Abstract
The 5′ 140 nucleotides of the mouse hepatitis virus (MHV) 5′ untranslated region (5′UTR) are predicted to contain three secondary structures, stem-loop 1 (SL1), SL2, and SL4. SL1 and SL2 are required for subgenomic RNA synthesis. The current study focuses on SL4, which contains two base-paired regions, SL4a and SL4b. A series of reverse genetic experiments show that SL4a is not required to be base paired. Neither the structure, the sequence, nor the putative 8-amino-acid open reading frame (ORF) in SL4b is required for viral replication. Viruses containing separate deletions of SL4a and SL4b are viable. However, deletion of SL4 is lethal, and genomes carrying this deletion are defective in directing subgenomic RNA synthesis. Deletion of 131ACA133 just 3′ to SL4 has a profound impact on viral replication. Viruses carrying the 131ACA133 deletion were heterogeneous in plaque size. We isolated three viruses with second-site mutations in the 5′UTR which compensated for decreased plaque sizes, delayed growth kinetics, and lower titers associated with the 131ACA133 deletion. The second-site mutations are predicted to change either the spacing between SL1 and SL2 or that between SL2 and SL4 or to destabilize the proximal portion of SL4a in our model. A mutant constructed by replacing SL4 with a shorter sequence-unrelated stem-loop was viable. These results suggest that the proposed SL4 in the MHV 5′UTR functions in part as a spacer element that orients SL1, SL2, and the transcriptional regulatory sequence (TRS), and this spacer function may play an important role in directing subgenomic RNA synthesis.
INTRODUCTION
Mouse hepatitis virus (MHV) and severe acute respiratory syndrome (SARS)-coronavirus (CoV) are closely related viruses in the genus Betacoronavirus (previously known as “coronavirus group 2”; see http://talk.ictvonline.org/media/g/vertebrate-2008/default.aspx) in the family Coronaviridae of the order Nidovirales (9). The coronaviruses comprise a group of single-stranded, positive-sense, nonsegmented, enveloped RNA viruses. CoV RNA genomes are 25 to 31 kb, the largest genomes of all known RNA viruses, and are infectious when introduced into permissive cells. MHV is the most extensively studied CoV and provides mouse models for several human diseases, including SARS, hepatitis, and multiple sclerosis (35).
The MHV genome is 5′ capped and 3′ polyadenylated and encodes nonstructural replication-related proteins in the 5′ two-thirds of the genome and structural proteins in the 3′ one-third of the genome. Infected cells contain seven MHV-specific mRNAs with coterminal 3′ ends, the largest of which is the genomic RNA (18, 33). These mRNAs all carry identical 72-nucleotide (nt) leader sequences at their 5′ ends (15, 16, 18, 33). The transcriptional regulatory sequences located at the 3′ end of the leader sequence (TRS-L) and also upstream of the coding sequences for each transcriptional unit in the genome (TRS-B [body]) act as cis-regulators of transcription (4). MHV RNA transcription occurs in the cytoplasm. In the most widely applied model of coronavirus transcription, discontinuous transcription of negative-strand subgenomic RNAs from a genome-length template leads to leader-body joining (27, 30, 31, 34). These negative-strand products subsequently serve as templates for subgenomic mRNA synthesis. In an elaboration of this model, viral and/or cellular factors bind to cis-acting RNA elements in the genomic RNA 5′ untranslated region (5′UTR) and 3′UTR, circularizing the genome and promoting template switching by topologically enabling base pairing between TRS-L and the nascent complementary TRS-Bs (32, 38).
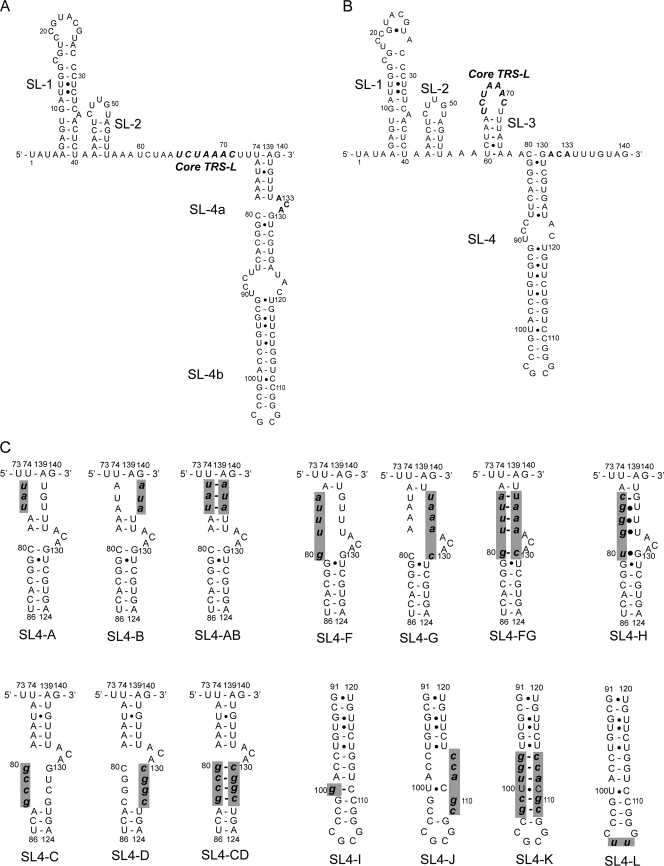
The MHV RNA 5′UTR and 3′UTR contain cis-acting sequences that are presumed to fold into secondary and higher-order structures important for RNA-RNA interactions and for the binding of viral and cellular proteins during RNA replication and translation (2, 22, 25). The first 140 nucleotides of the MHV genome are predicted to contain three cis-acting stem-loops (SL), SL1, SL2, and SL4, based on a consensus secondary structural model of nine representative CoVs representing all three CoV subgroups (Fig. 1A) (14, 24). Some CoVs, e.g., SARS-CoV and bovine coronavirus (BCoV), are also predicted to contain a third stem-loop, SL3, which folds the TRS-L into a hairpin loop. In MHV, a plausible base pairing scheme can be drawn which places the TRS into the loop of SL3 as well (Fig. 1B) (5), but the stem is not predicted to be stable at 37°C (24).
Fig. 1.
Predicted secondary structures of MHV A59 for the 5′ first 140 nucleotides, including SL1, SL2, and SL4, in our model (14, 24) (A) and including SL1, SL2, SL3, and SL4, in the Chen and Olsthoorn model (5) (B). The noncanonical base pairs shown at nt 94 to 117 and 96 to 115 were not present in our original model (14, 24) but were incorporated from the Chen and Olsthoorn model (5). (C) SL4-A, SL4-B, SL4-C, SL4-D, SL4-F, SL4-G, SL4-AB, SL4-CD, SL4-FG, and SL4-H represent individual mutant viruses containing clustered point mutations in the SL4a helix. SL4-I, SL4-J, SL4-K, and SL4-L represent the individual mutant viruses containing clustered point mutations in the SL4b helix. The lowercase letters with bold and italic in each mutant represent the introduced mutations. These mutations are highlighted by gray shading.
SARS-CoV SL1, SL2, and SL4 can separately replace their corresponding MHV counterparts in the MHV genome, and the constructed chimeric viruses are viable even though the sequences of the MHV 5′UTR and the SARS-CoV 5′UTR are significantly different (14). However, when the entire MHV 5′UTR is replaced by the SARS-CoV 5′UTR, the MHV chimera is not viable. The MHV 5′UTR SL1 has been shown to be functionally and structurally bipartite, and structural lability in SL1 is required to drive the interaction between the 5′UTR and the 3′UTR that stimulates subgenomic RNA synthesis (21). SL2 contains a highly conserved pentaloop (C/U)UUG(U/C) sequence in all nine examined CoVs (24). Subsequent nuclear magnetic resonance (NMR) determination of the solution structure and functional studies reveal that SL2 adopts a canonical uCUYG(U)a-like tetraloop stacked on a 5-base-pair stem with the 3′ U flipped out of the loop (17, 23). Mutational analysis demonstrates that SL2 is required for subgenomic RNA synthesis (23, 24).
SL4 is positioned just 3′ to the leader TRS (mapping at nt 74 through 139 in MHV) and is the first proposed structural RNA element of the 5′UTR that follows the leader. It is predicted to contain a bipartite stem-loop (SL4a and SL4b) separated by a bulge in our model (14, 24) (Fig. 1A). Chen and Olsthoorn (5) employed a phylogenetic approach to predict secondary structures of the 5′UTR and adjacent coding sequences in all CoVs. Their structural-phylogenetic analysis is largely consistent with our model and predicts that the core TRS-L and the flanking sequences are poorly structured in CoVs but strongly supports the existence of SL4 downstream of the TRS-L (Fig. 1B). The structure of the MHV-A59 SL4 predicted by Chen and Olsthoorn (5) differs primarily from SL4 in the model predicted by our group (14, 24) in that the proximal 6 base pairs of our predicted stem are not base paired in the Chen and Olsthoorn model (compare Fig. 1A to B). We hypothesize that the proposed SL4 in the MHV 5′UTR functions as a cis-acting sequence important for viral replication and viral RNA synthesis. In this study, we have used a reverse genetic approach to test this hypothesis and further investigated if the secondary structure or specific nucleotides in SL4 influence viral replication.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney cells expressing the MHV receptor CEACAM1a (BHK-R) (8, 37) were originally provided by Ralph Baric (University of North Carolina at Chapel Hill) and maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum, 10% tryptose phosphate broth, 800 μg/ml of Geneticin (G418 sulfate; Sigma) to select for cells expressing the MHV receptor, 100 IU/ml of penicillin, and 100 μg/ml of streptomycin. DBT cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% calf serum. L2 cells were maintained at 37°C and 3% CO2 in DMEM supplemented with 10% calf serum. MHV-A59-1000 (37) was used as our wild-type (WT) virus, and all mutant viruses were propagated in DBT cells.
Mutagenesis and plasmid construction.
Plasmid A, corresponding to the first 4,882 nt of the MHV-A59 genome, was used as a substrate for site-directed mutagenesis (37). Mutations were introduced into the MHV 5′UTR with the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions or by overlapping PCR. The sequences of the oligonucleotides used for mutagenesis are available on request. Mutagenized plasmids were sequenced between unique MluI-BamHI restriction sites to verify the introduced mutations. The MluI-BamHI fragments containing the introduced mutations were excised and ligated with MluI-BamHI-digested parental unmutagenized plasmid A to generate the plasmids used for generating mutant viruses. The sequences of these plasmids between the MluI and BamHI sites were all verified again by sequencing to minimize the possibility that the plasmids used to generate virus by reverse genetics contain unintended mutations inadvertently introduced during mutagenesis.
Assembly of full-length MHV-A59 cDNAs and recovery of mutant viruses.
cDNAs representing the entire MHV-A59 genome, carrying either the wild-type sequence or mutations in SL4, were constructed by sequential ligation of digested and gel-purified cDNAs A to G (37). The ligated cDNA was transcribed in vitro using the Ambion T7 mMESSAGE mMACHINE kit (Applied Biosystems), as described previously (13, 37). The transcription reactions were then electroporated into BHK-R cells that were then overlaid onto DBT cells. Cultures were incubated for up to 72 h and monitored by phase microscopy for the development of the cytopathic effect (CPE). Cultures that did not demonstrate CPE were frozen at −80°C, thawed, sonicated, and clarified, and the supernatant was blind passaged in DBT cells. A mutation was not considered to be lethal until at least three independent trials were performed, at least one of which was done at 34°C and 39°C to allow for recovery of temperature-sensitive viruses. Recovered viruses were plaque purified on L2 cell monolayers and propagated in DBT cells, and their 5′UTR and 3′UTR were amplified by reverse transcription (RT)-PCR and sequenced to confirm the presence of the introduced mutations and to identify any second-site mutations that might have arisen during virus recovery and propagation. For each mutant, usually 3 or 4 plaque-purified isolates at passage 1 (P1) were selected for sequencing. If second-site mutations were recovered from the P1 virus, serial passages P2 to P5 were performed for the plaque-purified isolates that did not contain second-site mutations, and the 5′UTR and 3′UTR were sequenced at each passage level to determine when second-site mutations present in the P5 virus became established during serial passage. Sequences at the extreme 5′ end of the 5′UTR were determined by 5′ rapid amplification of cDNA ends (5′ RACE) as described previously (21). Briefly, mutant plaque-purified virus isolates were propagated in DBT cells, total RNA extracted, and then reverse transcribed using primer JM19 [A59(−)531-513, ATGGCTTAACCAAGACGGC]. The cDNA products had poly(A) tails added to their 3′ ends with terminal transferase (Roche). The poly(A)-tailed RT products were repurified using the QIAquick PCR purification kit. Two rounds of nested PCR were performed using primers JM19 and A59(−)282-263 (ATGCGTTCGGAAGCATCCAT), respectively, with the adapter primer (AP-dT17) (GATCAGGACGTTCGTTTGAGTTTTTTTTTTTTTTTTT), and the PCR products were gel purified and sequenced.
Plaque size determination and growth curve assays.
Plaque assays were performed in L2 cells as described previously (13). Plaque diameters were calculated from 40 well-formed plaques for each virus by projecting the crystal violet-stained monolayers alongside a millimeter-scaled ruler, and the diameters of the projected plaques were measured. Plaque diameters were calculated after the enlargement factor was determined by measuring the projected millimeter ruler, thus allowing very accurate determinations of plaque diameter. Growth curves for each recovered mutant in comparison to that of the WT virus were also determined to establish the growth phenotype of each mutant. Triplicate wells of DBT cells in 96-well plates were infected with mutant or wild-type viruses at a multiplicity of infection (MOI) of 3. In experiments with low-titer viruses, an MOI of 0.1 or 1 was used. Infected cultures were harvested at 0, 4, 8, 12, 16, and 24 h postinfection (hpi). Virus titers were determined by plaque assays. The Kruskal-Wallis test and one-way analysis of variance (ANOVA) were used for statistical comparisons among mutant and WT viruses. Error bars represent the standard errors of the mean.
Detection of negative-strand genomic and subgenomic RNAs for nonviable mutants.
To determine if genome replication or subgenomic RNA transcription was taking place in cells electroporated with nonviable mutant genomes, we performed RT-PCR assays to detect the presence of negative-sense RNAs corresponding to these species. Total RNAs were extracted from cultures 4 h and 8 h postelectroporation and subsequently treated with DNase I to eliminate any cDNA that entered cells during electroporation. The total cellular RNAs were assayed by nested RT-PCR as described previously (13, 24) to detect negative-sense RNA corresponding to the genome and negative-sense subgenomic RNA3 and RNA6. Parallel reactions without an RT step to synthesize cDNA were carried out as controls to ensure that the PCRs did not detect residual DNA transcription templates taken up by the cells during electroporation. Previous work utilizing in vitro transcripts containing a frameshift mutation in nsp12, the gene encoding the RNA-dependent RNA polymerase (RdRP), indicated that these assays were not affected by any copy-back RNAs present in the in vitro transcription reactions (24).
RESULTS
The proximal region of SL4a is not required to be base paired.
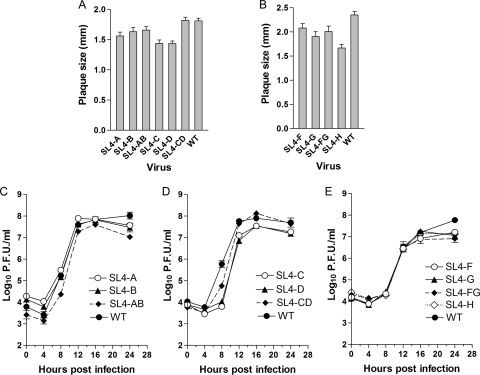
In our model (14, 24), SL4 in MHV-A59, positioned just 3′ to the leader TRS (nt 74 through 139), is predicted to contain a bipartite stem-loop (SL4a and SL4b) separated by a bulge (Fig. 1A). The Chen and Olsthoorn model (5) strongly supports the existence of SL4, but nt 74 to 79 are not paired with nt 134 to 139 (Fig. 1B). Therefore, we initiated our investigations of SL4 by focusing on the structure and function of SL4a. To determine the functional importance of structure and sequence in the helical regions of SL4a (nt 74 to 86 and nt 124 to 139), 10 viruses containing clustered point mutations (Fig. 1C, SL4-A through SL4-D, SL4-F through SL4-H, SL4-AB, SL4-CD, SL4-FG) in this region were generated as described in Materials and Methods. These mutations were predicted to open up the stem by separately altering the sequences on the left side (SL4-A, SL4-C, SL4-F) or the corresponding right side (SL4-B, SL4-D, SL4-G) of the SL4a helix or by changing the sequences on both sides of the helix (SL4-AB, SL4-CD, SL4-FG) while restoring canonical Watson-Crick base pairing (Fig. 1C). An additional mutant, SL4-H, changed the sequence UAAAC at nt 76 to 80 to CGGGU. This mutation potentially maintains base pairing via noncanonical UG base pairs while altering the sequence in this region (Fig. 1C). All 10 mutants were viable, and most had only small to moderate impairment of virus replication compared to that of the WT virus as assessed by plaque diameter and one-step growth curves (Fig. 2). The mutants with the largest replication deficits among these 10 mutants were the SL4-C and SL4-D viruses, containing mutations in the distal portion of SL4a in our model. These viruses had somewhat smaller plaque sizes and achieved a somewhat lower titer than the WT virus, although they grew with kinetics similar to those of the WT (Fig. 2A and D). However, the phenotype of the SL4-CD mutant, a mutant predicted to maintain the secondary structure while altering the sequences on both sides of the helix, more closely resembled that of the WT virus, with plaque size and growth kinetics essentially identical to those of the WT virus (Fig. 2A and D). The 5′ and 3′UTRs of four plaque-purified isolates of each mutant were sequenced at virus passage 1, and all contained the desired mutations, with no other substitutions in these regions of the genome. These results indicate that the proximal portion of SL4a in our model (nt 75 to 79 and 134 to 138) is likely not base paired, as proposed in the Chen and Olsthoorn model (5), or if it is base paired, this region of SL4a (Fig. 1A and C, mutants SL4-A, -B, -AB) has no functional significance. The upper portion (Fig. 1A and C, mutants SL4-C, -D, -CD) of the SL4a stem is also not required for viral replication but does contribute to optimal viral replication independently of the sequence.
Fig. 2.
Growth phenotypes of SL4a mutant viruses. (A and B) Average plaque sizes of mutant and wild-type viruses. (C, D, and E) Growth curves of mutant and wild-type viruses at an MOI of 3.
Neither the structure, the sequence, nor the putative 8-amino-acid ORF in SL4b has a critical role in viral replication.
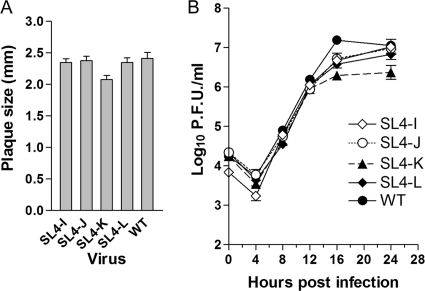
The Brian lab has previously shown that SL4b was necessary to continuously propagate BCoV defective interfering (DI) RNA replicons in that mutations which disrupted this stem reduced DI replication by greater than 99% (28). BCoV, as do most CoVs, including MHV, contains a short intra-5′UTR open reading frame (ORF) that resides in SL4b; there is a positive correlation between maintenance of the short intra-5′UTR ORF and maximal DI RNA accumulation in BCoV DI RNA (28). Guided by the results obtained by the Brian lab, we generated four mutants in SL4b (nt 91 through 120) to investigate the requirement for SL4b in the context of the complete genome. These mutations were predicted to destabilize the SL4b stem by altering the sequences on the right side of the SL4b helix (SL4-J) or to maintain base pairing by changing the sequences on both sides of the helix (SL4-K) (Fig. 1C). We also constructed additional mutants with substitutions of AUG→AGG (SL4-I) and CGCG→CUUG in the predicted terminal loop (SL4-L) (Fig. 1C). These mutations were designed to mutate the presumptive initiating methionine (SL4-I) and to alter the amino acid sequence (SL4-L) of this ORF. In contrast to the results obtained with the BCoV DI RNA replication system (28), the four mutant viruses that we constructed were viable and were only modestly impaired in their ability to replicate, as assessed by their relative plaque sizes and growth curves, compared to that of the WT (Fig. 3). Sequencing of the 5′ and 3′UTRs of four isolates of each mutant virus showed that each contained the desired mutations with no other substitutions, with the exception of SL4-K. For SL4-K, one of the four plaque-purified isolates contained a second-site mutation, U60C, in addition to the introduced substitutions, but was identical to the other three SL4-K mutants in terms of plaque size, growth kinetics, and titer achieved (data not shown). These data indicate that neither the structure, the sequence, nor the 8-amino-acid small ORF has a critical role in viral replication, in that none of the mutant viruses had peak titers less than 10% of that of the WT. This contrasts with the data obtained with a DI model replicon from the Brian lab, where the destruction of the helix that corresponds to SL4b reduced DI replication to less than 1% of that of wild-type DI RNA (28).
Fig. 3.
Growth phenotypes of SL4b mutant viruses. (A) Average plaque sizes of mutant and wild-type viruses. (B) Growth curves of mutant and wild-type viruses at an MOI of 3.
Deletion of SL4 is lethal.
The above-described results with multiple mutant viruses inspired us to explore whether SL4 is essential for MHV replication. To do this, we attempted to recover a virus containing a deletion of the entire SL4 (nt 75 to 138). In three independent experiments, including an attempt to recover temperature-sensitive virus by incubation at 34°C and 39°C, viral genomes with a deletion of SL4 failed to generate infectious virus, even after three sequential blind passages of each electroporation. This result indicates that some as-yet-unknown property of SL4 is required for virus replication.
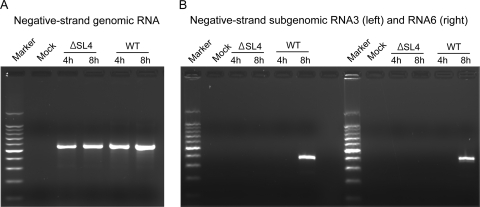
To determine the RNA species that might have been generated in cells electroporated with SL4-deleted genomes, we performed nested RT-PCR assays to detect any negative-strand genomic RNA and negative-strand RNAs complementary to subgenomic mRNA3 and mRNA6. These RNAs serve as templates for genomic and subgenomic mRNA synthesis (27, 32, 38). Nested RT-PCR results showed that negative-strand genomic RNAs were present in cells at 4 and 8 h after electroporation with in vitro-transcribed genomes deleted in SL4, similar to what we observed after electroporation of WT genomes (Fig. 4A). In contrast, neither negative-strand subgenomic RNA3 nor RNA6 was detected in cells electroporated with in vitro-transcribed SL4-deleted genomes, whereas cells electroporated with WT genomes contained negative-strand subgenomic RNA3 and RNA6 at 8 h incubation (Fig. 4B). For each sample, parallel RT-PCRs without an RT step were performed to ensure that residual DNA templates taken up by the cells during electroporation did not produce PCR signals (data not shown). Taken together, these data show that deletion of SL4 is lethal, and genomes carrying this deletion are defective in directing subgenomic RNA synthesis.
Fig. 4.
Analysis of negative-strand genomic and subgenomic RNAs for the nonviable ΔSL4 mutant by nested RT-PCR. (A) Negative-strand genomic RNA synthesis. Marker, 1-kb ladder; Mock, mock-infected cells. (B) Negative-strand subgenomic RNA3 (left) and RNA6 (right).
Mutant viruses containing separate deletions of SL4a and SL4b are viable.
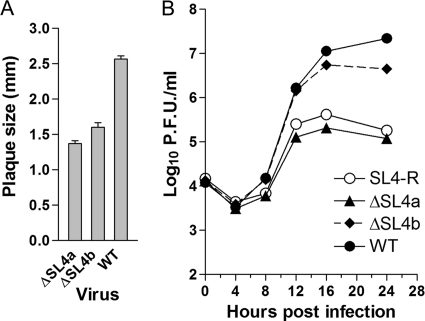
To explore the relative functional importance of SL4a and SL4b for MHV replication, mutant viruses containing separate deletions of either SL4a (nt 75 to 85 and nt 125 to 138) or SL4b (nt 92 to 119) (see Fig. 1) were constructed, and their replication phenotypes were examined. Both viruses were viable but did display impaired virus replication. The average plaque size of the ΔSL4a mutant was 53% of that of the WT, and the plaque size of the ΔSL4b mutant was 62% relative to the WT plaque size (Fig. 5A). Growth curves of these mutants revealed that although both mutants grew with kinetics similar to those of the WT virus, the peak titer achieved by the ΔSL4a mutant (2.05 × 105 PFU/ml) was approximately 100-fold less than that reached by the WT (2.20 × 107 PFU/ml) (Fig. 5B). The titer of the ΔSL4b mutant (5.67 × 106 PFU/ml) was also depressed relative to that of the WT virus, but this approximately 4-fold effect on titer was much less than that observed with the ΔSL4a mutant (Fig. 5B). Sequencing the 5′UTRs of four plaque-purified isolates of each mutant virus at viral passage 1 demonstrated that all contained the desired deletions with no other substitutions in these regions. These results suggest that neither the proposed SL4a nor SL4b is separately absolutely essential for viral replication.
Fig. 5.
Growth phenotypes of ΔSL4a, ΔSL4b, and SL4-R mutant viruses. (A) Average plaque sizes of ΔSL4a and ΔSL4b mutants and wild-type viruses. (B) Growth curves of ΔSL4a, ΔSL4b, and SL4-R mutants and wild-type viruses at an MOI of 1.
Deletion of 131ACA133 gives rise to second-site revertants in the 5′UTR.
To further investigate the functional importance of SL4a in our model, we characterized a mutant virus containing a less extensive 3-nt deletion of 131ACA133, a predicted 3-nt bulge in the middle of SL4a in our model (Fig. 1A) which lies just 3′ to SL4 in the Chen and Olsthoorn model (Fig. 1B). This mutant virus, named SL4-E, was viable as expected. However, viruses containing this bulge deletion were remarkably heterogeneous in phenotype (plaque size) and in some experiments contained second-site mutations in the 5′UTR in addition to the introduced bulge deletion (Table 1). Sequencing of the 5′ and 3′UTRs of eight plaque-purified isolates at passage 1 classifies the eight SL4-E mutant isolates into three groups: two isolates (SL4-E1 and SL4-E2) contained only the desired deletion of the ACA bulge and formed significantly smaller plaques than the WT (51% and 47% of that of the WT, respectively); two had a five-adenosine insertion between SL1 and SL2 in addition to the bulge deletion; the remaining four isolates contained an additional deletion of C at position 80 (ΔC80) just opposite the 131ACA133 bulge deletion in our model (Fig. 1). Interestingly, the six plaque-purified isolates harboring second-site mutations formed plaques that were only slightly smaller than the WT (from 82% to 93% of that of the WT) (Table 1) and were significantly larger than those formed by SL4-E1 and SL4-E2 mutant viruses containing no second-site mutations. There were no substitutions in the 3′UTR for all 8 plaque isolates. We then serially passaged SL4-E1 and -E2 in order to determine if viruses containing only the bulge deletion were genetically unstable, thus resulting in mutations with a compensatory adaptation taking over the culture. SL4-E1 serial passages from P1 to P5 were found to maintain the bulge deletion without additional mutations in their 5′UTRs, but the plaque sizes became significantly larger during serial passage (Table 1; Fig. 6A). In contrast, SL4-E2 serial passages, starting with P3, acquired a UCUAA insertion between SL2 and SL4 (preceding U60 or A70) while maintaining the bulge deletion (Table 1). There were no substitutions in the 3′UTRs for these two mutant viruses at any passage level.
Table 1.
Relative plaque sizes of 131ACA133 deletion-related mutantsa
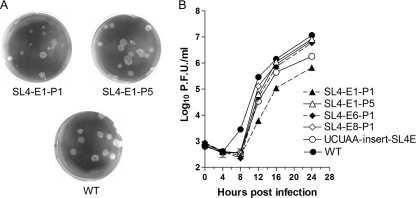
| Virus | Intended effect of mutation on SL4 | Viable | Sequencing of 5′UTR | Plaque size ± SD (mm) | Relative plaque sizeb |
|---|---|---|---|---|---|
| SL4-E1-P1 | Deletion of 131ACA133 | Yes | Deletion of 131ACA133 | 1.55 ± 0.11 | 0.51* |
| SL4-E1-P5 | Deletion of 131ACA133 | Yes | Deletion of 131ACA133 | 2.67 ± 0.10 | 0.88 |
| SL4-E2-P1 | Deletion of 131ACA133 | Yes | Deletion of 131ACA133 | 1.43 ± 0.07 | 0.47* |
| SL4-E6-P1 | Deletion of 131ACA133 | Yes | Deletion of 131ACA133 and insertion of AAAAA | 2.50 ± 0.10 | 0.82 |
| 5Ainsert-SL4E | Deletion of 131ACA133 and insertion of AAAAA | Yes | Deletion of 131ACA133 and insertion of AAAAA | 2.66 ± 0.09 | 0.88 |
| SL4-E8-P1 | Deletion of 131ACA133 | Yes | Deletion of 131ACA133 and deletion of C80 | 2.83 ± 0.08 | 0.93 |
| ΔC-SL4E | Deletion of 131ACA133 and deletion of C80 | Yes | Deletion of 131ACA133 and deletion of C80 | 2.98 ± 0.08 | 0.98 |
| SL4-E2-P3 | Deletion of 131ACA133 | Yes | Deletion of 131ACA133 and insertion of UCUAA | 1.83 ± 0.07 | 0.60* |
| UCUAAinsert-SL4E | Deletion of 131ACA133 and insertion of UCUAA | Yes | Deletion of 131ACA133 and insertion of UCUAA | 1.94 ± 0.10 | 0.64* |
| ACA-UGU | 131ACA133 change to 131UGU133 | Yes | 131ACA133 change to 131UGU133 | 2.90 ± 0.06 | 0.96 |
| WT | Yes | 3.03 ± 0.07 | 1.00 |
This table lists only prototypical plaque isolates for each genotype.
Asterisks represent statistical significance (P < 0.001).
Fig. 6.
Growth phenotypes of 131ACA133 deletion SL4-E mutant and related viruses. (A) Plaque morphologies of SL4-E1-P1 and SL4-E1-P5 mutants and wild-type viruses. (B) Growth curves of the representative mutants and wild-type viruses at an MOI of 0.1.
Since we obtained a diverse collection of second-site mutations in the 5′UTR upon deletion of 131ACA133, we performed two additional experiments starting from the assembly of the full-length cDNA in order to determine whether the diversity of these recovered mutant viruses was truly caused by the deletion. Eight plaque-purified isolates were recovered in each experiment, and their 5′ and 3′UTRs were sequenced. Surprisingly, all 16 plaque-purified virus isolates were found to contain only the desired 131ACA133 deletion with no other substitutions detected in the 5′ UTR, and all were characterized by plaque sizes very similar to those observed for the SL4-E1 and SL4-E2 viruses. Recombinant viruses (UCUAA insert-SL4E, 5A insert-SL4E, ΔC80-SL4E) were separately generated by constructing genomes containing these second-site mutations in addition to the bulge deletion and confirmed by sequencing. As anticipated, their plaque sizes and shapes were very similar to those of the originally recovered corresponding mutant viruses (Table 1). Moreover, one additional mutant virus was generated by changing the sequence of the bulge from 131ACA133 to UGU. Sequencing of four plaque-purified isolates of this mutant confirmed the presence of the desired UGU mutations without any additional changes in their 5′ and 3′UTR sequences. The plaque sizes of these mutants were almost as large as the WT plaques (Table 1), suggesting that any requirement for the predicted ACA bulge is not likely to be sequence specific.
Since the titer of SL4-E1 from passage 1 was approximately 1,000-fold lower than that of the WT virus, 7 × 105 PFU/ml (WT, 8.8 × 108 PFU/ml), we performed growth curves at an MOI of 0.1 with mutant isolates representing the four sequence classes of SL4-E viruses that we recovered and with serial passage 5 of the SL4-E1 virus (SL4-E1-P5) (Fig. 6B). The growth curves demonstrate that all of the representative mutants had delayed growth kinetics compared to those of the WT, lagging by 4 h. The mutants containing second-site mutations generally made up for the 4-h lag with somewhat more rapid growth from 8 to 16 hpi. The SL4-E6-P1 (contains an AAAAA insertion between SL1 and SL2 in addition to the ACA bulge deletion), SL4-E8-P1 (contains a second-site ΔC80 mutation in addition to the ACA bulge deletion), and SL4-E1-P5 viruses grew sufficiently well in this time period to reach near-WT titers (Fig. 6B).
SL4 can be replaced by a shorter sequence-unrelated stem-loop.
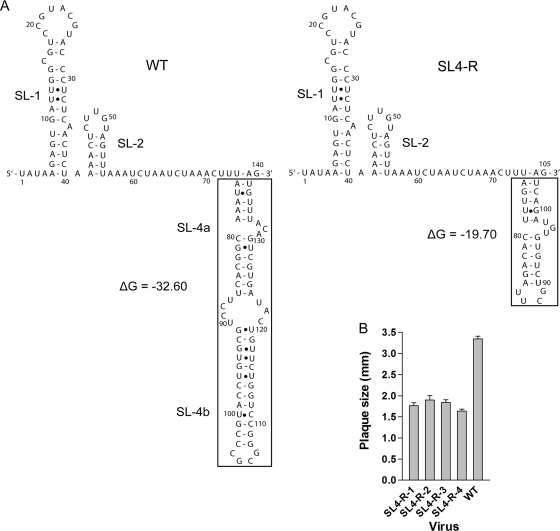
Since the viruses containing mutations designed to systematically disrupt the structure and sequence of SL4 were all viable with relatively modest effects on virus phenotype, but deletion of all of SL4 was lethal, we further explored whether the requirement for SL4 for MHV replication was based on the overall sequences or the structure of SL4. Thus, we constructed one additional mutant virus, SL4-R, which replaces SL4 with a shorter sequence-unrelated helical stem capped by a stable UUCG tetraloop (Fig. 7A). In addition, we replaced 131ACA133 with UGU. Remarkably, the SL4-R mutant virus was viable. Sequencing of the 5′ and 3′UTRs of four plaque-purified isolates revealed that all four isolates contained the intended mutation without other sequence alterations in their 5′UTRs but all exhibited plaque sizes about 50 to 60% that of the WT virus (Fig. 7B). Two contained WT 3′UTRs, while two contained C→A substitutions at position 31226 (SL4-R-2) or 31148 (SL4-R-4). The SL4-R2 and -R4 isolates were not studied further since their plaque sizes were not significantly different from the SL4-R1 and -R3 isolates with WT 3′UTR sequences. We next compared the growth kinetics of the SL4-R mutant to those of the ΔSL4a and ΔSL4b mutants and the WT virus (Fig. 5B). The growth curve of the SL4-R mutant was almost identical to that of the ΔSL4a mutant but did not replicate nearly as well as the WT virus and the ΔSL4b mutant.
Fig. 7.
Secondary structure and average plaque size of the SL4-R mutant. (A, left) The predicted secondary structures of the WT 5′ 140 nt (positions 1 through 140) including SL1, SL2, and SL4 in our model (14, 24). ΔG = −32.60 kcal/mol. (A, right) The secondary structures of SL4-R mutant (positions 1 through 105) predicted by Mfold 3.0, including SL1, SL2, and SL4, replaced by a shorter sequence-unrelated stem-loop. ΔG = −19.70 kcal/mol. The structures in the boxes represent the corresponding replacement. (B) Average plaque sizes of the four plaque-purified isolates of the SL4-R mutant.
DISCUSSION
The MHV RNA genome 5′UTR and 3′UTR contain cis-acting sequences, folding into secondary and higher-order structures, important for RNA-RNA interactions, for binding of viral and/or cellular proteins during RNA replication and translation (2, 22, 25). Therefore, mapping of RNA structures is important to understand RNA function, and such information is likely to assist understanding other closely related CoVs, such as the SARS-CoV. In studies of the MHV 5′UTR, our group has previously reported that SL1 adopts a bipartite structure that drives a 5′UTR-3′UTR interaction and that SL2 folds into a tetraloop structure that is required for subgenomic RNA synthesis (21, 23, 24). In the current study, we have performed an extensive mutational and functional analysis of SL4, a bulged stem-loop positioned just 3′ to the leader TRS and the first proposed structural RNA element of the 5′UTR following the leader. Our mutational analysis of SL4, particularly the replacement of SL4 with a sequence-unrelated stem-loop (mutant SL4-R), supports the hypothesis that SL4 functions in part as a “spacer element,” and this spacer function plays an important role in directing subgenomic RNA synthesis during virus replication.
SL4 in MHV-A59 is predicted to contain a bipartite stem-loop (SL4a and SL4b) separated by a bulge (14, 24). Our results, based on the comprehensive mutational and functional analyses of SL4a, are consistent with the Chen and Olsthoorn phylogenetic-based model (5), leaving nt 74 to 79 and 131 to 139 unpaired (Fig. 1A and B). However, our results with the four SL4b mutant viruses indicate that for SL4b, neither the structure, the sequence, nor the 8-amino-acid small ORF has a critical role in viral replication. This is contrary to the finding in BCoV DI RNA replication assays that these elements are necessary to continually passage BCoV DI RNA (28, 29). There are three possible explanations for the conflicting results from DI assays and assays done with similar mutants in the context of a complete viral genome. (i) By their very nature, DI replication assays are competition assays with helper wild-type virus and recombinant WT DI RNAs that arise during the experiment. Thus, the DI experiments may have detected subtle decreases in relative fitness in the BCoV DI RNA replication that are not detected in straightforward viral replication assays that focus on recovering viable viruses. (ii) The functional role of SL4b differs in MHV and BCoV and/or between DI replication assays and virus replication. (iii) The full-length RNA genome generates compensatory adaptive mutations outside the 5′UTR and 3′UTR of the genome during virus recovery and propagation, since we only sequenced the 5′UTR and adjacent nucleotides (nt 1 to 250) and 3′UTR (324 nt), whereas the entire genome is 31,357 nt. Currently, we favor the first interpretation as the most likely explanation for the discordant results, although we cannot formally exclude the third possibility. The competitive nature of the DI RNA assays resembles in some respects competition assays between wild-type and mutant viruses. We have previously found similar discordant results with MHV DI RNAs and recombinant viruses containing identical mutations in their 3′UTRs (13).
Our data demonstrate that deletion of the entire MHV 5′UTR SL4 is lethal, and the genome carrying this deletion is defective in directing subgenomic RNA synthesis. One possible explanation for this result is that SL4, as one of the cis-acting elements in the 5′UTR, could facilitate binding to viral and/or cellular factors leading to the formation of a 5′UTR and 3′UTR complex through protein-RNA and protein-protein interaction, bringing TRS-L in close proximity to the 3′ end of the genomic RNA, as described in the negative-strand discontinuous transcription model defined by Zuniga et al. (38). Subsequently, template switching occurs through the base pairing between TRS-L and the nascent individually complementary TRS-Bs. Pyrimidine tract-binding protein (PTB) has been reported to bind to the pentanucleotide repeat UCUAA in the positive-strand MHV RNA leader TRS, and heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) binds to the negative-strand leader and the complement of the TRS-B sequences (6, 19, 20). MHV N protein is also known to bind with high affinity and specificity to the TRS-L and possesses helix unwinding properties that suggest a role in template switching (1, 10). Another hnRNP, synaptotagmin-binding cytoplasmic RNA-interacting protein (SYNCRIP), binds to the MHV 5′UTR and to the complement of the 5′UTR (7). The BCoV nsp1-encoded protein has been determined to bind to three cis-acting stem-loops in the 5′UTR, including SLIII, which corresponds to SL4b in our model, and to regulate viral RNA translation and replication (3, 12). It is likely that nps1-encoded protein of MHV has a similar function.
Another possibility is that deletion of SL4 changes the spacing between crucial cis-acting structures, SL1, and SL2 and, as a result, the disposition of TRS-L sequences 5′ to SL4 and cis-acting elements downstream of SL4. Additionally, this mutation deletes all of the sequence 3′ to the core TRS that is within the BCoV 65-nt-wide 5′-proximal genomic acceptor hot spot for discontinuous transcription demonstrated by Wu et al. (36), possibly affecting subgenomic RNA transcription. These two explanations are not mutually exclusive, and both effects of the deletion might contribute to this mutant's failure to replicate.
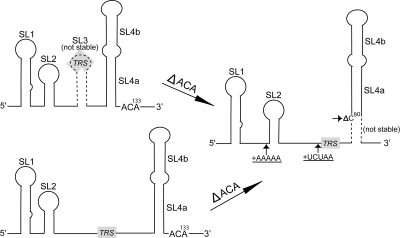
As depicted schematically in Fig. 8 by the dotted lines, both SL3 and the proximal portion of SL4a in our original model are likely to be unstable, and our mutational results are most consistent with a model in which the predominant structure is an extended single-stranded region (nt 57 to 79) between SL2 and SL4. Our mutational analysis demonstrated that deletion of 131ACA133 has a profound impact on viral replication (Table 1). Viruses carrying this deletion were heterogeneous in phenotype (plaque size) and were not genetically stable, with isolation of second-site revertants in a number of places in the 5′UTR, which compensates the decreased plaque sizes, delayed growth kinetics, and lower peak titer associated with 131ACA133 deletion. Deletion of these three nucleotides would favor a more extended SL4a conformation by increasing the stability of the potential SL4a helix originally predicted. This extension of the SL4a helix would change the nucleotide spacing between SL4 and the 5′ stem-loop structures, such as SL1 and SL2, and any 3′ cis-acting structures (3, 5, 11, 12, 29) by converting two single-stranded regions to a helix (Fig. 8). The second-site mutations within the 5′UTR are predicted to change either the spacing between SL1 and SL2 (insertion of five adenosines) or the spacing between SL2 and SL4 (insertion of UCUAA) or to destabilize the proximal portion of the extended SL4a by deletion of C at position 80, creating a 1-nt bulge, thus decreasing the stability of the helix in this region and likely restoring a structure similar to that of the wild type. Together, these suggest that the distance between SL4, or elements 3′ to SL4, and SL1 and/or SL2 is important for viral replication. Interestingly, Makino and Lai showed that viruses with more than two UCUAA repeats (the number of repeats present in wild-type MHV-A59) evolve the number of repeats downward with serial passage (26). This is consistent with the idea that there is an optimal spacing between SL4 and SL1. We also isolated viruses that did not have second-site mutations in the 5′UTR but were characterized by increased plaque size and growth curves upon subsequent serial passage; this suggests the presence of a second-site mutation(s) elsewhere in the genome outside the 5′UTR and 3′UTR, which reverses the functional deficits associated with the 131ACA133 deletion. We are in the process of sequencing the genomes of the SL4-E mutant viruses (with and without serial passages) to search for second-site mutations outside the 5′UTR and 3′UTR, in an effort to determine the structural origin of these effects.
Fig. 8.
A schematic cartoon representation of second-site mutations that result upon passage of MHV harboring 131ACA133 deletion and their effect on spacing of 5′UTR stem-loop structures. Three possible conformations are shown. Our data are most compatible with the possibility of the lower extended single-stranded region being the predominant conformation for the wild-type sequence. The approximate positions of second-site insertions (+) and the deletion (small Δ) that occur after deletion of 131ACA133 are indicated by small arrows. Inserted nucleotides are underlined. Deletion of 131ACA133 potentially stabilizes and extends SL4a. Note that SL3 and the extended SL4a are both predicted to be unstable at 37°C (indicated by broken lines) and are mutually exclusive structures. The extension of SL4a alters the spacing between SL4 and other stem-loop structures (SL1, SL2) in the 5′UTR. The position of the 5′ UCUAAAC core TRS-L is indicated with shading in the figure.
The MHV genome can tolerate the deletion of either SL4a or SL4b and yield viable viruses. Replacing SL4 with a sequence-unrelated shorter stem-loop structure (SL4-R) permits recovery of infectious recombinant virus, providing a further indication that the sequence of SL4 is relatively less important for virus replication than the spacing between SL4 and other 5′UTR stem-loops. It should be noted that care was taken in constructing SL4-R to approximate the low stability of the wild-type SL4a structure in our original model with the extended SL4a region, making it likely that the base pairing of the most proximal 6 bp of the SL4-R stem (nt 74 to 79/99 to 104; Fig. 7A) will be unstable, as we think to be the case for the wild-type SL4.
Taking the above data as a whole, we draw the conclusion that SL4 functions as a structural spacer element that properly orients SL1, SL2, TRS, and/or more distal structural elements of the 5′UTR. A schematic model that graphically illustrates this conclusion is shown in Fig. 8. The current data are consistent with our previous results in terms of SL1 and SL2 (21, 23, 24). These cis-acting elements provide the structural motifs that are crucial for negative-strand subgenomic RNA syntheses but are not required for full-length genomic negative-strand synthesis. Future work directed at further determining the cis-acting structural elements contained in the MHV 5′UTR and the 5′ end of the nsp1 coding sequences will enhance our understanding of MHV replication. We are also in an effort to investigate the interactions of these elements with viral and/or cellular proteins to further elucidate their function. Such information will assist understanding the common replication signals in other closely related CoVs, such as BCoV and SARS-CoV.
ACKNOWLEDGMENTS
We gratefully acknowledge support from U.S. National Institutes of Health grants AI067416 and AI051493.
Footnotes
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Baric R. S., et al. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 62:4280–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brian D. A., Baric R. S. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown C. G., Nixon K. S., Senanayake S. D., Brian D. A. 2007. An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J. Virol. 81:7716–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Budzilowicz C. J., Wilczynski S. P., Weiss S. R. 1985. Three intergenic regions of coronavirus mouse hepatitis virus strain A59 genome RNA contain a common nucleotide sequence that is homologous to the 3′ end of the viral mRNA leader sequence. J. Virol. 53:834–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen S. C., Olsthoorn R. C. 2010. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology 401:29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi K. S., Huang P., Lai M. M. 2002. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology 303:58–68 [DOI] [PubMed] [Google Scholar]
- 7. Choi K. S., Mizutani A., Lai M. M. 2004. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J. Virol. 78:13153–13162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dveksler G. S., et al. 1991. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 65:6881–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorbalenya A. E., Enjuanes L., Ziebuhr J., Snijder E. J. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grossoehme N. E., et al. 2009. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 394:544–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guan B. J., Wu H. Y., Brian D. A. 2011. An optimal cis-replication stem-loop 32 IV in the 5′ untranslated region of the mouse coronavirus genome extends 16 nucleotides into open reading frame 1. J. Virol. 85:5593–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gustin K. M., Guan B. J., Dziduszko A., Brian D. A. 2009. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J. Virol. 83:6087–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson R. F., et al. 2005. Effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 79:14570–14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kang H., Feng M., Schroeder M. E., Giedroc D. P., Leibowitz J. L. 2006. Putative cis-acting stem-loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J. Virol. 80:10600–10614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai M. M., Baric R. S., Brayton P. R., Stohlman S. A. 1984. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc. Natl. Acad. Sci. U. S. A. 81:3626–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lai M. M., Patton C. D., Baric R. S., Stohlman S. A. 1983. Presence of leader sequences in the mRNA of mouse hepatitis virus. J. Virol. 46:1027–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee C. W., Li L., Giedroc D. P. 2011. The solution structure of coronaviral stem-loop 2 (SL2) reveals a canonical CUYG tetraloop fold. FEBS Lett. 585:1049–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leibowitz J. L., Wilhelmsen K. C., Bond C. W. 1981. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-a59 and MHV-JHM. Virology 114:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H. P., Huang P., Park S., Lai M. M. 1999. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J. Virol. 73:772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H. P., Zhang X., Duncan R., Comai L., Lai M. M. 1997. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci. U. S. A. 94:9544–9549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L., et al. 2008. Structural lability in stem-loop 1 drives a 5′ UTR-3′ UTR interaction in coronavirus replication. J. Mol. Biol. 377:790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu P., Leibowitz J. L. 2010. RNA higher-order structures within the coronavirus 5′ and 3′ untranslated regions and their roles in viral replication, p. 47–61 In Lal S. K. (ed.), Molecular biology of the SARS-coronavirus. Springer, New York, NY [Google Scholar]
- 23. Liu P., et al. 2009. Mouse hepatitis virus stem-loop 2 adopts a uYNMG(U)a-like tetraloop structure that is highly functionally tolerant of base substitutions. J. Virol. 83:12084–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu P., et al. 2007. A U-turn motif-containing stem-loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA 13:763–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y., Wimmer E., Paul A. V. 2009. cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim. Biophys. Acta 1789:495–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Makino S., Lai M. M. 1989. Evolution of the 5′-end of genomic RNA of murine coronaviruses during passages in vitro. Virology 169:227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pasternak A. O., van den Born E., Spaan W. J., Snijder E. J. 2003. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 77:1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raman S., Bouma P., Williams G. D., Brian D. A. 2003. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 77:6720–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raman S., Brian D. A. 2005. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 79:12434–12446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sawicki S. G., Sawicki D. L. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sawicki S. G., Sawicki D. L. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215–219 [DOI] [PubMed] [Google Scholar]
- 32. Sola I., Moreno J. L., Zuniga S., Alonso S., Enjuanes L. 2005. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 79:2506–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spaan W. J., Rottier P. J., Horzinek M. C., van der Zeijst B. A. 1982. Sequence relationships between the genome and the intracellular RNA species 1, 3, 6, and 7 of mouse hepatitis virus strain A59. J. Virol. 42:432–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Marle G., et al. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. U. S. A. 96:12056–12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weiss S. R., Leibowitz J. L. 2007. Pathogenesis of murine coronavirus infections, p. 259–278 In Perlman S., Gallagher T., Snijder E. J. (ed.), Nidoviruses. ASM Press, Washington, DC [Google Scholar]
- 36. Wu H. Y., Ozdarendeli A., Brian D. A. 2006. Bovine coronavirus 5′-proximal genomic acceptor hot spot for discontinuous transcription is 65 nucleotides wide. J. Virol. 80:2183–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yount B., Denison M. R., Weiss S. R., Baric R. S. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zuniga S., Sola I., Alonso S., Enjuanes L. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980–994 [DOI] [PMC free article] [PubMed] [Google Scholar]