Abstract
Disulfide bonds reportedly stabilize the capsids of several viruses, including papillomavirus, polyomavirus, and simian virus 40, and have been detected in herpes simplex virus (HSV) capsids. In this study, we show that in mature HSV-1 virions, capsid proteins VP5, VP23, VP19C, UL17, and UL25 participate in covalent cross-links, and that these are susceptible to dithiothreitol (DTT). In addition, several tegument proteins were found in high-molecular-weight complexes, including VP22, UL36, and UL37. Cross-linked capsid complexes can be detected in virions isolated in the presence and absence of N-ethylmaleimide (NEM), a chemical that reacts irreversibly with free cysteines to block disulfide formation. Intracellular capsids isolated in the absence of NEM contain disulfide cross-linked species; however, intracellular capsids isolated from cells pretreated with NEM did not. Thus, the free cysteines in intracellular capsids appear to be positioned such that disulfide bond formation can occur readily if they are exposed to an oxidizing environment. These results indicate that disulfide cross-links are normally present in extracellular virions but not in intracellular capsids. Interestingly, intracellular capsids isolated in the presence of NEM are unstable; B and C capsids are converted to a novel form that resembles A capsids, indicating that scaffold and DNA are lost. Furthermore, these capsids also have lost pentons and peripentonal triplexes as visualized by cryoelectron microscopy. These data indicate that capsid stability, and especially the retention of pentons, is regulated by the formation of disulfide bonds in the capsid.
INTRODUCTION
Virus capsids have evolved mechanisms to protect viral genomes from the extracellular environment while maintaining the ability to release the viral genome upon entry into a new host cell during the next round of infection. The formation of capsids containing the herpes simplex virus type 1 (HSV-1) double-stranded DNA (dsDNA) genome is a complex process involving a preassembled protein shell (procapsid) containing the viral protease and scaffolding proteins (encoded by UL26 and UL26.5), concatemeric viral DNA, and seven cleavage and packaging proteins (4, 13, 24). The capsid shell is composed primarily of the major capsid protein (VP5), two triplex proteins (VP19C and VP23), and VP26, as well as a dodecameric UL6 portal ring located at a unique vertex through which DNA most likely is packaged and released.
During wild-type (WT) infection, three capsid forms are observed: A capsids, which have participated in an abortive encapsidation process and have lost both scaffold proteins and DNA; B capsids, which contain proteolytically processed forms of the internal scaffold proteins but no DNA; and DNA-containing mature C capsids, which have lost scaffold during the process of taking up DNA (18). DNA-containing C capsids undergo structural alterations which result in the acquisition of increased amounts of a heterodimer made up of UL25 and UL17 (81) believed to stabilize DNA-containing capsids (10, 12, 49, 56, 69, 78, 79, 82). The UL25/UL17 heterodimeric structure initially was termed the C capsid-specific complex (CCSC); however, it now has been observed on reconstructions of A, B, and C capsids and is referred to as the capsid vertex-specific component (CVSC) (81). In addition to stabilizing DNA-containing capsids, a role for UL25 has been recognized during the uncoating of the viral genome (2, 39, 65).
Capsid assembly and genome encapsidation in the herpesviruses are reminiscent of processes found in the double-strand DNA bacteriophages, such as T4, T7, P22, HK97, and λ, which have provided excellent model systems for HSV and the other herpesviruses (13, 76). As with the phage systems, HSV procapsid maturation involves dramatic structural remodeling, resulting in the conversion of a relatively fragile procapsid into a more stable form (65, 76). In the case of herpesviruses, spherical procapsids later are converted to a more angular icosahedron filled with DNA (7, 24, 73). DNA-filled capsids eventually bud from the nucleus and undergo a series of envelopment and deenvelopment steps prior to release into the extracellular environment (reviewed in reference 44). During the next round of infection, capsids are released into the cytoplasm, traffic to the nucleus, and dock at a nuclear pore, at which point they undergo a disassembly process resulting in the release of viral DNA into the nucleus (58, 71). The structural and biochemical changes that contribute to capsid stability and metastability in HSV-1 are poorly understood, and in this paper we explore the role of disulfide bonds in maintaining capsid stability.
It has been recognized for some time that extracellular papilloma- and polyomaviruses contain disulfide bonds that play an important role in the metastability of capsids (22, 25, 85). HSV also has been reported to contain disulfide bonds between capsid proteins. Zweig et al. first reported that the herpes simplex virus type 2 (HSV-2) VP19C, VP5, and scaffold proteins participate in disulfide linkages in the virion (93), and Yang et al. reported that the HSV scaffold protein could participate in both inter- and intramolecular disulfide bonds (92). Furthermore, the in vitro assembly of HSV capsids can be prevented in the presence of dithiothreitol (DTT) (50), and HSV nucleocapsids have been reported to be unstable in the presence of mercaptoethanol (38). In this paper, we confirm and extend these findings by showing that when disulfide bond formation was prevented, B and C capsids were unstable and lost scaffold and DNA, respectively. In addition, capsids that are unable to form disulfide bonds were shown to lack pentons and peripentonal triplexes.
MATERIALS AND METHODS
Cells, viruses, antibodies, and other reagents.
African green monkey kidney fibroblast (Vero) cells were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% fetal bovine serum and 0.1% penicillin-streptomycin. The KOS strain of herpes simplex virus type 1 (HSV-1) was used as the wild-type virus in all experiments. Monoclonal antibodies for VP5 (3E8) (6), UL6 (1C9 and 4G9) (9, 51), VP23 (1D2) (52), and VP19C (4A11) (52) were provided by Jay Brown, University of Virginia Health System. UL17-specific anti-chicken polyclonal antibody (89) was a gift from Joel Baines (Cornell University). An anti-UL25 monoclonal antibody (2D9) (39) was obtained from Fred Homa (University of Pittsburgh School of Medicine). A polyclonal antibody (ID1) against UL25 raised against a glutathione S-transferase-UL25 fusion protein was provided by Nels Pederson (East Carolina University School of Medicine). NEM (N-ethylmaleimide), trypsin, and soybean trypsin inhibitor were purchased from Sigma. NEM was prepared fresh every time as a 1 M stock in ethanol.
Isolation of extracellular virions.
Confluent monolayers of Vero cells were infected with KOS at a multiplicity of infection (MOI) of 3 PFU/cell. When cytopathic effects were observed in most cells, the media containing virions were collected and subjected to centrifugation at 200 × g for 10 min in a Beckman S4750 rotor to remove detached cells. The virion-containing supernatant then was subjected to centrifugation at 1,000 × g for 15 min in a Beckman S4750 rotor to remove cell debris. Virions in the supernatant were incubated with NP-40 at a final concentration of 0.5% for 15 min at room temperature and pelleted through 1.5 ml of a 30% (wt/vol) sucrose cushion for 1.5 h at 71,000 × g in an SW41 rotor. NP-40 removes the envelope and some tegument, resulting in the release of capsids. Released capsids were reconstituted in TNE buffer (20 mM Tris, pH 7.6, 500 mM NaCl, 2 mM EDTA) supplemented with protease inhibitors (Roche Complete EDTA-free protease inhibitor cocktail tablets), briefly sonicated in a cup horn sonicator (two 10-s bursts at 50% power), and stored at −80°C.
Isolation of NEM-treated virions.
Virions were isolated as described above, except that NEM was added to the virion-containing supernatant for 15 min on ice prior to NP-40 treatment (final concentration, 10 mM NEM). NEM also was present during subsequent steps performed as described above.
Intracellular capsid isolation.
Confluent monolayers of Vero cells were infected with KOS at an MOI of 3 PFU/cell. At 18 to 20 h postinfection, the medium was discarded, the monolayers were washed with phosphate-buffered saline (PBS), and the cells were scraped into 20 ml of PBS. Cells were pelleted at 200 × g in a Beckman S4750 rotor for 15 min, and the pellet was resuspended in 5 ml of 20 mM Tris (pH 7.6) buffer followed by the addition of 5 ml of 2× lysis buffer (2% Triton X-100, 20 mM Tris, pH 7.6, 1 M NaCl, 4 mM EDTA). For every 40 to 50 billion cells, 10 ml of final lysis buffer was used. Cell lysates were incubated on ice for 30 min, treated with DNase (0.1 mg/ml DNase and 20 mM MgCl) to reduce viscosity for 15 to 20 min at 37°C, and briefly sonicated in a cup horn sonicator (three 20-s bursts at 50% power). Insoluble material was removed by centrifugation at 10,000 × g for 15 min in a Beckman S4750 rotor. The supernatants containing intracellular capsids were spun through a 1.5-ml cushion of 30% (wt/vol) sucrose in TNE buffer at 71,000 × g for 1 h in an SW41 rotor. Each capsid pellet (crude capsids) was resuspended in 700 μl of TNE, briefly sonicated in a cup horn sonicator to break up clumps, and layered over a continuous gradient of 20 to 50% (wt/vol) sucrose in TNE. Gradients were centrifuged at 71,000 × g for 1 h in SW41 rotor. A, B, and C capsids were visualized by light scattering and removed with a syringe. For capsids isolated in the presence of NEM, cell monolayers were incubated at 37°C in Dulbecco's modified essential medium (DMEM) containing 10 mM NEM for 10 min prior to harvest. NEM at a final concentration of 10 mM also was added to all other buffers used during the process of capsid isolation. Capsid preparations removed from the gradient were diluted three to four times with TNE and centrifuged for 1.5 h at 71,000 × g in an SW41 rotor. Capsid pellets were reconstituted in TNE supplemented with protease inhibitors (except for capsids used for trypsin digestions).
Reducing and nonreducing SDS-PAGE and Western blotting.
Capsids and virions were reconstituted in reducing SDS buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 100 mM DTT, 5% [vol/vol] β-mercaptoethanol, 0.02% [wt/vol] bromophenol blue) or nonreducing SDS buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 20 mM NEM, 0.02% [wt/vol] bromophenol blue). Samples were heated at 95°C for 3 min, briefly sonicated, and resolved on an 8 to 16% Tris-glycine gradient gel. Proteins were electrotransferred to polyvinylidene difluoride (PVDF) membrane at 35 V overnight and blocked with 5% fat free milk, and membranes were incubated with primary antibody specific for various capsid proteins for 1.5 h. After washing with TBST (150 mM NaCl, 20 mM Tris, pH 7.5, and 0.1% Tween), blots were incubated with alkaline phosphatase-conjugated secondary antibodies at 1:5,000 for 1 h, washed again with TBST, and developed using Promega color detection reagents according to the manufacturer's instructions. Primary antibodies were used at the following dilutions: anti-VP5 (3E8), 1:1,000; anti-VP19C (4A11), 1:2,000; anti-VP23 (1D2), 1:10,000; anti-UL6 (4G9 and 1C9), 1:10,000; anti-UL17, 1:50,000; anti-UL25 (ID1), 1:10,000 (see Fig. 1 and Fig. 3); monoclonal anti-UL25, 1:2,000 (see Fig. 2).
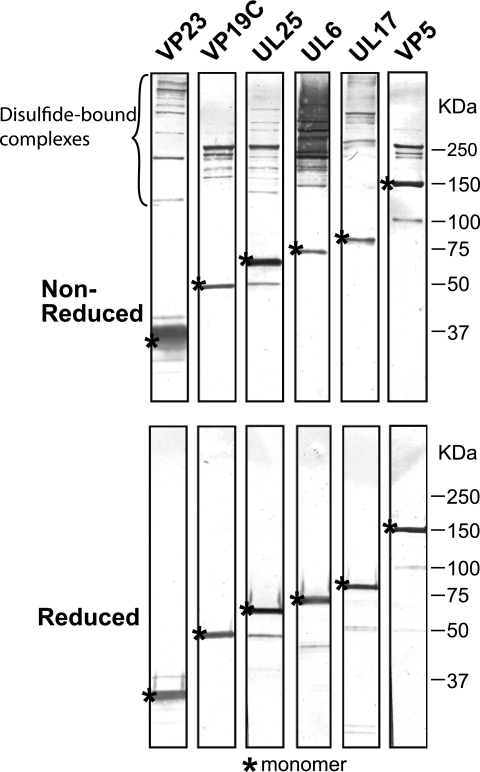
Fig. 1.
Disulfide-bonded complexes of capsid proteins present in mature virions. Virions were isolated as described in Materials and Methods and separated by SDS-PAGE under nonreducing and reducing conditions, followed by immunoblotting. Species migrating slower than the monomer size under nonreducing conditions that are not present in reducing gels are indicative of disulfide-bonded complexes.
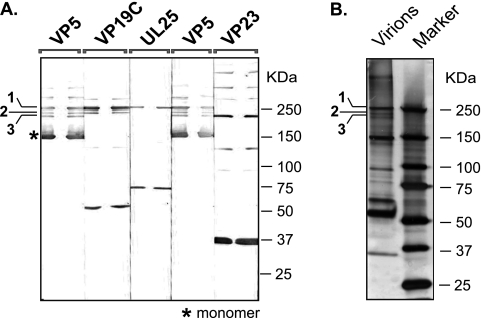
Fig. 3.
Composition of major disulfide-bonded complexes present in virions. Virions isolated as described in Materials and Methods were separated on 8 to 16% gradient SDS-PAGE under nonreducing conditions. (A) After electrotransfer, PVDF membrane was cut into strips in a way that cuts through the sample loading well; the strips were immunoblotted with antibody to various capsid proteins. After alkaline phosphates detection, strips were aligned back together. Bands that resolved at the same position for any protein were considered part of disulfide-bonded complex. (B) Silver-stained strip of SDS-PAGE gel.
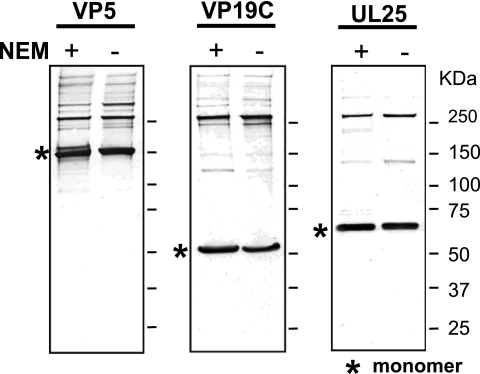
Fig. 2.
NEM does not effect the formation of slower-migrating protein complexes in extracellular virions. Nonreducing SDS-PAGE of extracellular virions isolated in the presence or absence of NEM as described in Materials and Methods. Immunoblotting was performed with various capsid protein antibodies.
Mass spectroscopy.
Extracellular virions were resolved under nonreducing conditions on a 1-mm-thick, 8 to 16% Tris-glycine gradient gel and silver stained as follows. The gel was fixed for 1 h in a 40% ethanol and 10% acetic acid solution and washed twice in 30% ethanol (10 min each wash), followed by one 20-min wash with water. The gel was sensitized in 0.1% sodium thiosulfate for 1 min, briefly rinsed with water, and incubated in cold 0.1% silver nitrate for 20 min at 4°C. Silver nitrate solution was removed; the gel was washed twice in water (1 min each wash) and developed with 3% potassium bicarbonate and 0.05% formaldehyde. Once the protein bands were detected, staining was terminated by replacing the developing solution with 5% acetic acid solution for 10 min. An identical gel was run in parallel and immunoblotted with various antibodies to support the identification of the bands of interest. Three silver-stained bands (as marked in Fig. 2) were excised from the gel and analyzed by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) on a Micromass Q-Tof API mass spectrometer at the Keck Biotechnology Resource Laboratory at Yale University.
Trypsin digestion.
Concentrated KOS capsids isolated in the presence or absence of NEM as described above were divided into five equal fractions. Each fraction was treated with increasing concentrations of trypsin, ranging from 10 to 80 μg/ml or no trypsin, and incubated at 37°C for 30 min. Digestion was terminated by the addition of soybean trypsin inhibitor at a 250 μg/ml final concentration and 1 mM phenylmethylsulfonyl fluoride (PMSF). Treated capsids were separated by SDS-PAGE, electrotransferred to PVDF membrane, and immunoblotted with monoclonal antibody specific for VP5 (3E8).
Cryo-EM and image analysis.
Frozen hydrated samples of purified capsids were prepared for cryoelectron microscopy (cryo-EM) by applying 3.5 μl of sample to Quantifoil 300-mesh R2/1 grids (Quantifoil Micro Tools GmbH, Jena, Germany), blotting, and plunge freezing with an FEI Vitrobot (FEI, Hillsboro, OR) into a 50:50 mixture of liquid ethane-propane (80) cooled in a bath of liquid nitrogen. Frozen grids were transferred onto a Gatan 626 cryoholder (Gatan, Pleasanton, CA) and imaged using low-dose techniques in an FEI T20 FEG microscope operating at 200 kV and equipped with a Gatan Ultrascan 4000 charge-coupled-device (CCD) camera. A magnification setting of 50,000× on the microscope, combined with a postcolumn magnification of 1.4× and a CCD camera pixel size of 15 μm, yielded a nominal sampling rate of 2.14 Å/pixel for the specimen. Images were further binned by a factor of 2 to yield a final pixel size of 4.28 Å. Capsid images were selected from the digital micrographs using x3dpreprocess software (14), and contrast transfer parameters were estimated manually for each micrograph using BSOFT (23). Three-dimensional reconstructions were calculated with the AUTO3DEM suite (91) using the RMC method (90) to generate starting models in each case, and density maps were visualized with the UCSF Chimera software (63). For the control KOS capsid sample, 412 empty capsid images were collected from 57 CCD micrographs, of which 370 capsid images were included in the final density map with a resolution estimated at 30 Å by the Fourier shell correlation method (83) at a cutoff of 0.5. The NEM-treated sample included 769 empty capsid images from 83 CCD micrographs, of which 690 capsid images were included in the final density map with an estimated resolution of 29 Å.
GuHCl and DTT treatment of B capsids.
Concentrated intracellular KOS capsids isolated in the absence of NEM as described above were suspended in TNE at 0.1 mg/ml and treated with 2 M guanidine hydrochloride (GuHCl) or 15 mM DTT alone or in combination. Treatment was carried out for 1 h at 4°C with occasional mixing. Capsids were recovered by centrifugation through 150 μl of a 25% (wt/vol) sucrose cushion at 33,000 rpm in a Beckman TLS55 for 1 h at 4°C. Pellets were reconstituted in TNE and analyzed by a 20 to 50% (wt/vol) sucrose gradient prepared in TNE. Gradients were centrifuged for 45 min at 23,000 rpm in a Beckman SW55Ti rotor at 4°C. Gradients were illuminated from the bottom to visualize capsid bands by light scattering. Gradients of treated or untreated samples were aligned with each other and compared to gradient-resolved A, B, and C capsids and photographed. Final images were aligned using Adobe Photoshop.
RESULTS
HSV virions contain complexes linked by disulfide bonds.
Disulfide-linked protein complexes involving VP19C, VP5, and scaffold proteins have been reported in herpes simplex virions (92, 93). To confirm the existence of disulfide-linked capsid proteins, extracellular virions from wild-type KOS-infected cells were isolated and subjected to SDS-PAGE under reducing and nonreducing conditions. Resolved proteins were transferred to a PVDF membrane, which subsequently was cut into strips. Each strip was immunoblotted with a different capsid protein antibody as shown in Fig. 1. The proteins in the nonreduced samples migrated differently from samples that had been reduced with ß-mercaptoethanol and DTT. For each of the known capsid proteins, VP23, VP19C, UL25, UL6, UL17, and VP5, a monomeric protein band of the expected size was observed; however, in addition, slower-migrating species also were detected in the nonreducing gel. The slower-migrating species disappeared when the samples were run under reducing conditions.
The results shown in Fig. 1 suggest that HSV capsid proteins within extracellular virions contain disulfide bonds; however, since the virions were treated with NP-40, it was possible that disulfide bonds only formed when the envelope was removed with NP-40 and were not actually present in capsids within extracellular virions. To distinguish between disulfides that preexist in virions and those that form only after exposure to oxidizing conditions, N-ethylmaleimide (NEM), a small hydrophobic molecule that efficiently passes through membranes, was added to extracellular virions before (and during) NP-40 treatment. NEM irreversibly modifies cysteine residues that are present in proteins as free thiols; thus, the presence of NEM can prevent de novo disulfide bond formation and allows us to distinguish between disulfides preexisting in extracellular virions and those that form only after envelope removal. Figure 2 shows capsids released from extracellular virions subjected to SDS-PAGE and immunoblotted for VP5, VP19C, and UL25. Bands migrating slower than the monomer were seen both in NEM-treated and untreated virion samples, suggesting that these bands represent cross-linked species preexisting in virions. Despite minor differences in the treated and untreated samples for a particular band, the patterns overall are remarkably similar.
To determine the composition of the major cross-linked bands, virion samples were subjected to SDS-PAGE under nonreducing conditions, transferred to PVDF membranes, and cut into strips such that each lane was bisected vertically. Strips were treated with antibodies to various capsid proteins, and after detection, strips were realigned. Consistently with the data shown in Fig. 1 and 2, slow-migrating species of VP5, VP19C, UL25, and VP23 were detected (Fig. 3A). Although slight differences in the number of slow-migrating bands were apparent between Fig. 1, 2, and 3, we repeatedly observed species that correspond to the bands labeled 1, 2, and 3 in Fig. 3A. Bands 1, 2, and 3 also were the major cross-linked species observed on silver-stained gels (Fig. 3B).
By aligning strips, we were able to make an initial determination of the composition of bands 1, 2, and 3. Band 1 reacted with antibodies to VP5, VP19C, and UL25 (Fig. 3A). Band 2 also reacted with VP5 and VP19C (Fig. 3A). In addition, UL25 was detected in band 2 in similar experiments that were developed with a UL25 polyclonal antibody rather than the monoclonal antibody used in this experiment (Fig. 1 and data not shown). Band 3 reacted strongly with VP5 and VP23. VP19C also was detected in band 3 after longer exposures (Fig. 1 and data not shown). To confirm the identity of bands 1, 2, and 3, a similar SDS-PAGE gel run under nonreducing conditions was silver stained, and bands equivalent to bands 1, 2, and 3 were cut out and subjected to mass spectrometry (MS). Table 1 confirmed that band 1 contained peptides that correspond to VP5, UL25, and VP19C, as expected from the immunoblot analysis. In addition, peptides corresponding to the tegument protein UL37 were identified. The MS analysis also confirmed that band 2 contained VP5, VP19C, and UL25. In addition, this analysis indicates that band 2 contained peptides corresponding to tegument proteins VP22, UL36, and UL37. However, the numbers of peptides from UL36 and UL37 in band 2 were low (1.3 and 1.2%, respectively), and as the large size of UL36 may cause it to migrate independently in that region of the gel, the assignment of UL36 and perhaps UL37 as part of the cross-linked complex is tentative and will require verification. Peptides corresponding to VP5, VP23, VP19C, UL25, and the tegument protein VP22 were identified in band 3. Thus, it appears that capsid proteins as well as VP22 are cross-linked in mature virions.
Table 1.
Mass spectrometry analysis of high-molecular-mass disulfide-bonded complexes
| Protein name | Mass (kDa) | % Coverage | Identified by Western blotting |
|---|---|---|---|
| Band 1 | |||
| Major capsid protein VP5 | 149 | 59.3 | Yes |
| DNA packaging tegument protein UL25 | 63 | 40.5 | Yes |
| Capsid triplex protein VP19C | 50 | 37.6 | Yes |
| Tegument protein UL37 | 120 | 2.0 | |
| Band 2 | |||
| Major capsid protein VP5 | 149 | 43.4 | Yes |
| Capsid triplex protein VP19C | 50 | 34.4 | Yes |
| Tegument protein VP22 | 32 | 16.9 | |
| DNA packaging tegument protein UL25 | 63 | 8.1 | Yes |
| Large tegument protein UL36 | 336 | 1.3 | |
| Tegument protein UL37 | 120 | 1.2 | |
| Band 3 | |||
| Major capsid protein VP5 | 149 | 28.7 | Yes |
| Tegument protein VP22 | 32 | 21.9 | |
| Capsid triplex protein VP23 | 34 | 20.4 | Yes |
| Capsid triplex protein VP19C | 50 | 14.6 | Yes |
| DNA packaging tegument protein UL25 | 63 | 6.4 | Yes |
NEM treatment prevents disulfide bonds in B and C capsids.
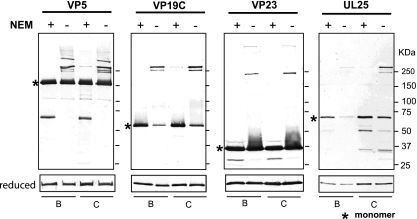
We next asked whether disulfide bonds could be detected in intracellular capsids. Although the cytoplasm and nucleus generally are considered to be reducing environments, many nuclear processes are controlled by the regulation of the redox state, including transcriptional regulation, nuclear protein trafficking, and DNA repair (20, 29). For instance, transcription factors such as NF-κB, AP-1, Nrf-2, glucocorticoid receptor, and p53 contain thiols that can undergo reversible oxidation to disulfides in response to signal transduction and thus can be regulated by redox state (77). To determine whether intracellular capsids also contained disulfide bonds, B and C capsids were isolated from NEM-treated or untreated cells and subjected to SDS-PAGE under nonreducing conditions. B and C capsids isolated in the absence of NEM display slower-migrating bands for VP5, VP19C, VP23, and UL25; however, if NEM was added prior to isolation, most of the slower-migrating bands were not detectable, and the monomeric form predominated. It should be noted that a small amount of slower-migrating material was detected in C but not B capsids isolated in the presence of NEM. The absence of detectable disulfide-cross-linked species in NEM-treated B capsids indicates that cross-linking probably occurs during or after packaging.
UL25 was previously reported to be present in both B and C capsids but at reduced levels in B capsids compared to those in C capsids (69). Consistently with this observation, the panel on the right in Fig. 4 shows that significantly less UL25 was detected in B capsids than in C capsids. The observation that B and C capsids are capable of forming disulfide bonds only if exposed to an oxidizing environment suggests that the reactive cysteines in capsid proteins are in close proximity to each other and can react upon cell lysis and exposure to oxidizing conditions. In summary, our results indicate that virions contain preexisting disulfide-bonded complexes between capsid proteins.
Fig. 4.
NEM blocks formation of slower-migrating protein complexes in intracellular capsids. Intracellular capsids isolated in the presence or absence of NEM as described in Materials and Methods were resolved on reducing and nonreducing SDS-PAGE, followed by immunoblotting with antibodies against various capsid proteins. Bands resolved above monomer represent disulfide-linked complexes. Immunoblots of reduced samples show total protein levels.
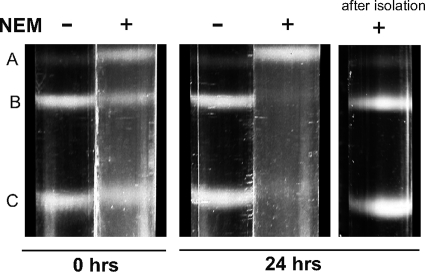
NEM-treated capsids are unstable.
In the course of our studies, we noticed that capsids isolated from cells that have been treated with NEM were unstable. In Fig. 5 capsids isolated either in the presence or absence of NEM were subjected to sucrose gradient centrifugation either immediately after cell lysis or 24 h later. Capsids isolated in the absence of NEM displayed the expected pattern of A, B, and C capsids at both 0 and 24 h after isolation. On the other hand, capsids isolated from NEM-treated cells displayed A, B, and C capsids at 0 h after isolation, but by 24 h the B and C capsids had disappeared, giving rise to a band that sedimented slightly slower than the 0-h A capsid band. Thus, within 24 h of isolation, NEM-treated B and C capsids lose their contents and sediment in a sucrose gradient at a position similar to that of A or empty capsids. Even at 0 h, there were more A and fewer B capsids in the NEM-treated sample than in the untreated sample. Thus, capsids isolated from NEM-treated cells appear to be unstable, while capsids isolated from untreated cells were stabilized possibly by the formation of disulfide bonds. We wanted to rule out the possibility that NEM itself was able to destabilize HSV capsids by reacting with some component other than a reactive cysteine. A preparation of capsids isolated from cells that had not been treated with NEM subsequently was exposed to NEM and subjected to sucrose gradient sedimentation. A, B, and C capsids can be seen in this preparation (Fig. 5, right), indicating that the NEM treatment of capsids whose disulfide bonds had already formed did not cause destabilization by reacting with some other component.
Fig. 5.
Capsids isolated in the presence of NEM are not stable. Intracellular capsids were isolated in the presence or absence of NEM and resolved on a 20 to 50% sucrose gradient (left panel). An aliquot of the crude capsid sample from each isolation was stored at 4°C and resolved on a sucrose gradient 24 h later (middle panel). An aliquot of the crude capsid sample isolated in the absence of NEM was exposed to NEM after isolation, stored at 4°C, and resolved on a sucrose gradient 24 h later (right panel).
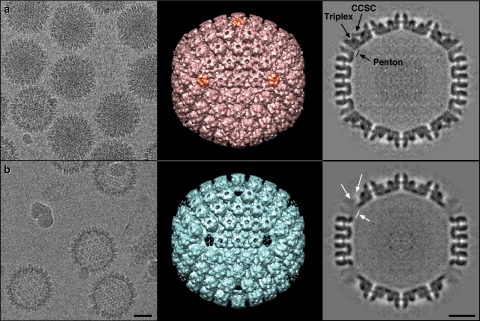
The loss of scaffold from B capsids and DNA from C capsids indicates that a significant conformational change has occurred in capsids isolated from NEM-treated cells. To address the nature of this alteration, capsids isolated from treated or untreated cells were visualized by cryoelectron microscopy (Fig. 6). C capsids from untreated cells (Fig. 6A) appeared to be predominantly DNA filled, and the few empty capsids likely resulted from the loss of a capsomer during preparation for electron microscopy. In contrast, the NEM-treated sample (Fig. 6b) included only empty capsids. Density maps calculated from the cryo-EM images show a full complement of capsid structural elements (hexons, pentons, triplexes, and CVSCs) for the untreated C capsids, while the structure of NEM-treated capsids is striking in its lack of almost all density corresponding to the pentons, the peripentonal triplexes, and the CVSC molecules. The consequent pores in the capsid would allow the leakage of any packaged DNA or scaffold, causing all capsids to appear empty.
Fig. 6.
Capsid structures from cryoelectron microscopy data. KOS C capsids used as a control (a) and NEM-treated capsids (b) are shown. The images at left show portions of cryoelectron micrographs revealing typical views of purified HSV capsids, some in panel a include packaged DNA while others have lost most or all DNA, whereas very few capsids in panel b contain detectable quantities of DNA. Bar, 500 Å. The central images show surface representations of density maps calculated from the corresponding cryoelectron micrographs, and at right are central gray-scale-encoded sections from the density maps. The comparison of the structures reveals that the pentons (orange in panel a) are absent from the NEM-treated sample, together with the CVSC molecules and the peripentonal triplexes. The remaining capsid components in panel b appear to be largely undisturbed at this resolution. Bar, 250 Å.
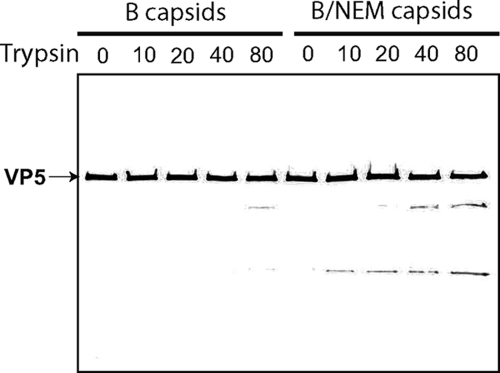
To further demonstrate that NEM-treated capsids were unstable, we subjected B capsids from NEM-treated and untreated cells to partial proteolysis by trypsin. Capsids then were analyzed by SDS-PAGE and immunoblot analysis for VP5. We found that VP5 in NEM-treated capsids was significantly more susceptible to trypsin digestion than B capsids from untreated cells (Fig. 7), which is consistent with the results presented in Fig. 5 and 6 showing that capsids isolated from NEM-treated cells are significantly less stable than those isolated from untreated cells.
Fig. 7.
VP5 protein of capsids isolated in the presence of NEM has different conformation. B capsids isolated in the presence or absence of NEM were partially digested with increasing concentrations of trypsin from 0 to 80 μg/ml and incubated for 30 min at 30°C. Digested samples were resolved on SDS-PAGE, followed by immunoblotting with VP5 monoclonal antibody (3E8).
Taken together, the data shown in Fig. 5, 6, and 7 suggest that cross-linking by disulfide bond formation contribute to proper capsid conformation and stability; however, other explanations for the observed instability are possible. For instance, the experiments were performed with capsids from cells that had been pretreated with NEM, which is known to irreversibly modify cysteines by binding directly to free sulfhydryls. It thus is possible that the observed capsid instability was due to NEM binding rather than the prevention of disulfide bond formation. To address this possibility, we isolated intracellular capsids in buffer at pH 5.5, which is expected to prevent the formation of disulfide bonds without the addition of the larger and hydrophobic NEM molecule. Capsids isolated at pH 5.5 contained much less of the slower-migrating cross-linked species, and the recovery of A, B, and C capsids was greatly diminished, suggesting instability (data not shown); however, the low-pH capsids also showed a tendency to aggregate, complicating the interpretation.
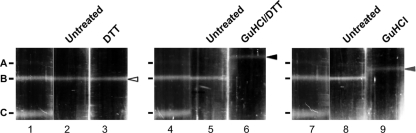
A third method was used to test the importance of disulfide bonds in capsid stability. We showed above (Fig. 4) that intracellular capsids isolated in the absence of NEM contained disulfide bonds. We next asked whether these capsids could be destabilized by the addition of reducing agents. B capsids were isolated in the absence of NEM, treated with DTT, or left untreated, and they were subjected to sucrose gradient sedimentation. A gradient containing A, B, and C capsids is shown to provide a reference for the sedimentation pattern of all three capsid types (Fig. 8, lane 1). DTT treatment alone had no effect on B capsid sedimentation (Fig. 8, lane 3) and was not able to reduce disulfide bonds as detected by nonreducing SDS-PAGE (data not shown). The only way that we have been successful in reducing the disulfide bonds in these capsids was treatment with the denaturing agents GuHCl and DTT in combination (data not shown). Figure 8 (lane 6) shows B capsids exposed to GuHCl and DTT shifted to a position that migrated slightly slower than A capsids, reminiscent of the 24-h NEM-treated capsids shown in Fig. 5 that have lost pentons (Fig. 6). This result indicates that disulfide bonds present in B capsids were buried in the protein such that they were not accessible to DTT without GuHCl treatment. GuHCl treatment alone resulted in B capsids that retain scaffold (data not shown). These B capsids sedimented slightly slower than untreated B capsids, indicating some conformational change but no loss of pentons. These data confirm a previous report showing that treatment with GuHCl and DTT together result in the loss of pentons (48). Thus, we conclude that disulfide bonds formed in capsids isolated in the absence of NEM can be reduced, leading to the loss of pentons. This experiment thus supports the argument that disulfide bonds are important for penton retention and capsid stability.
Fig. 8.
Treatment of B capsids with GuHCl and DTT. Sucrose gradient-purified and concentrated B capsids were exposed to DTT or GuHCl either alone or in combination. Treated and untreated capsids were separated on a 20 to 50% sucrose gradient and illuminated from the bottom to visualize capsids bands. Gradients of treated or untreated samples were aligned with each other as well as with gradient-resolved A, B, and C capsids and photographed. Final images were aligned using Adobe Photoshop.
DISCUSSION
In this paper, we have confirmed earlier reports that HSV virions contain disulfide cross-links involving structural proteins in the HSV capsid (41, 92, 93). In addition, immunoblotting and mass spectroscopy indicated that cross-linked species contain the major capsid protein VP5, triplex proteins VP19C and VP23, and both of the CVSC proteins UL17 and UL25. In addition, several tegument proteins were tentatively identified within cross-linked complexes in virions, including VP22, UL36, and UL37. When disulfide bond formation was blocked with NEM, intracellular capsid structures visualized by cryo-EM showed a dramatic loss of specific structural components, including pentons, peripentonal triplexes, and CVSC molecules, as well as a lack of internal DNA and scaffold protein. These results suggest that disulfide bonds contribute to capsid stability, which is reminiscent of other viruses shown to utilize cross-linking to stabilize capsids, including betanodavirus, foot and mouth virus, polyomavirus, retrovirus, and hepatitis B and C viruses (8, 17, 22, 25, 33, 37, 55, 61, 64, 85–87).
Several viruses have evolved mechanisms that exploit disulfide bonds to regulate aspects of capsid assembly, maturation, and penetration. Viral glycoproteins found in the membranes of enveloped viruses rely on disulfide bonds for proper conformation and infectivity. It has long been recognized that viral glycoproteins fold and mature in subcellular compartments in the trans-Golgi network (TGN) that are topologically equivalent to the extracellular environment (44). Thus, viral glycoproteins mature in compartments that provide an oxidative folding environment. For disulfides in proteins other than glycoproteins, it is not clear how and when the disulfide bonds are formed, since the assembly sites in the cytoplasmic and nuclear compartments are considered to be reducing due to the presence of thiol-containing molecules such as glutathione (GSH) and thioredoxin-1 (Trx1) (19, 20, 26, 29). In reoviruses, which assemble in the cytoplasm, disulfide bonds appear to form late in infection in dead or dying cells, perhaps after the breakdown of cytosolic reduction mechanisms (55). Interestingly, poxviruses, which assemble in the cytosol, encode three cytoplasmic thiol oxidoreductases conserved in all pox viruses that facilitate the formation of disulfide bonds in the otherwise unfavorable reducing environment of the cytosol (67, 68).
The timing and location of disulfide bond cross-links between HSV capsid proteins remain a mystery. Viral capsids are formed and take up DNA in the nucleoplasm, followed by budding into the perinuclear space, where they are not directly exposed to the otherwise oxidizing environment due to the membrane which surrounds them. After fusion with the outer nuclear membrane (70), the capsid is once again exposed to the reducing environment of the cytoplasm. Capsids acquire their final envelope by budding into TGN-derived vesicles and are released into the extracellular space via the secretory machinery of the cell (43, 44, 70). Thus, unlike glycoproteins that mature in subcellular compartments known to be oxidizing, HSV capsids are surrounded by the nucleoplasm or the cytoplasm, environments generally thought to be reducing. Thus, the observation that disulfide cross-linked capsid proteins are present in mature virions may be surprising considering that during all of these steps, capsids are never exposed to a cellular compartment that is topologically equivalent to the extracellular environment.
The interior of a virion could become more oxidizing and conducive to disulfide bond formation if oxygen was allowed to cross the membrane. This could theoretically occur in any of the oxidizing compartments that enveloped capsids are known to pass through (periplasm, TGN vesicle, or extracellular space); however, it is not clear how efficient this process would be. The level of cross-linking was higher in extracellular virions than in intracellular C capsids. This indicates that disulfides do not form efficiently until virus is released from cells, perhaps because in this environment membrane permeability to oxygen is enhanced. However, the observation that intracellular C capsids contained cross-linked capsid proteins, albeit at lower levels, suggests that some cross-linking occurs in the cell. Interestingly, we have recently observed that disulfide linkages are present in UL6, the HSV-1 portal protein (3). In contrast to the disulfide bonds detected between capsid proteins, those in UL6 can be detected in cells pretreated with NEM, suggesting that they are preexisting in infected cells (3). For these bonds to form in the reducing environment of the nucleus or cytoplasm, it may be necessary to create an oxidizing microenvironment. One scenario would involve the participation of viral or cellular chaperones reminiscent of the situation described above for the poxviruses (67, 68). Alternatively, HSV infection has been shown to cause a decrease in GSH levels (60), which would be expected to alter the redox state in cells, making it more conducive to disulfide bond formation. Interestingly, the treatment of cells with GSH, which would reinstate the reducing environment, was shown to inhibit virus growth (59, 60, 84). It is temping to speculate that the reason for the observed inhibition of HSV growth is that GSH treatment reinstates a reducing environment, thereby preventing disulfide bond formation. Thus, many scenarios are possible to explain the presence of disulfide bonds in both HSV portal rings and in capsids themselves, and experiments are in progress to distinguish among them.
Interestingly, several of the proteins that have been shown in this paper to contain disulfide cross-links have been implicated in various stages, including capsid assembly, nuclear egress, tegumentation, and virion maturation. UL25 is required for the primary envelopment of mature C capsids at the inner nuclear membrane and likely plays an important role in promoting the stability of DNA-containing capsids (31, 32). UL25 also plays an important role in the acquisition of tegument, as it has been reported to interact with the tegument protein UL36 (11, 40, 62). The process by which HSV capsids acquire tegument proteins is poorly understood. Some tegument proteins appear to be added prior to nuclear egress; however, others may be incorporated in the cytoplasm, and yet others are acquired during final envelopment as nucleocapsids bud into TGN-derived vesicles. The major tegument protein VP22 has been reported to associate with capsids in the perinuclear space (58a) as well as in the TGN (46). In both HSV and pseudorabies virus, UL36 is an important tegument protein that is required for the recruitment of other tegument proteins, including UL37 (30, 36, 45, 47). Interestingly, the tegument layer in HSV-1 virions has been shown to undergo dynamic time-dependent alterations that affect its symmetrical appearance by electron microscopy and its resistance to extraction by Triton X-100 (53). Reports from the Wills laboratory suggest that interactions between the tegument protein UL16 and the capsid are dynamic and can be regulated by pH and NEM by a mechanism that involves cysteines (41, 42). The finding that capsid and tegument appear to participate in covalent cross-links suggests that disulfide bond formation plays roles at several stages of the viral life cycle, including capsid assembly, the initiation of packaging, nuclear egress, tegument recruitment, and the maturation of extracellular virions.
We are also intrigued by the possibility that disulfide bonds also play a role during subsequent infections of new host cells. Following fusion with the plasma membrane, HSV-1 releases its capsid and tegument into the cytoplasm of a host cell, and the capsid is transported on microtubules (71), docking at the nuclear pore complexes (58). It is possible that the reduction of disulfide bonds either in the portal or the capsid play a role in this process. Since many of the proteins that participate in disulfide cross-linking, such as UL6, UL25, and preliminarily UL36, also have been implicated in the process of genome release (1, 5, 16, 27, 57, 65), it will be of interest to determine whether this process is regulated by the rearrangement of disulfide bonds. Interestingly, work with pseudorabies virus suggests that UL25 and at least two tegument proteins, UL36 and UL37, remain associated with the capsid following fusion (21, 28, 35).
HSV capsids are known to undergo structural changes as they mature from procapsid to C capsid during the encapsidation reaction and as they further develop into extracellular virions (34, 54, 66). In this report, we demonstrated that capsids lacking disulfide bonds are unstable and prone to the loss of pentons and peripentonal triplexes. This observation suggests that disulfide bonds are directly responsible for contacts between the penton and surrounding capsid or indirectly through maintaining structural elements in the correct conformations for stabilizing the penton-capsid interfaces. A recent study using atomic force microscopy (AFM) indicated that B capsids are less stable and break at lower forces than A and C capsids; however, after the release of pentons following GuHCl treatment, A, B, and C capsids demonstrated similar mechanical properties (66). Thus, the increased stability of A and C capsids may be related to the special constraints present at the vertices. Since the curvature of the capsid surface is greatest at the vertices and the intersubunit angular displacements are larger for pentons than hexons, the pentons and adjacent triplexes are likely to be the more strained and hence the most easily lost in the absence of stabilizing disulfide bonds. The need to enhance capsid integrity by bolstering the pentons is reminiscent of pathways used in the structurally related capsids of tailed bacteriophages. For instance, phages T4 and λ employ decoration proteins (72, 75) and phage HK97 covalent cross-links (88) to achieve stabilization following the encapsidation of viral genomes. The addition of decoration proteins or chemical cross-linking occur following expansion transformation from procapsid to capsid. Expansion brings binding sites or cross-linking residues into the correct conformation for the stabilization reactions to take place (15, 74). Furthermore, expansion is thought to be triggered by DNA packaging (15); thus, stabilization is precisely timed to strengthen the capsid against the internal pressure of the packaged genome. The degree to which herpesviruses mimic this aspect of bacteriophage capsid assembly remains to be seen.
ACKNOWLEDGMENTS
We thank the members of the Weller laboratory for helpful discussions and suggestions throughout the course of this study. Phil Greer and Douglas Bevan are gratefully acknowledged for help with computational analysis.
This work was supported by grants from the NIH (AI37549) to S.K.W. and from the Commonwealth of Pennsylvania (SAP 4100031302) to J.F.C.
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Abaitua F., O'Hare P. 2008. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J. Virol. 82:5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Addison C., Rixon F., Palfreyman J., O'Hara M., Preston V. 1984. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246–259 [DOI] [PubMed] [Google Scholar]
- 3. Albright B. S., Nellissery J., Szczepaniak R., Weller S. K. 2011. Disulfide bond formation in the herpes simplex virus 1 UL6 protein is required for portal ring formation and genome encapsidation. J. Virol. 85:8616–8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baines J., Weller S. K. 2005. Cleavage and packaging of herpes simplex virus 1 DNA. In Catalano C. (ed.), Viral genome packaging machines: genetics, structure and mechanism. Landes Bioscience, Austin, TX [Google Scholar]
- 5. Batterson W., Furlong D., Roizman B. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blewett E. L., Black D., Eberle R. 1996. Characterization of virus-specific and cross-reactive monoclonal antibodies to Herpesvirus simiae (B virus). J. Gen. Virol. 77:2787–2793 [DOI] [PubMed] [Google Scholar]
- 7. Brown J., McVoy M. A., Homa F. L. 2002. Packaging DNA into herpesvirus capsids, p. 111–155 In Bogner A. H. A. E. (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, New York, NY [Google Scholar]
- 8. Bruss V. 2007. Hepatitis B virus morphogenesis. World J. Gastroenterol. 13:65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burch A., Weller S. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during HSV-1 infection. J. Virol. 78:7175–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cockrell S. K., Huffman J. B., Toropova K., Conway J. F., Homa F. L. 2011. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J. Virol. 85:4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coller K. E., Lee J. I.-H., Ueda A., Smith G. A. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conway J., et al. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J. Mol. Biol. 397:575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conway J. F., Homa F. 2011. Nucleocapsid structure, assembly and DNA packaging of herpes simplex virus. In Weller S. K. (ed.), Alphaheresviruses: molecular virology. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 14. Conway J. F., Steven A. C. 1999. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128:106–118 [DOI] [PubMed] [Google Scholar]
- 15. Conway J. F., et al. 2001. Virus maturation involving large subunit rotations and local refolding. Science 292:744–748 [DOI] [PubMed] [Google Scholar]
- 16. Copeland A., Newcomb W., Brown J. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delmas O., Durand-Schneider A.-M., Cohen J., Colard O., Trugnan G. 2004. Spike protein VP4 assembly with maturing rotavirus requires a postendoplasmic reticulum event in polarized caco-2 cells. J. Virol. 78:10987–10994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gibson W., Roizman B. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilbert H. F. 1990. Molecular and cellular aspects of thiol-disulfide exchange. Adv. Enzymol. Relat. Areas Mol. Biol. 63:69–172 [DOI] [PubMed] [Google Scholar]
- 20. Go Y.-M., Jones D. P. 2008. Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta 1780:1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Granzow H., Klupp B. G., Mettenleiter T. C. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hare J., Chan J. 1968. Role of hydrogen and disulfide bonds in polyoma capsid structure. Virology 34:481–491 [DOI] [PubMed] [Google Scholar]
- 23. Heymann J. B., Belnap D. M. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157:3–18 [DOI] [PubMed] [Google Scholar]
- 24. Homa F., Brown J. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107–122 [DOI] [PubMed] [Google Scholar]
- 25. Huang E. S., Estes M. K., Pagano J. S. 1972. Structure and function of the polypeptides in simian virus 40. I. Existence of subviral deoxynucleoprotein complexes. J. Virol. 9:923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones D. P., Go Y.-M. 2010. Redox compartmentalization and cellular stress. Diabetes Obes. Metab. 12(Suppl. 2):116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jovasevic V., Liang L., Roizman B. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaelin K., Dezélée S., Masse M. J., Bras F., Flamand A. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kietzmann T. 2010. Intracellular redox compartments: mechanisms and significances. Antioxid. Redox. Signal. 13:395–398 [DOI] [PubMed] [Google Scholar]
- 30. Klupp B. G., Fuchs W., Granzow H., Nixdorf R., Mettenleiter T. C. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klupp B. G., Granzow H., Keil G. M., Mettenleiter T. C. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuhn J., et al. 2008. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J. Virol. 82:5725–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kushima Y., Wakita T., Hijikata M. 2010. A disulfide-bonded dimer of the core protein of hepatitis C virus is important for virus-like particle production. J. Virol. 84:9118–9127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liashkovich I., et al. 2008. Exceptional mechanical and structural stability of HSV-1 unveiled with fluid atomic force microscopy. J. Cell Sci. 121:2287–2292 [DOI] [PubMed] [Google Scholar]
- 35. Luxton G. W. G., et al. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luxton G. W. G., Lee J. I.-H., Haverlock-Moyns S., Schober J. M., Smith G. A. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Majhen D., Gabrilovac J., Eloit M., Richardson J., Ambriović-Ristov A. 2006. Disulfide bond formation in NGR fiber-modified adenovirus is essential for retargeting to aminopeptidase N. Biochem. Biophys. Res. Commun. 348:278–287 [DOI] [PubMed] [Google Scholar]
- 38. McCombs R. M., Williams G. A. 1973. Disruption of herpes virus nucleocapsids using lithium iodide, guanidine and mercaptoethanol. J. Gen. Virol. 20:395–400 [DOI] [PubMed] [Google Scholar]
- 39. McNab A., et al. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McNabb D. S., Courtney R. J. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221–232 [DOI] [PubMed] [Google Scholar]
- 41. Meckes D. G., Wills J. W. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 81:13028–13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meckes D. G., Wills J. W. 2008. Structural rearrangement within an enveloped virus upon binding to the host cell. J. Virol. 82:10429–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mettenleiter T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167–180 [DOI] [PubMed] [Google Scholar]
- 44. Mettenleiter T. C., Klupp B. G., Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 45. Mijatov B., Cunningham A. L., Diefenbach R. J. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26–31 [DOI] [PubMed] [Google Scholar]
- 46. Miranda-Saksena M., Boadle R. A., Armati P., Cunningham A. L. 2002. In rat dorsal root ganglion neurons, herpes simplex virus type 1 tegument forms in the cytoplasm of the cell body. J. Virol. 76:9934–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Newcomb W., Brown J. 2010. Structure and capsid association of the herpesvirus large tegument protein UL36. J. Virol. 84:9408–9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Newcomb W., Brown J. C. 1994. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J. Virol. 68:433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Newcomb W., Homa F., Brown J. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 80:6286–6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Newcomb W., Homa F., Thomsen D., Ye Z., Brown J. 1994. Cell-free assembly of the herpes simplex virus capsid. J. Virol. 68:6059–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Newcomb W., Thomsen D., Homa F., Brown J. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862–9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Newcomb W., et al. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Newcomb W. W., Brown J. C. 2009. Time-dependent transformation of the herpesvirus tegument. J. Virol. 83:8082–8089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Newcomb W. W., et al. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432–446 [DOI] [PubMed] [Google Scholar]
- 55. Odegard A., Chandran K., Liemann S., Harrison S., Nibert M. 2003. Disulfide bonding among micro 1 trimers in mammalian reovirus outer capsid: a late and reversible step in virion morphogenesis. J. Virol. 77:5389–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ogasawara M., Suzutani T., Yoshida I., Azuma M. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O'Hara M., et al. 2010. Mutational analysis of the herpes simplex virus type 1 UL25 DNA packaging protein reveals regions that are important after the viral DNA has been packaged. J. Virol. 84:4252–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ojala P. M., Sodeik B., Ebersold M. W., Kutay U., Helenius A. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20:4922–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58a. Padula M. E., Sydnor M. L., Wilson D. W. 2009. Isolation and preliminary characterization of herpes simplex virus 1 primary enveloped virions from the perinuclear space. J. Virol. 83:4757–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palamara A. T., et al. 2004. New synthetic glutathione derivatives with increased antiviral activities. Antivir. Chem. Chemother. 15:83–91 [DOI] [PubMed] [Google Scholar]
- 60. Palamara A. T., et al. 1995. Evidence for antiviral activity of glutathione: in vitro inhibition of herpes simplex virus type 1 replication. Antiviral Res. 27:237–253 [DOI] [PubMed] [Google Scholar]
- 61. Parry N., et al. 1990. Structural and serological evidence for a novel mechanism of antigenic variation in foot-and-mouth disease virus. Nature 347:569–572 [DOI] [PubMed] [Google Scholar]
- 62. Pasdeloup D., Blondel D., Isidro A., Rixon F. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pettersen E. F., et al. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 64. Pornillos O., Ganser-Pornillos B. K., Banumathi S., Hua Y., Yeager M. 2010. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J. Mol. Biol. 401:985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Preston V. G., Murray J., Preston C. M., McDougall I. M., Stow N. D. 2008. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J. Virol. 82:6654–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roos W. H., et al. 2009. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc. Natl. Acad. Sci. U. S. A. 106:9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Senkevich T. G., White C. L., Koonin E. V., Moss B. 2002. Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc. Natl. Acad. Sci. U. S. A. 99:6667–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Senkevich T. G., White C. L., Koonin E. V., Moss B. 2000. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl. Acad. Sci. U. S. A. 97:12068–12073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sheaffer A., et al. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Skepper J. N., Whiteley A., Browne H., Minson A. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment→deenvelopment→reenvelopment pathway. J. Virol. 75:5697–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sodeik B., Ebersold M., Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sternberg N., Weisberg R. 1977. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J. Mol. Biol. 117:733–759 [DOI] [PubMed] [Google Scholar]
- 73. Steven A., Spear P., Chiu W., Burnett R., Garcea R. 1997. Herpesvirus capsid assembly and envelopment, p. 312–351 In Chiu W., Burnett R. M., Garcea R. (ed.), Structural Biology of Viruses. Oxford University Press, New York, NY [Google Scholar]
- 74. Steven A. C., Bauer A. C., Bisher M. E., Robey F. A., Black L. W. 1991. The maturation-dependent conformational change of phage T4 capsid involves the translocation of specific epitopes between the inner and the outer capsid surfaces. J. Struct. Biol. 106:221–236 [DOI] [PubMed] [Google Scholar]
- 75. Steven A. C., Greenstone H. L., Booy F. P., Black L. W., Ross P. D. 1992. Conformational changes of a viral capsid protein. Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, co-operativity, and super-stabilization by soc binding. J. Mol. Biol. 228:870–884 [DOI] [PubMed] [Google Scholar]
- 76. Steven A. C., Heymann J. B., Cheng N., Trus B. L., Conway J. F. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 15:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Su D. G. T., et al. 2010. Interactions of APE1 with a redox inhibitor: evidence for an alternate conformation of the enzyme. Biochemistry 50:82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thurlow J., et al. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Thurlow J. K., Murphy M., Stow N. D., Preston V. G. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tivol W. F., Briegel A., Jensen G. J. 2008. An improved cryogen for plunge freezing. Microsc. Microanal. 14:375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Toropova K., Huffman J., Homa F. L., Conway J. 1 June 2011. The HSV-1 UL17 protein is the second constituent of the capsid vertex specific component (CVSC) required for DNA packaging and retention. J. Virol. [Epub ahead of print.] doi:10.1128/JVI.00837–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Trus B., et al. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol. Cell 26:479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Van Heel M. 1987. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21:111–123 [DOI] [PubMed] [Google Scholar]
- 84. Vogel J. U., et al. 2005. Effects of S-acetylglutathione in cell and animal model of herpes simplex virus type 1 infection. Med. Microbiol. Immunol. 194:55–59 [DOI] [PubMed] [Google Scholar]
- 85. Walter G., Deppert W. 1975. Intermolecular disulfide bonds: an important structural feature of the polyoma virus capsid. Cold Spring Harb. Symp. Quant. Biol. 39:255–257 [DOI] [PubMed] [Google Scholar]
- 86. Wang C.-H., et al. 2010. Roles of cysteines Cys115 and Cys201 in the assembly and thermostability of grouper betanodavirus particles. Virus Genes 41:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Whitehurst C. B., et al. 2007. Location and role of free cysteinyl residues in the Sindbis virus E1 and E2 glycoproteins. J. Virol. 81:6231–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wikoff W. R., et al. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129–2133 [DOI] [PubMed] [Google Scholar]
- 89. Wills E., Scholtes L., Baines J. 2006. Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid. J. Virol. 80:10894–10899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yan X., Dryden K. A., Tang J., Baker T. S. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 157:211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yan X., Sinkovits R. S., Baker T. S. 2007. AUTO3DEM–an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yang C. C., Yang Y. Y., Lin K. L., Lin S. J. 2000. Different forms of HSV-1 VP22a within purified virion and infected cells. J. Microbiol. Immunol. Infect. 33:141–148 [PubMed] [Google Scholar]
- 93. Zweig M., Heilman C. J., Hampar B. 1979. Identification of disulfide-linked protein complexes in the nucleocapsids of herpes simplex virus type 2. Virology 94:442–450 [DOI] [PubMed] [Google Scholar]