Abstract
Noroviruses are the most common cause of epidemic gastroenteritis. Genotype II.3 is one of the most frequently detected noroviruses associated with sporadic infections. We studied the evolution of the major capsid gene from seven archival GII.3 noroviruses collected during a cross-sectional study at the Children's Hospital in Washington, DC, from 1975 through 1991, together with capsid sequence from 56 strains available in GenBank. Evolutionary analysis concluded that GII.3 viruses evolved at a rate of 4.16 × 10−3 nucleotide substitutions/site/year (strict clock), which is similar to that described for the more prevalent GII.4 noroviruses. The analysis of the amino acid changes over the 31-year period found that GII.3 viruses evolve at a relatively steady state, maintaining 4% distance, and have a tendency to revert back to previously used residues while preserving the same carbohydrate binding profile. In contrast, GII.4 viruses demonstrate increasing rates of distance over time because of the continued integration of new amino acids and changing HBGA binding patterns. In GII.3 strains, seven sites acting under positive selection were predicted to be surface-exposed residues in the P2 domain, in contrast to GII.4 positively selected sites located primarily in the shell domain. Our study suggests that GII.3 noroviruses caused disease as early as 1975 and that they evolve via a specific pattern, responding to selective pressures induced by the host rather than presenting a nucleotide evolution rate lower than that of GII.4 noroviruses, as previously proposed. Understanding the evolutionary dynamics of prevalent noroviruses is relevant to the development of effective prevention and control strategies.
INTRODUCTION
Noroviruses (NoVs) are a major cause of acute gastroenteritis in children and adults worldwide. In developing countries, NoVs have been estimated to cause around 1.1 million hospitalizations and more than 200,000 deaths in children annually (33). In addition, antibodies to NoVs can be detected almost universally in children between the ages of 5 to 15 years worldwide, demonstrating the widespread distribution within human populations (19, 20, 30, 32).
NoVs are a genetically diverse group of small (approximately 27 nm in diameter) icosahedral viruses belonging to the family Caliciviridae, which includes the genera Norovirus, Sapovirus, Vesivirus, Lagovirus, and Nebovirus. They have a positive-sense, single-stranded RNA genome of approximately 7.6 kb (20). The genome is comprised of three open reading frames (ORFs). ORF1 encodes the six nonstructural proteins, ORF2 encodes the major capsid protein (VP1), and ORF3 encodes a minor capsid protein (VP2) (20, 25). The genus Norovirus is further classified into five genogroups (GI to GV), with genogroups I, II, and IV having the capability to infect humans (8, 11, 48). Additionally, these genogroups are further classified into genotypes (48).
It has been reported that one of the host factors involved in norovirus infection is histo-blood group antigens (HBGAs) (23). HBGAs are carbohydrates expressed on cell surfaces and are the determinants of ABO, Lewis, and secretor blood types. The major capsid protein, VP1, contains sites that are predicted to interact with HBGAs, and mutations occurring in this region may be driven by the evasion of the host immune response (11).
Despite the large number of NoV genotypes cocirculating in human populations, specific genotypes have predominated over time. The majority of acute gastroenteritis outbreaks due to NoV infection are caused by GII.4 NoVs (28, 34, 41), while GII.3 NoVs are one of the most common genotypes associated with sporadic NoV infection, particularly in children, where they often are identified as the dominant genotype (1, 2, 9, 13, 34). In addition, GII.3 NoVs also have been implicated in food-borne outbreaks in developed and developing countries (44, 45). Although both GII.3 and GII.4 genotypes have an important role in human NoV infections, few studies have addressed the genetic basis of the epidemiological differences between these NoVs (5, 39). It has been suggested that GII.4 NoVs predominate over time through cluster replacement with intermediate periods of stasis and the ability of the clusters to shift the interactions between different HBGAs (28). In contrast, it was proposed that GII.3 NoVs had an evolutionary disadvantage compared to GII.4 NoVs due to a lower rate of evolution and a less processive polymerase (5).
NoVs were discovered in 1972 (24); however, the oldest GII.3 strain identified to date is from a 1983 sample isolated in the Goulburn Valley of Australia (42). It has been proposed that the incidence of GII.3 NoVs increased in the 1990s following GII.4 pandemic periods (6), but little is known about GII.3 NoVs prior to this time.
The Children's Hospital National Medical Center of Washington, DC (CHDC), study was initiated in 1974 to determine the etiology of severe acute gastroenteritis in infants and children (2, 4). The current study aims to characterize the molecular evolution of GII.3 NoVs from 1975 through 2006 by analyzing the sequence of the VP1 gene of these viruses from archival clinical samples collected from the CHDC study in combination with GII.3 sequences available in GenBank. The additional comparison of the evolution of the GII.3 NoVs with GII.4 NoVs over the same time period was performed to examine potential differences that may have led to the predominance of the GII.4 genotype over the GII.3 genotype. A better understanding of how GII.3 NoVs evolve over time may help clarify how these viruses evade the host immune response and persist in the human population and determine whether they indeed have an evolutionary disadvantage compared to GII.4 NoVs. These insights will aid in the development of prevention and control strategies, including vaccination and drug therapies.
MATERIALS AND METHODS
Sample collection.
Seven of 23 NoV GII.3 strains previously identified in archive stool materials from infants and children admitted to the Children's Hospital National Medical Center, in Washington, DC (CHDC), were selected for analysis in this study. These archival samples originally were collected as part of a cross-sectional study which included patients with diarrhea, vomiting, or both, as well as controls, and was conducted between January 1974 and July 1982 and from 1987 through 1991 (2, 4). Samples were selected based on the quality and quantity of RNA available, and to span the whole time frame of sample collection, selected samples were collected in 1975, 1976, 1979, 1988, 1990, and 1991.
The CHDC study originally was supported by several grants (including AI01528-17/10) and contracts (NIH-NIAID-71-2091) with the NIAID, NIH, and has been described previously (2, 4). This research was reviewed by the Office of Human Subjects Research at the National Institutes of Health and was determined to be exempt from institutional review board approval.
RNA extraction.
RNA extraction for NoV capsid gene amplification and sequencing was performed as described previously (2). Briefly, stool samples were reconstituted into 10% suspensions in phosphate-buffered saline (PBS) and then further clarified using Genetron (1,1,2-trichloro-1,2,2-trifluoroethane; Aqua Solutions, Deer Park, TX). RNA was extracted using an RNeasy kit (Qiagen, Valencia, CA) by following the manufacturer's recommendations. RNA was stored at −70°C.
VP1 gene sequencing.
Previously, NoVs were provisionally genotyped by submitting the partial polymerase sequence to a BLAST search (2, 29). Selected samples positive for GII.3 NoVs based on the polymerase screening were subjected to full VP1 gene sequencing. The seven samples selected were Hu/NoV/GII.3/CHDC2005/1975/US, Hu/NoV/GII.3/CHDC32/1976/US, Hu/NoV/GII.3/CHDC4671/1979/US, Hu/NoV/GII.3/CHDC4031/1988/US, Hu/NoV/GII.3/CHDC4090/1988/US, Hu/NoV/GII.3/CHDC5261/1990/US, and Hu/NoV/GII.3/CHDC5365/1991/US. Full-length VP1 sequences were obtained using nested or one-step reverse transcription-PCR (RT-PCR) by primer walking (see Fig. S1 in the supplemental material). cDNA for use in nested PCRs was generated using the Superscript III first-strand synthesis super mix (Invitrogen), using either oligo(dT) or random hexamers (Invitrogen) as primers, by following the manufacturer's recommendations. PCR was carried out using the elongase system (Invitrogen) by following the manufacturer's recommendations. One-step RT-PCR was carried out using the Superscript III one-step RT-PCR system HiFi (Invitrogen) by following the manufacturer's recommendations. PCR products were run on 1.2% agarose gels and were purified using a QIAquick gel extraction kit (Qiagen). The nucleotide sequence was determined directly from the purified DNA amplicon using the BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Carlsbad, CA), and the sequencing products were resolved on an ABI PRISM 3730 automated DNA sequencer (Applied Biosystems).
Phylogeny.
Evolutionary analysis was performed on seven full-length VP1 nucleotide sequences corresponding to GII.3 NoVs from the CHDC study, together with 56 sequences available in the GenBank database as of 1 April 2010 that reported a reliable date of sample collection (see Fig. S2 in the supplemental material). Sequence alignments were performed using Clustal X 1.8, and the alignments were manually edited in MegAlign version 8.0 (Lasergene, Madison, WI). The parameter values for the best-fit model of nucleotide substitutions were determined to be GTR+I+G (general time reversible model; proportion of invariable sites [I], 0.4381; gamma distribution shape parameter [G], 1.0671) using Akaike information criterion (AIC) as implemented in MODELTEST (36). A phylogenetic tree was inferred by maximum-likelihood reconstruction based on the nucleotide alignment of full-length VP1 sequences as implemented in PhyML software (21). The statistical significance of phylogenies constructed was estimated by bootstrap analysis with 100 pseudoreplicate data sets. The tree was displayed with the Fig Tree program (http://tree.bio.ed.ac.uk/software/figtree/) (37).
Evolutionary analysis.
The parameter values for the best-fit model of nucleotide substitutions were determined, as described above, using AIC as implemented in MODELTEST (36). The corresponding sample collection dates were assigned using the BEAUTi application, which is part of the BEAST package (15, 16). Rates of nucleotide substitution per site and the time to the most recent common ancestor (TMRCA) were estimated using Bayesian Markov chain Monte Carlo (MCMC) as implemented in the BEAST software package (15, 16). The model of substitution utilized was the GTR, and the data set was partitioned by codon positions. Since no demographic model is available from organisms showing cyclic annual/seasonal behavior, the Bayesian skyline model for population growth was selected, and the corresponding plots were observed in a graph (17). BEAST files were run assuming a constant rate of evolution across the tree (strict molecular clock) or using relaxed-clock models (log normal and exponential), which model a varying molecular rate among lineages (14). In all cases, statistical uncertainty in parameter values across the sampled trees is given by the 95% highest probability density (HPD) values. For all models, chains were run until convergence was achieved (as assessed using the TRACER program, which is part of the BEAST package), and BEAST files were run three times independently for each model.
The nucleotide and amino acid variations within and between clusters observed in the phylogenetic tree were examined by applying the Tamura-Nei model and Poisson correction, respectively (MEGA version 4) (43). Amino acids of all GII.3 NoV and GII.4 NoV VP1 sequences available in GenBank were aligned with their respective genotype in Clustal X 1.8. The amino acid distance between each strain and the most ancestral strain in the respective genotype, Hu/NoV/GII.3/CHDC2005/1975/US (GII.3) or Hu/NoV/GII.4/CHDC2094/1974/US (GII.4), was calculated using MEGA version 4 (43) and plotted against the year the strain was collected.
Positive selection.
Site-specific positive selection was evaluated as rates of nonsynonymous versus synonymous substitutions among sequences present in the GII.3 alignment. Our analysis was performed using the single-likelihood ancestor counting (SLAC) method available in the HyPhy package (35). The GTR substitution model was chosen and a neighbor-joining tree was used as an input, with a limit of significance of 0.25.
Residue mapping on protein structure.
A homology model of the Hu/NoV/GII.3/CHDC2005/1975/US P domain was derived by comparing the amino acid sequence to sequences existing in the Protein Data Bank. The homology model was created using the I-TASSER web server and was modeled after Hu/NoV/GII.4/VA387/1998/US (2OBT) (and similar) templates (7, 38, 46, 47). The amino acids evolving under positive selection were mapped onto the Hu/NoV/GII.3/CHDC2005/1975/US P domain minimized homology model. The GII.4 (Hu/NoV/GII.4/VA387/1998/US (2OBT) (7) amino acid sites that corresponded to the GII.3 (Hu/NoV/GII.3/CHDC2005/1975/US)-positive selection sites were highlighted in complex with blood group trisaccharide B. Additionally, the model ribbon structures of Hu/NoV/GII.4/VA387/1998/US and Hu/NoV/GII.3/CHDC2005/1975/US were superimposed for comparison. The resulting P2 protein structures were visualized using PyMOL (DeLano Scientific, LLC, San Francisco, CA).
Virus-like particle expression and HBGA binding/blocking assay.
The VP1 genes of Hu/NoV/GII.3/CHDC2005/1975/US, Hu/NoV/GII.3/CHDC32/1976/US, Hu/NoV/GII.3/CHDC4031/1988/US, and Hu/NoV/GII.3/CHDC5261/1990/US were amplified and cloned into a pENTR plasmid (Invitrogen). The VP1 genes of Hu/NoV/GII.3/Arg320/1995/AG and Hu/NoV/GII.3/Maizuru010524/2001/JPN were synthesized (GenScript, Piscataway, NJ). The recombination of plasmid DNA with baculovirus DNA was performed using the Baculodirect kit (Invitrogen), and recombinant virus-like particles (rVLPs) were amplified and purified as previously described (2).
The ability of VLPs to bind to synthetic oligosaccharides and of antibodies to block the binding of VLPs to synthetic HBGA H type 3 (H3) carbohydrate was determined as previously described (2, 3). Briefly, neutravidin-coated plates (Pierce, Rockford, IL) were treated with 1 mg/ml biotinylated carbohydrate (Glycotech, Gaithersburg, MD) for 2 h and washed with PBS containing 0.05% Tween 20 and 0.1% bovine serum albumin (BSA). Wells were coated for 1 h with 1.5 μg/ml of either VLP alone or VLP combined with VLP-specific sera, which were generated in rVLP-immunized guinea pigs. The binding of captured VLP was determined by incubation with guinea pig anti-VLP-specific hyperimmune sera (1:5,000 dilution), followed by incubation with a peroxidase-conjugated goat anti-guinea pig serum (1:2,000 dilution; KPL, Gaithersburg, MD). The reaction was revealed with peroxidase substrate 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS; KPL) and read at 405 nm by a Dynex Technologies Revelation 4.25 plate reader (Dynex Technologies, Chantilly, VA). All incubations were conducted at room temperature.
Nucleotide sequence accession numbers. The complete VP1 gene sequences from the seven CHDC GII.3 NoVs sequenced were submitted to the GenBank database under the following accession numbers: HM072045 (CHDC2005), HM072046 (CHDC32), HM072042 (CHDC4671), HM072044 (CHDC4031), HM072043 (CHDC4090), HM072041 (CHDC5261), and HM072040 (CHDC5365).
RESULTS
Identification of noroviruses in Children's Hospital stool collection.
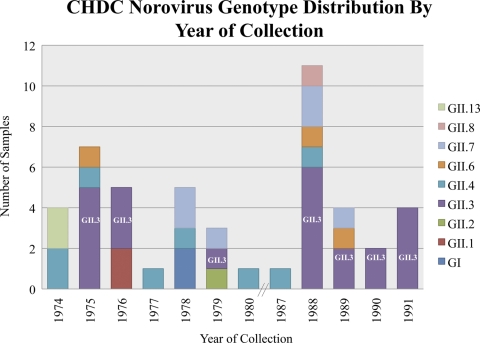
The GII.3 strains selected for analysis were among the 48 NoV samples identified during the recent examination of 5,424 archival stool materials from the CHDC cross-sectional study (2, 4). The distribution of NoV genotypes in the 48 positive CHDC samples was found to be the following: GII.3, 48%; GII.4, 17%; GII.7, 13%; GII.6, 6%; GII.1, 4%; GI, 4%; GII.13, 4%; GII.2, 2%; and GII.8, 2% (2). GII.3 viruses represented the most common genotype identified within the norovirus-positive samples, accounting for 23 of the 48 samples, and they were identified in 7 of the 12 years available for study (Fig. 1). It is interesting that 2 of the 23 GII.3 NoV-positive samples were found in stool material collected from controls. GII.3 NoVs were identified as the most common NoV strains isolated in 1975, 1976, and 1988 to 1991; only one sample tested positive in 1979, and GII.3 viruses were not detected in any of the remaining years sampled. The GII.3 viruses studied here were isolated in 1975, 1976, 1979, 1988, 1990, and 1991.
Fig. 1.
Distribution of genotypes in the NoV-positive samples of the CHDC study by year of collection.
Evolutionary dynamics of GII.3 noroviruses.
The seven GII.3 NoV strains initially characterized using a region of the polymerase gene were confirmed as GII.3 by the comparison of the VP1 sequences to NoV VP1 sequences representing GI, GII, GIII, GIV, and GV strains available in the GenBank database. All CHDC GII.3 sequences clustered with known GII.3 strains, confirming that they were GII.3 NoVs (see Fig. S3 in the supplemental material).
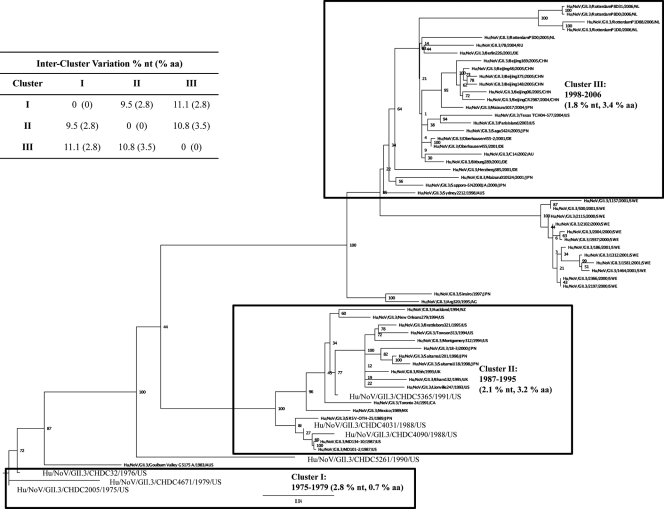
We studied the evolutionary dynamics of GII.3 VP1 sequences over time, comparing them to previous similar studies of GII.4 NoVs from the same time period (2). Sixty-three GII.3 NoV VP1 sequences from 1975 through 2006 were included in a maximum-likelihood phylogenetic analysis that was used to map out the evolution of GII.3 NoVs over time (Fig. 2). Clusters were defined as a minimum of three sequences of monophyletic origin. The tree showed that three distinct clusters (I, II, and III) have emerged over time. An additional group of sequences that was not included in the cluster classification was comprised of strains isolated from an immunocompromised patient in Sweden (SWE in Fig. 2) with a chronic NoV infection from 2000 to 2001 (31). Cluster I comprised only three sequences, the oldest CHDC strains from the 1970s (Hu/NoV/GII.3/CHDC2005/1975/US, Hu/NoV/GII.3/CHDC32/1976/US, and Hu/NoV/GII.3/CHDC4671/1979/US). Cluster II was comprised of strains that emerged in the 1980s and 1990s, including Hu/NoV/GII.3/CHDC4031/1988/US, Hu/NoV/GII.3/CHDC4090/1988/US, Hu/NoV/GII.3/CHDC5365/1991/US, and the prototype GII.3 strain Hu/NoV/GII.3/Toronto 24/1991/CA. Finally, cluster III included strains isolated in the late 1990s through 2006 (the most recent VP1 sequence available in GenBank). Four strains did not fit into any specific cluster. Two of these strains, one derived from Japan in 1997 (Hu/NLV/GII.3/Sinsiro/1997/JPN) and one derived from Argentina in 1995 (Hu/NoV/GII.3/Arg320/1995/AG), were most closely related to each other and were predicted to be most similar to the cluster III sequences. An Australian isolate from 1983 (Hu/NoV/GII.3/Goulburn Valley G5175A/1983/AUS) did not cluster with any defined group and seemed to have emerged out of the cluster I strains from the 1970s. Interestingly, one CHDC strain (Hu/NoV/GII.3/CHDC5261/1990/US) did not fall into any of the three clearly defined clusters.
Fig. 2.
Maximum-likelihood analysis of VP1 region sequences from GII.3 NoVs. GII.3 VP1 sequences from 1975 to 2006 (for GenBank accession numbers, see Fig. S2 in the supplemental material), including the seven new CHDC sequences, are shown, circled within proposed clusters as a result of the analysis. The percent nucleotide (nt) and percent amino acid (aa) variation within each cluster are shown in parentheses, and between-cluster variation is shown in the inset table.
The mean nucleotide variation within and between the three clusters was examined. The clusters had similar intracluster nucleotide variation values of 2.8, 2.1, and 1.8% for clusters I, II, and III, respectively (Fig. 2). Interestingly, while the level of nucleotide variation within clusters I, II, and III was very similar, the variation at the amino acid level was much lower in cluster I (0.7%) than in cluster II (3.2%) or III (3.4%) (Fig. 2). The level of intercluster variation among the amino acid sequences was lower between cluster I and III (2.8%), which are furthest apart in the tree, than the variation between clusters II and III (3.5%), which are closer in the tree. Not surprisingly, the variation within the SWE samples, isolated over the course of a year from a single immunocompromised patient with a chronic infection, was very low, 0.4 and 0.6% for nucleotide and amino acid, respectively, compared to the variation seen within the clusters. Additionally, the nucleotide variation between the SWE group and the defined clusters was much higher than what was seen between the clusters, with values of 13, 12.2, and 6.3% between the SWE samples and clusters I, II, and III, respectively, and 6.3, 7.2, and 5.7% amino acid variation between the SWE samples and clusters I, II, and III, respectively.
A Bayesian coalescent method was used to infer the rates of evolutionary change expressed as nucleotide substitutions per site per year, using three different molecular clock models, a strict-clock and two relaxed-clock estimations, an uncorrelated exponential derivation model (UCED), and an uncorrelated log-normal model (UCLN). The most conservative clock, the strict model, estimated that GII.3 NoVs evolved at a rate of 4.16 × 10−3 nucleotide substitutions/site/year (Table 1), which is comparable to the nucleotide evolution rate previously calculated for GII.4 NoVs (2). The relaxed-clock estimations calculated similar rates of evolution, 7.39 × 10−3 and 5.8 × 10−3 nucleotide substitutions/site/year for the UCED and UCLN clocks, respectively. The same Bayesian approach was used to estimate the time to the most recent common ancestor using each clock model. The mean age of the GII.3 VP1 most recent ancestor derived from the population analyzed was 35.5 years (32.3 to 38.13) by the strict clock, which would date the most recent ancestor to 1970, while the relaxed-clock models dated the ancestor to 1972 to 1973 (Table 1).
Table 1.
Nucleotide substitution rates and divergence times over a 31-year period for the VP1 gene of GII.3 NoV strainsa
| Molecular clock | Nucleotide substitution rateb (10−3 substitutions/site/yr) | TMRCA by: |
|
|---|---|---|---|
| No. of yrb | Date (range) | ||
| Strict | 4.16 (3.5–4.8) | 35.5 (32.3–38.1) | 1970 (1967–1973) |
| UCED | 7.39 (5–9.96) | 33.1 (31–37.6) | 1972 (1968–1975) |
| UCLN | 5.8 (4.4–7.4) | 32.9 (31–35.9) | 1973 (1970–1975) |
The nucleotide substitution rate is the mean rate for the three individual determinations. UCED, uncorrelated exponential deviation. UCLN, uncorrelated log normal. TMRCA, time to most recent common ancestor.
Values in parentheses are 95% HPDs.
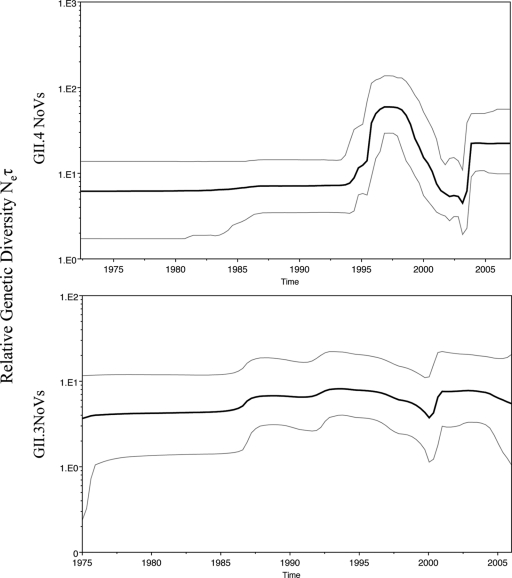
It has been suggested previously that GII.4 NoVs present an epochal pattern of evolution with periodic bursts of GII.4 activity (28). A plot comparing the relative genetic diversity over time of GII.4 and GII.3 NoV sequences (Fig. 3) shows a marked difference in the evolutionary dynamics of these two genotypes. These plots show the relative genetic diversity of a population over time, where Ne is the effective population size and τ is the generation. These relative genetic diversity plots can help illustrate a demographical history by showing increases and decreases in genetic diversity within a defined time frame. The overall genetic diversity among strains is lower for GII.3 than for GII.4. In addition, GII.3 strains do not show an increased peak of activity after 1995 but rather a constant circulation of genetic variants. The observed differences could be due partially to the smaller number of GII.3 sequences catalogued over time and might be confirmed by analyzing the Bayesian skyline plot of polymerase sequences which have been reported to better reflect epidemiological observations (40).
Fig. 3.
Bayesian skyline plots of GII.4 and GII.3 NoV VP1 sequences. The plots illustrate the relative genetic diversity of circulating GII.4 and GII.3 NoVs through time. The black line represents the median posterior value and the gray lines the 95% highest probability density (HPD) intervals.
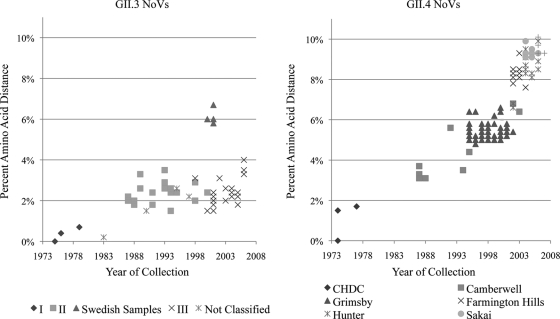
To investigate if the nucleotide variability of these two genotypes is reflected in amino acid fixation on the viral population over time, the percent amino acid distance was calculated between the available VP1 sequences in GenBank (GII.3, n = 56; GII.4, n = 185 [2]), the CHDC samples (GII.3, n = 7; GII.4, n = 5), and their respective CHDC most ancestral strains (GII.3, Hu/NoV/GII.3/CHDC2005/1975/US; GII.4, Hu/NoV/GII.4/CHDC2094/1974/US) (Fig. 4). The time span for each analysis was 1975 to 2006 and 1974 to 2005 for GII.3 and GII.4, respectively. The amino acid distance for GII.3 NoVs demonstrated a relatively constant level of change (∼4%) compared to the CHDC reference strain, with the exception of the Swedish sequences (∼6%), all of which were isolated from an immunocompromised patient and may have evolved under different pressures. In contrast, the GII.4 NoVs demonstrated a linear increase in amino acid distance over time when strains were compared to the ancestral reference strain, reaching more than 10% accumulated amino acid variation from 1974 to 2008.
Fig. 4.
Amino acid evolution of GII.3 and GII.4 NoV sequences compared to sequences of the most ancestral CHDC strains. Amino acid distance from most ancestral GII.3 (Hu/NoV/GII.3/CHDC2005/1975/US) and GII.4 (Hu/NoV/GII.4/CHDC2094/1974/US) strains was calculated for all VP1 sequences available in GenBank (GenBank accession numbers for GII.3 are in Fig. S2 in the supplemental material; those for GII.4 are in reference 2). Samples are marked by cluster from the phylogenetic analysis.
Amino acid variation and positive selection in VP1 capsid protein.
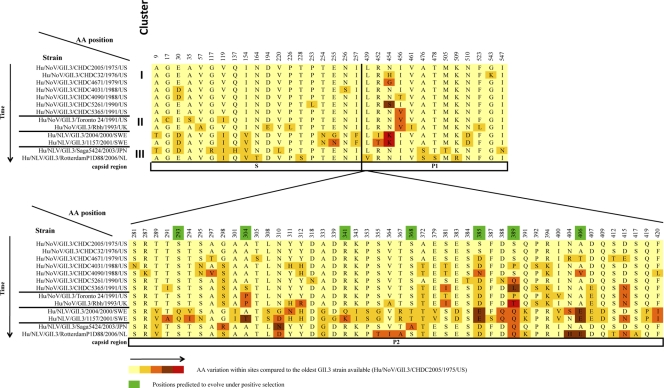
The VP1 amino acid sequence from 13 GII.3 NoVs representative of the three clusters and the Swedish samples (spanning 1975 to 2006), including the seven CHDC sequences from this study, were aligned. Eighty variable sites were detected, representing 14.6% of the total VP1 amino acid sequence of 548 residues (Fig. 5). For more than half the variable positions, 48 sites (60%), were located in the P2 region of the capsid. When comparing the most ancestral sequence available (Hu/NoV/GII.3/CHDC2005/1975/US) with the most recent sequence available (Hu/NoV/GII.3/RotterdamP1D88/2006/NL), only 24 amino acid (aa) residues were different between the two strains, representing only 30% of the total variable sites (24/80). This also accounts for a change in only 4% (24 out of 548) of the complete VP1 amino acid sequence between these two NoVs, which is comparable to the percent distance observed between other GII.3 samples and the most archival CHDC GII.3 strain (Hu/NoV/GII.3/CHDC2005/1975/US) (Fig. 4).
Fig. 5.
Amino acid variation of the VP1 capsid region of GII.3 NoVs over time. The VP1 amino acid sequences of the GII.3 strains from the CHDC study were compared to sequences of representative strains from all three clusters predicted for GII.3 evolution. Amino acids predicted to be under positive selective pressure are highlighted in green and were determined using the single-likelihood ancestor counting (SLAC) method available in the HyPhy package.
The VP1 amino acid sequences from the 63 GII.3 sequences used in the alignment spanning the years 1975 to 2006 (see Fig. S2 in the supplemental material) were used for positive selection analysis. Positive selection or selective pressure is defined as the evolutionary force that increases the frequency of a beneficial mutation until it becomes fixed in the population (26). An analysis of the VP1 amino acid residues with evidence for positive selective pressure (P < 0.25) identified seven sites, all in the P2 domain. Positions are highlighted in green in Fig. 5 (aa 293, 304, 341, 368, 385, 389, and 406).
Protein structure modeling.
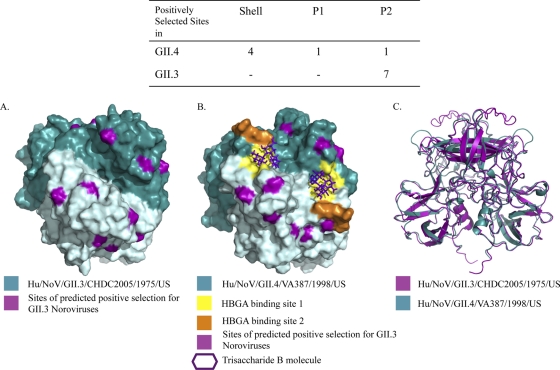
A homology model was determined by comparison of the amino acid sequence of Hu/NoV/GII.3/CHDC2005/1975/US P domain to sequences existing in the Protein Data Bank. All seven amino acids evolving under positive selection were highlighted in purple on the Hu/NoV/GII.3/CHDC2005/1975/US P domain dimer minimized structure (Fig. 6A). Interestingly, all seven of the positively selected sites mapped to surface-exposed residues in the homology model. The capsid structure for GII.3 NoVs has not yet been solved and can only be estimated by computer generation based on sequence, but the structure of the P2 capsid region has been reported for a GII.4 strain, Hu/NoV/GII.4/VA387/1998/US (7). Using the GII.4 model to map selected GII.3 residues of interest onto the solved GII.4 structure allows for preliminary insight into the locations of these amino acids on the viral structure. A solved model of the GII.3 capsid structure will help clarify the true locations of these residues on the viral capsid. To better understand the locations of the amino acids under positive selection in GII.3 NoVs, these sites were highlighted on the minimized structure of the Hu/NoV/GII.4/VA387/1998/US (2OBT) P domain (7) in complex with blood group trisaccharide B (Fig. 6B). Six out of seven of the positively selected sites could be mapped to the GII.4 structure. All of the mapped sites corresponded to surface-exposed residues near the two HBGA binding sites (7). Moreover, there is a marked difference between the location of positively selected sites in GII.3 and that in GII.4 NoVs. Using the same parameters in both determinations, we found that all positively selected sites were located in the P2 region of GII.3 NoVs, while most of the positively selected sites were found in the shell region in GII.4 NoVs (Fig. 6) (2). Ribbon structures of the Hu/NoV/GII.4/VA387/1998/US and Hu/NoV/GII.3/CHDC2005/1975/US minimized structures were superimposed for comparison (Fig. 6C). The ribbon models illustrated that the majority of the P region is structurally similar between the two models.
Fig. 6.
Structural modeling of GII.3 P2 dimer. Homology modeling was performed to predict the P2 protein structure of the GII.3 P region using the amino acid sequence of Hu/NoV/GII.3/CHDC2005/1975/US. (A) Sites predicted to undergo positive selection (highlighted in green in Fig. 5) were highlighted in purple on this minimized structure. (B) The same residues were mapped on the protein structure of the Hu/NoV/GII.4/VA387/1998/US dimer and are highlighted in purple. The two HBGA binding sites are shown in yellow (site 1) and orange (site 2), with the trisaccharide B molecule shown in purple. (C) An overlay of the two minimized ribbon structures predicted for Hu/NoV/GII.3/CHDC2005/1975/US P domain (purple) and Hu/NoV/GII.4/VA387/1998/US P domain (teal) was performed. The inset table shows the number and locations of positively selected sites.
HBGA binding profile of GII.3 NoV over time.
It has been proposed that GII.4 NoVs evolve through cluster replacement over time, and that each emerging cluster is able to escape the immune response and successfully infect susceptible individuals partially due to their ability to bind different types of HBGAs over time (28). We studied the binding profile of the GII.3 clusters established in this analysis. Consistent with their low and constant level of amino acid variation over a 27-year period, GII.3 NoV clusters differ only slightly in their ability to bind different carbohydrates over time (Table 2), as all clusters were able to bind to HBGAs Leb and Lex, and H3 was bound only by VLPs representing clusters I and II.
Table 2.
Interactions of GII.3 NoVs with synthetic oligosaccharides spanning a 27-year perioda
| Strainb | Date | GII.3 cluster | Interaction result with: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | A | B | Lea | Leb | Lex | Ley | |||
| CHDC2005 | 1975 | I | − | − | + | − | − | − | + | + | − |
| CHDC32 | 1976 | I | − | − | + | − | − | − | − | − | − |
| CHDC5261 | 1990 | NC | − | − | + | − | − | − | − | − | − |
| CHDC4031 | 1988 | II | − | − | − | − | − | − | ± | + | − |
| TV* | 1991 | II | − | − | + | − | − | − | − | − | − |
| Arg320 | 1996 | NC | − | − | ± | − | − | − | + | + | − |
| Maizuru | 2001 | III | − | − | − | − | − | − | + | + | − |
NC, not classified; +, positive interaction; −, no interaction; ±, weak interaction.
*TV HBGA binding results were not assayed in this study (22).
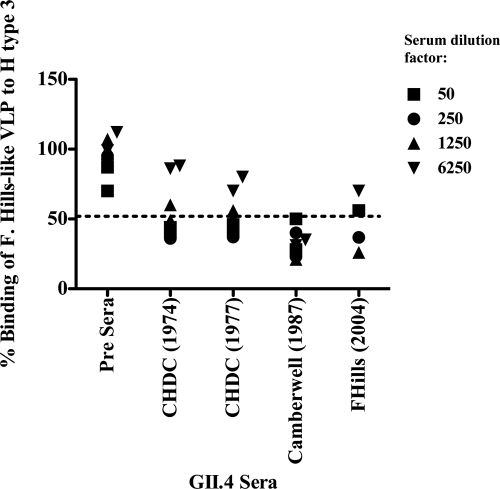
A surrogate neutralization assay also was performed to test the hypothesis that sera raised against an individual cluster would not be able to block the interaction of a heterologous NoV VLP with a specific HBGA. This “blockade” assay has been shown to correlate well with protection against infection (3, 27). Guinea pig sera raised against three different GII.4 clusters spanning 30 years (CHDC, Camberwell, and Farmington Hills [F. Hills]) were tested for their ability to block the binding of an F. Hills-like VLP to H type 3. All sera, including sera raised against a VLP generated from a GII.4 virus circulating 30 years earlier, were able to block the binding of the F. Hills NoV VLP to H type 3, whereas preimmune sera did not interfere with binding (Fig. 7). The same assay was attempted between GII.3 NoV clusters (data not shown), but we encountered technical problems with poor binding to HBGAs. Therefore, it was not possible to observe a significant optical density (OD) difference between pre- and postimmunization sera. However, we hypothesize that hyperimmune sera raised against the different GII.3 clusters (I, II, and III) block the HBGA binding of heterologous VLPs, since their amino acid variability over time is much lower than that of GII.4 NoVs and all GII.3 NoV clusters interact with the same types of carbohydrates.
Fig. 7.
Detection of HBGA blocking antibodies in sera from GII.4 clusters. Sera from three different GII.4 clusters (CHDC, Camberwell, and F. Hills) were tested for the ability to block the interaction between recombinant F. Hills VLPs and H3 carbohydrates.
DISCUSSION
Noroviruses are important causative agents of acute viral gastroenteritis in both children and adults worldwide. Their persistence in the human population may be due, in part, to the genetic diversity of these viruses. The majority of studies on the evolution of NoVs have focused broadly on all genotypes or on a few specific genotypes, such as GII.4, because of their high prevalence in outbreaks (28). However, GII.3 NoVs, which are responsible for a large number of infections, particularly in settings in which it is endemic and in children, have not been thoroughly studied (6, 39). The goal of this study was to compare differences in the evolutionary dynamics of these viruses to inform the development of control strategies to prevent NoV infection and dissemination.
In this study, we have identified and described the VP1 region of the earliest known GII.3 NoV strains isolated from children hospitalized in Washington, DC. Prior to this study, the oldest GII.3 full-length VP1 sequence (Hu/NoV/GII.3/Goulburn Valley G5175A/1983/AUS) reported was collected in 1983 (42). GII.3 NoVs were present and caused acute gastroenteritis in the human population at least 8 years earlier, in 1975. Previously, and in unpublished data, we showed that GII.3 NoVs were the most prevalent genotype detected from children hospitalized with acute gastroenteritis at the Children's Hospital National Medical Center of Washington, DC, from 1974 to 1991 (2). It has been proposed that the most successful NoV genotypes, which currently are responsible for the majority of gastroenteritis cases (GII.4 and GII.3), emerged during the first peak detected in the 1980s (GII.4) (28) and 1990s (GII.3) (6). Our studies of norovirus samples collected prior to those years in the Children's Hospital study now have shown that both GII.4 and GII.3 viruses have been circulating for at least several decades (unpublished data) (2 and K. L. Shumansky, E. J. Abente, S. V. Sosnovtsev, A. Z. Kapikian, K. Y. Green, and K. Bok, unpublished data). This suggests that regardless of the mechanism of evolution of different NoV genotypes and the cumulative host immunity acquired after each gastroenteritis episode, certain genotypes, such as GII.4 and GII.3 remain predominant. Unlocking the key to this evolutionary advantage might be essential for the development of adequate control strategies.
One of the key features of RNA viruses that have allowed them to persist in human populations is their ability to undergo genetic changes which may lead to a more fit generation of viruses (10). It has been reported that RNA viruses evolve at an approximate rate of 10−3 nucleotide substitutions/site/year (18). Consistently with this, it was previously reported that GII.4 NoVs evolved at a rate of 4.3 to 6.5 × 10−3 nucleotide substitutions/site/year (2). The GII.3 NoVs in our study evolved at a rate of 4.16 × 10−3 to 7.39 × 10−3 nucleotide substitutions/site/year (strict- and relaxed-molecular-clock models), which is comparable to the rate for GII.4 viruses. These data differ from a previous study that calculated a lower rate of evolution for GII.3 compared to that of GII.4 (5). Our data suggest that the overall lower prevalence of GII.3 compared to GII.4 NoVs cannot be attributed to differences in the rate of nucleotide evolution in the VP1 region, since the rates of evolution between the two genotypes were remarkably similar for both strict- and relaxed-molecular-clock models. The addition of the CHDC samples to both the GII.3 and the GII.4 (2) evolutionary analyses, together with the use of more statistically advanced algorithms (Bayesian estimation), may have afforded better estimations of evolutionary rates.
The nucleotide and amino acid variations both within and between the three clusters identified in our GII.3 phylogenetic tree were analyzed and compared to values previously described for GII.4. The GII.3 clusters I, II, and III had intracluster nucleotide variation of 2.8, 2.1, and 1.8%, respectively. These values are lower than the range of intracluster nucleotide variation seen in GII.4 NoVs, which demonstrated nucleotide variations ranging from 0.8 to 7% (2). However, this difference might also be due to the availability of a larger number of GII.4 VP1 sequences in the public databases. Intercluster nucleotide variation increased with increasing distance between clusters in the tree, with clusters I and II, II and III, and I and III demonstrating variation of 9.5, 10.8, and 11.1%, respectively. It was hypothesized that intercluster amino acid variation would follow a similar pattern; however, the intercluster variation between clusters II and III (3.5%) was higher than the intercluster variation between clusters I and II (2.8%) and I and III (2.8%). These results indicate that while the nucleotide composition of GII.3 NoVs continues to change over time, viruses in the earliest cluster are more similar to viruses in the most recent cluster in their amino acid composition than the two most modern clusters are to each other.
The percent amino acid distances between the two most distant GII.3 strains in time (Hu/NoV/GII.3/CHDC2005/1975/US, cluster I, and Hu/NoV/GII.3/RotterdamP1D88/2006/NL, cluster III) shown in Fig. 5 were analyzed. Only 30% of the amino acids in VP1 changed over time, whereas 67.1% of amino acids changed in a similar analysis of GII.4 NoVs (Hu/NoV/GII.4/CHDC2094/1974/US compared to Hu/NoV/GII.4/Sakai/2005/US) (2). Additionally, the percent distance of all GII.3 NoVs individually compared to that of Hu/NoV/GII.3/DC2005/1975/US demonstrated a stable rate of approximately 4 to 6% amino acid distance over time, with sample isolation dates ranging from 1975 to 2006 (Fig. 4). In contrast, GII.4 NoVs showed a linear rate of amino acid change that continually increased over time, from 1974 to 2005, to greater than 10%, which appeared to correlate with the emergence of the GII.4 clusters. Taken together, these data demonstrate a striking difference in the evolution of GII.3 and GII.4 NoVs at the amino acid level, despite their similar rates of nucleotide substitution. It seems that while certain amino acid residues have undergone change over time in GII.3 NoVs, many residues in cluster III that had undergone mutation between clusters I and II are the same as the residues in cluster I. While we cannot determine if these mutations were fixed from cluster I, we did not have a large enough sample size to demonstrate this, and could not demonstrate that these mutations have actually mutated twice, this phenomenon of similar amino acids between only clusters I and III may reflect an advantage in viral fitness in relation to these particular residues. Alternatively, perhaps only a small repertoire of amino acids can be tolerated by the virus at these positions, which may have undergone mutation due to pressure to evade the host immune response or because of an error-prone RNA-dependent RNA polymerase (RdRp). Since these residues were present in the viral population in the 1970s, and assuming only short periods of antibody protection are elicited after infection, reverting back to those particular residues may allow these viruses to infect current populations, which may have gained immunity to more recently circulating strains.
In contrast, GII.4 NoVs seem to evolve in a linear fashion without much reversion to previously utilized amino acids. This difference may reflect the difference in hosts that these two genotypes tend to infect. For GII.4 to cause epidemics in adult populations, it needs to continue to reinfect previously exposed hosts, which requires constant changes in the capsid protein to evade the host immune response developed in response to previous infections. Conversely, GII.3 NoVs have been shown to be prevalent in young children and infants (1, 9, 13, 34). These young naïve hosts may not have established immune-associated protection from previous infections, and therefore GII.3 NoVs may not need to continually adapt to evade the immune response, as the pool of young children and infants is continually renewed. Why these two genotypes have a tendency to infect different age groups in the population has yet to be determined, but it may be due to a variety of factors, including seroprotection from heterologous strains or the minimal infectious dose necessary to cause disease.
The alignment of VP1 sequences representative of all three GII.3 clusters revealed that greater than 50% of the amino acid changes occurred in the P2 domain. Bull and colleagues identified 15 evolutionary hotspots on the GII.4 NoV capsid protein (5). These hotspots vary between each pandemic cluster within this genotype. Interestingly, they found that six (aa 310, 312, 389, 392, 395, and 404) of these GII.4 hotspots also were hypervariable sites in GII.3 sequences, and that they clustered onto four exposed loops in the P2 domain (5). Consistent with those results, our analysis found variation over time at the same positions with the exception of residue 395 (Fig. 5). Residue 312 was identified as a residue which had undergone genetic mutation over time but had reverted back to the most ancestral amino acid in the most recent isolates. Residue 389 was predicted, in our analysis, to be under positive selective pressure. Our data confirmed that except for amino acid 395, these hotspots might actually vary between GII.3 clusters.
Our analysis on positive selection sites within the VP1 region of GII.3 NoVs resulted in the identification of only seven residues; however, all seven were located in the P2 domain (Fig. 5). Previous analysis of the GII.4 NoVs over the same time period identified six positively selected sites; however, only one was located in the P2 domain, while four were identified in the shell (S) domain and one in the P1 domain (2). These differences in the location of sites under positive selection between GII.3 and GII.4 NoVs indicate that the two genotypes are under differing host selection pressures, such as possible coreceptors or immune-driven selection.
The structural modeling of the Hu/NoV/GII.3/CHDC2005/1975/US P domain sequence demonstrated that all seven of the positively selected amino acid sites were surface-exposed residues on the GII.3 minimized structure (Fig. 6). It is important that while there is no capsid structure solved for GII.3 viruses, the ability to model with the solved GII.4 structure may give interesting preliminary insight into the location of certain residues. By mapping the same sites on the better-understood GII.4 minimized model, Hu/NoV/GII.4/VA387/1998/US, we showed that out of the six residues that could be mapped, all remained in surface-exposed locations. Additionally, these residues mapped to regions surrounding the predicted HBGA binding sites. This suggests that these sites directly interact with host ligands, including HBGAs. Moreover, these residues may be undergoing positive selection as a direct evasion strategy of the host immune response and neutralizing antibodies. However, we cannot differentiate between mutations occurring due to host-driven pressure and mutations occurring due to RdRp error. Mutations that remain fixed in the population, such as those predicted to be undergoing positive selection, can be hypothesized to be due to host pressure, but further investigation will be necessary to better describe this dynamic process.
The role of the immune-driven evolution of GII.3 NoVs merits further study, particularly with the investigation of HBGA binding in relationship to the evolutionary changes of the P2 domain over time. Previous studies have found that GII.4 NoVs bind HBGA types A, B, H3, Leb, and Ley in oligosaccharide-based binding assays, while GII.3 NoVs bound only types A and B strongly and Leb weakly (23). The fewer HBGAs bound by GII.3 NoVs in comparison to GII.4 NoVs may explain why GII.3 NoVs are not as prevalent as GII.4 NoVs in adults (28), in that there may be fewer hosts who are susceptible to GII.3 NoV infection. The studies on HBGA binding in the GII.3 NoVs were performed on more contemporary clusters II and III (Hu/NoV/GII.3/Mexico/1989/MX, cluster II; Hu/NoV/GII.3/ParisIsland/2003/US, cluster III). Our binding results agree with the lower level of amino acid variation found on GII.3 NoVs, since all of the clusters analyzed typically bound the same types of carbohydrates, contrasting with a wider and continually changing repertoire of HBGAs bound by GII.4 NoVs over time. Regardless of the binding pattern differences between GII.4 clusters, cluster-specific sera were able to block the interaction of heterologous GII.4 VLPs to HBGAs, even between clusters three decades apart. These data suggest the presence of several conserved antigenic sites within strains of a particular genotype. Several studies show evidence for GII.4 NoVs escaping the immune response through cluster replacement (2, 6, 11, 12, 27), but our HBGA-blocking binding assay data did not support these predictions. If GII.4 NoVs are indeed escaping the immune response by cluster replacement, the “blockade” assay might not adequately predict major antigenic shifts within a genotype.
Our study provides novel insight into the evolution of the VP1 protein of GII.3 NoVs over a 31-year period and includes the characterization of three sequences older than any previously described. These data demonstrate that GII.3 NoVs were circulating and causing severe disease in humans prior to 1983, the date of the oldest GII.3 NoV VP1 sequence previously available in public databases, and that the currently circulating GII.3 strains probably have a common ancestor between 1970 and 1973. Our study also provided insight into the evolution of GII.3 NoVs compared to that of their more prevalent relative, the GII.4 NoVs. While both of these genotypes had similar rates of nucleotide evolution, the fixation of amino acid substitutions over time was strikingly different, with GII.3 NoVs demonstrating more similarity in amino acid sequence between samples isolated three decades ago than samples isolated in the same decade. This, together with the difference in location of the positively selected amino acids, suggests that these two genotypes are under differing host-specific selective pressures. A clear and thorough characterization of the various patterns of evolution employed by different NoV genotypes is critical for the development of suitable vaccine candidates which need to be formulated to target the most likely strains circulating at a given point in time.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
We thank Norma Santos and Taka Hoshino (LID, NIAID) for their contributions to this study. We also thank Michael Dolan for his assistance with structure modeling. We also thank the Children's Hospital National Medical Center Research Group, led by H. W. Kim, which originally obtained the specimens and the clinical data.
We have no conflicts of interest to disclose.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Barreira D. M., et al. 2010. Viral load and genotypes of noroviruses in symptomatic and asymptomatic children in southeastern Brazil. J. Clin. Virol. 47:60–64 [DOI] [PubMed] [Google Scholar]
- 2. Bok K., et al. 2009. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83:11890–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bok K., et al. 2011. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc. Natl. Acad. Sci. U. S. A. 108:325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brandt C. D., et al. 1983. Pediatric viral gastroenteritis during eight years of study. J. Clin. Microbiol. 18:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bull R. A., Eden J. S., Rawlinson W. D., White P. A. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6:e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cannon J. L., et al. 2009. Herd immunity to GII.4 noroviruses is supported by outbreak patient sera. J. Virol. 83:5363–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao S., et al. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. CDC 23 February 2010, posting date Norovirus: technical fact sheet. CDC, Atlanta, GA: http://www.cdc.gov/ncidod/dvrd/revb/gastro/norovirus-factsheet.htm [Google Scholar]
- 9. Dey S. K., et al. 2010. Seasonal pattern and genotype distribution of norovirus infection in Japan. Pediatr. Infect. Dis. J. 29:e32–34 [DOI] [PubMed] [Google Scholar]
- 10. Domingo E., Holland J. J. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151–178 [DOI] [PubMed] [Google Scholar]
- 11. Donaldson E. F., Lindesmith L. C., Lobue A. D., Baric R. S. 2008. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 225:190–211 [DOI] [PubMed] [Google Scholar]
- 12. Donaldson E. F., Lindesmith L. C., Lobue A. D., Baric R. S. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. Microbiol. 8:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dove W., et al. 2005. Detection and characterization of human caliciviruses in hospitalized children with acute gastroenteritis in Blantyre, Malawi. J. Med. Virol. 77:522–527 [DOI] [PubMed] [Google Scholar]
- 14. Drummond A. J., Ho S. Y., Phillips M. J., Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drummond A. J., Nicholls G. K., Rodrigo A. G., Solomon W. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161:1307–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drummond A. J., Rambaut A., Shapiro B., Pybus O. G. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185–1192 [DOI] [PubMed] [Google Scholar]
- 18. Duffy S., Shackelton L. A., Holmes E. C. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9:267–276 [DOI] [PubMed] [Google Scholar]
- 19. Gray J. J., Jiang X., Morgan-Capner P., Desselberger U., Estes M. K. 1993. Prevalence of antibodies to Norwalk virus in England: detection by enzyme-linked immunosorbent assay using baculovirus-expressed Norwalk virus capsid antigen. J. Clin. Microbiol. 31:1022–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Green K. Y. 2007. Caliciviridae: the noroviruses, p. 949–979 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 21. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 22. Hansman G., Jiang X., Green K. Y. 2010. Caliciviruses. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 23. Huang P., et al. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 188:19–31 [DOI] [PubMed] [Google Scholar]
- 24. Kapikian A. Z., et al. 1972. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 10:1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar S., Ochoa W., Kobayashi S., Reddy V. S. 2007. Presence of a surface-exposed loop facilitates trypsinization of particles of Sinsiro virus, a genogroup II.3 norovirus. J. Virol. 81:1119–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemey P., Salemi M., Vandamme A. 2009. The Phylogenetic handbook, 2nd ed Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 27. Lindesmith L. C., Donaldson E. F., Baric R. S. 2011. Norovirus GII.4 strain antigenic variation. J. Virol. 85:231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lindesmith L. C., et al. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McGinnis S., Madden T. L. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakata S., Estes M. K., Chiba S. 1988. Detection of human calicivirus antigen and antibody by enzyme-linked immunosorbent assays. J. Clin. Microbiol. 26:2001–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nilsson M., et al. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 77:13117–13124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Numata K., Nakata S., Jiang X., Estes M. K., Chiba S. 1994. Epidemiological study of Norwalk virus infections in Japan and southeast Asia by enzyme-linked immunosorbent assays with Norwalk virus capsid protein produced by the baculovirus expression system. J. Clin. Microbiol. 32:121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patel M. M., et al. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Phan T. G., et al. 2005. Etiologic agents of acute gastroenteritis among Japanese infants and children: virus diversity and genetic analysis of sapovirus. Arch. Virol. 150:1415–1424 [DOI] [PubMed] [Google Scholar]
- 35. Pond S. L., Frost S. D. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 36. Posada D., Crandall K. A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 37. Rambaut A. 2009. Tree figure drawing tool Fig Tree, 1.3.1 ed University of Edinburgh, Edinburgh, United Kingdom [Google Scholar]
- 38. Roy A., Kucukural A., Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siebenga J. J., et al. 2008. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: a model for in vivo molecular evolution. J. Infect. Dis. 198:994–1001 [DOI] [PubMed] [Google Scholar]
- 40. Siebenga J. J., et al. 2010. Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants. PLoS Pathog. 6:e1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Siebenga J. J., et al. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J. Infect. Dis. 200:802–812 [DOI] [PubMed] [Google Scholar]
- 42. Symes S. J., Gunesekere I. C., Marshall J. A., Wright P. J. 2007. Norovirus mixed infection in an oyster-associated outbreak: an opportunity for recombination. Arch. Virol. 152:1075–1086 [DOI] [PubMed] [Google Scholar]
- 43. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 44. Tan M., et al. 2008. Outbreak studies of a GII-3 and a GII-4 norovirus revealed an association between HBGA phenotypes and viral infection. J. Med. Virol. 80:1296–1301 [DOI] [PubMed] [Google Scholar]
- 45. Xerry J., Gallimore C. I., Iturriza-Gomara M., Gray J. J. 2009. Tracking the transmission routes of genogroup II noroviruses in suspected food-borne or environmental outbreaks of gastroenteritis through sequence analysis of the P2 domain. J. Med. Virol. 81:1298–1304 [DOI] [PubMed] [Google Scholar]
- 46. Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y. 2009. I-TASSER: fully automated protein structure prediction in CASP8. Proteins 77(Suppl. 9):100–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zheng D. P., et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.