Abstract
PG9 and PG16 are two recently isolated quaternary-specific human monoclonal antibodies that neutralize 70 to 80% of circulating HIV-1 isolates. The crystal structure of PG16 shows that it contains an exceptionally long CDR H3 that forms a unique stable subdomain that towers above the antibody surface to confer fine specificity. To determine whether this unique architecture of CDR H3 itself is sufficient for epitope recognition and neutralization, we cloned CDR H3 subdomains derived from human monoclonal antibodies PG16, PG9, b12, E51, and AVF and genetically linked them to a glycosyl-phosphatidylinositol (GPI) attachment signal. Each fusion gene construct is expressed and targeted to lipid rafts of plasma membranes through a GPI anchor. Moreover, GPI-CDR H3(PG16, PG9, and E51), but not GPI-CDR H3(b12 and AVF), specifically neutralized multiple clades of HIV-1 isolates with a great degree of potency when expressed on the surface of transduced TZM-bl cells. Furthermore, GPI-anchored CDR H3(PG16), but not GPI-anchored CDR H3(AVF), specifically confers resistance to HIV-1 infection when expressed on the surface of transduced human CD4+ T cells. Finally, the CDR H3 mutations (Y100HF, D100IA, and G7) that were previously shown to compromise the neutralization activity of antibody PG16 also abolished the neutralization activity of GPI-CDR H3(PG16). Thus, we conclude that the CDR H3 subdomain of PG16 neutralizes HIV-1 when targeted to the lipid raft of the plasma membrane of HIV-1-susceptible cells and that GPI-CDR H3 can be an alternative approach for determining whether the CDR H3 of certain antibodies alone can exert epitope recognition and neutralization.
INTRODUCTION
During human immunodeficiency virus type 1 (HIV-1) infection, a proportion of individuals develop broadly neutralizing sera over time (32). From a few such individuals, a number of potent and broadly cross-neutralizing monoclonal antibodies (MAbs) have also been isolated (36, 38, 40). Among them, PG9 and PG16 are recently isolated quaternary-specific neutralizing MAbs from a subtype A HIV-1-infected individual in Africa that neutralize 70 to 80% of circulating HIV-1 isolates (36). PG9 and PG16 bind to overlapping, but distinct, gp120 epitopes composed of conserved elements from the second and third variable regions (V2 and V3, respectively). The quaternary epitopes are glycosylated (6) and are preferentially displayed on envelope trimers on the surface of virions and transfected cells but not on recombinant monomeric gp120 or soluble trimers (36).
To gain insight into the molecular features of antibody binding and neutralizing activities, Pancera et al. (23) and Pejchal et al. (24) recently determined the crystal structures of the Fab fragment of PG16. Antibodies PG9 and PG16 were found to be sulfated (24). The fine specificity of the antibodies is conferred by an exceptionally long third-heavy-chain complementarity-determining region (CDR H3) that forms a unique stable subdomain towering above the antibody surface (23, 24).
The lipid raft is a specialized dynamic microdomain of the plasma membrane that is rich in cholesterol, sphingolipids, and glycerophospholipids (31). The lipid raft has been shown to be a gateway for HIV-1 budding (4, 17) as well as for HIV-1 entry into T cells and macrophages (2, 26, 27). Interestingly, CD4, the receptor for HIV-1 entry, was found to be located in the lipid raft of the plasma membrane (14, 25). Previously, we showed that by genetically linking single-chain Fv (scFv) of human anti-HIV-1 envelope antibodies with a glycosyl-phosphatidylinositol (GPI) attachment signal derived from decay-accelerating factor (DAF) (18), scFvs are targeted into the lipid raft of the plasma membrane. GPI-anchored scFvs (X5, 48d, and 4E10) exhibit greater neutralization against diverse HIV-1 strains than do their soluble counterparts (37).
Therefore, the exceptionally long and unique structure of the CDR H3 subdomain of PG16 led us to postulate that the CDR H3 subdomain itself may bind to the epitope of gp120 and that the targeting of this subdomain to the lipid raft of the plasma membrane of HIV-1-susceptible cells could neutralize HIV-1 infection efficiently.
To test this hypothesis, we constructed CDR H3 derived from five human monoclonal antibodies, PG16, PG9, b12, E51, and AVF. Antibody AVF recognizes the influenza virus hemagglutinin, which is used here as a negative control (33). Antibody b12 is a well-known broadly neutralizing antibody with a protruding, fingerlike, long CDR H3 that penetrates the recessed CD4 binding site of gp120 (1, 29, 41). In addition, a Tyr residue in the CDR H2 loop and a number of Arg residues in CDR L1 are also important for b12 binding (42). Nevertheless, a soluble b12 CDR H3 peptide exhibits relatively weak neutralization (42). Antibody E51 is another sulfated antibody that recognizes the CCR5 binding site of gp120 (39). A sulfated peptide derived from CDR H3 of E51 binds gp120 and inhibits HIV-1 infection (7). In addition, we constructed three CDR H3 mutants (Y100HF, D100IA, and G7) of PG16. These CDR H3 mutants were previously shown to compromise the neutralization activity of antibody PG16 (24). Here, we report that by genetically linking the CDR H3 of PG16, PG9, AVF, b12, and E51 and the CDR H3 mutants of PG16 with a GPI attachment signal of DAF, CDR H3 and the CDR H3 mutants are targeted to lipid rafts of plasma membranes through a GPI anchor. Moreover, GPI-CDR H3 derived from PG16, PG9, and E51 [GPI-CDR H3(PG16, PG9, and E51)], but not GPI-CDR H3(AVF and b12), neutralizes multiple clades of HIV-1 isolates with a high degree of potency. Furthermore, the transduction of human CD4+ T cells with GPI-CDR H3(PG16), but not GPI-CDR H3(AVF), confers resistance to HIV-1 infection. Finally, the CDR H3 mutations (Y100HF, D100IA, and G7) that were previously shown to compromise the neutralization activity of antibody PG16 also abolished the neutralization activity of GPI-CDR H3(PG16). Thus, we conclude that the CDR H3 subdomain of PG16, when targeted to the lipid raft of the plasma membrane of HIV-1-susceptible cells, efficiently neutralizes HIV-1 infection.
MATERIALS AND METHODS
Gene construction and cells.
Fusion gene fragments encoding the CDR H3 subdomain of antibodies PG16, PG9, E51, b12, and AVF and the CDR H3 mutants (Y100HF, D100IA, and G7) of PG16 along with an IgG3 hinge, a His tag, and a GPI attachment signal were generated by overlapping PCR, ligated into the TA vector system (Invitrogen Life Technologies, San Diego, CA), and sequenced as described previously (28). The gene fragments with the correct sequences were religated between the BamHI and SalI sites of a third-generation lentiviral transfer vector, pRRLsin-18.PPT.hPGK.Wpre (8). The resulting lentiviral transfer constructs were designated pRRL-CDR H3(PG16, PG9, E51, b12, and AVF)/IgG3 hinge/His tag/DAF (Fig. 1A) and pRRL-CDR H3 mutants (Y100HF, D100IA, and G7) of PG16/IgG3 hinge/His tag/DAF (see Fig. 4A), respectively.
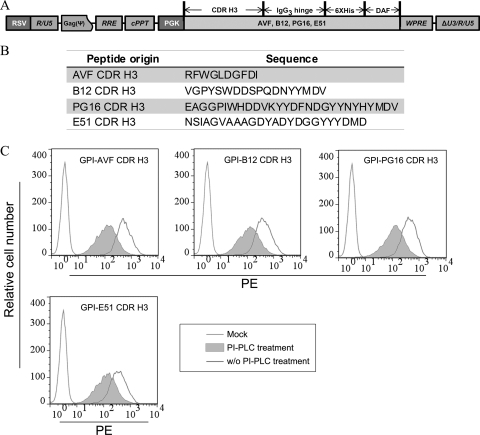
Fig. 1.
Expression of GPI-anchored CDR H3 in transduced TZM-bl cells. (A) Schematic diagram of the lentiviral vectors pRRL-CDR H3/hinge/His tag/DAF. CDR H3s were derived from four human monoclonal antibodies, PG16, b12, E51, and AVF. Hinge, a human IgG3 hinge region; His tag, a six-histidine-residue tag; DAF, the C-terminal 34 amino acid residues of decay-accelerating factor; RSV, Roys sarcoma virus; PGK, human phosphoglycerate kinase gene promoter; RRE, Rev-responsive element; cPPT, central polypurine tract and termination sequence. (B) List of CDR H3 peptide sequences of PG16, b12, E51, and AVF. (C) FACS analysis of cell surface expression of CDR H3/hinge/His tag/DAF in mock- and CDR H3(PG16, b12, E51, and AVF)/hinge/His tag/DAF-transduced TZM-bl cells with or without PI-PLC treatment.
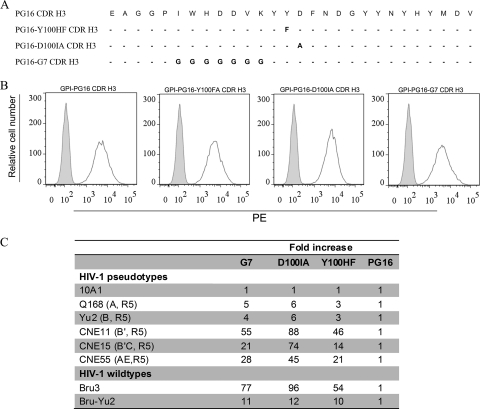
Fig. 4.
Relative susceptibilities to 10A1 and HIV-1 pseudotypes and replication-competent HIV-1 of TZM-bl cells transduced with GPI-CDR H3(PG16) and with GPI-CDR H3 mutants (G7, D100HI, and Y100HF). (A) Amino acid sequence comparison of wild-type CDR H3(PG16) and its mutants. (B) FACS analysis of cell surface expression of TZM-bl cells transduced with GPI-CDR H3(PG16) and with GPI-CDR H3 mutants (G7, D100HI, and Y100HF). (C) Relative susceptibility to pseudotypes and replication-competent HIV-1 of TZM-bl cells transduced with GPI-CDR H3 mutants (G7, D100HI, and Y100HF) compared to cells transduced with GPI-CDR H3(PG16).
The packaging cell line 293T was purchased from Invitrogen Life Technologies and was maintained in complete Dulbecco's modified Eagle's medium (DMEM) (i.e., high-glucose DMEM supplemented with 10% fetal bovine serum [FBS], 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin [100 U/ml], and streptomycin [100 μg/ml]) plus G418 (500 μg/ml) (Invitrogen Life Technologies). The human CD4+ T cell line CEMss-CCR5 was generated as described previously (37). TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP; Germantown, MD), contributed by J. Kappes and X. Wu (16). CEMss-CCR5 and TZM-bl cells were maintained in complete DMEM.
Generation of recombinant lentiviral vectors.
Recombinant lentiviral vectors were generated as described previously (35, 37). Briefly, 4 × 106 293T cells were seeded onto a P-100 dish in 10 ml complete DMEM. After culturing overnight, cells were cotransfected with 20 μg transfer construct (one of the above-mentioned pRRL-CDR H3/IgG3 hinge/His tag/DAF or pRRL-CDR H3 mutants of PG16/IgG3 hinge/His tag/DAF constructs), 10 μg packaging construct encoding HIV-1 Gag/Pol (pLP1), 7.5 μg plasmids encoding the vesicular stomatitis virus G protein (VSV-G) envelope (pLP/VSVG), and 7.5 μg HIV-1 Rev protein (pLP2) (Invitrogen), using a calcium phosphate precipitation method. Sixteen hours later, culture supernatants were removed and replaced with fresh complete DMEM plus 1 mM sodium butyrate (Sigma). Eight hours later, supernatants were again removed and replaced with fresh DMEM plus 4% FBS. After another 20 h, the culture supernatants were harvested and concentrated by ultracentrifugation as described previously (35, 37). The vector pellets were resuspended in a small volume of DMEM and stored in aliquots in a −80°C freezer. Vector titers were determined as previously described (35, 37). The amount of HIV-1 Gag p24 in concentrated vector stocks was determined by an enzyme-linked immunosorbent assay (ELISA) (see below).
Generation of stable cell lines expressing GPI-anchored CDR H3.
To transduce CEMss-CCR5 cells, 1 × 105 CEMss-CCR5 cells and 2 × 106 transducing units (TU) of one of the lentiviral vectors containing pRRL-CDR H3(PG16 and AVF)/IgG3 hinge/His tag/DAF were added to 24-well tissue culture plate in the presence of 8 μg/ml of Polybrene (Sigma). Twenty-four hours later cells were extensively washed and cultured in complete DMEM.
To transduce TZM-bl cells, 5 × 104 TMZ-bl cells per well were seeded onto a 24-well plate. After culturing overnight, 2 × 106 TU of one of the lentiviral vectors containing pRRL-CDR H3(PG16, PG9, E51, b12, and AVF)/IgG3 hinge/His tag/DAF or pRRL-CDR H3 mutants (Y100 HF, D100IA, and G7) of PG16/IgG3 hinge/His tag/DAF was added onto a 24-well tissue culture plate in the presence of 8 μg/ml of Polybrene. Twenty-four hours later cells were extensively washed and cultured in complete DMEM. The expressions of pRRL-CDR H3/IgG3 hinge/His tag/DAF and pRRL-CDR H3 mutant/IgG3 hinge/His tag/DAF constructs were measured by fluorescence-activated cell sorter (FACS) analysis (see below). We usually found that after a single round of transduction, over 98% of cells express transgenes (data not shown). After transduced cells were generated, cells were continuously cultured in complete DMEM and split every 3 or 4 days. Periodically, the expression of transgenes was measured. We found that the level of transgene expression was very stable in the transduced cell lines (data not shown).
FACS analysis.
To study the cell surface expression of pRRL-CDR H3/IgG3 hinge/His tag/DAF and pRRL-CDR H3 mutant/IgG3 hinge/His tag/DAF, 2 × 105 mock-transduced and pRRL-CDR H3(PG16, PG9, E51, b12 and AVF)/IgG3 hinge/His tag/DAF-transduced or pRRL-CDR H3 mutant (Y100 HF, D100IA, and G7) of PG16/IgG3 hinge/His tag/DAF-transduced TZM-bl cells and mock-transduced and pRRL-CDR H3(PG16 and AVF)/IgG3 hinge/His tag/DAF-transduced CEMss-CCR5 cells were incubated with a mouse anti-His tag antibody (Sigma) for 45 min on ice. Cells were then washed twice with FACS buffer (phosphate-buffered saline [PBS] containing 1% bovine serum albumin [BSA] and 0.02% NaN3) and stained with phycoerythrin (PE)-conjugated goat anti-mouse IgG antibody (Sigma) for another 45 min on ice. Cells then were washed twice with FACS buffer and fixed with 1% formaldehyde in 0.5 ml of FACS buffer. FACS analysis was performed with a FACScan instrument (Becton Dickinson, Mountain View, CA).
To determine whether the expression of pRRL-CDR H3/IgG3 hinge/His tag/DAF or pRRL-CDR H3 mutant/IgG3 hinge/His tag/DAF is truly targeted through a GPI anchor, 8 × 105 mock- and pRRL-CDR H3(PG16, E51, b12, and AVF)/IgG3 hinge/His tag/DAF- or pRRL-CDR H3 mutant (Y100HF, D100IA, and G7) of PG16/IgG3 hinge/His tag/DAF-transduced TMZ-bl cells were first incubated with or without 6 U/ml phosphatidylinositol-specific phospholipase C (PI-PLC) (Invitrogen) in 0.5 ml 1× PBS and rocked at 4°C for 20 min. After the incubation, cells were washed twice to remove the remaining PI-PLC and then stained with a mouse anti-His tag antibody as described above.
Generation of pseudotypes of the HIV-1 vector and a single-cycle infectivity assay.
To generate pseudotypes with the HIV-1 vector, 4 × 106 293T packaging cells were cotransfected with 10 μg of an HIV-1–luciferase transfer vector (10) and 1 μg of a DNA plasmid encoding one of several HIV-1 envelopes (Q168, AD8, ADA, Yu2, JRFL, HXBc2, Mj4, consensus C, CNE3, CNE5, CNE6, CNE8, CNE11, CNE15, CNE17, CNE30, CNE50, CNE55, CH091.9, CH110.2, CH114.8, CH119.10, CH120.6, and CH181.12) or control retroviral envelope 10A1 (19) and VSV-G (37) by using a calcium phosphate precipitation method. DNA plasmids encoding HIV-1 envelopes Q168, AD8, ADA-M, JRFL, HXBc2, and Mj4 were obtained from the ARRRP. The DNA plasmid encoding consensus C was a generous gift of B. H. Hahn at the University of Alabama. Q168 is derived from an R5-tropic clade A virus (22). AD8, Yu2, JRFL, and ADA-M are derived from R5-tropic clade B viruses (12, 13). HXBc2 is derived from an X4-tropic clade B virus (9). Consensus C is an artificial HIV-1 envelope (12). Mj4 is derived from R5-tropic clade C viruses (21). CNE3, CNE5, CNE8, and CNE55 are derived from R5-tropic CRF01_AE viruses. The HIV-1 envelopes CNE6 and CNE11 are derived from a R5-tropic clade B′ viruses. HIV-1 envelopes CNE15, CNE17, CNE30, CNE50, CH091.9, CH110.2, CH114.8, CH119.10, CH120.6, and CH181.12 are derived from CRF07_B′C viruses (30). The pseudotype-containing supernatants were harvested and stored in aliquots in a freezer at −80°C. The amount of HIV-1 p24 in collected supernatants was measured by an ELISA.
In a single-cycle assay to measure the infectivity of pseudotypes, 10,000 mock-, pRRL-CDR H3/IgG3 hinge/His tag/DAF-, or pRRL-CDR H3 mutant/IgG3 hinge/His tag/DAF-transduced TZM-bl or CEMss-CCR5 cells were transduced with HIV-1, 10A1, or VSV-G pseudotype-containing supernatants equivalent to a relative luciferase activity (RLA) of 179,000 to 1,800,000 overnight. Cells were then washed twice with PBS and cultured in complete DMEM for 2 days. Cells were then washed once with PBS and lysed in 100 μl of lysis buffer. The luciferase activity in 50-μl cell suspensions was measured by a BrightGlo luciferase assay according to the manufacturer's instructions (Promega).
SIV and HIV-1 infections and luciferase and p24 assays.
Mock-, pRRL-CDR H3/IgG3 hinge/His tag/DAF-, or pRRL-CDR H3 mutant/IgG3 hinge/His tag/DAF-transduced TZM-bl cells at 5,000 cells per well were seeded onto a 96-well plate. After culturing overnight, cells were infected with HIV-1 strains Bru-3, Bru-Yu2, and AD8 (34) as well as a simian immunodeficiency virus (SIV) strain SIVMne027 as a control (11) at a multiplicity of infection (MOI) of 2 in a final volume of 0.2 ml overnight. Cells were then washed three times with Hanks' balanced salt solution (HBSS), cultured in 0.2 ml of complete DMEM, and incubated at 37°C for 2 days. The infectivity of HIV-1 was determined by a BrightGlo luciferase assay (see above).
To test the effect of GPI-CDR H3(PG16) on HIV-1 infection and replication in human CD4+ T cells, 1 × 106 CEMss-CCR5 cells transduced with GPI-CDR3(PG16 and AVF) were infected with HIV-1 strains Bru-3 and Bru-Yu2 at an MOI of 0.01 in a final volume of 0.5 ml overnight. Cells were then extensively washed with HBSS, resuspended in 6 ml of complete DMEM, and cultured for 15 days. Every 3 days, 4.5 ml of cell suspensions was harvested and replaced with fresh medium. The supernatants were then collected. HIV-1 p24 in the supernatants was measured by an ELISA (Beckman Coulter) according to the manufacturer's instructions.
Immunofluorescent staining and confocal analysis.
Parental and TZM-bl-GPI-CDR3(PG16 and AVF) cells were seeded (5,000 cells per well) onto a tissue culture-treated glass slide (BD Biosciences) and incubated at 37°C in 5% CO2 for 2 days. Cells were then washed twice with 500 μl PBS and fixed by fixation buffer (4% formaldehyde in PBS plus 1% BSA) for 15 min. Cells were washed twice with 500 μl PBS and blocked with blockage buffer (5% goat serum in PBS plus 1% BSA) for 1 h. Cells were stained with Alexa 555-conjugated cholera toxin subunit B (CtxB) (Invitrogen Life Technologies) at 4°C for 45 min. After being washed 3 times with PBS, cells were stained with mouse anti-His tag antibody (Sigma) at 4°C for 45 min and then stained with Alexa 488-conjugated goat anti-mouse IgG antibody (Invitrogen) at 4°C. After cells were washed 3 times with PBS, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) in permeabilization buffer (blockage buffer plus 0.5% saponin) for 5 min. The slides were mounted before being analyzed under a confocal fluorescence microscope (Zeiss model LSM 510).
RESULTS
Expression of the CDR H3 subdomain in the lipid raft of the plasma membrane through a GPI anchor.
To generate GPI-anchored CDR H3, the sequences encoding CDR H3 derived from four different human antibodies, PG16, b12, E51, and AVF, were genetically linked with the sequence encoding a His-tagged IgG3 hinge region and the sequence encoding a GPI attachment signal of DAF (18). The CDR H3/IgG3 hinge/His tag/DAF (PG16, b12, E51, and AVF) fusion genes were inserted into a third-generation lentiviral vector, pRRL (Fig. 1A). The peptide sequences of human antibodies PG16, b12, E51, and AVF CDR H3 are shown in Fig. 1B. The recombinant viruses were then generated as described previously (35) and used to transduce TZM-bl cells and human CEMss-CCR5 CD4+ T cells (see below).
To determine if CDR H3/hinge/His tag/DAF was expressed on the cell surface through a GPI anchor, four TZM-bl cells—one transduced with lentiviral vector expressing CDR H3(PG16)/hinge/His tag/DAF, one transduced with lentiviral vector expressing CDR H3(b12)/hinge/His tag/DAF, one transduced with lentiviral vector expressing CDR H3(E51)/hinge/His tag/DAF, and one transduced with lentiviral vector expressing CDR H3(AVF)/hinge/His tag/DAF—were treated with or without phosphatidylinositol-specific phospholipase C (PI-PLC) and stained with an anti-His tag antibody, followed by FACS analysis. Figure 1C shows that all 4 CDR H3/hinge/His tag/DAFs were highly expressed on the surface of the cells, and their expression was substantially reduced with PI-PLC treatment, indicating that the attachment of CDR H3/hinge/His tag/DAF to the cell surface is indeed through a GPI anchor. Thus, for the sake of simplicity, in the remaining text we will refer to CDR H3/hinge/His tag/DAF as GPI-CDR H3.
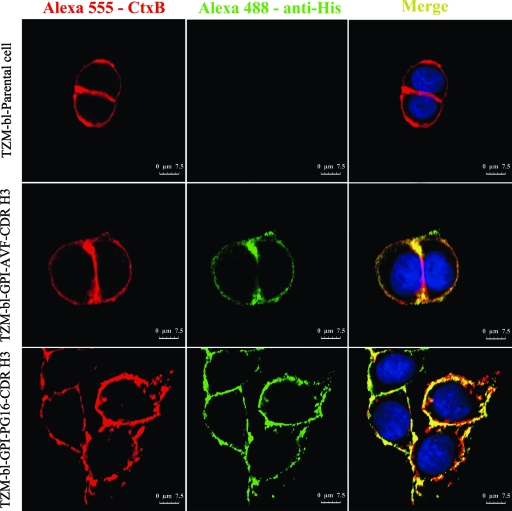
To determine if GPI-CDR H3 is located in the lipid rafts of plasma membranes, mock- and GPI-CDR H3(PG16 and AVF)-transduced TZM-bl cells were cultured overnight and costained with (i) mouse anti-His tag antibody followed by Alexa 488-conjugated goat anti-mouse IgG antibody, (ii) Alexa 555-conjugated cholera toxin subunit B (CtxB), and (iii) DAPI. CtxB interacts with GM1 (a lipid raft marker) on the cell surface. Figure 2 shows that both GPI-CDR H3(PG16) and GPI-CDR H3(AVF) are colocalized with GM1 on the cell surface, implying that they are located in the lipid raft of the plasma membrane.
Fig. 2.
Localization of GPI-anchored CDR H3 in transduced TZM-bl cells. Shown are data for confocal analysis of mock- or GPI-CDR H3(PG16 and AVF)-transduced TZM-bl cells. Cells were stained with Alexa 555-conjugated cholera toxin B subunit (CtxB), and cells were stained with a mouse anti-His tag antibody followed by Alexa 488-conjugated goat anti-mouse IgG antibody.
GPI-CDR H3(PG16) exhibits a remarkable degree of breadth and potency against HIV-1.
Next, we tested the neutralization activity of GPI-CDR H3 against HIV-1. Because the ability of GPI-CDR H3 to neutralize virus could be impacted by differences in the level of expression of CD4, CXCR4, or CCR5 on the surface of transduced TZM-bl cells, we first assessed the expression levels of these receptors in mock-transduced parental TZM-bl cells as well as GPI-CDR H3-transduced TZM-bl cells. In all cases, the values were similar (see Fig. S1 in the supplemental material), suggesting that the expression of the GPI-CDR H3 transgenes does not alter the expression of the receptor and the coreceptors for HIV-1 in transduced cells. In addition, the expression of the transgenes did not alter cell growth (data not shown).
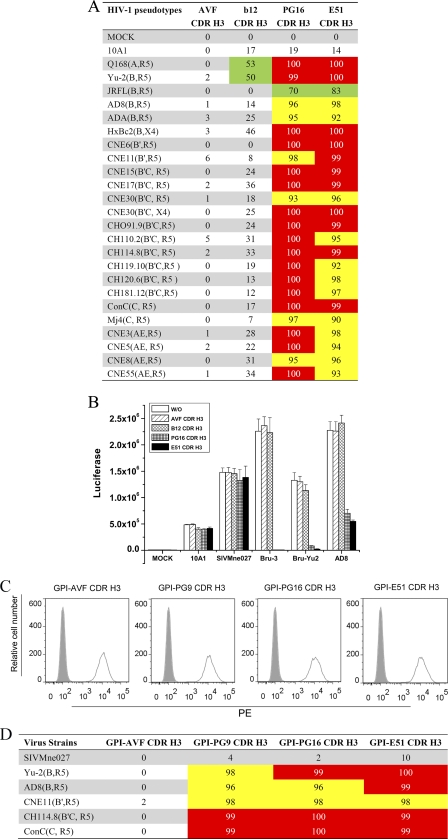
To test the neutralization activity of GPI-CDR H3 against HIV-1, a panel of 24 virions pseudotyped with envelopes representing different HIV-1 clades or a control retroviral envelope, 10A1, was used to infect GPI-CDR H3(PG16, b12, E51, and AVF)-transduced TZM-bl cells in a single-round infection experiment (37). The retroviral envelope 10A1 recognizes either Ram-1 or Glvr-1 as a receptor for cell entry (19) and was used here as negative control. The panel of HIV-1 pseudotypes consists of HIV-1 envelopes derived from clade A (Q168), clade B (ADA-M, AD8, HXBc2, JRFL, and Yu2), clade B′ (CNE6 and CNE11), clade C (Mj4 and consensus C), CRF07_B′C (CNE15, CNE17, CNE30, CNE50, CH091.9, CH110.2, CH114.8, CH119.10, CH120.6, and CH181.12), and CRF01_AE (CNE3, CNE5, CNE8, and CNE55). Figure 3A shows the percentage of the reduction of relative luciferase activity (RLA) in four GPI-CDR H3(PG16, b12, E51, and AVF)-transduced cells infected with these pseudotypes compared to mock-transduced parental cells infected with the same panel of pseudotypes. Compared to mock-transduced parental TZM-bl cells, cells transduced with GPI-CDR H3(AVF, PG16, b12, and E51) did not show any significant neutralization activity against the 10A1 pseudotyped control virus. Furthermore, compared to mock-transduced parental TZM-bl cells, cells transduced with GPI-CDR H3(AVF) did not show any significant neutralization activity against any of the 24 HIV-1 pseudotypes, and cells transduced with GPI-CDR H3(b12) had minimum neutralization activity against 2 (Q168 and Yu2) of the 24 HIV-1 pseudotypes with a low degree of potency. In contrast, cells transduced with both GPI-CDR H3(PG16) and GPI-CDR H3(E51) neutralized all 24 HIV-1 pseudotypes with remarkable degrees of potency. GPI-CDR H3(PG16) reduced the infection of 17 HIV-1 pseudotypes by over 99%, inhibited the infection of the other 6 HIV-1 pseudotypes by over 90%, and reduced the infection of JRFL by 70%. GPI-CDR H3(E51) conferred over 99% inhibition of 11 HIV-1 pseudotypes, over 90% inhibition of the other 12 HIV-1 pseudotypes, and 83% inhibition of JRFL. Both GPI-CDR H3(PG16) and GPI-CDR H3(E51) inhibited HIV-1 pseudotypes expressing envelopes derived from clades A, B, and B′ with similar degrees of potency, whereas for most of the HIV-1 pseudotypes derived from clades B′C, C, and AE, GPI-CDR H3(PG16) inhibited infection better than did GPI-CDR H3(E51).
Fig. 3.
Effect of GPI-CDR H3(PG16, PG9, b12, E51, and AVF) on infection of HIV-1 viruses and pseudotypes. (A) Effect of GPI-CDR H3s on efficiency of transduction of HIV-1 and 10A1 pseudotypes into GPI-CDR H3-transduced TZM-bl cells. Shown are percentages of reduction of the relative luciferase activities in TZM-bl cells transduced with GPI-CDR H3(AVF, b12, PG16, and E51) compared with mock-transduced parental TZM-bl cells. Green, ≥50% inhibition; yellow, ≥90% inhibition; red, ≥99% inhibition. The percentage of inhibition was based on the following calculation: (RLA in virus alone in a given transduced cell − RLA in no virus in the same transduced cell)/(RLA in virus alone in the parental cell − RLA in no virus in the parental cell). (B) Effect of GPI-CDR H3s on wild-type HIV-1 and SIVMne027 infection in GPI-CDR H3-transduced TZM-bl cells. w/o, parental TZM-bl cells. (C) FACS analysis of cell surface expression of CDR H3/hinge/His tag/DAF in CDR H3(PG16, PG9, E51, and AVF)/hinge/His tag/DAF-transduced TZM-bl cells. (D) Percentage of the reduction of relative luciferase activity in TZM-bl cells transduced with GPI-CDR H3(AVF, PG9, PG16, and E51) compared with mock-transduced parental TZM-bl cells. Yellow, ≥90% inhibition; red, ≥99% inhibition.
We next tested the neutralization activity of GPI-CDR H3 against three replication-competent HIV-1 strains (Bru-3, Bru-Yu2, and AD8) as well as a SIVMne027 control. Figure 3B shows means and standard deviations of RLA in parental TZM-bl cells and GPI-CDR H3(AVF, b12, PG16, and E51)-transduced TZM-bl cells infected with these HIV-1 and SIV strains. As expected, compared to mock-transduced parental TZM-bl cells, cells transduced with GPI-CDR H3(AVF, b12, PG16, and E51) did not show any significant neutralization activity against the SIVMne027 control. Compared to mock-transduced parental TZM-bl cells, cells transduced with GPI-CDR H3(AVF and b12) did not show any significant neutralization activity against any of 3 HIV-1 strains. In contrast, similar to what was observed with HIV-1-pseudotyped virions, cells transduced with GPI-CDR H3(PG16 and E51) neutralized all 3 viruses.
Since antibodies PG9 and PG16 are somatic variants and a 7-amino-acid region in CDR H3 is responsible for the fine specificity of these antibodies (24), we also constructed GPI-CDR H3(PG9) and transduced the construct into TZM-bl cells. We then compared the cell surface expressions and anti-HIV-1 activities of GPI-CDR H3(PG9) and GPI-CDR H3(PG16). Figure 3C shows similar degrees of cell surface expression of GPI-CDR H3(PG9) compared with those of the other GPI-CDR H3 constructs (PG16, AVF, and E51). Figure 3D shows that GPI-CDR H3(PG9) exhibited the same degree of inhibition against 5 representative HIV-1 pseudotypes as that of GPI-CDR H3(PG16 and E51). Again, none of these GPI-CDR H3 constructs exhibited any neutralization activity against the SIVMne027 control.
In addition, we synthesized three CDR H3 peptides (AVF, E51, and PG16) and constructed four new lentiviral vectors expressing the secretory form of CDR H3 [CDR H3(AVF), CDR H3(PG16), CDR H3(PG9), and CDR H3(E51)]. We then compared the anti-HIV-1 activities of TZM-bl cells expressing GPI-CDR H3(AVF, PG9, PG16, and E51) and TZM-bl cells expressing the secretory form of CDR H3(AVF, PG9, PG16, and E51). We found that only cells expressing GPI-CDR H3(PG9, PG16, and E51) but not cells expressing the secretory form of CDR H3(PG9, PG16, and E51) exhibited significant inhibition against five representative HIV-1 pseudotypes tested (data not shown). Similarly, soluble AVF, PG16, and E51 CDR H3 peptides at concentrations of 0.1, 1, and 10 μM also did not have any inhibitory effect on HIV-1 (data not shown). At this time, it is not clear whether the lack of an inhibitory effect of secreted CDR H3(PG9 and PG16) is due to a limited amount of CDR H3 peptides present in the culture supernatants and whether the lack of an inhibitory effect of synthetic CDR H3 peptides (E51 and PG16) is due to the lack of sulfation of these synthetic peptides.
Pejchal et al. showed previously that CDR H3 mutants (Y100HF, D100IA, and G7) of antibody PG16 reduced neutralization activity against JR-CSF, resulting in 2.2-, 200-, and >500-fold increases in the 50% inhibitory concentration (IC50) compared to that of wild-type antibody PG16 (24). Therefore, to test whether such CDR H3 mutants would also affect the neutralization activity of GPI-anchored CDR H3 of PG16, we constructed transfer vectors expressing three GPI-CDR H3 mutants (Y100HF, D100IA, and G7) of PG16 (Fig. 4A). Recombinant lentiviruses generated from these transfer vectors were used to transduce TZM-bl cells. Figure 4B shows that the level of cell surface expression of three GPI-CDR H3 mutants is similar to that of GPI-CDR H3(PG16). Figure S2 in the supplemental material shows that the expression of GPI-CDR H3 mutants did not alter the cell surface expressions of CD4, CCR5, and CXCR4. We then compared the susceptibilities of TZM-bl cells transduced with GPI-CDR H3 mutants (Y100 HF, D100IA, and G7) with those of TZM-bl cells transduced with GPI-CDR H3(PG16) by infecting these cells with HIV-1 pseudotypes generated with envelope proteins from Q168 (clade A), Yu-2 (clade B), CNE11 (clade B′), CNE15 (CRF07_B′C), and CNE55 (CRF01_AE) as well as replication-competent HIV-1 Bru-3 and Bru-Yu2. A pseudotyped virus expressing retroviral envelope 10A1 was also used as a negative control. Figure 4C shows the fold increase in RLA in TZM-bl cells transduced with these GPI-CDR H3 mutants compared with GPI-CDR H3(PG16) after the infection. As expected, no significant difference in susceptibility to the 10A1 pseudotype control was found among TZM-bl cells transduced with the wild type and mutants of GPI-CDR H3(PG16). In contrast, TZM-bl cells transduced with GPI-CDR H3 mutants (G7, D100IA, and Y100HF) were significantly more susceptible to all HIV-1 pseudotypes and replication-competent HIV-1 than cells transduced with GPI-CDR H3(PG16). Among the three mutants, TZM-bl cells transduced with GPI-CDR H3 mutant D100IA were more susceptible (i.e., higher fold increases in infection) than cells transduced with GPI-CDR H3 mutant G7 or Y100HF, and cells transduced with GPI-CDR H3 mutant Y100HF was less susceptible (i.e., smaller fold increases) than cells transduced with GPI-CDR H3 mutant G7.
GPI-anchored CDR H3(PG16) renders human CD4+ T cells resistant to HIV-1 but not to transduction of VSV-G-pseudotyped lentivirus.
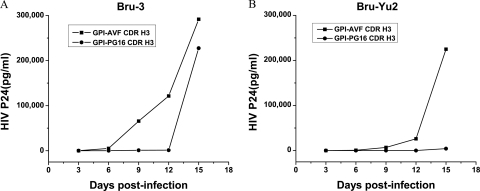
Next, we evaluated whether GPI-CDR H3(PG16) would confer resistance to HIV-1 in human CD4+ T cells. Human CD4+ CEMss-CCR5 cells (37) were transduced with GPI-CDR H3(AVF and PG16). The expression of GPI-CDR H3(AVF and PG16) as well as the expressions of CD4, CCR5, and CXCR4 in transduced CEMss-CCR5 cells were tested by immunostaining followed by FACS analysis as described above. In all cases, the levels of expression of GPI-CDR H3(AVF and PG16) as well as the levels of expression of CD4, CCR5, and CXCR4 between mock-transduced parental CEMss-CCR5 cells and GPI-CDR H3-transduced CEMss-CCR5 cells were similar (see Fig. S3 in the supplemental material). Transduced CEMss-CCR5 cells were then infected with HIV-1 strains Bru-3 and Bru-Yu2 at a multiplicity of infection of 0.01. As shown in Fig. 5A, the replication of HIV-1 Bru-3 was significantly inhibited in cells transduced with GPI-CDR H3(PG16) compared to cells transduced with GPI-CDR H3(AVF). At 6, 9, and 12 days postinfection there was a 2-log reduction in the amounts of HIV-1 Gag p24 produced by cells transduced with GPI-CDR H3(PG16) compared to the amounts produced by cells transduced with GPI-CDR H3(AVF). However, at 15 days postinfection, there was no significant difference in the amounts of HIV-1 Gag p24 produced by cells transduced with GPI-CDR H3(PG16) and cells transduced with GPI-CDR H3(AVF). As shown in Fig. 5B, compared to that in cells transduced with GPI-CDR H3(AVF), the replication of HIV-1 Bru-Yu2 was also significantly inhibited in cells transduced with GPI-CDR H3(PG16) throughout the experiments. At 6, 9, and 12 days postinfection there was a 1- or 2-log reduction in the amounts of HIV-1 Gag p24 produced by cells transduced with GPI-CDR H3(PG16) compared to cells transduced with GPI-CDR H3(AVF). At 15 days postinfection, there was still a 98% reduction in the amount of HIV-1 Gag p24 produced by cells transduced with GPI-CDR H3(PG16) compared to that produced by cells transduced with GPI-CDR H3(AVF).
Fig. 5.
Effect of GPI-CDR H3(PG16) on anti-HIV-1 activity of transduced human CD4+ T cells. (A) GPI-CDR H3(PG16) confers resistance to HIV-1 Bru-3 in human CD4+ T cells. (B) GPI-CDR H3(PG16) confers resistance to HIV-1 Bru-Yu2 in human CD4+ T cells.
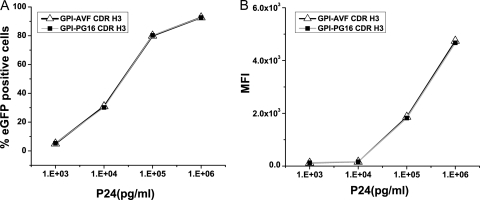
Finally, we transduced parental CEMss-CCR5 cells and CEMss-CCR5-GPI-CDR H3(AVF and PG16) cells with a VSV-G-pseudotyped HIV-1 vector expressing enhanced green fluorescent protein (EGFP) as described previously (37). The VSV-G envelope interacts with the lipid moiety in the lipid bilayer of the plasma membrane. Because of this, the VSV-G-pseudotyped lentiviral vector bypasses the requirement for the interaction between the HIV-1 envelope and its receptor and coreceptor to enter cells. We found that at all doses tested, the transduction of parental CEMss-CCR5 cells and CEMss-CCR5-GPI-CDR H3(AVF and PG16) cells with a VSV-G-pseudotyped lentiviral vector results in similar vector dose-dependent transduction efficiencies and transgene expressions (Fig. 6). These results demonstrate that GPI-CDR H3(PG16) does not inhibit VSV-G envelope-mediated viral entry, reverse transcription, integration, or the postintegration protein expression of the HIV-1 vector, indicating that the potent inhibition of HIV-1 replication seen for GPI-CDR H3(PG16)-transduced CEMss-CCR5 cells (Fig. 5) is HIV-1 envelope specific and at the level of viral entry.
Fig. 6.
EGFP expression in parental CEMss-CCR5 cells and CEMss-CCR5 cells expressing GPI-CDR H3(PG16 and AVF) transduced with a VSV-G-pseudotyped HIV-1 vector. (A) Percent EGFP-positive cells; (B) MFI (mean fluorescence intensity).
DISCUSSION
The crystal structure of quaternary-specific antibody PG16 reveals a unique stable hammerhead structure of the CDR H3 subdomain that rises above the antibody surface to confer fine specificity (23, 24). Thus, a crucial question is whether this unique architecture of CDR H3 itself is sufficient to recognize the quaternary epitope on the trimeric HIV-1 spike (gp1203/gp413) and exert neutralization. In this study, we constructed CDR H3s derived from five human monoclonal antibodies, PG16, PG9, b12, E51, and AVF, and genetically linked them with a GPI attachment signal of DAF. We demonstrate that fusion gene constructs are expressed and targeted to lipid rafts of plasma membranes through a GPI anchor (Fig. 1 and 2). GPI-anchored CDR H3s derived from PG16, PG9, and E51, but not GPI-anchored CDR H3s derived from AVF and b12, on the surface of transduced TZM-bl cells specifically neutralize multiple clades of HIV-1 isolates with a great degree of potency (Fig. 3). Moreover, GPI-anchored CDR H3(PG16), but not GPI-anchored CDR H3(AVF), on the surface of transduced human CD4+ T cells specifically confers resistance to HIV-1 infection (Fig. 5). Finally, the CDR H3 mutants (Y100HF, D100IA, and G7) that were previously shown to compromise the neutralization activity of antibody PG16 also abolish the neutralization activity of GPI-CDR H3(PG16) (Fig. 4). Thus, we conclude that the CDR H3 subdomain of PG16 itself, when targeted to the lipid raft of the plasma membrane of HIV-1-susceptible cells, exerts HIV-1 neutralization.
Changela et al. (3) recently determined the crystal structure of another quaternary-specific human antibody, 2909, which has limited neutralization breadth against HIV-1. To test the role of CDR H3 in neutralization, the CDR H3 sequences between 2909 and PG16 were swapped, and the chimeras were assayed for neutralization activity. Intriguingly, a chimera composed of antibody 2909 with PG16-derived CDR H3 does not neutralize any HIV-1 isolates tested, while a reciprocal chimera composed of antibody PG16 with the 2909-derived CDR H3 does not express neutralization activity. However, when the CDR H3 from each chimera was paired with its native light chain, neutralization activity was recovered although at a much reduced level and at a limited breadth (3). It was not clear what role the light chain plays. However, in light of the finding in our present study that the CDR H3 of PG16 itself, when targeted to the lipid raft of the plasma membrane of HIV-1-susceptible cells, exerts HIV-1 neutralization, it is unlikely that the light chain of PG16 could play a functional role in antigen recognition. Its structural role in maintaining the conformation of CDR H3, if any, may be found only at the whole-antibody level.
Although in transduced TZM-bl cells GPI-CDR H3(PG16) exhibited remarkable breadth and potency against diverse HIV-1 isolates tested (Fig. 3), in transduced CEMss-CCR5 cells GPI-CDR H3(PG16) showed significant inhibition only in the first 12 or 15 days postinfection (Fig. 5). This level of neutralization is quite different from what we previously reported for CEMss-CCR5 cells transduced with GPI-scFv (X5) (37). The transduction of CEMss-CCR5 cells with GPI-scFv (X5) conferred long-term resistance to HIV-1 infection (37). Although the difference may be attributed to the different forms, scFv versus CDR H3, it is more likely due to the different epitopes that the two antibodies (PG16 and X5) direct. In fact, we compared the anti-HIV-1 activities of TZM-bl cells transduced with GPI-CDR H3(PG16) and TZM-bl cells transduced with GPI-scFv(PG16) and found that cells transduced with GPI-scFv(PG16) exhibited less inhibition against HIV-1 pseudotypes than did cells transduced with GPI-CDR H3(PG16) (data not shown). The X5 epitope is a soluble CD4-inducible epitope, which resides within a conserved region (amino acid residues 417 to 434) of the gp120 core, particularly in the vicinity of amino acid residues at positions 423 and 432, which is in proximity to the CD4 and coreceptor binding sites (20). It was previously shown that scFv (X5) neutralizes HIV-1 better than Fab and whole IgG (15). The epitope recognized by PG16, however, is a glycosylated quaternary epitope (6) in a region on gp120 that includes the V2 and V3 loops. It forms, appropriately, only in the trimeric HIV-1 spike (36). Because of this, antibody PG16 can simultaneously interact with two epitopes present only in the oligomeric (gp1203/gp413) HIV-1 spike. As a result, antibody PG16 may have a much better avidity of binding to gp120 than GPI-CDR H3(PG16), which interacts with only one epitope at a time, even though the latter is present in the lipid raft domain of the plasma membrane. Therefore, a redesigning of fusion gene constructs that express dimeric and trimeric forms of GPI-CDR H3(PG16) may result in more potent and broad entry inhibitors of HIV-1.
GPI-CDR H3(PG16, PG9, and E51), with such a remarkable breadth of neutralization activity, should have potential either alone or in combination with other anti-HIV-1 gene constructs, such as GPI-scFv (X5), to be developed into antiviral agents for HIV-1 prevention and therapy. For example, GPI-CDR H3(PG16) and GPI-scFv (X5), due to their different specificities, could be codelivered into hematopoietic progenitor cells of HIV-1 patients ex vivo through a lentiviral vector, and transduced cells could then be transfused into patients, as recently described by DiGiusto et al. (5). However, in order to achieve clinical efficacy with this gene therapy approach, many details, such as efficient transduction and engraftment, maintenance of self-renewal, hematopoietic linage cell differentiation of transduced hematopoietic progenitor cells, sustainable transgene expression, and avoidance of potential insertion mutagenesis, have to be worked out.
Finally, it was previously shown that a fusion protein comprised of a sulfated peptide derived from CDR H3 of antibody E51 with the Fc domain of human IgG1 binds gp120 and inhibits HIV-1 infection (7). In the present study we show that in transduced TZM-bl cells, GPI-CDR H3(E51), like GPI-CDR H3(PG16 and PG9), also specifically neutralizes diverse HIV-1 isolates with a remarkable potency. Thus, it appears that GPI-CDR H3 can be an alternative approach for determining whether CDR H3 of certain antibodies alone can exert epitope recognition and neutralization.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Naldini at the University Torino Medical School, Torino, Italy, for the lentiviral transfer vector and B. H. Hahn at the University of Alabama for the DNA plasmid encoding the consensus C HIV-1 envelope protein. The cell line TZM-bl; HIV-1 molecular clones pMJ4, pBru-3, pBru-Yu2, and pAD8; as well as expression vectors pADA, pAD8, pQ168ENVa2, and pNL4-3.luc.R−E− were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Germantown, MD. These reagents were originally developed and contributed by J. Kappes, X. Wu, N. Landau, R. Risser, J. Overbaugh, E. Freed, M. Essex, T. Ndung'u, A. Adachi, M. A. Martin, I. R. Chen, G. W. Shaw, and B. H. Hahn.
This work was supported by research grants from the Chinese National Science Foundation (grant no. 30740008), the Chinese Science and Technology Ministry 973 Program Project (grant no. 2006CB504308), National Science and Technology Major Projects (grant no. 2008ZX10001-010, 2009ZX10004-105, and 2009ZX10004-016), the Shanghai Pasteur Foundation (grant no. SPHRF2007001), and French Energy Company Areva to P.Z. and by research grants from the National Institutes of Health to J.T.K. (grant AI47725) and the Baylor-UTHouston CFAR (grant P30AI036211).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 29 June 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Burton D. R., et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 2. Carter G. C., et al. 2009. HIV entry in macrophages is dependent on intact lipid rafts. Virology 386:192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Changela A., et al. 2011. Crystal structure of human antibody 2909 reveals conserved features of quaternary structure-specific antibodies that potently neutralize HIV-1. J. Virol. 85:2524–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chazal N., Gerlier D. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67:226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DiGiusto D. L., et al. 2010. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2:36ra43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doores K. J., Burton D. R. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84:10510–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dorfman T., Moore M. J., Guth A. C., Choe H., Farzan M. 2006. A tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J. Biol. Chem. 281:28529–28535 [DOI] [PubMed] [Google Scholar]
- 8. Follenzi A., Ailles L. E., Bakovic S., Geuna M., Naldini L. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217–222 [DOI] [PubMed] [Google Scholar]
- 9. Freed E. O., Myers D. J., Risser R. 1989. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J. Virol. 63:4670–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He J., et al. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kimata J. T., Kuller L., Anderson D. B., Dailey P., Overbaugh J. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535–541 [DOI] [PubMed] [Google Scholar]
- 12. Kothe D. L., et al. 2007. Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology 360:218–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koyanagi Y., et al. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819–822 [DOI] [PubMed] [Google Scholar]
- 14. Kozak S. L., Heard J. M., Kabat D. 2002. Segregation of CD4 and CXCR4 into distinct lipid microdomains in T lymphocytes suggests a mechanism for membrane destabilization by human immunodeficiency virus. J. Virol. 76:1802–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Labrijn A. F., et al. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y., et al. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liao Z., Cimakasky L. M., Hampton R., Nguyen D. H., Hildreth J. E. 2001. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum. Retroviruses 17:1009–1019 [DOI] [PubMed] [Google Scholar]
- 18. Medof M. E., Kinoshita T., Nussenzweig V. 1984. Inhibition of complement activation on the surface of cells after incorporation of decay-accelerating factor (DAF) into their membranes. J. Exp. Med. 160:1558–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller A. D., Chen F. 1996. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J. Virol. 70:5564–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moulard M., et al. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. U. S. A. 99:6913–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen D. H., Giri B., Collins G., Taub D. D. 2005. Dynamic reorganization of chemokine receptors, cholesterol, lipid rafts, and adhesion molecules to sites of CD4 engagement. Exp. Cell Res. 304:559–569 [DOI] [PubMed] [Google Scholar]
- 22. Overbaugh J., Anderson R. J., Ndinya-Achola J. O., Kreiss J. K. 1996. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res. Hum. Retroviruses 12:107–115 [DOI] [PubMed] [Google Scholar]
- 23. Pancera M., et al. 2010. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 84:8098–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pejchal R., et al. 2010. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 107:11483–11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Percherancier Y., et al. 2003. HIV-1 entry into T-cells is not dependent on CD4 and CCR5 localization to sphingolipid-enriched, detergent-resistant, raft membrane domains. J. Biol. Chem. 278:3153–3161 [DOI] [PubMed] [Google Scholar]
- 26. Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Popik W., Alce T. M., Au W. C. 2002. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4(+) T cells. J. Virol. 76:4709–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prodromou C., Pearl L. H. 1992. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. 5:827–829 [DOI] [PubMed] [Google Scholar]
- 29. Saphire E. O., et al. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155–1159 [DOI] [PubMed] [Google Scholar]
- 30. Shang H., et al. 2011. Genetic and neutralization sensitivity of diverse HIV-1 ENV clones from chronically infected patients in China. J. Biol. Chem. 286:14531–14541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simons K., Ikonen E. 1997. Functional rafts in cell membranes. Nature 387:569–572 [DOI] [PubMed] [Google Scholar]
- 32. Stamatatos L., Morris L., Burton D. R., Mascola J. R. 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15:866–870 [DOI] [PubMed] [Google Scholar]
- 33. Sun L., et al. 2009. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS One 4:e5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Theodore T. S., et al. 1996. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retroviruses 12:191–194 [DOI] [PubMed] [Google Scholar]
- 35. Tsai C., et al. 2009. Measurement of neutralizing antibody responses against H5N1 clades in immunized mice and ferrets using pseudotypes expressing influenza hemagglutinin and neuraminidase. Vaccine 27:6777–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walker L. M., et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wen M., et al. 2010. GPI-anchored single chain Fv—an effective way to capture transiently-exposed neutralization epitopes on HIV-1 envelope spike. Retrovirology 7:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu X., et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiang S. H., et al. 2003. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology 315:124–134 [DOI] [PubMed] [Google Scholar]
- 40. Zhou T., et al. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou T., et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zwick M. B., et al. 2003. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 77:5863–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.