Abstract
Respiratory syncytial virus (RSV) is a major cause of bronchiolitis and pneumonia in infants, the immunocompromised, and the elderly in both developed and developing countries. Reinfections are common, and G protein variability is one mechanism to overcome herd immunity. This is illustrated by the appearance of the BA genotype with a 60-nucleotide duplication dominating the subtype B genotypes in epidemics worldwide. To investigate the evolution of subtype B in South Africa since 2002, the genetic variability of the G protein was analyzed in all recent strains isolated over 4 years (2006 to 2009) in South African hospitals. Bayesian analysis revealed a replacement of all subtype B genotypes previously identified in South Africa with the BA genotype since 2006, while subtype A genotypes identified in previous years are still circulating. Compared to BA strains from other countries, the evolutionary rate of the South African BA genotype was shown to be 2.305 × 10−3 nucleotide substitutions/site/year and drift was evident. The most recent common ancestor (MRCA) of the South African BA viruses was determined to date back to 1996. All South African BA isolates clustered with the BA-IV subgenotype, and the appearance of new subgenotypes within this branch may occur if drift continues. Sequencing of the complete G protein of selected South African strains revealed an additional 6-nucleotide deletion. Acquisition of the 60-nucleotide duplication appeared to have improved the fitness of this virus, and more recent subtype B strains may need to be included in experimental vaccines to evaluate their efficacy in the current setting of evolved circulating strains.
INTRODUCTION
Respiratory syncytial virus (RSV), a member of the genus Pneumovirus in the Paramyxoviridae family, is a major cause of severe pediatric respiratory tract disease in infants, the immunocompromised, and the elderly (3, 10, 41). In moderate climates, RSV epidemics occur yearly in the winter months, whereas outbreaks are associated with the rainy season in humid climates (5). Although the mortality rate for RSV infections has decreased significantly over the past 20 years, approximately 500 deaths still occur annually in the United States, of which 80% occur in children <1 year old (29). Globally, the World Health Organization estimates that RSV causes 64 million infections and 160,000 deaths annually (44). A few studies have characterized the disease burden of RSV in developing countries. A study in Kilifi, Kenya, estimated that 85,000 infant cases of severe lower respiratory tract infection (LRTI) were due to RSV per year (23). Reinfection is known to occur throughout life. Children are infected in the presence of maternal antibodies, and natural infection affords only partial protection (31, 43).
RSV has a single-stranded, negative-sense RNA genome containing 10 genes encoding 11 proteins (6). Two antigenic subtypes (A and B) exist, with little cross protection (2). Major antigenic differences between subtypes are a feature of the attachment protein G, a type II transmembrane glycoprotein with a conserved central region with four cysteines postulated to be a receptor binding site. Variability is concentrated in two hypervariable regions of the ectodomain (15). Several G protein genotypes within the two subtypes have been identified, including GB1 to GB4 (24). SAA1, SAB1, SAB2, and SAB3 were identified in South Africa (39) and, subsequently, in various other geographic locations (26, 30). Venter et al. (38) also showed that identical RSV genotypes were identified in different regions in South Africa during one season. A new BA genotype has been identified in Buenos Aires in 1999 that is characterized by a 60-nucleotide duplication starting after residue 791 of the G protein (36). Subsequently, strains with this duplication have been found in clinical specimens from distantly related places in the world (16, 21, 27, 28, 46, 47), including Kenya in East Africa (28). This BA genotype was first detected in South Africa during the investigation of a nosocomial outbreak in Pretoria in 2006, which motivated us to reevaluate the current RSV molecular epidemiology in South Africa (40).
It is unclear why some children experience severe disease and others develop milder disease. It may be due to host factors, maternal immunity, or differences in the virus itself. Genotypes show complex circulation patterns probably facilitated by herd immunity to certain genotypes which might influence disease severity (24, 25).
Despite the importance of RSV as a respiratory pathogen, there is currently no vaccine or effective therapy available. Experimental vaccines have been based on prototype strains identified shortly after its discovery in the 1960s (7, 8, 20). Protective vaccines may, however, have to take recent strains into account based on drift demonstrated relative to these strains. To determine the evolution of the subtype B genotype in South African hospitals since the emergence of the BA genotype, the molecular epidemiology of RSV epidemics in children in South African hospitals was investigated. The correlation between disease severity and infecting strains was also investigated.
MATERIALS AND METHODS
Study population.
The study population consisted of all children <1 year of age who were diagnosed between February 2006 and May 2009 with RSV using the respiratory panel 1 immunofluorescence assay (Chemicon, Hampshire, United Kingdom) or the Directigen RSV rapid test (Becton Dickinson Microbiology Systems, Franklin Lakes, NJ) at the Department of Medical Virology, University of Pretoria, National Health Laboratory Service (NHLS), Tshwane Academic Division, and confirmed to have RSV by reverse transcription (RT)-PCR at our laboratory. The NHLS serves three secondary and tertiary hospitals which serve the public sector in the Pretoria region and act as referral hospitals for the Northern Parts of South Africa: Kalafong Secondary Hospital, Steve Biko Academic Hospital, and 1-Military Hospital. For the analysis of association with disease severity, patients were divided into two classes: mild disease was classified as outpatients not requiring hospitalization and severe disease as patients requiring hospitalization with severe LRTI or requiring intensive care treatment. This study was reviewed, approved, and monitored by the human ethics committee, University of Pretoria (25/2006).
Laboratory analysis.
RSV-positive specimens were confirmed by RT-PCR and screened for coinfections by multiplex PCR (17). Specimens were labeled according to the following system: SA for South Africa, isolate number, OP for outpatient/H for hospitalized, and year of isolation.
RNA extraction.
RNA was extracted directly from nasopharyngeal aspirates with a QIAamp viral RNA minikit according to the manufacturer's recommendations (Qiagen, Valencia, CA).
Subtype identification: multiplex RT-PCR and nested PCR.
The specimens were subtyped with a multiplex nested RT-PCR method as described before (38). Full-length G protein RT-PCR products were amplified with primers G1-21 (37) and F164 (32). In brief, 10 μl RNA was added to 10 μl 10× reaction buffer, 10 mM each deoxynucleoside triphosphate (dNTP), 20 pmol each primer (G1-21 and F164), 5 mM dithiothreitol solution, 10 U RNase inhibitor, and 1 μl Titan enzyme mix; the 50-μl volume was made up with distilled water. The following cycling conditions were used: 50°C for 30 min, 94°C for 2 min; 35 cycles of 94°C for 10 s, 53°C for 30 s, 68°C for 1 min; 68°C for 7 min. This was followed by nested PCR for weak specimens with primers G32B (32) and F1 (25). The nested PCR was conducted in a 100-μl reaction mixture volume. Two microliters of the RT-PCR product was mixed with 10 mM each dNTP, 20 pmol each primer (G32B and F1), 2.6 U Expand High-Fidelity PCR enzyme mix, and 10× Expand High-Fidelity buffer with 15 mM MgCl2 according to the following conditions: 94°C for 2 min; 35 cycles of 94°C for 30 s, 53°C for 30 s, 72°C for 1 min; 72°C for 7 min. PCR products were analyzed on a 1.5% agarose gel against a 100-bp ladder as molecular weight marker (DNA molecular marker XIV; Roche Diagnostics, Mannheim, Germany).
Nucleotide sequencing.
The Wizard SV gel and PCR cleanup system was used for PCR product purification (Promega, Madison, WI). Cycle sequencing was performed with a BigDye terminator 3.1 cycle sequencing kit as recommended by the manufacturer (Applied Biosystems, Foster City, CA). Primers were as described by Venter et al. (38). Nucleotide sequencing was carried out on both strands, and the editing was performed with Sequencher version 4.6 (Gene Codes Corporation, Ann Arbor, MI).
Phylogenetic analysis.
A region spanning 270 nucleotides (330 nucleotides for the BA strains) representing the second hypervariable region of the G protein gene was used for phylogenetic analysis as described before (25). This region corresponds to nucleotides 649 to 918 of prototype strain A2 for subtype A and 652 to 921 of prototype strain 18537 for subtype B (15). The South Africa sequences were compared against reference sequences of each genotype, available in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html). Nucleotide sequences of subtype A and B viruses were aligned separately with ClustalX 1.81, using the multiple alignment option (35). Midpoint-rooted maximum-likelihood trees were constructed under the HKY codon position substitution model using PhyML (12). Confidence estimates were based on bootstrap resampling carried out with 1,000 replicates.
Bayesian evolutionary analysis was carried out with BEAST (Bayesian Evolutionary Analysis by Sampling Trees) version 1.4.8 (9) using the Bayesian Markov chain Monte Carlo method (MCMC) to generate maximum-clade-credibility trees with the Tree Annotator software and showing the highest posterior probability values for each node of the tree for the BA genotype identified in South Africa from 2006 to 2009 together with 63 unique BA sequences from the rest of the world since 1999. According to Trento et al. (37), the BA genotype could be further divided into six different branches (subgenotypes), BA-I to -VI. These guidelines were used to assign South African BA strains to specific subgenotypes. All the trees were plotted using Figtree version 1.1.2 (http://tree.bio.ed.ac.uk).
Nucleotide and amino acid sequence analysis.
Pairwise distances (P-distance) were calculated between individual genotypes, as well as within each genotype, using Mega version 4 (34). Positive selection was calculated by the Nei-Gojobori method and is indicated by a Ka/Ks ratio of >1, where Ka is the nonsynonymous substitution rate and Ks the synonymous substitution rate (22). BioEdit Sequence Alignment Editor version 7.0.4.1 was used for amino acid analysis (13).
Evolutionary rate and date of MRCA.
The evolutionary rate was calculated using a Bayesian MCMC method available in the BEAST package, assuming an uncorrelated relaxed lognormal molecular clock that assumes independent rates on different branches (9). MCMC chains were run for sufficient time to achieve convergence (as assessed using the TRACER program). Statistical uncertainty in parameter estimates is given by the 95% highest probability density (95% HPD) values. The root-to-tip regression plot displays the correlation between phylogenetic branch length and the time of sampling of the viral strains. This was performed with Path-O-Gen version 1.1.2 (http://tree.bio.ed.ac.uk/software/pathogen). The crossing point was taken as the date of the most recent common ancestor (MRCA) for all the sequences under analysis.
Statistical analysis.
The association between genetic variability and disease severity was calculated with Fisher exact using STATA 10 (College Station, TX). P values of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
Sequences of partial G protein genes were deposited in GenBank under accession numbers HQ711628 to HQ711839, and sequences of full-length G protein genes were deposited under accession numbers JF704219 to JF704234.
RESULTS
Frequency of RSV subtypes and genotypes over four consecutive years in South Africa.
Of 621 specimens that tested positive for RSV at the NHLS Tshwane laboratory over the 4 years (2006 to 2009) of the study period, 245 specimens met the criteria for RT-PCR and 209 were confirmed as RSV positive. Of the 209 patients, 81 (39%) were HIV seropositive (54 were DNA PCR negative and 27 DNA PCR positive for HIV), 26 (12%) were HIV negative, and 102 (49%) had unknown HIV status. One hundred eighty-five (89%) patients had single RSV infections, while 24 (11%) had coinfections with other viruses as tested with a respiratory multiplex PCR (17).
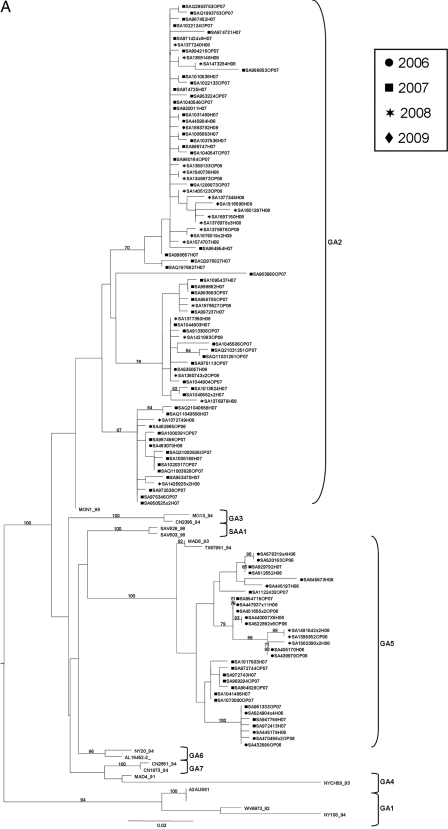
As shown in Table 1, subtype A was the more prevalent subtype in 2006, 2007, and 2008, whereas subtype B was more prevalent in 2009. Several dual infections with both subtypes were also seen over 2 of the 4 years. Two genotypes within subtype A were detected, GA2 and GA5 (Fig. 1A and Table 2). Genotype GA5 predominated among the subtype A genotypes in 2006 at 88%, whereas GA2 predominated in 2007 and 2008 at 82% and 83%, respectively. All of our subtype B isolates clustered within the BA genotype and had the characteristic 60-nucleotide duplication (Fig. 1B and Table 2). All previously identified subtype B genotypes in South Africa were replaced by the BA genotype for all 4 years investigated.
Table 1.
Frequencies of respiratory syncytial virus subtypes A and B over four consecutive years in South Africa
| Subtype(s) | No. (%) of specimens positive for subtype(s) in: |
|||
|---|---|---|---|---|
| 2006 (76 specimens) | 2007 (74 specimens) | 2008 (32 specimens) | 2009 (25 specimens) | |
| A | 45 (59) | 67 (90) | 29 (91) | 4 (16) |
| B | 27 (36) | 5 (7) | 3 (9) | 21 (84) |
| Dual infections | 4 (5) | 2 (3) | 0 | 0 |
Fig. 1.
Midpoint-rooted maximum-likelihood trees for subtype A (A) and subtype B (B) constructed under the HKY codon position substitution model using PhyML(12), drawn to scale with the bars indicating 0.03 nucleotide substitutions. Estimates were based on bootstrap resampling carried out with 1,000 replicates. Only bootstrap values of >70 are shown. The virus designations refer to the place of isolation (SA, South Africa)/isolate number/disease severity for strains isolated in this study (OP, outpatient; H, hospitalized)/year of isolation. The number of identical sequences is indicated as ×n. The genotypes assigned are indicated at the right by brackets. The sources of the reference sequences obtained from GenBank were as follows: NY, New York; AL, Alabama; MO, Missouri; TX, Texas; CN, Canada; CH, Rochester, NY (24, 25); WV, West Virginia (33); MON, Montevideo, Uruguay; MAD, Madrid, Spain (11, 18); BE, Belgium; Ken, Kenya; QUE, Quebec; SA, South Africa. The prototype strain for subtype A is strain A2 (A2AUS61, Australia) (42), and prototype strains for subtype B are Sw8-60 (Sweden) (33) and 18537 (CH18537_62, United States) (15).
Table 2.
Frequencies of respiratory syncytial virus genotypes over four consecutive years in South Africa
| Subtype and genotype | No. (%) or total no.a of strains sequenced in: |
|||
|---|---|---|---|---|
| 2006 | 2007 | 2008 | 2009 | |
| Subtype A (total no.) | 49 | 69 | 29 | 4 |
| GA2 | 4 (8) | 56 (81) | 24 (83) | 4 (100) |
| GA5 | 43 (88) | 13 (19) | 5 (17) | 0 |
| Untypeable | 2 (4) | 0 | 0 | 0 |
| Subtype B (total no.) | 31 | 7 | 3 | 23 |
| BA | 30 (97) | 7 (100) | 3 (100) | 23 (100) |
| Untypeable | 1 (3) | 0 | 0 | 0 |
Total number of strains sequenced. Due to dual infections in some patients, these totals are higher than the number of specimens tested.
Characterization of the South African subtype A strains.
Subtype A strains identified in previous epidemics are still circulating in South Africa, although the previously identified SAA1 genotype was not detected. The intragenotypic P-distances were 0.040 and 0.031 for GA2 and GA5, respectively, and the average intergenotypic P-distance between the different genotypes within subtype A fell in the range of 5 to 16%, with 11% difference between the two dominant genotypes, GA2 and GA5. The percentage of nonsynonymous changes among South African subtype A isolates within the C-terminal end of the G protein was 74%. Genotype-specific amino acid substitutions could also be identified. The Nei-Gojobori method identified positive selection within genotype GA2 with a Ka/Ks ratio of 1.08. Using BEAST, the evolutionary rate of South African subtype A strains was estimated as 3.382 × 10−3 (95% HPD, 1.911 × 10−3 to 4.954 × 10−3) substitutions/site/year. A root-to-tip transgression plot estimation of South African subtype A strains indicated the MRCA to date back to 1980. Subtype A isolates utilized four termination codons. In total, 18% used the UAG stop codon, 36% used UGA, and 2% used UAA at position 909, while 44% used stop codon UAG at position 912.
Characterization of the South African subtype B strains.
All subtype B genotypes previously identified in South Africa have been replaced by the BA genotype since 2006. The overall nucleotide and amino acid identity levels were shown to be 97.7% and 96.6%, respectively, for all the South African BA strains. It was shown that the same sequence in different patients was stable for three successive years. Using the reference strains in Fig. 1B, differences of 4 to 15% were observed between the subtype B genotypes. The BA strains currently circulating in South Africa differ by 8%, 11%, and 5% from SAB1, SAB2, and SAB3, respectively, that were previously identified in South Africa.
Except for the 60-nucleotide duplication, only substitutions and no deletions, insertions, or frameshift mutations were observed in the C-terminal end of the G protein. The percentage of nonsynonymous substitutions among South African subtype B isolates was 52% within the C-terminal end of the G protein. The full-length G protein gene was sequenced for selected BA strains for each of the 4 years, including five sequences for 2006, three sequences for 2007, two for 2008, and six for 2009. Compared to other complete G protein nucleotide sequences in GenBank, all 16 South African BA strains sequenced here had a 6-nucleotide (2-amino-acid) deletion at position 490 that resulted in the deletion of a proline and a lysine. South African BA isolates used the UAA stop codon at nucleotide position 946 (79%), resulting in a protein length of 315 amino acids, while 21% of the isolates used the UAG stop codon at position 967, resulting in a length of 322 amino acids.
Evolutionary relationship between South African BA strains and BA strains isolated in other countries.
Figure 2 illustrates Bayesian analysis of BA strains isolated worldwide since 1999 together with all the BA strains isolated during this study period (2006 to 2009) in South Africa. It was shown that all South African BA strains isolated during the 4 years clustered within group four (BA-VI). Differences ranging from 0.3% to 6.4% were seen between South African BA-IV strains. All the South African BA strains were unique with respect to the other strains within this group from other countries. South African strains were most closely related to strains from Brazil, Argentina, and India. Strains from Belgium, Canada, and Kenya within group IV clustered separately from the South African strains. Differences of 1.7% to 4.7% were seen between the different BA branches. Positive selection was identified in BA-II, BA-III, and BA-VI with Ka/Ks ratios of 1.25, 1, and 2, respectively. Positive selection could not be identified within BA-IV.
Fig. 2.
A midpoint-rooted BEAST phylogenetic tree of BA genotypes isolated worldwide since 1999 together with all the BA genotypes isolated during this study (2006 to 2009) (9). The lengths of the horizontal lines are proportional to the genetic distance between viruses. The bar represents 2 substitutions per site. The numbers on the branches represent the highest posterior probability value for each node of the tree. The virus designations refer to the place of isolation (SA, South Africa)/isolate number/disease severity for strains isolated in this study (OP, outpatient; H, hospitalized)/year of isolation. South African strains were compared to strains from Buenos Aires (BA), Japan, Kenya (Ken), Quebec (QUE), Jundiai, Brazil (JU), Sau Paulo, Brazil (SP), Ribeirao Preto, Brazil (RP), India (PN) and Belgium (BE) (28, 37, 48). The number of identical sequences is indicated as ×n. Branches are indicated at the right by brackets. The scale at the bottom indicates years since first isolation.
Genetic drift is visible within the BA genotype since its emergence 11 years ago, as can be seen by the dates of isolation and the location on the tree. The older strains are closer to the root than the more recently isolated strains. Drift is visible within the South African BA-IV strains. When the BA viruses were first isolated in 1999, the 60-nucleotide duplication was an exact replicate of the preceding 60 nucleotides (37). However, numerous changes were observed between these two regions in the South African BA strains (Fig. 3). Using BEAST, the average nucleotide substitution rate across all sites in the alignment was calculated as 2.305 × 10−3 (95% HPD, 1.112 to 3.665 × 10−3) substitutions/site/year for the South African BA strains. With the use of the program Path-O-Gen, the MRCA of the South African BA genotype was determined to date back to 1996.
Fig. 3.
Amino acid alignment of the duplicated region within the BA genotype. The alignments are shown relative to the BE12445 strain isolated in Belgium in 1999.
DISCUSSION
Our study showed that the BA genotype has replaced all previously identified B genotypes in South Africa and illustrates the ability of RSV to evolve and overcome herd immunity. Such variability complicates human RSV vaccine development. Strong evidence was shown for a common ancestry of all BA viruses with this 60-nucleotide duplication (37). Since its discovery, strains with this duplication have been found in clinical specimens from distantly related places (16, 21, 27, 28, 46, 47).
In the first 3 years studied in South Africa, subtype A predominated, whereas subtype B predominated in the 4th year. Two genotypes (GA2 and GA5) dominated among the subtype A specimens that have previously been isolated in South Africa (39). The SAA1 genotype was not detected again, suggesting that GA2 and GA5 are the current stable genotypes. All of the subtype B specimens clustered with the BA genotype and had the characteristic 60-nucleotide duplication replacing all B genotypes previously identified in South Africa. Referring to Venter et al. (38), identical genotypes were previously found in different regions in South Africa during one season and, thus, strains identified in one region probably represent the strains circulating in South Africa. The fact that only BA subtype B strains were identified over all 4 years in major public sector referral hospitals in the capital of the country is also strong evidence that the previously identified genotypes are not circulating any more. Investigations of strains from the rest of Africa will determine whether this is true for the whole continent. The BA genotype has also been detected in Kenya in 2006 but cocirculated with the SAB1 genotype in that epidemic (28). Compared to other genotypes, South African BA strains were more closely related to the SAB3 genotype previously detected in South Africa. It is not certain whether new genotypes in South Africa occurred as a result of spontaneous mutations or importation. Apart from the 60-nucleotide duplication, no deletions, insertions or frameshift mutations were detected in the C-terminal region of the G protein gene, although sequencing of the full-length G protein gene identified a 6-nucleotide (2-amino-acid) deletion at position 490 resulting in a missing proline and lysine. This deletion has also been detected in strains from Kenya and Buenos Aires (1). The dominance of this genotype indicates that the duplication might give this genotype an evolutionary advantage. Sequencing of the full gene suggests that further changes in the G gene may occur. Whole-genome sequencing of the new BA strains would elucidate changes in other proteins.
To determine the evolutionary rate of the South African BA genotype, as well as the MRCA of these strains, Bayesian evolutionary analysis was conducted on all South African BA isolates, as well as BA isolates from all over the world since it was first discovered in 1999. First, it was shown that all the strains isolated in South Africa clustered within branch IV. According to Trento et al. (37), this is the most heterogeneous branch regarding the date and the place of isolation. Thus, it is possible that this 60-nucleotide duplication changed the antigenic structure of these BA genotypes, giving them an evolutionary advantage to reinfect individuals previously exposed to subtype B.
When the BA viruses were first isolated in 1999, the 60-nucleotide duplication was an exact replicate of the preceding 60 bp (37). This region accumulated further nucleotide substitutions over time, and certain nucleotide positions were shown to be under positive selection (4, 19, 45–47). These changes might enhance the fitness of these viruses, which would explain why they are now becoming dominant worldwide and replacing all other B genotypes. This region is believed to be involved in changing/replacing the antigenic epitopes which contribute to the evasion of the immune response of the population. Certain amino acids in this region have been shown to be under positive selection, as well as the change of 952CAA to UAA (Q313 to STOP) that was present in the most recent lineages of the BA-IV branch (4, 46). Most of the South African BA strains (79%), all from BA-IV, had this stop codon change. However, it was at position 946 because of the 6-nucleotide deletion. Changes in stop codon usage are thought to be associated with antigenic variation that allows immune evasion, especially in the subtype B isolates that are less variable than the subtype A isolates (18).
Positive selection was shown to occur within some of the different BA branches. The dates when the strains were isolated and their placement on the tree indicate how much this genotype changed over time, especially the South African strains (BA-IV). The strains first isolated in 1999 lie closer to the root of the tree than the strains isolated in 2009. The appearance of new subgenotypes within the BA-IV branch may occur as drift continues, with four different clusters already existing within BA-IV (Fig. 2). Substitution rates have been estimated for only a limited number of viruses. In general, values lie close to 1 × 10−3 substitutions/site/year, although considerable variation exists. This variation is partly due to their error-prone replication, which causes mutations to accumulate quickly (14). The rate of evolution for South African BA strains across all sites in the alignment was calculated as 2.305 × 10−3 substitutions/site/year, which correlates with findings by Trento et al. (37). According to them, the origin of the MRCA was dated between 1998 and 1999, shortly before it was discovered in 1999. Our data indicate that the MRCA of the South African BA sequences dates back to 1996.
This study did have some limitations and was not able to fully identify possible associations between disease severity and genotype. Although information on hospitalization was available, the number of outpatients was small and no significant associations between subtype/genotype and disease severity were found. Nevertheless, most other studies only evaluated this during one epidemic season, whereas the present study investigated this over four consecutive seasons. The accumulation of herd immunity may determine the disease association of different strains in a community from season to season. Also, information on HIV infection and immune status for most study participants was lacking, which limited our ability to address the role of HIV in the association of disease severity with the different RSV genotypes.
To conclude, G protein variability may play a significant role in RSV pathogenesis by allowing immune evasion. Certain substitutions or alterations may enhance the fitness of viruses, as is evident with the BA strains that replaced all other B genotypes previously identified in South Africa. The G protein's ability to accommodate such substantial changes and facilitate immune evasion may complicate vaccine development. It remains to be seen whether the BA genotype will remain dominant or whether dominance will eventually fade because of herd immunity. Our study suggests that subtype B strains may need to be updated with recent strains in experimental vaccines to reevaluate their efficacy.
ACKNOWLEDGMENTS
This study was reviewed, approved, and monitored by the human ethics committee, University of Pretoria (25/2006).
The study was funded by the Poliomyelitis Research Foundation.
A special thanks to Perry Smith and Kariuki Njenga of the CDC for critical reviews of this paper.
Footnotes
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Agoti C. N., et al. 2010. Intrapatient variation of the respiratory syncytial virus attachment protein gene. J. Virol. 84:10425–10428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson L. J., et al. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626–633 [DOI] [PubMed] [Google Scholar]
- 3. Boeck K. D. 1996. Respiratory syncytial virus bronchiolitis: clinical aspects and epidemiology. Monaldi Arch. Chest Dis. 51:210–213 [PubMed] [Google Scholar]
- 4. Botosso V. F., et al. 2009. Positive selection results in frequent reversible amino acid replacements in the G protein gene of human respiratory syncytial virus. PLoS Pathog. 5:e1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cane P. A. 2001. Molecular epidemiology of respiratory syncytial virus. Rev. Med. Virol. 11:103–116 [DOI] [PubMed] [Google Scholar]
- 6. Collins P. L., Crowe J. E., Jr 2007. Respiratory syncytial virus and metapneumovirus, p. 1601–1646 In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed., vol. 2 Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 7. Collins P. L., Murphy B. R. 2002. Respiratory syncytial virus: reverse genetics and vaccine strategies. Virology 296:204–211 [DOI] [PubMed] [Google Scholar]
- 8. Crowe J. E., Jr 2001. Respiratory syncytial virus vaccine development. Vaccine 20(Suppl. 1):S32–S37 [DOI] [PubMed] [Google Scholar]
- 9. Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Falsey A. R., Hennessey P. A., Formica M. A., Cox C., Walsh E. E. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352:1749–1759 [DOI] [PubMed] [Google Scholar]
- 11. Garcia O., et al. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J. Virol. 68:5448–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guindon S., Lethiec F., Duroux P., Gascuel O. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 14. Jenkins G. M., Rambaut A., Pybus O. G., Holmes E. C. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:156–165 [DOI] [PubMed] [Google Scholar]
- 15. Johnson P. R., Spriggs M. K., Olmsted R. A., Collins P. L. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. U. S. A. 84:5625–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuroiwa Y., et al. 2005. A phylogenetic study of human respiratory syncytial viruses group A and B strains isolated in two cities in Japan from 1980-2002. J. Med. Virol. 76:241–247 [DOI] [PubMed] [Google Scholar]
- 17. Lassauniére R., Kresfelder T., Venter M. 2010. A novel multiplex real-time RT-PCR assay with FRET hybridization probes for the detection and quantification of 13 traditional and newly identified respiratory viruses. J. Virol. Methods 165:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martinez I., et al. 1999. Evolutionary pattern of the G glycoprotein of human respiratory syncytial viruses from antigenic group B: the use of alternative termination codons and lineage diversification. J. Gen. Virol. 80:125–130 [DOI] [PubMed] [Google Scholar]
- 19. Melero J. A., Garcia-Barreno B., Martinez I., Pringle C. R., Cane P. A. 1997. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J. Gen. Virol. 78(Pt. 10):2411–2418 [DOI] [PubMed] [Google Scholar]
- 20. Murata Y. 2009. Respiratory syncytial virus vaccine development. Clin. Lab. Med. 29:725–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagai K., Kamasaki H., Kuroiwa Y., Okita L., Tsutsumi H. 2004. Nosocomial outbreak of respiratory syncytial virus subgroup B variants with the 60 nucleotides-duplicated G protein gene. J. Med. Virol. 74:161–165 [DOI] [PubMed] [Google Scholar]
- 22. Nei M., Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 23. Nokes D. J., et al. 2008. Respiratory syncytial virus infection and disease in infants and young children observed from birth in Kilifi District, Kenya. Clin. Infect. Dis. 46:50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peret T. C., et al. 2000. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J. Infect. Dis. 181:1891–1896 [DOI] [PubMed] [Google Scholar]
- 25. Peret T. C., Hall C. B., Schnabel K. C., Golub J. A., Anderson L. J. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 79:2221–2229 [DOI] [PubMed] [Google Scholar]
- 26. Riccetto A. G., et al. 2009. Genotypes and clinical data of respiratory syncytial virus and metapneumovirus in Brazilian infants: a new perspective. Braz. J. Infect. Dis. 13:35–39 [DOI] [PubMed] [Google Scholar]
- 27. Sato M., et al. 2005. Molecular epidemiology of respiratory syncytial virus infections among children with acute respiratory symptoms in a community over three seasons. J. Clin. Microbiol. 43:36–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scott P., et al. 2004. Molecular epidemiology of respiratory syncytial virus in Kilifi district, Kenya. J. Clin. Microbiol. 74:344–354 [DOI] [PubMed] [Google Scholar]
- 29. Shay D. K., Holman R. C., Roosevelt G. E., Clarke M. J., Anderson L. J. 2001. Bronchiolitis-associated mortality and estimates of respiratory syncytial virus-associated deaths among US children, 1979–1997. J. Infect. Dis. 183:16–22 [DOI] [PubMed] [Google Scholar]
- 30. Shobugawa Y., et al. 2009. Emerging genotypes of human respiratory syncytial virus subgroup A among patients in Japan. J. Clin. Microbiol. 47:2475–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sullender W. M. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 13:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sullender W. M., Sun L., Anderson L. J. 1993. Analysis of respiratory syncytial virus genetic variability with amplified cDNAs. J. Clin. Microbiol. 31:1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sullender W. M., Wertz G. W. 1991. Synthetic oligonucleotide probes differentiate respiratory syncytial virus subgroups in a nucleic acid hybridization assay. J. Clin. Microbiol. 29:1255–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 35. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trento A., et al. 2003. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J. Gen. Virol. 84:3115–3120 [DOI] [PubMed] [Google Scholar]
- 37. Trento A., et al. 2006. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J. Virol. 80:975–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Venter M., Collinson M., Schoub B. D. 2002. Molecular epidemiological analysis of community circulating respiratory syncytial virus in rural South Africa: comparison of viruses and genotypes responsible for different disease manifestations. J. Med. Virol. 68:452–461 [DOI] [PubMed] [Google Scholar]
- 39. Venter M., Madhi S. A., Tiemessen C. T., Schoub B. D. 2001. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J. Gen. Virol. 82:2117–2124 [DOI] [PubMed] [Google Scholar]
- 40. Visser A., Delport S., Venter M. 2008. Molecular epidemiological analysis of a nosocomial outbreak of respiratory syncytial virus associated pneumonia in a kangaroo mother care unit in South Africa. J. Med. Virol. 80:724–732 [DOI] [PubMed] [Google Scholar]
- 41. Welliver R. C. 2003. Review of epidemiology and clinical risk factors for severe respiratory syncytial virus (RSV) infection. J. Pediatr. 143:S112–S117 [DOI] [PubMed] [Google Scholar]
- 42. Wertz G. W., et al. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. U. S. A. 82:4075–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wertz G. W., Moudy R. M. 2004. Antigenic and genetic variation in human respiratory syncytial virus. Pediatr. Infect. Dis. J. 23:S19–S24 [DOI] [PubMed] [Google Scholar]
- 44. WHO Department of Vaccines Biologicals 2001. Report of a workshop on the epidemiology of respiratory syncytial virus (RSV) in developing countries, Johannesburg, South Africa, 30 November—1 December 2000. Document V&B/VAM/SCEFR/00.01 WHO, Geneva, Switzerland [Google Scholar]
- 45. Woelk C. H., Holmes E. C. 2001. Variable immune-driven natural selection in the attachment (G) glycoprotein of respiratory syncytial virus (RSV). J. Mol. Evol. 52:182–192 [DOI] [PubMed] [Google Scholar]
- 46. Zlateva K. T., Lemey P., Moes E., Vandamme A. M., Van Ranst M. 2005. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J. Virol. 79:9157–9167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zlateva K. T., Lemey P., Vandamme A. M., Van Ranst M. 2004. Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup A: positively selected sites in the attachment G glycoprotein. J. Virol. 78:4675–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zlateva K. T., Vijgen L., Dekeersmaeker N., Naranjo C., Van Ranst M. 2007. Subgroup prevalence and genotype circulation patterns of human respiratory syncytial virus in Belgium during ten successive epidemic seasons. J. Clin. Microbiol. 45:3022–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]