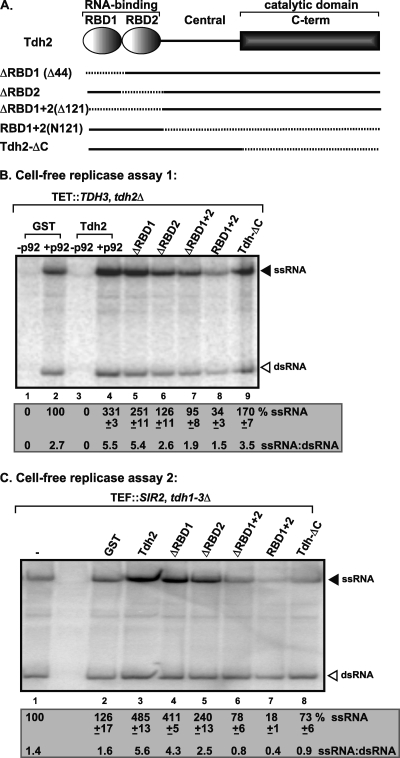
Fig. 2.
Effect of GAPDH mutants on TBSV repRNA replication based on CFE assay. (A) Schematic representation of GAPDH (Tdh2p) mutants expressed as GST fusion proteins in E. coli and affinity purified. Deleted regions are indicated by dotted lines. (B and C) Nondenaturing PAGE analysis of the 32P-labeled repRNA products obtained using GAPDH-depleted CFE (B) or CFE containing E. coli GAPDH (C) (all three yeast TDH genes are deleted, and E. coli GAPDH is expressed). Equal quantities of purified recombinant GAPDH mutants were added to each assay. Each experiment was repeated three times. See further details in Fig. 1A.
To test which domains of GAPDH are important for the stimulatory effect on positive-strand synthesis, we expressed and purified various truncated versions of the yeast GAPDH (Tdh2p, Fig. 2A) and applied an equal quantity of proteins to CFE with depleted GAPDH (Fig. 2B and C). The in vitro data obtained revealed that deletion of the RNA-binding domains (RBDs) in GAPDH (ΔRBD2, Fig. 2B, lane 6, and Fig. 2C, lane 5; ΔRBD1+2, Fig. 2B, lane 7, and Fig. 2C, lane 6) led to a reduced stimulatory effect on TBSV (+)RNA synthesis. Notably, the second RBD demonstrated a greater effect on TBSV repRNA replication than the first RBD. Overall, both RBDs in GAPDH seem to play a role in TBSV replication.
Purified dominant negative GAPDH mutant inhibits positive-strand RNA synthesis in the yeast cell extract.
Additional analyses of GAPDH deletion mutants revealed that the N-terminal RNA-binding domains (RBD1+2, Fig. 2A) in the absence of the catalytic domain acted as a dominant negative mutant, resulting in 3- to 5-fold lower level of TBSV replication in the CFE-based assay (Fig. 2B, lane 8, and Fig. 2C, lane 7) than those containing the purified recombinant GST controls (Fig. 2B, lane 2, and Fig. 2C, lanes 1 and 2). The dominant negative RBD1+2 mutant strongly inhibited positive-strand synthesis, as well as slightly inhibited negative-strand synthesis, altogether changing the ssRNA/dsRNA ratio (i.e., a positive-strand/negative-strand ratio to close to or below 1 (Fig. 2B, lane 8, and Fig. 2C, lane 7).
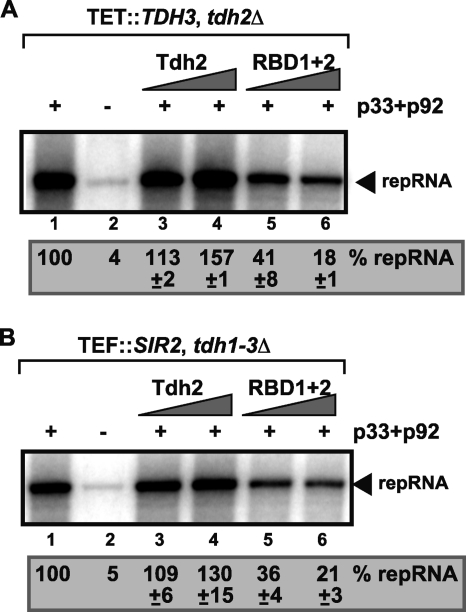
Since RBD1+2 mutant affected (−)RNA synthesis, which also depends on previous steps of replication, such as RNA recruitment and replicase assembly; therefore, we performed an RNA recruitment assay based on CFE (24). The viral RNA recruitment to the cellular membrane required p33 and p92pol replication proteins (Fig. 3, lanes 1 and 2), and in the presence of the full-length recombinant GAPDH (Tdh2p) there was a slight increase in RNA recruitment (up to 57%, lane 4). In contrast, RBD1+2 inhibited RNA recruitment by ∼5-fold (Fig. 3, lane 6) in a CFE assay. Altogether, these data demonstrate that RBD1+2 inhibits negative-strand synthesis in the CFE assay (Fig. 2), likely due to inhibition of viral RNA recruitment into the membrane fraction of CFE. Nevertheless, the effect of RBD1+2 on negative-strand synthesis is much less than that on positive-strand synthesis.
Fig. 3.
Inhibition of viral (+)RNA recruitment by a dominant negative mutant of GAPDH based on CFE assay. The CFE assay was based on yeast strain TDH3/THC (TET::TDH3 tdh2Δ) or (B) SIR2 (tdh1-3Δ) expressing the E. coli GAPDH. A denaturing PAGE analysis of the 32P-labeled repRNA template bound to the cellular membranes was performed. The full-length repRNA is indicated at by an arrowhead. Each experiment was repeated three times.
GAPDH stimulates positive-strand RNA synthesis by the purified tombusvirus replicase.
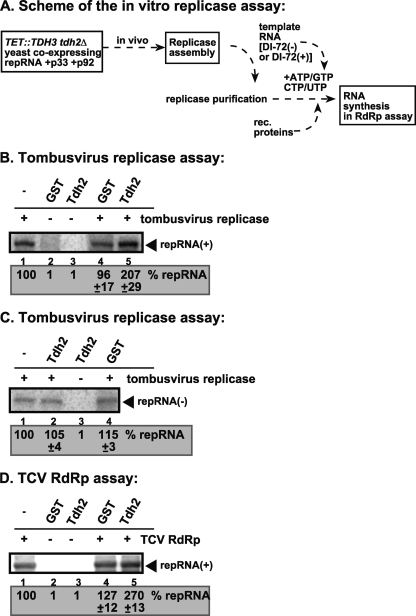
To obtain additional direct evidence that GAPDH stimulates viral positive-strand synthesis, we utilized the affinity-purified tombusvirus replicase, which can utilize TBSV negative-strand RNA as a template in vitro (41, 42). The purified tombusvirus replicase was obtained from GAPDH-depleted yeast, as shown in Fig. 4A. The addition of purified recombinant GAPDH (Tdh2, Fig. 4B, lane 5) increased the RdRp activity of the tombusvirus replicase preparation by ∼2-fold in vitro. Since this preparation can only produce positive-strand RNA products on the (−)RNA template (33, 41, 42), the enhanced level of the viral RNA product in the above assay supports the model that GAPDH is directly involved in positive-strand RNA synthesis.
Fig. 4.
GAPDH directly stimulates positive-strand synthesis by the purified tombusvirus replicase. (A) A scheme for obtaining the purified replicase and the in vitro replicase assay. (B) Denaturing PAGE analysis of the in vitro TBSV replicase assay in the presence of negative-strand DI-72 repRNA. The assay contained the purified template-dependent tombusvirus-replicase obtained from yeast coexpressing p33 and p92pol and the (+)repRNAs (as shown in panel A). The activity of the affinity-purified replicase was tested by using the same amount of RNA template. The replicase product is marked with an arrowhead. Note that the endogenous template was removed during replicase purification (data not shown). (C) An in vitro TBSV replicase assay was performed in the presence of positive-strand DI-72 repRNA. Further details were as described in panel B. (D) GAPDH promotes positive-strand synthesis by the TCV RdRp in vitro. Purified GAPDH was added to the TCV RdRp assay as shown. The template was the negative-strand DI-72 repRNA.
In contrast, the addition of recombinant GAPDH to the purified tombusviral replicase in the presence of positive-stranded RNA template did not increase RdRp activity (Fig. 4C). These data further support previous findings that GAPDH does not significantly stimulate negative-strand synthesis.
The purified tombusvirus replicase contains several host proteins in addition to p33/p92pol viral proteins (54); therefore, GAPDH might stimulate the activity of any one of these components during positive-strand RNA synthesis. To identify which component in the replicase complex is being specifically stimulated by GAPDH, we utilized the purified TCV RdRp (a relative of TBSV) expressed in E. coli (48). This RdRp preparation, which consists only of the recombinant p88 viral protein, is active on added TBSV (−)RNA templates in the absence of the auxiliary viral protein and host proteins present in the tombusvirus replicase (48). Nevertheless, the recombinant GAPDH stimulated the RdRp activity of the TCV RdRp preparation by 2.7-fold in vitro (Fig. 4D, lane 5), suggesting that the activity of the viral RdRp protein is directly stimulated by GAPDH in vitro.
The dominant negative GAPDH mutant inhibits tombusvirus replication in yeast and in plants.
To demonstrate if the above in vitro findings are also relevant in living cells, we investigated whether RBD1+2 dominant negative mutant could also inhibit TBSV replication in yeast. Therefore, we overexpressed RBD1+2 from both low- and high-copy-number plasmids in yeast supporting TBSV repRNA replication, as shown in Fig. 5. Approximately 40 to 85% inhibition of TBSV replication was observed depending on the expression level of RBD1+2 and the growth temperature (Fig. 5B and C). The amounts of p33 and p92 replication proteins were comparable in the yeast transformants described above, suggesting that the primary target of RBD1+2 is likely the RNA recruitment and RNA synthesis steps, as shown by the in vitro data (Fig. 2 and 3).
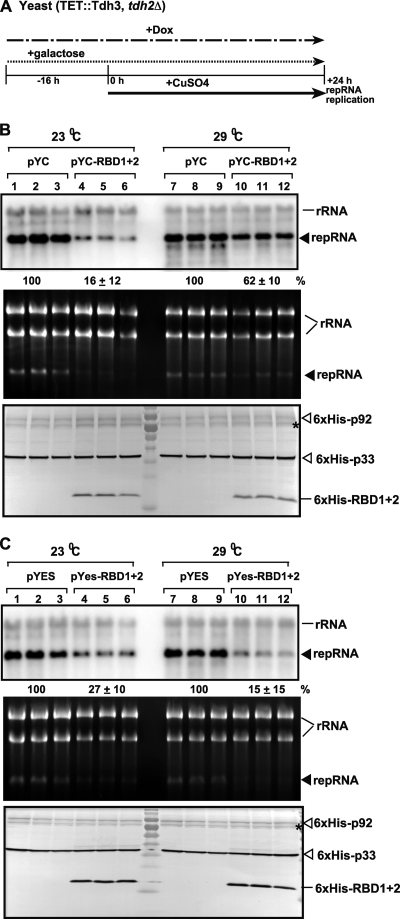
Fig. 5.
Inhibitory effect of GAPDH RBD1+2 dominant negative mutant overexpression on TBSV repRNA accumulation in yeast. (A) For these experiments, TET::TDH3 tdh2Δ yeast was used, which contains a doxycycline regulatable promoter replacing the native TDH3 promoter. Therefore, the addition of doxycycline to the culture media can downregulate GAPDH (Tdh3p) expression at the mRNA level. To launch TBSV repRNA replication, we expressed 6×His-p33 and 6×His-p92 from the copper-inducible CUP1 promoter and DI-72(+) repRNA from the galactose-inducible GAL1 promoter. First, the GAPDH level was reduced by adding doxycycline- and galactose-containing media to induce the expression of RBD1+2 at 16 h prior to expressing the viral components. Then, the yeast cells were cultured for 24 h at either 23 or 29°C on 2% galactose SC minimal medium containing doxycycline plus 50 μM CuSO4. TBSV repRNA accumulation was tested at the two different temperatures since 23°C is the most favorable for TBSV repRNA replication, while the expression of GAPDH is highest at 29°C. (B) For the top panel, Northern blot analysis was used to detect DI-72(+) repRNA accumulation in a yeast strain overexpressing RBD1+2 dominant negative mutant from the low-copy-number pYC plasmid as shown. For the middle panel, an ethidium-bromide stained gel of total RNA extracts of the samples was used for the Northern blotting described above. For the bottom panel, a Western blot analysis of the accumulation level of 6×His-tagged p33, 6×His-tagged p92, and 6×His-tagged RBD1+2 using anti-6×His antibody was performed. The asterisk (*) indicates an SDS-resistant p33 dimer. (C) The same experiments as in panel B, except that RBD1+2 was expressed from the high-copy-number pYES plasmid, as shown.
Constitutive expression of RBD1+2 in N. benthamiana also resulted in significant inhibition in the accumulation of CNV, a close relative of TBSV (Fig. 6A and B). Surprisingly, expression of the yeast RBD1+2 showed more inhibitory effect than that of the RBD1+2 from the GAPDH gene of N. benthamiana (Fig. 6A). The inhibition of tombusvirus accumulation was even detectable in systematically infected younger leaves (Fig. 6B), suggesting the spread of CNV was delayed from the inoculated leaf expressing RBD1+2. Accordingly, the symptoms caused by CNV infection were greatly attenuated in plants expressing RBD1+2 (Fig. 6C). Altogether, the yeast and the plant data of RBD1+2 further support a significant role of GAPDH in tombusvirus replication.
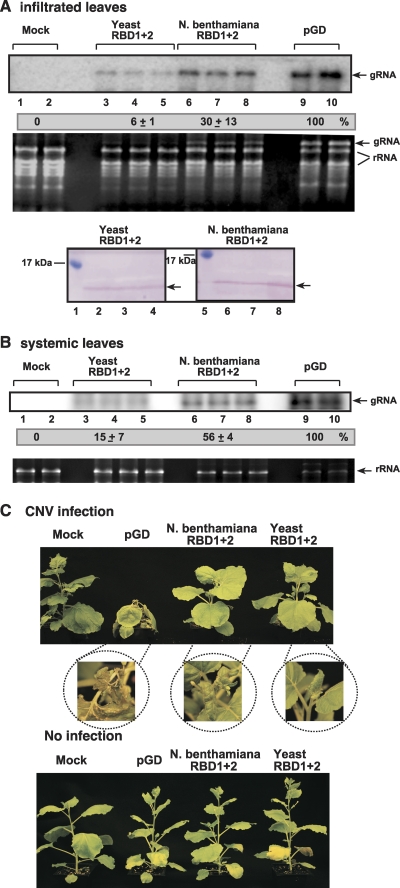
Fig. 6.
Inhibition of tombusvirus RNA accumulation in plants by overexpression of RBD1+2 dominant negative GAPDH mutant. (A) Expression of yeast or N. benthamiana RBD1+2 was evaluated in N. benthamiana leaves, which were coinfiltrated with Agrobacterium carrying a plasmid to launch CNV replication from the 35S promoter. The control samples were obtained from leaves expressing no proteins (pGD, lanes 9 to 10). Total RNA was extracted from leaves 4 days after agroinfiltration. The accumulation of CNV gRNA in the infiltrated N. benthamiana leaves was measured by Northern blotting (top panel). The rRNA was used as a loading control and is shown in an agarose gel stained with ethidium bromide (second panel). The bottom panel shows the accumulation of the yeast or N. benthamiana RBD1+2 in N. benthamiana leaves. (B) Northern blot analysis of the accumulation of CNV gRNA in the systemically infected N. benthamiana leaves, measured as shown in panel A. (C) Symptoms of CNV-infected or mock-infected N. benthamiana plants expressing different RBD1+2 proteins are shown 11 days after infection. Note that the yellow color of the older leaves are due to the damaging effect of agroinfiltration.
GAPDH is recruited by the p92pol replication protein to the site of replication.
Previous work has shown that a large portion of the cytosolic GAPDH is recruited into tombusvirus replicase complexes located on the host peroxisomal membrane surface (64). Moreover, the recruitment of GAPDH was most likely independent from that of p33 replication protein (64). To determine whether p92pol might be involved in the recruitment of GAPDH, we coexpressed Tdh2-YFP with CFP-p92 in yeast cells. Confocal laser microscopy revealed that Tdh2-YFP colocalized with CFP-p92 forming the characteristic punctate structures (Fig. 7A). Similar punctate structures with colocalized Tdh2-YFP and CFP-p92 were also visible in yeast cells supporting TBSV replication (Fig. 7B). In contrast, coexpression of Tdh2-YFP and CFP-p33 did not result in colocalization of the two proteins to the punctate structures formed by CFP-p33 (Fig. 7C). These data suggest that p33 is unlikely to be responsible for the robust recruitment of GAPDH, while p92pol is the more likely candidate that performs most of GAPDH recruitment.
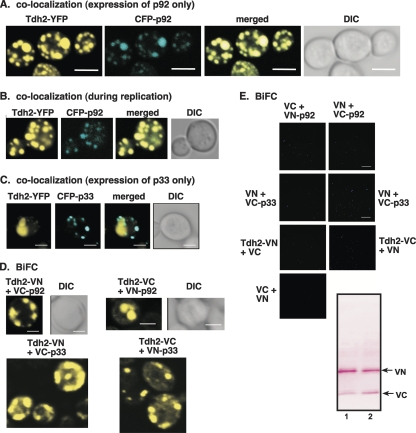
Fig. 7.
Redistribution of cytoplasmic GAPDH to the yeast peroxisomal membranes is driven by p92. (A) Confocal laser microscopy images show the colocalization of Tdh2p-YFP with CFP-p92 expressed from the ADH1 promoter in the BY4741 yeast strain. The merged images show the colocalization of Tdh2p-YFP with CFP-p92. Differential interference contrast (DIC) images are shown on the right. (B) Colocalization of Tdh2p-YFP with CFP-p92 is also observed in yeast supporting TBSV repRNA replication. (C) Absence of colocalization of Tdh2p-YFP with CFP-p33. (D) BiFC analysis of interaction between Tdh2p and p92. (E) Control BiFC experiments that show no interaction between the tags and p33 and p92 in the absence of Tdh2. Western blotting shows the expression of tags in pESC-VC and pYES-VN. The expression of p33, p92, and Tdh2p derivatives is not shown. Yeast was grown under similar conditions, and images were obtained as described in panel D.
To test whether GAPDH interacts with p92pol, we performed BiFC analysis in yeast cells. These experiments revealed that p92pol and Tdh2p interacted in punctate structures in yeast cells (Fig. 7D). Interestingly, p33 also interacted with Tdh2p (Fig. 7D), suggesting that direct association could occur between these molecules, possibly in the replicase complex following p92pol-driven GAPDH recruitment. The interaction between p33 and GAPDH, however, does not seem to be efficient enough to lead to robust recruitment of GAPDH to the site of replication, as shown above in the colocalization experiment (Fig. 7A and B).
To further test the binding between the replication proteins and GAPDH (Tdh2p), we performed pulldown experiments using affinity-purified recombinant p33 and p92pol, which are immobilized to beads. The results showed that both p33 and p92pol bound to GAPDH expressed in yeast (Fig. 8A and B). When tested in the pulldown assay, the C-terminal portion of p33 (termed p33C) and the C-terminal half of p92pol (termed p92C) also showed interaction with GAPDH (Fig. 8B). The binding between the replication proteins and GAPDH was also confirmed in a membrane-based split-ubiquitin assay (Fig. 8D).
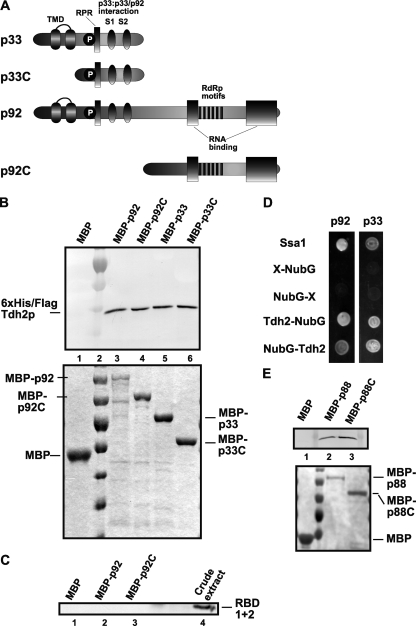
Fig. 8.
Binding of GAPDH (Tdh2p) to TBSV p33 and p92 proteins. (A) Schematic representation of viral proteins and their derivatives used in a binding assay. The various domains include the following: TMD, the transmembrane domain; RPR,the arginine-proline-rich RNA-binding domain; P; phosphorylated serine and threonine; and the S1 and S2 subdomains involved in p33:p33/p92 interaction. The two RNA binding regions in p92 are shown with gray boxes. (B) In the top panel, a Western blot affinity-binding (pull-down) assay was performed to detect interaction between 6×His/Flag-Tdh2p and the MBP-tagged viral proteins. The MBP-tagged viral proteins and MBP produced in E. coli were immobilized on amylose-affinity columns. The yeast lysate containing 6×His-tagged Tdh2p was then passed through amylose-affinity columns with immobilized MBP-tagged proteins. The affinity-bound proteins were specifically eluted with maltose from the columns. The eluted 6×His-tagged Tdh2p was analyzed by Western blotting with anti-6×His antibody. For the bottom panel, the amounts of MBP-tagged proteins bound to the amylose columns were analyzed by Coomassie blue staining of SDS-PAGE gels. (C) Pulldown assay to detect interaction between 6×His-RBD1+2 (the N-terminal RNA-binding domains of Tdh2p) and the MBP-tagged p92 viral protein. The MBP-tagged viral proteins and MBP were immobilized on amylose affinity columns as described for panel B. The yeast lysate with 6×His-tagged RBD1+2 was then passed through the amylose affinity columns with immobilized MBP-tagged proteins. The eluted 6×His-tagged RBD1+2 was analyzed by Western blotting with anti-6×His antibody. Total sample represents the crude yeast cell lysate. (D) A split-ubiquitin assay was used to test binding between p33/p92 and Tdh2p in yeast. The bait p33 or p92 was coexpressed with the indicated prey proteins. Ssa1p (HSP70 chaperone) and the empty prey vector (NubG) were used as positive and negative controls, respectively. (E) Pulldown assay to detect interaction between 6×His-Tdh2p and the TCV MBP-p88 and MBP-p88C proteins. See further details in panel B.
To test the interaction between the dominant negative RBD1+2 mutant carrying the RNA-binding domains and TBSV p92 replication protein, we also performed pulldown experiments (Fig. 8C) wherein Tdh2p was used as a positive control (data not shown). The lack of interaction between RBD1+2 and p92 or p92C suggests that RBD1+2 likely manifests its dominant negative function via binding to the viral RNA.
Furthermore, we also used pulldown experiments with affinity-purified recombinant TCV p88 and the N-terminally truncated p88C to show interaction between these RdRp proteins and Tdh2p (GAPDH) (Fig. 8E).
DISCUSSION
The host metabolic enzyme, GAPDH, is a permanent resident in the tombusvirus replicase complex (54, 64). In the present study, we used in vitro approaches in combination with yeast and plant experiments to dissect GAPDH function in TBSV replication. The data obtained provide several lines of evidence that the co-opted GAPDH plays a direct role in TBSV RNA replication. First, the GAPDH-depleted CFE supported TBSV replication resulting in a positive-strand/negative-strand ratio of ∼1:1 (Fig. 1), similar to the previously observed phenomenon in yeast with suppressed expression of GAPDH (64). Second, the addition of recombinant GAPDH to the GAPDH-depleted CFE increased TBSV positive-strand synthesis. Third, the RdRp activity of the purified tombusvirus replicase from GAPDH-depleted yeast on viral negative-strand template was stimulated by the recombinant GAPDH. Finally, the recombinant GAPDH also increased the RdRp activity of the recombinant TCV RdRp protein, demonstrating that GAPDH can directly stimulate RdRp activity in the absence of the viral auxiliary replication protein (TCV p28, which is similar to the TBSV p33 protein) or additional subverted host factors. More importantly, GAPDH selectively enhanced positive-strand synthesis, confirming its direct role in the regulation of asymmetric viral RNA replication (64).
The results obtained with the RBD1+2 dominant negative mutant also support the role of GAPDH during positive-strand synthesis. The inhibitory effect of RBD1+2 on in vitro RNA recruitment and negative-strand synthesis could be due to less-selective binding of RBD1+2 to the (+)repRNA or p92pol compared to the full-length GAPDH (Tdh2p). Altogether, the strong inhibitory effect of RBD1+2 on tombusvirus replication in yeast and plants supports the idea that GAPDH is a major functional component of the tombusvirus replicase. A similar conclusion can also be drawn from the reduced level of tombusvirus replication in yeast expressing low levels of GAPDH and GAPDH-knockdown plants (64).
Furthermore, the robust recruitment of GAPDH to the site of replication, which is the peroxisomal membrane or ER under some conditions (18, 26, 37, 43), is likely performed by the p92pol replication protein (Fig. 7). Accordingly, we show direct binding between p92pol and GAPDH based on BiFC experiments in live-yeast, split-ubiquitin, and in vitro assays. Based on our data, the efficient interaction between p92pol and GAPDH seems to be sufficient to recruit GAPDH in the absence of p33 replication protein and the viral RNA. However, since p92pol also binds to eEF1A, which interacts with GAPDH (14) and Pex19p peroxisomal shuttle protein (43), it is possible that a multiprotein complex might have assembled to facilitate host factor recruitment to the site of replication. The viral (+)RNA and p33 replication protein are also likely present in the complex described above, since purified GAPDH slightly increased the recruitment of repRNA to the membrane in vitro (Fig. 3). The detailed mechanism of the recruitment of GAPDH or the proposed riboprotein complex remains to be dissected.
The emerging picture based on our data is that the co-opted GAPDH plays several roles in TBSV replication. Previous work demonstrated that GAPDH affects the ability of the viral replicase to retain the RNA template (64). GAPDH was proposed to play a role in the selective retention of negative strands in the replicase complex due to its specific binding to an AU-rich segment in the negative-strand RNA. Here, we discovered an additional function of GAPDH based on in vitro experiments using GAPDH-depleted CFE, which showed that GAPDH directly stimulates viral (+)RNA synthesis. GAPDH might function in (+)RNA synthesis by binding to the viral p92pol RdRp protein and facilitating the recruitment of p92pol to the negative-strand RNA intermediate, as shown in Fig. 9. In addition, GAPDH likely stimulates the polymerase activity of the p92pol RdRp protein by facilitating the correct positioning of p92pol over the replication promoter and enhancer sequences at the 3′ end of the negative-strand RNA template (Fig. 9). These proposed functions of GAPDH are similar to a transcription factor that recruits polymerase II to DNA prior to transcription. Indeed, GAPDH was reported to activate transcription as part of the OCA-S transcription coactivator complex (8, 71).
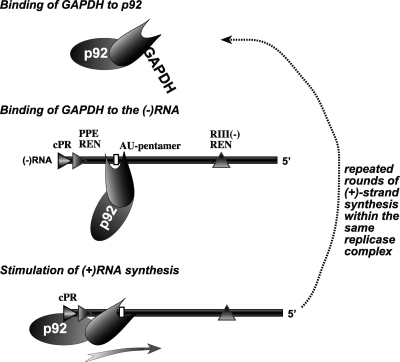
Fig. 9.
Model for the role GAPDH in stimulation of tombusvirus positive-strand synthesis. GAPDH likely binds to p92 and recruit p92 to the (−)RNA intermediate within the viral replicase complex through binding to an AU-pentamer sequence as shown. Note that the reverse order of binding GAPDH to the (−)RNA first and then to p92 is also equally possible. This is followed by the GAPDH-mediated proper positioning of p92 over the negative-strand template, thus facilitating positive-strand synthesis. The GAPDH-driven binding/positioning of p92 onto the (−)RNA template could occur many times, leading to the production of abundant positive-strand progeny and resulting in asymmetric RNA synthesis.
Similar to its role in TBSV replication, GAPDH likely plays important roles during replication of other (+)RNA viruses. Indeed, GAPDH is known to affect the replication of HAV, HCV, Japanese encephalitis virus, transmissible gastroenteritis coronavirus, hepatitis delta virus RNA, and human parainfluenza virus type 3 (9, 10, 13, 25, 44, 50, 53, 58, 64, 68). Based on these observations, we suggest that GAPDH deserves further studies to unravel its functions during (+)RNA virus replication.
ACKNOWLEDGMENTS
We thank Daniel Barajas, Jing-Yi Lin, Zhenghe Li, and Judit Pogany for valuable comments. We appreciate the yeast strains provided by Hans Lehrach (Max Planck Institute, Germany). We thank Markus Ralser (MPI, Germany) for suggestions on using a yeast strain expressing the E. coli GAPDH.
This study was supported by the National Science Foundation (IOB-0517218), NIH-NIAID (5R21AI072170-02), and the Kentucky Tobacco Research and Development Center (P.D.N.).
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Ahlquist P., Noueiry A. O., Lee W. M., Kushner D. B., Dye B. T. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barajas D., Jiang Y., Nagy P. D. 2009. A unique role for the host ESCRT proteins in replication of tomato bushy stunt virus. PLoS Pathog. 5:e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barajas D., Li Z., Nagy P. D. 2009. The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J. Virol. 83:11751–11764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barajas D., Nagy P. D. 2010. Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein. Virology 397:358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonafe N., et al. 2005. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) mRNA in human ovarian cancer cells: possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 65:3762–3771 [DOI] [PubMed] [Google Scholar]
- 6. Brunner J. E., et al. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 79:3254–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng C. P., Jaag H. M., Jonczyk M., Serviene E., Nagy P. D. 2007. Expression of the Arabidopsis Xrn4p 5′-3′ exoribonuclease facilitates degradation of tombusvirus RNA and promotes rapid emergence of viral variants in plants. Virology 368:238–248 [DOI] [PubMed] [Google Scholar]
- 8. Dai R. P., et al. 2008. Histone 2B (H2B) expression is confined to a proper NAD+/NADH redox status. J. Biol. Chem. 283:26894–26901 [DOI] [PubMed] [Google Scholar]
- 9. De B. P., Gupta S., Zhao H., Drazba J. A., Banerjee A. K. 1996. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and LA protein with cis-acting RNAs of human parainfluenza virus type 3. J. Biol. Chem. 271:24728–24735 [DOI] [PubMed] [Google Scholar]
- 10. Dollenmaier G., Weitz M. 2003. Interaction of glyceraldehyde-3-phosphate dehydrogenase with secondary and tertiary RNA structural elements of the hepatitis A virus 3′ translated and non-translated regions. J. Gen. Virol. 84:403–414 [DOI] [PubMed] [Google Scholar]
- 11. Duclos-Vallee J. C., Capel F., Mabit H., Petit M. A. 1998. Phosphorylation of the hepatitis B virus core protein by glyceraldehyde-3-phosphate dehydrogenase protein kinase activity. J. Gen. Virol. 79(Pt. 7):1665–1670 [DOI] [PubMed] [Google Scholar]
- 12. Emara M. M., Liu H., Davis W. G., Brinton M. A. 2008. Mutation of mapped TIA-1/TIAR binding sites in the 3′ terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J. Virol. 82:10657–10670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galan C., et al. 2009. Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology 391:304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gavin A. C., et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631–636 [DOI] [PubMed] [Google Scholar]
- 15. Herold J., Andino R. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jaag H. M., Stork J., Nagy P. D. 2007. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology 368:388–404 [DOI] [PubMed] [Google Scholar]
- 17. Jiang Y., Serviene E., Gal J., Panavas T., Nagy P. D. 2006. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J. Virol. 80:7394–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jonczyk M., Pathak K. B., Sharma M., Nagy P. D. 2007. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362:320–330 [DOI] [PubMed] [Google Scholar]
- 19. Laliberte J. F., Sanfacon H. 2010. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48:69–91 [DOI] [PubMed] [Google Scholar]
- 20. Li W., et al. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989–12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Z., Barajas D., Panavas T., Herbst D. A., Nagy P. D. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 82:6911–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z., Nagy P. D. 2011. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 8:305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Z., et al. 2009. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication cofactor. Virology 385:245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z., et al. 2010. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 6:e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin S. S., Chang S. C., Wang Y. H., Sun C. Y., Chang M. F. 2000. Specific interaction between the hepatitis delta virus RNA and glyceraldehyde 3-phosphate dehydrogenase: an enhancement on ribozyme catalysis. Virology 271:46–57 [DOI] [PubMed] [Google Scholar]
- 26. McCartney A. W., Greenwood J. S., Fabian M. R., White K. A., Mullen R. T. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mendu V., Chiu M., Barajas D., Li Z., Nagy P. D. 2010. Cpr1 cyclophilin and Ess1 parvulin prolyl isomerases interact with the tombusvirus replication protein and inhibit viral replication in yeast model host. Virology 406:342–351 [DOI] [PubMed] [Google Scholar]
- 28. Mnaimneh S., et al. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118:31–44 [DOI] [PubMed] [Google Scholar]
- 29. Moradpour D., Penin F., Rice C. M. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 30. Nagy P. D. 2008. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 46:217–242 [DOI] [PubMed] [Google Scholar]
- 31. Nagy P. D., Carpenter C. D., Simon A. E. 1997. A novel 3′-end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. U. S. A. 94:1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagy P. D., Pogany J. 2010. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against tomato bushy stunt virus. Adv. Virus Res. 76:123–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nagy P. D., Pogany J. 2000. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology 276:279–288 [DOI] [PubMed] [Google Scholar]
- 34. Nagy P. D., Pogany J. 2006. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344:211–220 [DOI] [PubMed] [Google Scholar]
- 35. Nagy P. D., Wang R. Y., Pogany J., Hafren A., Makinen K. 2011. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411:374–382 [DOI] [PubMed] [Google Scholar]
- 36. Noueiry A. O., Ahlquist P. 2003. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41:77–98 [DOI] [PubMed] [Google Scholar]
- 37. Panavas T., Hawkins C. M., Panaviene Z., Nagy P. D. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 338:81–95 [DOI] [PubMed] [Google Scholar]
- 38. Panavas T., Nagy P. D. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314:315–325 [DOI] [PubMed] [Google Scholar]
- 39. Panavas T., Serviene E., Brasher J., Nagy P. D. 2005. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 102:7326–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Panavas T., Serviene E., Pogany J., Nagy P. D. 2008. Genome-wide screens for identification of host factors in viral replication. Methods Mol. Biol. 451:615–624 [DOI] [PubMed] [Google Scholar]
- 41. Panaviene Z., Panavas T., Nagy P. D. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 79:10608–10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Panaviene Z., Panavas T., Serva S., Nagy P. D. 2004. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 78:8254–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pathak K. B., Sasvari Z., Nagy P. D. 2008. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 379:294–305 [DOI] [PubMed] [Google Scholar]
- 44. Petrik J., Parker H., Alexander G. J. 1999. Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly(U) tract of the 3′ non-coding region of hepatitis C virus genomic RNA. J. Gen. Virol. 80(Pt. 12):3109–3113 [DOI] [PubMed] [Google Scholar]
- 45. Pogany J., Nagy P. D. 2008. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J. Virol. 82:5967–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pogany J., Stork J., Li Z., Nagy P. D. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc. Natl. Acad. Sci. U. S. A. 105:19956–19961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rajendran K. S., Nagy P. D. 2003. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 77:9244–9258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rajendran K. S., Pogany J., Nagy P. D. 2002. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76:1707–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ralser M., Zeidler U., Lehrach H. 2009. Interfering with glycolysis causes Sir2-dependent hyper-recombination of Saccharomyces cerevisiae plasmids. PLoS One 4:e5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Randall G., et al. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roehl H., Semler B. 1995. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J. Virol. 69:2954–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sasvari Z., Nagy P. D. 2010. Making of viral replication organelles by remodeling interior membranes. Viruses (Basel) 2:2436–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schultz D. E., Hardin C. C., Lemon S. M. 1996. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′-nontranslated RNA of hepatitis A virus. J. Biol. Chem. 271:14134–14142 [DOI] [PubMed] [Google Scholar]
- 54. Serva S., Nagy P. D. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 80:2162–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Serviene E., Jiang Y., Cheng C. P., Baker J., Nagy P. D. 2006. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 80:1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Serviene E., et al. 2005. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc. Natl. Acad. Sci. U. S. A. 102:10545–10550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi S. T., Lai M. M. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 287:95–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sikora D., Greco-Stewart V. S., Miron P., Pelchat M. 2009. The hepatitis delta virus RNA genome interacts with eEF1A1, p54(nrb), hnRNP-L, GAPDH, and ASF/SF2. Virology 390:71–78 [DOI] [PubMed] [Google Scholar]
- 59. Sirover M. A. 1999. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432:159–184 [DOI] [PubMed] [Google Scholar]
- 60. Sirover M. A. 2005. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J. Cell Biochem. 95:45–52 [DOI] [PubMed] [Google Scholar]
- 61. Sparkes I. A., Runions J., Kearns A., Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1:2019–2025 [DOI] [PubMed] [Google Scholar]
- 62. Tristan C., Shahani N., Sedlak T. W., Sawa A. 2011. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal 23:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Walter B. L., Parsley T. B., Ehrenfeld E., Semler B. L. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang R. Y., Nagy P. D. 2008. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3:178–187 [DOI] [PubMed] [Google Scholar]
- 65. Wang R. Y., Stork J., Nagy P. D. 2009. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 83:3276–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang R. Y., Stork J., Pogany J., Nagy P. D. 2009. A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology 394:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. White K. A., Nagy P. D. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acids Res. Mol. Biol. 78:187–226 [DOI] [PubMed] [Google Scholar]
- 68. Yang S. H., Liu M. L., Tien C. F., Chou S. J., Chang R. Y. 2009. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) interaction with 3′ ends of Japanese encephalitis virus RNA and colocalization with the viral NS5 protein. J. Biomed. Sci. 16:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yi M., Schultz D. E., Lemon S. M. 2000. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J. Virol. 74:6459–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zang W. Q., Fieno A. M., Grant R. A., Yen T. S. 1998. Identification of glyceraldehyde-3-phosphate dehydrogenase as a cellular protein that binds to the hepatitis B virus posttranscriptional regulatory element. Virology 248:46–52 [DOI] [PubMed] [Google Scholar]
- 71. Zheng L., Roeder R. G., Luo Y. 2003. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114:255–266 [DOI] [PubMed] [Google Scholar]