Abstract
Biliary atresia (BA) is a devastating disease of childhood for which increasing evidence supports a viral component in pathogenesis. The murine model of BA is induced by perinatal infection with rhesus rotavirus (RRV) but not with other strains of rotavirus, such as TUCH. To determine which RRV gene segment(s) is responsible for pathogenesis, we used the RRV and TUCH strains to generate a complete set of single-gene reassortants. Eleven single-gene “loss-of-function” reassortants in which a TUCH gene replaced its RRV equivalent and 11 single-gene “gain-of-function” reassortants in which an RRV gene replaced its TUCH equivalent were generated. Newborn BALB/c mice were inoculated with the reassortants and were monitored for biliary obstruction and mortality. In vitro, the ability to bind to and replicate within cholangiocytes was analyzed. Infection of mice with the “loss-of-function” reassortant RT(VP4), where gene 4 from TUCH was placed on an RRV background, eliminated the ability of RRV to cause murine BA. In a reciprocal fashion, the “gain-of-function” reassortant TR(VP4) resulted in murine BA with 88% mortality. Compared with those for RRV, RT(VP4) binding and titers in cholangiocytes were significantly attenuated, while TR(VP4) binding and titers were significantly increased over those for TUCH. Reassortants RT(VP3) and TR(VP3) induced an intermediate phenotype. RRV gene segment 4 plays a significant role in governing tropism for the cholangiocyte and the ability to induce murine BA. Gene segment 3 did not affect RRV infectivity in vitro but altered its in vivo effect.
INTRODUCTION
Biliary atresia (BA) is a progressive inflammatory cholangiopathy of infancy leading to obstruction of the biliary tract. Despite current therapy, BA results in cirrhosis and end-stage liver disease. Among children, it is the most common indication for liver transplantation (23, 35). Although the etiology of BA is unclear, evidence from both human and murine studies supports the hypothesis that biliary atresia is induced by a perinatal viral infection that triggers a host inflammatory immune response (25, 35). The evidence includes patient-based investigations in which reovirus, cytomegalovirus, human papillomavirus, Epstein-Barr virus, and rotavirus (7, 9, 25, 31–34, 36, 39) were found in the livers of infants diagnosed with biliary atresia, as well as murine studies in which newborn BALB/c pups infected with rhesus rotavirus (RRV) developed inflammatory cholangiopathy and bile duct obstruction in a manner paralleling the disease process that occurs in infants (33). In this invaluable mouse model, the initiating event is RRV infection of the biliary epithelial cell (cholangiocyte) (1).
The molecular basis of RRV tropism for cells of hepatobiliary origin has not been defined. Previously, we showed that the tropism for the biliary epithelial cell is strain specific. Among five strains studied, only RRV and SA11-FM (a simian/bovine reassortant) were found to directly infect cholangiocytes and induce extrahepatic bile duct inflammation and obstruction (1). Interestingly, SA11-SM, the parent strain of SA11-FM, could be found in hepatobiliary tissue but did not cause direct cholangiocyte injury. In vitro, only RRV and SA11-FM could infect cholangiocytes, mirroring the in vivo findings. Since that study, we tested many other strains of rotavirus and identified another simian strain, TUCH, that could be found in liver extracts of inoculated mice but did not cause murine BA and could not infect cholangiocytes in vitro.
Rotaviruses are members of the family Reoviridae. A rotaviral particle consists of 3 concentric protein layers surrounding a genome of 11 double-stranded RNA (dsRNA) segments encoding 6 structural viral proteins (VP1 to VP4, VP6, and VP7) and 6 nonstructural proteins (NSP1 to NSP6). Reverse genetics is difficult to perform in rotavirus; however, rotavirus can undergo genetic reassortment after mixed infection in vivo or in cell culture (11, 18, 19, 28, 30, 37, 38). When a host cell is coinfected with two strains of rotavirus, progeny viruses, termed reassortants, that contain different combinations of the parental genes are generated. Single-gene reassortants (i.e., reassortants in which all genes but one are derived from one parent) potentially allow the determination of the function of that gene. Previous studies utilizing reassortants have reported that gene segments encoding VP3, VP4, VP7, NSP1, NSP2, and NSP4 are associated with virulence (2–4, 15, 26, 27) and that NSP3 is a determinant of the extraintestinal spread of RRV (26).
The goal of this study was to determine the specific RRV gene(s) that governs the induction of murine BA. We utilized the property of reassortment to systematically generate a complete set of 22 single-gene reassortants derived from the parental strains RRV and TUCH: 11 loss-of-function “knockout” reassortants that contain 10 genes derived from RRV and 1 gene replaced by its TUCH equivalent, and 11 reciprocal gain-of-function “knock-in” reassortants that contain 1 gene derived from RRV and 10 genes derived from TUCH. Administration of these reassortants to neonatal mice and characterization of their disease phenotypes indicated that gene segment 4, encoding rotavirus protein VP4, was a primary determinant for biliary injury. In vitro, gene segment 4 governed RRV attachment to and infection of the cholangiocyte. In vivo, gene segment 3 was found to affect the ability of RRV to cause BA. The mechanism by which these genes and their translated proteins determine RRV tropism for bile duct epithelial cells requires further study.
MATERIALS AND METHODS
Cells, viruses, and animals.
MA104 cells (BioWhittaker, Walkersville, MD) were grown in Dulbecco's modified Eagle's medium (DMEM) (Cellgro) supplemented with 10% fetal bovine serum (FBS) (Gibco/BRL, Gaithersburg, Md.), 0.01% penicillin-streptomycin (Gibco/BRL), 0.01% l-glutamine (Gibco/BRL), and 0.005% amphotericin B (Cellgro). The mouse cholangiocyte cell line (mCL) generously provided by the laboratory of James Boyer (Yale Liver Care Center, Hartford, CT) was cultured as described previously (16). We used two rotavirus strains: (i) RRV, a simian strain of genotype G3P[3] (kindly provided by H. Greenberg, Stanford University, Palo Alto, CA), and (ii) TUCH (named after the locations where the strain was isolated: Tulane National Primate Research Center and Cincinnati Children's Hospital), a simian strain of genotype G3P[24] (and subgroup 1 [24]).
Breeding pairs of BALB/c mice (Harlan Labs, Indianapolis, IN) were kept in microisolator cages in a virus-free environment with free access to sterilized chow and water. The mice were bred, and pups in litters of >4 pups were used.
Generation, purification and analysis of reassortants.
Reassortants were generated by coinfection of MA104 cells, the standard kidney epithelial cell line used to maintain rotavirus strains in cell culture. MA104 cells were seeded in polystyrene tubes in 2 ml DMEM plus 10% FBS. Monolayers were coinfected with RRV and TUCH at varying multiplicities of infection (MOI). After 1 h of absorption, serum-free DMEM plus 4 μg/ml trypsin (1:250) (Invitrogen, Carlsbad, CA) was added. At 24 and 48 h, the cultures were evaluated for cytopathic effect (CPE), and they were frozen when CPE was complete. Several single-gene reassortants were derived by backcrossing double- or multigene-segment reassortants with parental or other multigene reassortants.
Reassortants were plaque purified in confluent cell culture plates containing MA104 cells in DMEM plus 10% FBS. The cells were infected with 0.2 ml of serially diluted virus supernatants for 1 h, overlaid with 5 ml of medium containing 0.2% agarose (Lonza, Rockland, ME), and incubated for 3 to 4 days at 37°C. Plaques were picked and amplified in MA104 cells. Viral RNA was extracted, and the dsRNA segments were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver staining as described previously (14). The migration patterns of the reassortants' dsRNA genes were compared with those for the parental strains, and gene assignments were made based on how RRV gene segments migrated. Progeny with single-gene substitutions were identified and reamplified. The parental origins of the single-gene reassortants were reconfirmed by gel electrophoresis and sequencing after amplification.
Sequencing of the reassortant genes using RT-PCR.
To verify the findings based on gel electrophoresis, all 11 genes found in each of the 22 single-gene reassortants were amplified and underwent partial sequence analysis. Genomic dsRNA was isolated from infected-cell lysates as described above. The RNA was used as a template for the preparation of viral cDNAs using a one-step reverse transcription-PCR (RT-PCR) kit (Invitrogen) and appropriate segment-specific primers (listed in file S1 in the supplemental material). PCRs were performed in 50-μl reaction mixtures containing 10 mM Tris buffer (pH 8.3), 3 mM MgSO4, a 0.2 mM concentration of each deoxynucleoside triphosphate (dNTP), a 0.25 μM concentration of each primer, and 2 U of Platinum Taq polymerase (Invitrogen). PCR conditions were 35 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min 30 s, using a PTC-200 thermocycler (MJ Research). PCR products were confirmed by separation on 1.5% agarose gels stained with ethidium bromide. DNA sequencing of RT-PCR products was performed according to the dideoxynucleotide chain termination method, using an ABI Prism BigDye Terminator cycle-sequencing ready reaction kit and an ABI Prism 3730 XL DNA analyzer (Applied Biosystems) according to the manufacturer's instructions (for sequence data, see file S1 in the supplemental material).
Viral inoculation of newborn mice for phenotypic characterization.
Newborn pups were injected intraperitoneally (i.p.) with single-gene reassortants at a dose of 1.5 × 106 focus-forming units (FFU) per mouse within 24 h of birth. Saline-injected pups served as controls. Clinical signs of hepatobiliary injury (i.e., jaundice in non-fur-covered skin, acholic stools, and bilirubinuria) and survival rates were recorded. The presence of bilirubin in the urine was detected quantitatively using commercially available urine dipsticks (Bayer Co., Elkhart, IN). A subset of injected mice was sacrificed 7 days postinjection, and the liver and extrahepatic biliary tract were harvested, preserved in formalin, and analyzed histologically as described previously (1). For another subset of mice, the extrahepatic biliary tract was weighed (wet weight) and homogenized in Earle's balanced salt solution (EBSS). Tissue samples were analyzed for the presence of infectious rotavirus by a focus-forming assay (FFA), and quantities of infectious virus were reported as focus-forming units (FFU) per milliliter per milligram (wet weight) of tissue as described previously (1).
Measurement of viral binding using an attachment assay.
The ability of the virus to attach to mCL cells was assessed by binding assays as described previously (16, 17). In brief, cultured cells were grown to confluence in 24-well plates. Attachment assays were performed in triplicate. At the time of assay, the cells, medium, and inoculating virus were cooled to 4°C. Cells were inoculated with varying amounts of virus and were incubated for 1 h at 4°C. The inoculum was removed, and the cells were washed twice to remove any unbound virus. The wash fluids and the residual inoculum were combined to account for all unbound viruses. The cells underwent 2 freeze-thaw cycles, and any virus found within the final cell fraction reflected bound virus. The amounts of bound and unbound virus were determined by FFA analysis. The amount of bound virus was expressed as a percentage of the total amount of virus used to inoculate the cells.
In vitro assay for infectious virus in cholangiocytes.
For the assay for infectious virus, mCL cells were seeded in 24-well plates and were grown to confluence. Wells were washed with EBSS and were inoculated with reassortants at an MOI of 1 at 37°C for 1 h. The cultures were washed and were incubated with serum-free DMEM plus 4 μg trypsin/ml at 37°C for 48 h. Cultures were monitored for the development of CPE, and the viral yield was assessed by an FFA using MA104 cells as described below.
Focus-forming assay.
The focus-forming assay was performed by seeding 96-well plates with MA104 cells, which were grown for 4 days. Once confluent, the cells were exposed to serially diluted virus samples for 1.5 h. The cells were washed with DMEM and were incubated at 37°C for 14 to 16 h with DMEM containing 4 μg of trypsin/ml. The medium was aspirated, and the cells were fixed with cold 80% acetone for 15 min at 20°C. Following a wash with phosphate-buffered saline (PBS), guinea pig anti-rotavirus immunoglobulin G (IgG) (1:1,000) was added as the primary antibody and was incubated for 30 min. Wells were washed with PBS, and fluorescein isothiocyanate (FITC)-tagged goat anti-guinea pig IgG (1:500) was added as the secondary antibody and was incubated for 30 min at 37°C. Wells were washed twice and were allowed to dry completely. Plates were scored using a UV microscope (10× objective), and quantities of infectious virus were reported as focus-forming units per milligram (wet weight) of tissue.
Statistical analysis.
Our assessment of the development of symptoms and of mortality rates following rotavirus inoculation were based on experimental groups of at least 12 pups each. The findings are presented as percentages of pups expressing at least two symptoms and percentages of survival. These noncontinuous variables were analyzed using an arcsine square root transformation to make comparisons between control and treatment groups. A multiple-testing adjustment was made by calculating the Bonferroni adjusted P values due to the comparison of the control to the 11 treatment groups (10). Each subset utilized for the FFAs consisted of at least 5 pups. The results of these continuous variables were expressed as arithmetic means ± standard errors (SE) and were analyzed by analysis of variance (ANOVA) with post hoc testing where appropriate. A P value of <0.05 was considered significant.
RESULTS
Perinatal infection with TUCH does not cause BA in the murine model.
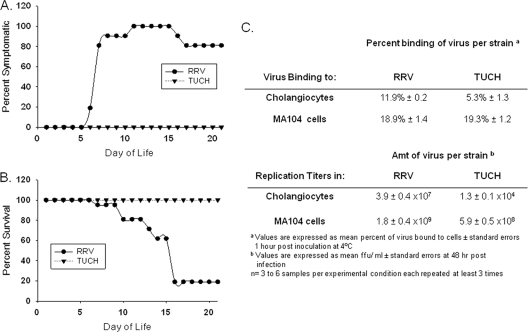
In contrast to RRV, which induces BA in the murine model, i.p. inoculation of newborn BALB/c pups with the TUCH virus caused no signs of hepatobiliary injury, even though both RRV and TUCH were detected in liver and common bile duct extracts within 2 days of inoculation (Table 1). None of the pups displayed jaundice during the 21 days after inoculation, and the mortality rate was 0% (Fig. 1A and B). In vitro, TUCH had poor binding capacity and virtually no ability to replicate within cholangiocytes, correlating with its in vivo effect. In contrast, TUCH bound to, and replicated in, MA104 cells similarly to RRV (Fig. 1C). These qualities made TUCH an ideal virus for the generation of single-gene reassortants when used for coinfection with RRV.
Table 1.
Titers of live virus of each strain in organ extracts following inoculationa
| Organ and day postinjection | Arithmetic mean titer ± SEb |
|
|---|---|---|
| RRV | TUCH | |
| Intestines | ||
| 2 | 28.5 ± 7.7* | 1.7 ± 0.9 |
| 5 | 409.6 ± 44.9* | 33.5 ± 24.5 |
| 7 | 1,199.7 ± 298.1* | —c |
| 10 | 25.5 ± 16.0* | — |
| 14 | — | — |
| Liver | ||
| 2 | 47.7 ± 28.9 | 15.6 ± 5.1 |
| 5 | 26,369.6 ± 12,112.0* | 37.8 ± 19.0 |
| 7 | 24,566.6 ± 13,445.0* | 512.1 ± 418.6 |
| 10 | 33.4 ± 24.0 | 23.1 ± 13.5 |
| 14 | — | — |
| Bile duct | ||
| 2 | 750.6 ± 395.2* | 94.4 ± 31.9 |
| 5 | 97,943.8 ± 36,176.8 | 26,622.9 ± 11,252.7 |
| 7 | 234,949.2 ± 76,252.7* | 29,011.6 ± 6,053.2 |
| 10 | 4,890.8 ± 3,515.5* | — |
| 14 | — | — |
Groups of ≥10 pups per time point were injected within 24 h of birth with 1.5 × 106 FFU of rotavirus. Organs were harvested on the day indicated and were homogenized, and the amount of virus was determined by a focus-forming assay in MA104 cells.
Expressed as focus-forming units per milliliter per milligram of tissue harvested from mice. Asterisks indicate significant differences (P ≤ 0.05) between RRV and TUCH.
—, value below the limit of detection.
Fig. 1.
Effects of RRV and TUCH in vivo in the murine model of BA and in vitro in MA104 and mCL cells. (A and B) Pups were infected within the first 24 h of birth with RRV or TUCH at 1.5 × 106 FFU/pup. RRV injection leads to symptoms of cholestasis in mice between 6 and 15 days (A) and to decreased survival (B). The onset of cholestasis was very mild, and survival was 100%, in mice infected with TUCH. The total numbers of animals at the time of injection were 20 for RRV and 16 for TUCH. During the course of the study, all mice injected with TUCH showed mild symptoms, remained healthy, and survived. Eighteen of 20 RRV-injected mice had died by day 16. (C) RRV bound to cholangiocytes and replicated within them at levels almost 100-fold higher than those of TUCH, but the two strains bound to, and replicated in, MA104 cells almost similarly.
Generation of viral reassortants.
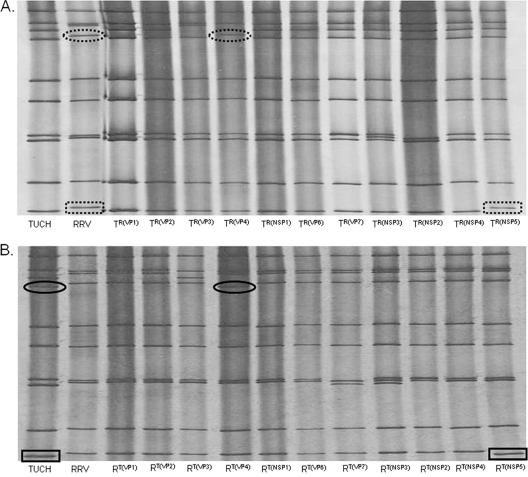
To generate single-gene reassortants, MA104 cells were coinfected with the parental strains RRV and TUCH at varying MOI. The resulting progeny underwent plaque purification followed by polyacrylamide gel electrophoresis and sequencing to determine the parental origin of the genetic content. Reassortants were classified according to the sources of their individual genes (Fig. 2). A total of 1,226 plaques were picked. Seven hundred three plaques were generated by coinfections with different MOI combinations of RRV and TUCH. Two hundred twenty-five of these 703 plaques contained copies of both parental genes, indicating that the plaques were not pure; these plaques were not used further. Of the 478 pure plaques, 337 clones had a parental phenotype and 141 were reassortants. Among the reassortants, 71 were on a RRV background and 70 were on a TUCH background. The background was determined by which parent contributed 6 or more genes to the reassortant strain. A total of 59 plaques contained single-segment reassortants, 39 were double, 18 were triple, and 25 had 4 or 5 gene segment substitutions. Because we could not isolate all single-gene reassortants resulting from coinfection with the parental strains, we backcrossed first-generation double- or multiple-gene segment reassortants with a parental strain. Under these conditions, 523 plaques were picked. From these backcrosses, six single-gene reassortants were generated: TR(VP2), TR(VP3), TR(VP4), TR(NSP2), TR(NSP5), and RT(VP6).
Fig. 2.
Gel electrophoresis of dsRNA from the reassortants. dsRNAs extracted from the 22 single-gene reassortants were separated by SDS-PAGE and were silver stained to reveal the migration rates of the gene segments. The dashed ovals and rectangles mark RRV gene shifts on the TUCH background. The solid ovals and rectangles mark TUCH gene shifts on the RRV background.
Initially, reassortants were identified by gel electrophoresis. In addition, upon identification of the complete set of 22 gene reassortants, all 11 genes within each of the 22 reassortants underwent partial sequence analysis using RT-PCR. The sequence data (see file S1 in the supplemental material) verified the single-gene change within the background of 10 genes derived from the parent strain RRV or TUCH and validated the findings of gel electrophoresis.
The 22 single-gene reassortants generated were labeled according to the following nomenclature: RT(xxxx) (reassortant containing 10 genes derived from RRV with the xxxx gene segment derived from TUCH) or TR(xxxx) (the reciprocal reassortant containing 10 genes derived from TUCH with the xxxx gene derived from RRV). For example, the single-gene reassortant in which the TUCH VP1 gene was placed on an RRV background is designated RT(VP1). The reciprocal reassortant in which the RRV VP1 was placed on the TUCH background is designated TR(VP1).
Gene segment 4 governs the RRV-induced murine model of BA.
Groups of newborn pups were inoculated with the 22 single-gene reassortants. The abilities of the reassortants to cause clinical manifestations of biliary obstruction were determined, as were mortality rates (Table 2). Dramatic changes were found with reassortants RT(VP4) and TR(VP4). The reassortant RT(VP4) (containing gene segment 4 from TUCH placed on an RRV background) elicited no manifestation of biliary injury, and 100% of injected pups survived, completely eliminating the RRV parental effect (Table 2). In contrast, reassortant TR(VP4) (containing gene segment 4 from RRV on a TUCH background) induced signs of biliary obstruction in 100% of pups and produced a mortality rate of 88.2%, reversing the TUCH parental effect (Table 2). An intermediate phenotype was seen in RT(VP3)-infected mice. Both the proportion of mice with symptoms and the mortality rate were significantly lower than those of RRV-injected mice (P < 0.05 [Table 2]). Among mice infected with TR(VP3), significantly more pups developed signs of obstructive jaundice than those infected with TUCH, but the mortality rate remained low.
Table 2.
Signs of biliary obstruction, mortality rates, and replication-competent virus levels in bile ducts of pups after infection with reassortantsa
| Panel and reassortant | % of mice with symptomsb | Mortality rate (%)c | Amt of infectious virus in bile ducts (104 FFU/ml/mg of tissue)d |
|---|---|---|---|
| Panel A | |||
| RRV | 100.0 | 80.0 | 29.5 ± 4.5 |
| RT(VP1) | 100.0 | 100.0 | 9.0 ± 2.1* |
| RT(VP2) | 100.0 | 85.7 | 8.4 ± 3.3* |
| RT(VP3) | 58.4* | 20.8* | 4.5 ± 1.0* |
| RT(VP4) | 0* | 0* | 1.1 ± 0.1* |
| RT(NSP1) | 85.0 | 85.0 | 9.4 ± 3.2* |
| RT(VP6) | 95.0 | 55.0 | 6.3 ± 0.9* |
| RT(VP7) | 91.7 | 87.5 | 7.7 ± 1.9* |
| RT(NSP3) | 89.5 | 89.5 | 18.7 ± 4.7 |
| RT(NSP2) | 76.9 | 53.9 | 19.9 ± 6.4 |
| RT(NSP4) | 95.2 | 81.0 | 8.5 ± 1.3* |
| RT(NSP5) | 95.5 | 59.1 | 5.8 ± 1.4* |
| Panel B | |||
| TUCH | 0 | 0 | 2.9 ± 0.6 |
| TR(VP1) | 0 | 0 | 0.5 ± 0.0* |
| TR(VP2) | 0 | 0 | 0.7 ± 0.2* |
| TR(VP3) | 88.9** | 5.6 | 12.5 ± 4.4** |
| TR(VP4) | 100.0** | 88.2** | 25.9 ± 4.0** |
| TR(NSP1) | 27.8 | 0 | 4.7 ± 0.6 |
| TR(VP6) | 68.8** | 0 | 10.4 ± 2.7** |
| TR(VP7) | 0 | 0 | 0.2 ± 0.0* |
| TR(NSP3) | 84.2** | 5.3 | 8.7 ± 2.7 |
| TR(NSP2) | 43.5** | 0 | 2.2 ± 0.6 |
| TR(NSP4) | 43.8** | 0 | 7.6 ± 0.6 |
| TR(NSP5) | 52.6** | 0 | 5.5 ± 1.4 |
Groups of 15 to 20 pups per strain were injected within 24 h of birth with 1.5 × 106 FFU of rotavirus and were monitored for symptoms of BA for 21 days. A subset of these mice (6 to 10 pups per strain) had their bile ducts harvested on day 7 of life for determination of viral contents. Single asterisks indicate values significantly lower (P < 0.05) than those for RRV (panel A) or TUCH (panel B). Double asterisks indicate values significantly higher (P < 0.05) than those for RRV (panel A) or TUCH (panel B).
Showing two or more signs of biliary obstruction.
Over 21 days.
Bile ducts were harvested 7 days postinfection, and the amount of virus was determined by a focus-forming assay. Values are means ± standard errors.
Of the remaining nine “knockout” strains, all behaved similarly to RRV, causing biliary obstruction in 76.9 to 100% of mice, with mortality rates ranging from 53.9 to 89.5%. There were only subtle differences in the onset and duration of clinical symptoms (data not shown). Among the remaining nine “gain-of-function” reassortants, TR(NSP1), TR(VP6), TR(NSP2), TR(NSP4), and TR(NSP5) caused some symptoms of hepatobiliary injury, but the rates were significantly lower than that with RRV. All pups injected with TR(NSP1), TR(VP6), TR(NSP2), and TR(NSP5) survived (P < 0.05), while there was a slight increase in mortality among pups infected with TR(NSP3). Reassortants TR(VP1), TR(VP2), and TR(VP7) did not elicit any sign of hepatobiliary injury or cause mortality.
The yield of virus in extrahepatic biliary samples mirrored the development of clinical symptoms of BA. Although “knockouts” of gene segments 1 through 7, 10, and 11 all reduced the yield of virus in the bile ducts from that for RRV (P < 0.05 [Table 2]), the most dramatic reduction was seen with reassortant RT(VP4), with a viral yield 27-fold less than that of RRV ([1.1 ± 0.1] × 104 FFU/ml/mg versus [29.5 ± 4.5] × 104 FFU/ml/mg, respectively; P < 0.05). In contrast, the yield of virus following infection with TR(VP4) was 9 times higher than that of its parent strain TUCH ([25.9 ± 4.0] × 104 FFU/ml/mg versus [2.9 ± 0.6] × 104 FFU/ml/mg, respectively; P < 0.05 [Table 2]). Among the “knock-in” strains that induced signs of obstructive jaundice [TR(VP3), TR(NSP1), TR(VP6), TR(NSP3), TR(NSP2), TR(NSP4), and TR(NSP5)], viral titers were slightly higher than that of the parental strain TUCH, but only the titers after infection with TR(VP6) and TR(VP3) were statistically significant (P < 0.05). Consistent with the observation that mice injected with TR(VP1), TR(VP2), or TR(VP7) showed no signs of disease, viral titers in the bile ducts were low.
Histological assessment of the liver and extrahepatic biliary tract.
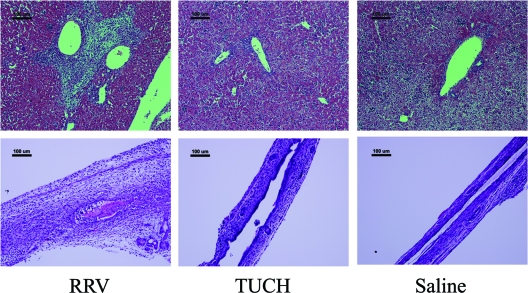
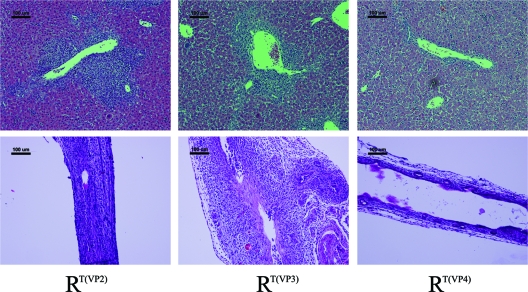
The livers and extrahepatic bile ducts harvested from mice at 7 days after inoculation with reassortants were histologically analyzed, and the findings were compared to those for specimens obtained from mice inoculated with the parental strains (Fig. 3). The histologic appearance of the portal area and the extrahepatic bile duct after the infection of mice with different monoreassortant strains was consistent with the symptoms. The histology of livers from mice injected with the reassortants on an RRV background revealed marked infiltration of inflammatory cells within the area of the portal tract, except for those inoculated with RT(VP4) (Fig. 4). Periductal inflammatory infiltration, epithelial sloughing, stromal proliferation, and lumen obstruction were universally seen in the extrahepatic bile ducts of these groups. In contrast, the histology of the extrahepatic bile ducts and livers of pups injected with RT(VP4) was indistinguishable from that for TUCH- or saline-injected mice (Fig. 3 and 4).
Fig. 3.
Histology of the liver and extrahepatic bile ducts in RRV-, TUCH-, and saline-injected mice. (Top) RRV induced massive inflammation of the liver with remarkable portal expansion at 7 days, containing primarily lymphocytes in the portal area. In contrast, TUCH-injected mice, like saline-injected controls, showed no liver inflammation. There were 15 to 20 mice in each group. Bars, 100 μm. (Bottom) Injection of RRV led to obstruction of the lumen by inflammatory cells and by sloughed epithelial cells by day 7, while TUCH- and saline-injected mice showed normal epithelia and unobstructed lumens at 7 days. Bars, 100 μm. All the slides were stained with hematoxylin and eosin.
Fig. 4.
Histology of the liver and extrahepatic bile ducts in mice injected with selected RRV reassortants. (Top) RT(VP2) and RT(VP3), like the parent strain RRV, induced massive inflammation of the liver with remarkable portal expansion at 7 days, but RT(VP4)-injected mice showed no liver inflammation. There were 15 to 20 mice in each group. Bars, 100 μm. (Bottom) Injection of RT(VP2), like that of the parent strain RRV, led to complete obstruction of the lumen, while RT(VP4)-injected mice showed unobstructed lumens at 7 days. RT(VP3) showed an intermediate phenotype, with only a partially obstructed bile duct. Bars, 100 μm. All the slides were stained with hematoxylin and eosin.
Minimal [TR(VP1), TR(VP2), TR(VP7)] to medium [TR(VP3), TR(NSP1), TR(VP6), TR(NSP3), TR(NSP2), TR(NSP4), and TR(NSP5)] levels of inflammatory infiltration in the portal area of the liver were observed in all the mice infected with TUCH background reassortants, except for those inoculated with TR(VP4), in which portal tract infiltration was rather extensive. The histological changes in the extrahepatic bile ducts of TR(VP4)-injected mice were indistinguishable from those in RRV-injected mice (Fig. 3 and 5). Epithelial edema and submucosal inflammation were commonly seen in the extrahepatic bile ducts of mice infected with other TUCH-based clones. In addition, epithelial cell sloughing was also found in the TR(VP3), TR(NSP3), and TR(NSP5) groups, but intraluminal infiltration and stromal proliferation were rarely seen, and the lumens of bile ducts were patent, as demonstrated by the findings for the TR(VP2) group (Fig. 5).
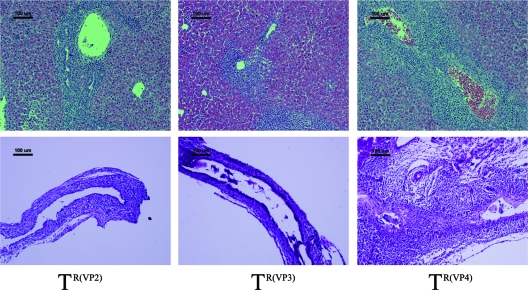
Fig. 5.
Histology of the liver and extrahepatic bile ducts in mice injected with selected TUCH reassortants. (Top) TR(VP2) and TR(VP3) induced very mild inflammation of the liver, while TR(VP4)-injected mice, like RRV-injected mice, showed massive inflammation with remarkable portal expansion at 7 days. There were 15 to 20 mice in each group. Bars, 100 μm. (Bottom) Injection of TR(VP4) led to complete obstruction of the lumen, while TR(VP2)- or TR(VP3)-injected mice showed unobstructed lumens at 7 days. Bars, 100 μm. All the slides were stained with hematoxylin and eosin.
The binding of RRV and TUCH to cholangiocytes was significantly altered by manipulation of gene segment 4.
To establish a mechanistic basis for the in vivo findings, we tested the reassortants in our in vitro model of BA, in which RRV, but not TUCH, binds to, and replicates within, cholangiocytes. We found that gene segment 4 of TUCH, when inserted into an RRV background, reduced viral binding from that of the parental RRV strain (4.0% ± 0.5% versus 11.9% ± 0.1%; P < 0.05 [Table 3]). While the binding ratios of reassortants RTVP1, RT(VP2), RT(VP3), and RT(NSP1) were similar to that of RRV, those of reassortants RT(VP6), RT(VP7), RT(NSP3), RT(NSP2), RT(NSP4), and RT(NSP5) were all slightly higher (Table 3).
Table 3.
In vitro binding and viral yield after infection of cholangiocytes with reassortantsa
| Panel and reassortant | % Bindingb | Amt of infectious virus in immortalized cholangiocytes (104 FFU/ml)c |
|---|---|---|
| Panel A | ||
| RRV | 11.9 ± 0.1 | 3,850.0 ± 449.2 |
| RT(VP1) | 13.6 ± 1.6 | 4,568.7 ± 321.7 |
| RT(VP2) | 11.6 ± 1.4 | 3,882.7 ± 298.7 |
| RT(VP3) | 15.3 ± 3.2 | 3,929.3 ± 291.9 |
| RT(VP4) | 4.0 ± 0.5* | 0.8 ± 0.1* |
| RT(NSP1) | 15.3 ± 2.4 | 5,012.0 ± 42.0 |
| RT(VP6) | 15.2 ± 0.3** | 2,884.0 ± 163.4 |
| RT(VP7) | 16.2 ± 0.2** | 4,652.7 ± 426.7 |
| RT(NSP3) | 17.6 ± 0.8** | 3,388.0 ± 809.6 |
| RT(NSP2) | 16.4 ± 0.6** | 3,168.7 ± 172.9 |
| RT(NSP4) | 17.3 ± 0.6** | 3,313.3 ± 382.0 |
| RT(NSP5) | 15.6 ± 0.7** | 4,265.3 ± 264.4 |
| Panel B | ||
| TUCH | 5.3 ± 0.6 | 1.3 ± 0.1 |
| TR(VP1) | 3.1 ± 0.7 | 0.8 ± 0.0* |
| TR(VP2) | 1.4 ± 0.2* | 0.3 ± 0.0* |
| TR(VP3) | 6.5 ± 0.6 | 65.9 ± 0.8** |
| TR(VP4) | 12.4 ± 0.5** | 226.3 ± 50.4** |
| TR(NSP1) | 6.2 ± 0.6 | 13.3 ± 1.7** |
| TR(VP6) | 5.9 ± 1.4 | 10.5 ± 2.0** |
| TR(VP7) | 7.2 ± 1.5 | 0.2 ± 0.0* |
| TR(NSP3) | 2.9 ± 0.2* | 20.3 ± 6.3** |
| TR(NSP2) | 4.9 ± 0.8 | 4.1 ± 0.9 |
| TR(NSP4) | 7.6 ± 0.7 | 12.5 ± 2.3** |
| TR(NSP5) | 5.3 ± 0.4 | 17.4 ± 0.8** |
Cholangiocytes were inoculated with reassortants in order to determine binding and replication rates. Six samples per strain were used. Single asterisks indicate values significantly lower (P < 0.05) than those for RRV (panel A) or TUCH (panel B). Double asterisks indicate values significantly higher (P < 0.05) than those for RRV (panel A) or TUCH (panel B).
Expressed as mean percentages of virus bound to cholangiocytes ± standard errors.
Values are means ± standard errors.
Placement of RRV gene segment 4 on a TUCH background enhanced viral attachment. Reassortant TR(VP4) exhibited a significantly higher binding ratio than the parental strain TUCH (12.4% ± 0.5% versus 5.3% ± 0.6%; P < 0.01 [Table 3]). None of the other reassortants showed binding abilities different from that of TUCH, except for strains TR(VP2) and TR(NSP3), whose binding ratios (1.37% ± 0.2% and 2.87% ± 0.2%, respectively) were significantly lower (P < 0.05).
Reassortant replication in cholangiocytes in vitro.
Infection of cholangiocytes in vitro with “knockout” reassortants resulted in viral replication rates that mirrored the in vivo findings. RT(VP4) dramatically lowered the yield of virus from that for RRV ([0.8 ± 0.1] × 104 FFU/ml versus [3,850.0 ± 449.2] × 104 FFU/ml, respectively; P < 0.05 [Table 3]). The remaining “knockout” reassortants had viral yields similar to that of the parental strain RRV.
The titers of the “knock-in” clones TR(VP3), TR(VP4), TR(NSP1), TR(VP6), TR(NSP2), TR(NSP4), and TR(NSP5) were significantly higher than that of TUCH (P < 0.05 [Table 3]). Consistent with the in vivo findings, the reassortant TR(VP4) replicated to the highest titer, 174 times greater than that of TUCH ([226.3 ± 50.4] × 104 FFU/ml versus [1.3 ± 0.1] × 104 FFU/ml) and 3 times greater than that of the second highest reassortant, TR(VP3). Reassortants TR(VP1), TR(VP2), and TR(VP7) had significantly reduced titers (P < 0.05 [Table 3]).
DISCUSSION
Previously, we found that among five rotavirus strains studied, RRV and SA11-FM had tropism for the biliary epithelial cell and could induce biliary atresia in the murine model; SA11-SM could be found in the hepatobiliary system but caused hepatitis instead of biliary obstruction; and EDIM and Wa caused no hepatobiliary disease (1). To determine the molecular basis of RRV tropism for the biliary epithelial cell, we utilized the rotavirus property of reassortment to determine which RRV gene(s) governs its ability to induce biliary atresia in the murine model. Initially, we attempted to generate reassortants with the parental strains RRV and EDIM, but because EDIM does not replicate well within MA104 cells, the generation of new reassortants was challenging. To overcome this challenge, we tested other strains of rotavirus in the murine model of BA and identified the simian strain TUCH, which, after injection into newborn BALB/c pups, could be found within hepatobiliary tissue but did not cause BA. Because it replicated well in MA104 cells, we used it for the generation of reassortants. By using RRV and TUCH, we generated a complete set of 11 loss-of-function “knockout” single-gene reassortants and 11 reciprocal gain-of-function “knock-in” single-gene reassortants. The gain-of-function “knock-in” reassortants were important, confirming that the presence of a specific RRV gene was by itself capable of causing biliary atresia in the murine model. These rotavirus monoreassortants permit the identification of the RRV gene(s) that governs the ability to induce BA.
Using these single-gene reassortants, we identified RRV gene segments 3 and 4 as important determinants in the pathogenesis of murine BA. We found that when gene segment 4 of RRV was replaced by the corresponding gene derived from TUCH, the manifestations of biliary obstruction were totally abolished. In a reciprocal fashion, when newborn mice were infected with TR(VP4) (the TUCH background clone with the VP4 gene derived from RRV), BA was induced. Supportive results were recently found using rotavirus strains RRV and UK (8). Experiments performed by Feng et al. demonstrated the requirement of both genes VP4 and NSP1 from RRV for replication in the mouse biliary tract. RRV and TUCH share 91% amino acid homology in NSP1, which could explain why we did not observe a role for NSP1 in our experiments. Our in vitro studies revealed that RRV gene segment 4 governed cholangiocyte binding and infectivity, establishing a mechanistic basis for the in vivo results. The protein product of gene segment 4, VP4, has been found to be a major determinant of rotavirus pathogenicity in several systems. Using heterologous bovine × simian viral reassortants, Offit et al. linked gene segment 4 to tropism for the intestine in mice (27). Two porcine rotavirus variants (4f and 4s) with different VP4 genes showed distinct pathogenicity profiles during serial passage in gnotobiotic piglets. Insertion of the pathogenic parental (4f) gene segment 4 into the nonpathogenic virus (4s) genome by reassortment caused the latter to develop tropism for the intestine and caused diarrhea in piglets (2). VP4 is essential for early virus-cell interactions, because it participates in receptor binding and cell penetration (12, 13).
The early rotavirus-cell interactions constitute a multistep process (21). In the multistep model, the initial contact of a neuraminidase-sensitive virus strain with the cell surface occurs through a sialic acid (SA)-containing cell receptor, using the VP8* subunit of VP4, which is positioned at the surfaces of rotavirus particles. The initial interaction of the virus with SA induces a subtle conformational change in VP4, which allows the virus to interact with a second cell receptor (currently proposed to be the α2β1 integrin) through a DGE-binding motif. After the second interaction, the virus interacts with integrins αXβ2, αVβ3, and hsc70. These interactions increase the permeability of the cell membrane, facilitating the penetration of the virus into the cell. Previously, we found that expression of the α2β1 integrin on the cell surface confers susceptibility to RRV infection on cholangiocytes (16). The different mechanisms by which TUCH VP4 and RRV VP4 interact with the cell surface require further study. The VP4 proteins from these simian rotaviruses share 87% homology, so it is probable that the mechanistic basis for their differing abilities to infect cholangiocytes lies within the nonhomologous regions.
Interestingly, VP7, the other major constituent of the outer protein layer, did not, under these conditions, govern hepatobiliary tropism. As an outer capsid protein, VP7 plays a role in viral entry; however, reassortants RT(VP7) and TR(VP7) behaved similarly to their parents. The function of VP7 is to facilitate cell entry and infection by interacting with αXβ2 and αVβ3 (12) in a postbinding step. Because the VP7 proteins of RRV and TUCH were both G3 types and had 90.5% homology, it is likely that manipulation of this gene segment did not change the protein structure of VP7 and thus did not affect cell entry/replication.
The mechanism by which gene segment 3 and its translated protein VP3 govern the induction of murine BA is unknown. At least one function of VP3 is to act as a guanylyltransferase and methyltransferase, enabling capping of the 5′ end of the mRNA synthesized in viroplasms by the RNA-dependent RNA polymerase, VP1 (5, 20, 29). The cap stabilizes viral mRNA, potentially protecting it from degradation by nucleases (6), and enables its translation by interaction with cellular translation protein complexes. Association of VP3 with three other genes (VP4, VP7, NSP4) in rotavirus virulence and host range restriction and attenuation has been suggested in reassortant studies (27). In these studies, piglets failed to develop diarrhea when challenged with a single-gene reassortant that derived its 3rd gene from an avirulent background, while diarrhea was induced only when virulence-associated genes encoding VP3, VP4, VP7, and NSP4 were present together on an avirulent-rotavirus background. Our results are somewhat different from these observations in that reassortant RT(VP3) did not abolish biliary injury; rather, it elicited an attenuated phenotype compared with that of RRV. Reassortant TR(VP3) caused a significant increase in the proportion of animals with biliary injury symptoms, similar to that with RRV, but mortality was minimal. We speculate that gene segment 3 might affect viral replication rates, thereby decreasing mortality, but this was not observed in infectivity assays. The basis for the intermediate phenotype induced by gene segment 3 is currently unclear, but it might result from the interactions of multiple factors, including the host immune response.
It was noteworthy that TUCH reassortants containing the RRV VP6, NSP2, NSP3, NSP4, or NSP5 gene increased symptoms of hepatobiliary injury without eliciting bile duct obstruction or mortality. The reverse effect was not seen in the reciprocal RRV reassortants containing the same TUCH genes. The TUCH reassortants produced larger amounts of replication-competent virus than did the TUCH parent both in vivo and in vitro. The increase in symptoms encountered following infection with these reassortants could be a consequence of viral loads causing hepatobiliary inflammation rather than obstruction. Further study will be required to determine how these genes contribute to pathogenesis.
The basis for the contributions of these gene segments to pathogenesis may extend beyond the mechanisms discussed. Several recent investigations have established both a cell- and an antibody-mediated immunological role in biliary obstruction. A recent study by Mack and colleagues showed that a component of VP4 results in the generation of an antibody that recognizes α-enolase expressed on cholangiocytes (22). This mimicry may contribute to BA pathogenesis. Interestingly, within the segment of interest identified in that study lie several amino acid differences between RRV and TUCH that may cause conformational changes in the peptide structure which could contribute to antibody recognition. Site-directed mutagenesis will be necessary to confirm these findings.
In summary, a set of 22 single-gene reassortants was generated that allowed the identification of RRV gene segments 3 and 4 as important determinants of murine BA. RRV gene segment 4, through its translated protein VP4, determined RRV tropism for the biliary system. The mechanism of binding to receptors on the surfaces of cholangiocytes remains to be characterized. Replacement of the 3rd gene segment of RRV did not affect viral infectivity in vitro; however, it caused an intermediate phenotype in vivo. Although rotavirus has been refractory to direct genetic manipulation, a reverse-genetics system has recently been developed which permits the rescue of a viral RNA segment produced from cDNA in vitro into replication-competent progeny virus (18, 19). In the future, this approach will be utilized to determine the precise molecular mechanism by which RRV gene segments 3 and 4 contribute to the pathogenesis of BA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Harry Greenberg for critical review of the manuscript, Marepalli Rao for statistical support, and Jelle Matthijnssens for help with primer design.
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants K08-DK-728858, R03-DK-087974 and R01-DK-091566 to G. Tiao.
No conflicts of interest exist.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Allen S. R., et al. 2007. Effect of rotavirus strain on the murine model of biliary atresia. J. Virol. 81:1671–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bridger J. C., Tauscher G. I., Desselberger U. 1998. Viral determinants of rotavirus pathogenicity in pigs: evidence that the fourth gene of a porcine rotavirus confers diarrhea in the homologous host. J. Virol. 72:6929–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Broome R. L., Vo P. T., Ward R. L., Clark H. F., Greenberg H. B. 1993. Murine rotavirus genes encoding outer capsid proteins VP4 and VP7 are not major determinants of host range restriction and virulence. J. Virol. 67:2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burke B., Desselberger U. 1996. Rotavirus pathogenicity. Virology 218:299–305 [DOI] [PubMed] [Google Scholar]
- 5. Chen D., Luongo C. L., Nibert M. L., Patton J. T. 1999. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology 265:120–130 [DOI] [PubMed] [Google Scholar]
- 6. Coutts M., Krowczynska A., Brawerman G. 1993. Protection of mRNA against nucleases in cytoplasmic extracts of mouse sarcoma ascites cells. Biochim. Biophys. Acta 1173:49–56 [DOI] [PubMed] [Google Scholar]
- 7. Domiati-Saad R., et al. 2000. Cytomegalovirus and human herpesvirus 6, but not human papillomavirus, are present in neonatal giant cell hepatitis and extrahepatic biliary atresia. Pediatr. Dev. Pathol. 3:367–373 [DOI] [PubMed] [Google Scholar]
- 8. Feng N., et al. 2011. Roles of VP4 and NSP1 in determining the distinctive replication capacities of simian rotavirus RRV and bovine rotavirus UK in the mouse biliary tract. J. Virol. 85:2686–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fischler B., Ehrnst A., Forsgren M., Orvell C., Nemeth A. 1998. The viral association of neonatal cholestasis in Sweden: a possible link between cytomegalovirus infection and extrahepatic biliary atresia. J. Pediatr. Gastroenterol. Nutr. 27:57–64 [DOI] [PubMed] [Google Scholar]
- 10. Foulkes A. S. 2009. Applied statistical genetics with R: for population-based association studies. Springer Verlag, New York, NY [Google Scholar]
- 11. Gombold J. L., Ramig R. F. 1986. Analysis of reassortment of genome segments in mice mixedly infected with rotaviruses SA11 and RRV. J. Virol. 57:110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graham K. L., et al. 2003. Integrin-using rotaviruses bind α2β1 integrin α2 I domain via VP4 DGE sequence and recognize αXβ2 and αVβ3 by using VP7 during cell entry. J. Virol. 77:9969–9978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graham K. L., Takada Y., Coulson B. S. 2006. Rotavirus spike protein VP5* binds α2β1 integrin on the cell surface and competes with virus for cell binding and infectivity. J. Gen. Virol. 87:1275–1283 [DOI] [PubMed] [Google Scholar]
- 14. Greenberg H. B., Vo P. T., Jones R. 1986. Cultivation and characterization of three strains of murine rotavirus. J. Virol. 57:585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoshino Y., et al. 1995. Identification of group A rotavirus genes associated with virulence of a porcine rotavirus and host range restriction of a human rotavirus in the gnotobiotic piglet model. Virology 209:274–280 [DOI] [PubMed] [Google Scholar]
- 16. Jafri M., et al. 2008. Cholangiocyte expression of α2β1-integrin confers susceptibility to rotavirus-induced experimental biliary atresia. Am. J. Physiol. Gastrointest. Liver Physiol. 295:G16–G26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jafri M., Donnelly B., McNeal M., Ward R., Tiao G. 2007. MAPK signaling contributes to rotaviral-induced cholangiocyte injury and viral replication. Surgery 142:192–201 [DOI] [PubMed] [Google Scholar]
- 18. Komoto S., Kugita M., Sasaki J., Taniguchi K. 2008. Generation of recombinant rotavirus with an antigenic mosaic of cross-reactive neutralization epitopes on VP4. J. Virol. 82:6753–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Komoto S., Taniguchi K. 2006. Establishment of a reverse genetics system for rotavirus. Uirusu 56:183–192 (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 20. Liu M., Mattion N. M., Estes M. K. 1992. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology 188:77–84 [DOI] [PubMed] [Google Scholar]
- 21. Lopez S., Arias C. F. 2004. Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol. 12:271–278 [DOI] [PubMed] [Google Scholar]
- 22. Lu B. R., Brindley S. M., Tucker R. M., Lambert C. L., Mack C. L. 2010. α-Enolase autoantibodies cross-reactive to viral proteins in a mouse model of biliary atresia. Gastroenterology 139:1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mack C. L., Sokol R. J. 2005. Unraveling the pathogenesis and etiology of biliary atresia. Pediatr. Res. 57:87R–94R [DOI] [PubMed] [Google Scholar]
- 24. McNeal M. M., et al. 2005. Development of a rotavirus-shedding model in rhesus macaques, using a homologous wild-type rotavirus of a new P genotype. J. Virol. 79:944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morecki R., Glaser J. H., Cho S., Balistreri W. F., Horwitz M. S. 1982. Biliary atresia and reovirus type 3 infection. N. Engl. J. Med. 307:481–484 [DOI] [PubMed] [Google Scholar]
- 26. Mossel E. C., Ramig R. F. 2002. Rotavirus genome segment 7 (NSP3) is a determinant of extraintestinal spread in the neonatal mouse. J. Virol. 76:6502–6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Offit P. A., Blavat G., Greenberg H. B., Clark H. F. 1986. Molecular basis of rotavirus virulence: role of gene segment 4. J. Virol. 57:46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Offit P. A., Clark H. F., Blavat G., Greenberg H. B. 1986. Reassortant rotaviruses containing structural proteins vp3 and vp7 from different parents induce antibodies protective against each parental serotype. J. Virol. 60:491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pizarro J. L., Sandino A. M., Pizarro J. M., Fernandez J., Spencer E. 1991. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. J. Gen. Virol. 72(Pt 2):325–332 [DOI] [PubMed] [Google Scholar]
- 30. Ramig R. F. 1997. Genetics of the rotaviruses. Annu. Rev. Microbiol. 51:225–255 [DOI] [PubMed] [Google Scholar]
- 31. Rauschenfels S., et al. 2009. Incidence of hepatotropic viruses in biliary atresia. Eur. J. Pediatr. 168:469–476 [DOI] [PubMed] [Google Scholar]
- 32. Riepenhoff-Talty M., et al. 1996. Detection of group C rotavirus in infants with extrahepatic biliary atresia. J. Infect. Dis. 174:8–15 [DOI] [PubMed] [Google Scholar]
- 33. Riepenhoff-Talty M., et al. 1993. Group A rotaviruses produce extrahepatic biliary obstruction in orally inoculated newborn mice. Pediatr. Res. 33:394–399 [DOI] [PubMed] [Google Scholar]
- 34. Rosenthal P. 1995. The association of reovirus 3 and biliary atresia: finally resolved? Am. J. Gastroenterol. 90:1895–1896 [PubMed] [Google Scholar]
- 35. Sokol R. J., Mack C. 2001. Etiopathogenesis of biliary atresia. Semin. Liver Dis. 21:517–524 [DOI] [PubMed] [Google Scholar]
- 36. Szavay P. O., Leonhardt J., Czech-Schmidt G., Petersen C. 2002. The role of reovirus type 3 infection in an established murine model for biliary atresia. Eur. J. Pediatr. Surg. 12:248–250 [DOI] [PubMed] [Google Scholar]
- 37. Trask S. D., Taraporewala Z. F., Boehme K. W., Dermody T. S., Patton J. T. 2010. Dual selection mechanisms drive efficient single-gene reverse genetics for rotavirus. Proc. Natl. Acad. Sci. U. S. A. 107:18652–18657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Troupin C., et al. 2010. Rearranged genomic RNA segments offer a new approach to the reverse genetics of rotaviruses. J. Virol. 84:6711–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tyler K. L., et al. 1998. Detection of reovirus RNA in hepatobiliary tissues from patients with extrahepatic biliary atresia and choledochal cysts. Hepatology 27:1475–1482 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.