Abstract
In response to mammalian orthoreovirus (MRV) infection, cells initiate a stress response that includes eIF2α phosphorylation and protein synthesis inhibition. We have previously shown that early in infection, MRV activation of eIF2α phosphorylation results in the formation of cellular stress granules (SGs). In this work, we show that as infection proceeds, MRV disrupts SGs despite sustained levels of phosphorylated eIF2α and, further, interferes with the induction of SGs by other stress inducers. MRV interference with SG formation occurs downstream of eIF2α phosphorylation, suggesting the virus uncouples the cellular stress signaling machinery from SG formation. We additionally examined mRNA translation in the presence of SGs induced by eIF2α phosphorylation-dependent and -independent mechanisms. We found that irrespective of eIF2α phosphorylation status, the presence of SGs in cells correlated with inhibition of viral and cellular translation. In contrast, MRV disruption of SGs correlated with the release of viral mRNAs from translational inhibition, even in the presence of phosphorylated eIF2α. Viral mRNAs were also translated in the presence of phosphorylated eIF2α in PKR−/− cells. These results suggest that MRV escape from host cell translational shutoff correlates with virus-induced SG disruption and occurs in the presence of phosphorylated eIF2α in a PKR-independent manner.
INTRODUCTION
The nonfusogenic mammalian orthoreoviruses (MRV) are members of a large family of animal and plant viruses (Reoviridae) that includes many members that are of considerable importance in human, animal, and plant disease. Following infection with many viruses from this family, including MRV, the host cell initiates a stress response that culminates in shutoff of protein translation (10, 34, 43). Viral mRNAs are able to escape this inhibition and continue to be translated in the shutoff environment (40, 41, 52). In the case of MRV, the innate immune response has been implicated in host translational shutoff via activation of the double-stranded RNA (dsRNA) kinase PKR (12, 37, 40, 45). PKR is activated by binding dsRNA, at which point it homodimerizes and undergoes autophosphorylation (51). Activated PKR phosphorylates serine 51 on the alpha subunit of the cellular translation initiation factor eIF2 (23). In the absence of cellular stress, eIF2α binds to GTP and initiator methionyl-tRNA (met-tRNAi) to form a ternary complex, which subsequently binds to the 40S ribosomal complex to form the 43S preinitiation complex. As translational initiation proceeds, eIF2-bound GTP is hydrolyzed to release initiation factors from the ribosome. The released GDP bound to eIF2 must be exchanged for GTP in a reaction catalyzed by the guanine nucleotide exchange factor, eIF2B. Upon this exchange, eIF2-GTP can again bind met-tRNAi to initiate a new round of translation. When phosphorylated, eIF2α changes from a substrate to a competitive inhibitor of eIF2B, preventing the exchange of GDP for GTP. This results in global inhibition of protein synthesis (reviewed in references 33 and 35). There is an excess of eIF2 relative to eIF2B in the cell; therefore, phosphorylation of as little as 30% of cellular eIF2α can completely inhibit protein synthesis (18, 20, 25).
Previous data suggested that some strains of MRV are able to prevent cellular induction of PKR activation, eIF2α phosphorylation, and subsequent host translational shutoff (22, 40, 41, 44). Prevention of translational shutoff was mapped to the σ3-encoding S4 gene by reassortant genetics (41). σ3 from all MRV strains is a sequence-independent dsRNA binding protein that has been shown to functionally replace adenovirus VAI RNA and vaccinia virus E3L protein, both known PKR inhibitors (1, 22). Based on this, it was proposed that σ3 from MRV strains that prevent host shutoff bind dsRNA in a manner that interferes with PKR activation, preventing eIF2α phosphorylation and subsequent translation inhibition. The differences between strains that interfere with PKR activity and those that do not may be dependent on the levels and localization of free σ3 protein in the infected cell (40). While strong evidence supports a role for σ3 in modulating the ability of MRV to prevent the host cell from shutting off protein translation in response to infection, the mechanism behind the ability of viral mRNAs to escape translational shutoff when PKR activation and eIF2α phosphorylation are not prevented remains poorly understood.
A number of studies have illustrated additional consequences of eIF2α phosphorylation on mRNA and the cellular translation machinery. The reduction in available ternary complex that results from eIF2α phosphorylation leads to an increase in 48S preinitiation complexes unable to recruit 60S ribosomal complexes for translation initiation. This destabilization of polysomes leads to the rapid localization of mRNAs, translation initiation factors, and small, but not large, ribosomal subunits to structures in the cytoplasm called stress granules (SGs) (13, 14, 17). A number of proteins, such as TIAR/TIA-1 and G3BP, all of which have RNA binding and self-aggregation domains, play key roles in the formation and recruitment of protein and RNA components to SGs (8, 47). eIF2α phosphorylation is sufficient to induce SGs; however, drugs that interfere with translation initiation (hippouristinol, pateamine A, 15d-PGJ2, and NSC119893) and small interfering RNAs (siRNAs) targeted to many translation initiation factors [eIF4B, 4H, or poly(A) binding protein] induce SG formation independently of eIF2α phosphorylation (3, 16, 26, 30). SGs are thought to function as sites of mRNA triage where mRNAs are held in a translationally silent state until the cell either recovers from stress or undergoes apoptosis (13).
We have recently shown that MRV infection induces formation of SGs in an eIF2α phosphorylation-dependent manner early during infection at a step following virus uncoating but preceding viral-gene expression. We found that as MRV infection proceeds, SGs are disrupted in a manner that is dependent on viral translation, suggesting that a viral protein or protein complex may be involved in SG disruption (36). Initial induction and subsequent disruption of SGs during viral infection occur in a number of viral systems. SG disruption following West Nile virus infection occurs by TIAR/TIA-1 binding to the 3′ stem-loop of negative-strand viral RNA (6, 21). Poliovirus prevents SG formation by cleaving G3BP with the viral 3C proteinase (49). Other viruses, including Semliki Forest virus and rotavirus, also interfere with SG formation, although the mechanisms for this interference remain to be identified (28, 31). In this study, we found that MRV disruption of SGs occurs downstream of eIF2α phosphorylation and examined the impact of SGs on viral and cellular translation in the absence and presence of eIF2α phosphorylation. We found that SG disruption correlates with release of viral, but not cellular, mRNA from host translational inhibition and that MRV translation occurs in the presence of high levels of phosphorylated eIF2α in a manner independent of PKR inhibition.
MATERIALS AND METHODS
Cells and reagents.
HeLa, Cos-7, and PKR−/− cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen Life Technologies) containing 10% fetal calf serum (Atlanta Biologicals) and penicillin-streptomycin (100 IU/ml; Mediatech). Spinner-adapted L929 cells were maintained in Joklik's minimal essential medium (c-MEM) (Irvine Scientific) containing 2% fetal calf serum, 2% bovine calf serum (HyClone Laboratories, Atlanta Biologicals), 2 mM l-glutamine (Mediatech), and penicillin-streptomycin (100 IU/ml; Mediatech). The primary antibodies used in immunofluorescence and immunoblotting assays were as follows: goat polyclonal anti-TIAR (α-TIAR) antibody (sc-1749; Santa Cruz Biotechnology), rabbit polyclonal α-tubulin antibody (11H10; Cell Signaling Technologies), rabbit polyclonal α-phospho-eIF2α (Ser51) antibody (9721; Cell Signaling Technologies), and rabbit anti-β-actin (4967; Cell Signaling Technologies). Rabbit polyclonal antiserum against μNS was made by the Iowa State University hybridoma facility by injection of rabbits with peptides corresponding to μNS amino acids (aa) 1 to 20 and μNS aa 21 to 40 synthesized on a Multiple Antigen Peptide System (MAPS). The secondary antibodies used in immunofluorescence and immunoblotting experiments were as follows: Alexa 488-, Alexa 594-, or Alexa 350-conjugated donkey α-mouse, α-rabbit, or α-goat IgG antibodies (Invitrogen) and alkaline phosphatase (AP)-conjugated goat α-rabbit IgG antibodies (Bio-Rad). Sodium arsenite (SA) (Sigma-Aldrich) was used where indicated at a final concentration of 0.5 mM. Cycloheximide (Sigma-Aldrich) was used where indicated at a final concentration of 10 μg/ml. 15D-PGJ2 (Sigma-Aldrich, Caymen Biochemicals) was used where indicated at a final concentration of 50 μM. NSC119893 was obtained from the NIH/NCI Developmental Therapeutics Program and used where indicated at a final concentration of 10 μM. AP-conjugated streptavidin (Invitrogen) and Alexa 488-conjugated streptavidin (Invitrogen) were used to stain nascent proteins in l-azidohomoalanine (l-AHA) labeling experiments.
Virions.
Purified MRV virions (T1L, T2J, and T3D strains) are our laboratory stocks. Purified virions were prepared as described previously (29), using Vertrel reagent (DuPont) in place of Freon, and stored in dialysis buffer (150 mM NaCl, 10 mM Tris, pH 7.4, 10 mM MgCl2) at 4°C.
Viral infection.
Cells (2.0 × 105 or 4.0 × 105) were seeded onto 35- or 60-mm dishes the day before infection. The cells were infected with MRV strain T2J or T3D at 1 or 100 PFU as determined by standard plaque assay on L929 cells (7) or 1 to 5 cell-infecting units (CIU) based on titers that were determined on each cell line (9, 39). To determine CIU, each purified virus stock was used to infect individual cell lines using 1, 10, and 100 μl of virus. The infected cells were incubated overnight and processed for immunofluorescence assay using μNS antiserum. The numbers of infected cells per microscope field were determined in 10 independent fields, and CIU/ml were calculated by multiplying the average number of infected cells per field by the number of fields per well, the reciprocal of dilution, and the volume factor. Using this calculation, 1 CIU resulted in infection of 75 to 95% of cells. Virus was diluted in phosphate-buffered saline (PBS) (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4 [pH 7.5]) plus 2 mM MgCl2 and adsorbed to cells for 1 h. The cells were then refed with DMEM and incubated at 37°C until they were harvested.
l-AHA protein labeling.
For measuring MRV translation and viral replication in the presence of l-AHA (Click-iT AHA; Invitrogen), L929 cells were seeded in 60-mm dishes and infected with 1 PFU of T3D in the absence and presence of 50 μM l-AHA. Cells were harvested at the indicated time points and processed for plaque assay on L929 cells (7) or for biotin labeling as described below, followed by blotting using streptavidin-conjugated AP to detect de novo-translated proteins. For Western blot assays, Cos-7 cells were seeded on 6-well dishes at a density of 2 ×105 cells/well and then incubated overnight at 37°C and infected with MRV or mock infected. At the indicated times postinfection (p.i.), the medium was replaced with prewarmed methionine-deficient medium containing SA, cycloheximide, or 15D-PGJ2 for 1 h, at which point l-AHA was added at a final concentration of 50 μM and the cells were incubated an additional 1 h. For biotin labeling, cells were harvested and lysed with 1% sodium dodecyl sulfate (SDS) in 50 mM Tris-HCl, pH 8.0, and then l-AHA-labeled proteins were conjugated with biotin as described by the manufacturer with the following changes. For each reaction, 30 μl of components A and B was added to 50 μl cell lysate and vortexed for 5 s; 5 μl of component C was then added to the cell lysate, followed by 5 μl of component D and 10 μl of component E. The treated lysates were incubated with rotation for 20 min at room temperature. Following labeling, the proteins were precipitated as described by the manufacturer, resuspended in protein-loading buffer, and separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For in situ translation, Cos-7 cells were seeded on 12-well dishes containing 12-mm-diameter round coverslips at a density of 1 × 105 cells/well, incubated overnight at 37°C, and then infected with MRV or mock infected. At the indicated times p.i., the medium was replaced with prewarmed methionine-minus medium containing SA, cycloheximide, or 15D-PGJ2 for 45 min, at which point l-AHA was added at a final concentration of 50 μM, and the cells were incubated an additional 30 min. The cells were fixed with 100% ice-cold methanol at −20°C for 3 min and then washed three times with PBS. The fixed cells were permeabilized by incubation with 0.2% Triton X-100 in PBS for 5 min and washed three times with PBS. The coverslips were transferred, cell side up, to Parafilm and incubated with reagents provided for biotin labeling from Invitrogen. Alterations to the protocol provided by the manufacturer were as follows: 25 μl of components A and B was added to 15 μl PBS, and 40 μl of this solution was added to each coverslip; 2.5 μl of component C was added to the coverslips, followed by 2.5 μl of component D and 5 μl of component E. The coverslips were incubated in this solution for 20 min and then rinsed and processed for immunofluorescence (IF) assay.
IF assay.
At the indicated times, cells were fixed at room temperature for 10 min with 2% paraformaldehyde in PBS or at −20°C with 100% methanol for 3 min. The fixed cells were washed three times with PBS, permeabilized by incubation with 0.2% Triton X-100 in PBS for 5 min, and then washed three times with PBS. Samples were blocked by a 10-min incubation with 2% bovine serum albumin (BSA) in PBS. Primary and secondary antibodies were diluted in 2% BSA in PBS. After blocking, the cells were incubated for 1 h with primary antibodies or streptavidin-conjugated Alexa 488, washed three times with PBS, and then incubated for an additional hour with secondary antibodies. The immunostained cells were washed a final three times with PBS and mounted on slides with Prolong reagent with or without DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (Invitrogen). The immunostained samples were examined with a Zeiss Axiovert 200 inverted microscope equipped with fluorescence optics or an Olympus laser scanning confocal microscope augmented with spectral deconvolution hardware and equipped with four variable-voltage color lasers. Images were prepared using Photoshop and Illustrator software (Adobe Systems). Following IF processing, quantification of SG formation and translation activity in SA- and 15D-PGJ2-treated cells was done by counting cells based on their SG and translation phenotypes. More than 100 cells were counted in each treatment group, and the averages of two independent experiments were subjected to an unpaired Student's t test using GraphPad software to determine the statistical significance of differences between mock- and MRV-infected samples in each phenotype group. Differences in groups that were found to be statistically different (P < 0.005) are indicated.
Immunoblotting.
Samples were harvested at the indicated times and lysed with 100 μl lysis buffer (50 mM Tris, pH 8.0, 1% SDS). Proteins were transferred to a nitrocellulose membrane by electroblotting in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol [pH 8.3]). The nitrocellulose membrane containing transferred proteins was blocked for 15 min with 5% BSA in Tris-buffered saline (20 mM Tris, 137 mM NaCl [pH 7.6]) containing 0.1% Tween (TBS-T) and then incubated overnight with primary antibodies in TBS-T containing 1% milk or AP-conjugated streptavidin in PBS containing 5% BSA. Blots incubated with primary antibodies were washed three times for 15 min with TBS-T, followed by 4 h of incubation with AP-conjugated secondary antibodies in TBS-T containing 1% milk. The blots were washed a final three times, and AP conjugate substrates (Bio-Rad [colorimetric] or Millipore [chemiluminescent]) were applied. Images were collected using a Chemi-doc XRS camera (Bio-Rad), and protein bands were quantified using Quantity One software (Bio-Rad). All experiments were independently performed between 3 and 5 times, and representative results are shown.
Quantification of phosphorylated eIF2α.
In all experiments where quantification of eIF2α phosphorylation is shown, the Quantity One-determined amount of eIF2α protein was divided by the amount of the in-gel loading control protein α-tubulin to control for gel-loading differences. For time course experiments, the quantity of phosphorylated eIF2α at time zero was arbitrarily set to 1 within each time course. The increase in eIF2α phosphorylation over time was determined by dividing the quantity present at each time point by the quantity present at time zero. For SA induction experiments, the increase in eIF2α phosphorylation in the presence of SA was determined by dividing the quantity of phosphorylated eIF2α present in the presence of SA by the quantity of phosphorylated eIF2α present in the absence of SA. Quantification was performed on at least two independent experiments, and representative results or mean values with standard deviations are shown.
RESULTS
eIF2α is phosphorylated in MRV-infected cells in a strain- and cell-type-specific manner.
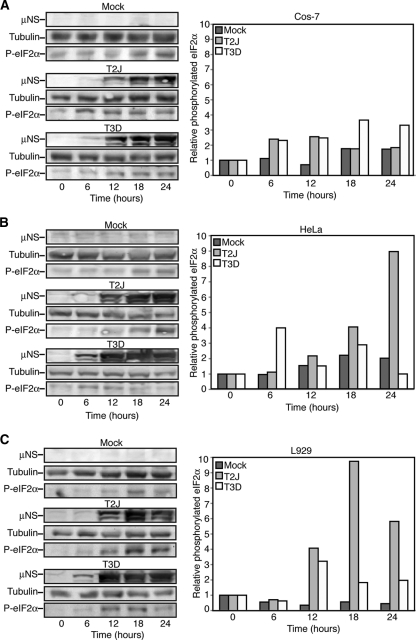
There is ample evidence that infection with MRV strain T2J elicits a cellular innate immune response that includes activation of PKR and subsequent phosphorylation of eIF2α in L929 cells, whereas MRV strain T3D prevents this activation (12, 37, 40, 44, 45). To further examine the impact of MRV on cellular eIF2α phosphorylation, we measured the levels of phosphorylated eIF2α over a 24-h time course in Cos-7, HeLa, and L929 cells infected with MRV strains T2J and T3D. Immunoblot analysis was performed on cell lysates harvested at 6-h intervals from mock-infected cells and cells infected with T2J or T3D at 1 CIU/cell. We found that infection with MRV led to an increase in eIF2α phosphorylation compared with mock-infected cells in a viral strain- and cell-type-dependent manner (Fig. 1). In Cos-7 cells, T3D induced eIF2α phosphorylation to levels above mock throughout infection, whereas T2J induced eIF2α phosphorylation at early times in infection that diminished by late times in infection (Fig. 1A). In HeLa and L929 cells, T2J infection induced increased levels of phosphorylated eIF2α over time, whereas T3D did not induce substantial levels of phosphorylated eIF2α compared to mock-infected cells (Fig. 1B and C). There was no observable change in the levels of total eIF2α in these experiments (data not shown). Although phosphorylation of eIF2α induced by MRV strains in Cos-7 and HeLa cells have not to our knowledge been previously investigated in the absence of exogenous interferon (32), the L929 data agree with previously published reports (44). Taken together, these data suggest that MRV infection results in induction of eIF2α phosphorylation in both strain- and cell-type-specific manners.
Fig. 1.
MRV infection induces eIF2α phosphorylation in a strain- and cell-type-specific manner. Cos-7 (A), HeLa (B), or L929 (C) cells were mock infected or infected with MRV T2J or T3D. At 0, 6, 12, 18, and 24 h p.i., cells were harvested and proteins were separated on SDS-PAGE and transferred to nitrocellulose. The membranes were blotted with rabbit α-μNS polyclonal antibody, rabbit α-tubulin polyclonal antibody, and rabbit α-phosphorylated (P)-eIF2α polyclonal antibody, followed by goat α-rabbit IgG conjugated with AP. Bound AP conjugates were detected by chemiluminescence staining and quantified with Quantity-One software. Quantified amounts of phosphorylated eIF2α were divided by quantified amounts of α-tubulin to adjust for differences in gel loading, and then increases in eIF2α phosphorylation relative to time zero were calculated and are shown on the right.
SGs are not present in MRV-infected cells at late times p.i.
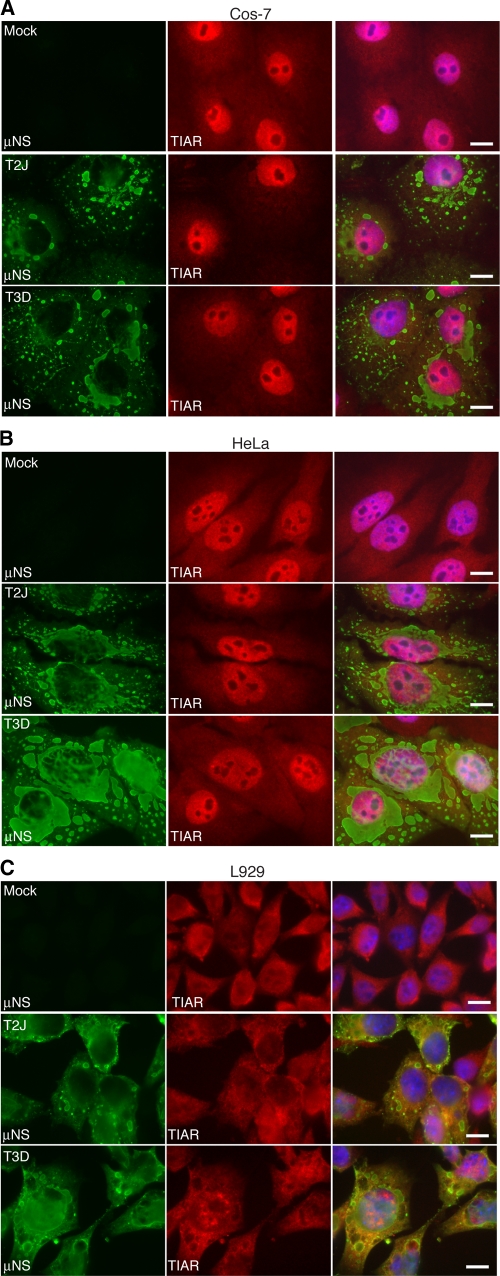
eIF2α phosphorylation is sufficient to induce SGs (15); therefore, based on the phosphorylation data in Fig. 1, we predicted that Cos-7 cells infected with MRV strain T3D and HeLa and L929 cells infected with MRV strain T2J would contain SGs. We previously showed that at later times in infection (10 to 24 h p.i.) only between 2 and 5% of T3D-infected HeLa cells contained SGs (36). We have now expanded these studies to determine if the absence of SGs late in infection is limited to MRV strains that do not induce eIF2α phosphorylation or whether MRV has the capacity to interfere with SGs in the presence of virus-induced phosphorylation of eIF2α. L929, Cos-7, and HeLa cells were infected with MRV strain T2J or T3D at 1 CIU/cell and at 24 h p.i. were fixed and stained with antibodies against the MRV nonstructural protein μNS and the SG component protein TIAR. The infected cells were then examined by immunofluorescence microscopy to determine whether MRV-infected cells contained SGs. We found that in Cos-7 (Fig. 2A), HeLa (Fig. 2B), or L929 (Fig. 2C) cells there were no SGs present in T2J (middle rows)- or T3D (bottom rows)-infected cells, and that localization of the SG-associated protein TIAR did not appear to be substantially different from that in mock-infected cells (top rows). In contrast, in mock-infected cells treated with sodium arsenite (SA), an oxidative substance that induces high levels of eIF2α phosphorylation via activation of the heme-regulated inhibitor (HRI) kinase (24, 27), 99% of cells contained SGs (Fig. 3A, B, and C, top rows). These data suggest that the lack of SGs at late times in MRV-infected cells is independent of the viral strain and cell type and is not dictated by whether the strain induces phosphorylation of eIF2α. Moreover, these data suggest that cellular signals connecting eIF2α phosphorylation and SG formation are uncoupled in MRV-infected cells.
Fig. 2.
MRV-infected cells do not contain SGs at late times p.i. Cos-7 (A), HeLa (B), and L929 (C) cells were mock infected (top row) or infected with MRV T2J (middle row) or T3D (bottom row). At 24 h p.i., the cells were fixed and immunostained with rabbit α-μNS poly- clonal antiserum (left column) and goat α-TIAR polyclonal antibody (middle column), followed by Alexa 594-conjugated donkey α-rabbit IgG and Alexa 488-conjugated donkey α-goat IgG. Merged images containing DAPI-stained nuclei (blue) are shown (right columns). Bars = 10 μm.
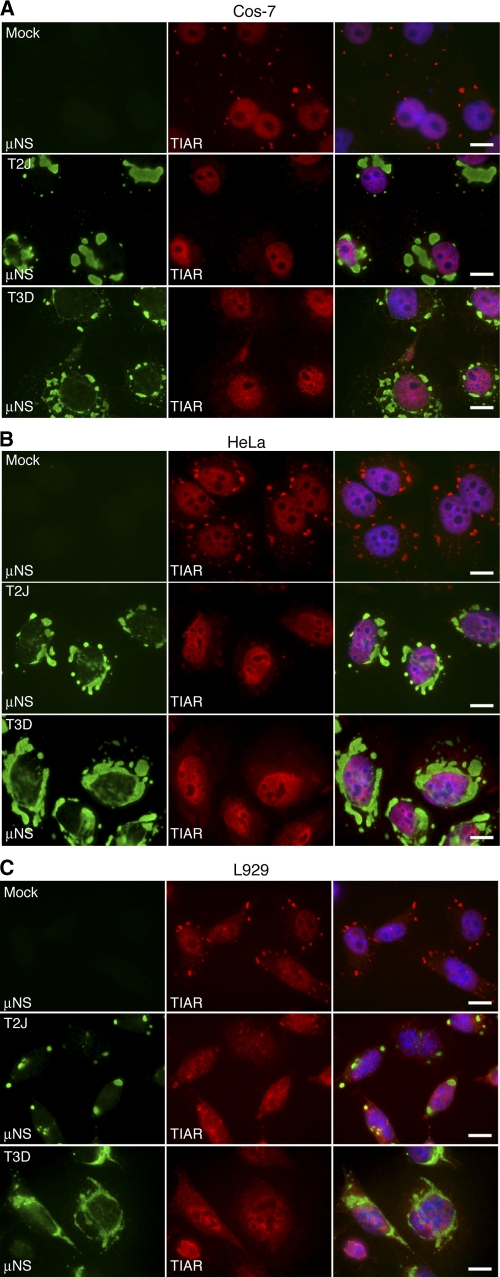
Fig. 3.
MRV infection renders cells unable to form SGs in response to SA. Cos-7 (A), HeLa (B), or L929 (C) cells were mock infected (top rows) or infected with MRV T2J (middle rows) or T3D (bottom rows). At 24 h p.i., the cells were treated with SA for 1 h and then fixed and immunostained with rabbit α-μNS polyclonal antiserum (left columns) and goat α-TIAR polyclonal antibody (middle columns), followed by Alexa 594-conjugated donkey α-rabbit IgG and Alexa 488-conjugated donkey α-goat IgG. Merged images containing DAPI-stained nuclei (blue) are shown (right columns). Bars = 10 μm.
MRV infection renders cells unable to form SGs in response to external stress signals.
The absence of SGs in cells where MRV induced eIF2α phosphorylation suggested that the virus is able to interfere with SG formation following virus-induced eIF2α phosphorylation. To examine the capacity of MRV to prevent SG formation in response to other cellular stress signals that induce eIF2α phosphorylation, we examined uninfected and infected cells for SG formation following SA treatment. Cos-7 cells were infected with MRV strain T2J or T3D at 1 CIU/cell and at 24 h p.i. were treated with SA for 1 h and then fixed and stained for immunofluorescence microscopy to visualize infected cells and SGs. Remarkably, while SGs were apparent in most uninfected cells following SA treatment (Fig. 3A, B, and C, top rows), infected cells did not contain SGs (Fig. 3A, B, and C, middle and bottom rows). This experiment was repeated using other known SG-localized protein markers, including TIA-1, eIF3, eIF4E, and eIF4G, with identical results (data not shown). This finding was both viral strain and cell type independent, as all three prototype strains (T1L, T2J, and T3D) interfered with SG formation in a number of cell types, including Cos-7, L929, HeLa, CV-1, and MEFs (Fig. 3A, B, and C and data not shown). Additionally, examination of SA-treated infected Cos-7, HeLa, and L929 cells by confocal microscopy using a sequential scan of Kalman-averaged planes with identical exposures and a Z-plane thickness of 0.33 μm confirmed that SGs were not obscured by viral factories in these experiments and that they did not form in response to SA treatment in MRV-infected cells (data not shown). These results strengthen the findings in Fig. 2 that suggest MRV uncouples the cellular signals between eIF2α phosphorylation and SG formation and also support a hypothesis that interference with SG formation is independent of the specific ability of the virus strain to interfere with PKR activation and eIF2α phosphorylation.
MRV SG interference is independent of eIF2α phosphorylation.
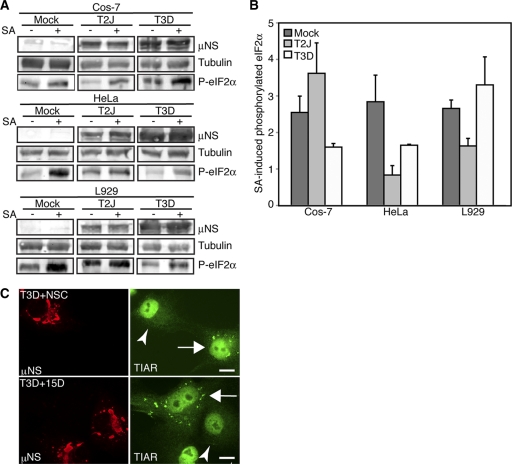
Because both MRV infection and SA treatment can induce eIF2α phosphorylation (24, 37, 40, 41, 44) (Fig. 1A, B, and C), our data suggested that MRV interference with SG formation at late times in infection with strains that induce eIF2α phosphorylation and following SA treatment occurs in the presence of phosphorylated eIF2α. However, it remained possible that the levels of eIF2α phosphorylation found in MRV-infected cells were not sufficient to induce SGs and that SG formation following SA treatment was also prevented via MRV interference with eIF2α phosphorylation. We used two assays to rule out this possibility. First, to determine if MRV infection led to a decrease in SA-induced eIF2α phosphorylation, we examined the levels of phosphorylated eIF2α in MRV-infected versus uninfected cells upon addition of SA. Cos-7 cells were mock infected or infected with T2J or T3D at 1 CIU/cell. At 24 h p.i., the cells were subjected to SA for 1 h and then harvested and immunoblotted with antibodies against viral nonstructural protein μNS, α-tubulin, and phosphorylated eIF2α (Fig. 4A). The amounts of phosphorylated eIF2α in the absence and presence of SA in uninfected and infected cells were quantified by chemiluminescence to determine the fold induction of phosphorylated eIF2α following SA treatment in each sample (Fig. 4B). We found that in T2J-infected Cos-7 cells and T3D-infected HeLa and L929 cells, SA induced eIF2α phosphorylation to levels similar to those in uninfected cells (Fig. 4B), suggesting MRV does not inhibit SA induction of eIF2α phosphorylation in these viral strain/cell type combinations. This quantification method was complicated by the fact that T3D infection of Cos-7 cells and T2J infection of HeLa and L929 cells leads to the induction of phosphorylated eIF2α in the absence of SA (Fig. 1A, B, and C and Fig. 4A). As a result, the fold increase in eIF2α phosphorylation in the presence of SA is artificially lower than the fold increase measured in mock-infected cells in these samples. However, there does appear to be a qualitative induction of eIF2α phosphorylation in the presence of SA in these infected samples compared to untreated mock-infected cells (Fig. 4A), suggesting that SA induces eIF2α phosphorylation in all MRV-infected cells. Because SA did not induce SG formation in any viral strain/cell type combination (Fig. 3), these data suggest MRV is able to interfere with SGs in the presence of levels of phosphorylated eIF2α that are sufficient to induce SGs in uninfected cells.
Fig. 4.
MRV interferes with SGs downstream of eIF2α phosphorylation. (A) Cos-7, HeLa, or L929 cells were mock infected or infected with MRV T2J or T3D. At 24 h.p.i., the cells were left untreated or treated with SA for 1 h. Samples were harvested, and proteins were separated on SDS-PAGE and electroblotted to nitrocellulose. The nitrocellulose membranes were immunoblotted with rabbit α-μNS polyclonal antiserum, rabbit α-tubulin polyclonal antibodies, and rabbit α-phosphorylated (P) eIF2α polyclonal antibodies, as indicated, followed by goat α-rabbit IgG conjugated with AP, and bound AP conjugates were detected by chemiluminescence. (B) AP conjugates from panel A were quantified with Quantity-One software. Quantified amounts of phosphorylated eIF2α were divided by quantified amounts of α-tubulin to adjust for differences in gel loading. Fold increases in levels of phosphorylated eIF2α in SA-treated cells relative to untreated cells were calculated, and the means and standard deviations of two experimental replicates are shown. (C) Cos-7 cells were infected with MRV T3D, and at 24 h p.i., the cells were treated with either NSC119893 (top row) or 15D-PGJ2 (bottom row) for 1 h and then fixed and immunostained with rabbit α-μNS polyclonal antiserum (left column) and goat α-TIAR polyclonal antibody (right column), followed by Alexa 594-conjugated donkey α-rabbit IgG and Alexa 488-conjugated donkey α-goat IgG. The arrowheads identify infected cells that do not contain SGs. The arrows identify uninfected cells that do contain SGs. Bars = 10 μm.
To more directly detect whether MRV interference with SG formation occurs independently of eIF2α phosphorylation, we also examined the ability of MRV to interfere with SGs induced by NSC119893 and 15D-PGJ2, both of which induce SGs independently of eIF2α phosphorylation (16, 30). NSC119893 binds to eIF2α and inhibits ternary-complex formation (30), while 15D-PGJ2 binds to eIF4A and inhibits translation initiation (16). Cos-7 cells were infected with T3D, and at 24 h p.i., the cells were treated with NSC119893 or 15D-PGJ2 for 1 h. An immunofluorescence assay was then performed to visualize infected cells and SGs. We found that SGs did not form in response to these drugs in any MRV-infected cells (Fig. 4C, arrowheads), while SGs were found following drug treatment in most uninfected cells (Fig. 4C, arrows), confirming that interference with SG formation by MRV occurs independently of inhibition of eIF2α phosphorylation. The ability to interfere with SGs induced by these drugs was virus strain independent, as MRV T1L and T2J also prevented SG formation in identical experiments (data not shown). Taken together, the data in Fig. 3 and 4 show that at late times in infection, MRV prevents SG formation in the presence of cellular stress signals that are sufficient to induce SGs in uninfected cells in a manner independent of cellular eIF2α phosphorylation status. These data strongly suggest that MRV infection uncouples cellular signals between eIF2α phosphorylation and SG formation.
A nonradioactive labeling strategy for detection of newly translated proteins does not interfere with viral replication.
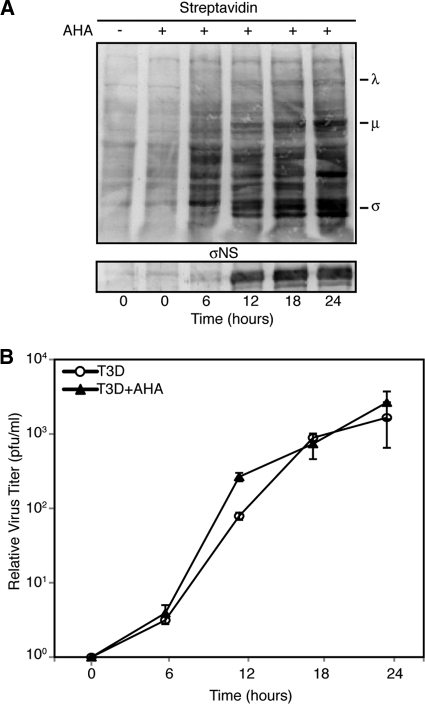
It is possible that disruption of SGs plays an important role in the MRV replication cycle. We hypothesized that SG disruption may be necessary for MRV escape from host translational shutoff. To examine this possibility, we were interested in measuring de novo synthesis of viral proteins in the presence and absence of SGs. To perform these experiments, we wanted to utilize a newly described technology that involves incorporation of the methionine analog l-AHA into proteins as they are synthesized for nonradioactive protein labeling. Following l-AHA incorporation, proteins are labeled with biotin-alkyne and are detected by immunoblotting or immunofluorescence assay using streptavidin-conjugated chemiluminescent or fluorescent substrates. This labeling strategy has been extensively tested and found to be nontoxic to cells, to specifically label newly synthesized proteins to levels comparable to 35S, and to have no impact on protein degradation or global protein synthesis rates (5). Nonetheless, to address the impact of this labeling strategy on MRV infection, we examined both viral protein translation and replication in the presence of l-AHA. L929 cells were infected with T3D at 1 PFU/cell for 1 h, at which point the cells were left untreated or treated with 50 μM l-AHA. At 6-h intervals, cells were harvested and prepared for immunoblot analysis or plaque assay. When examined by immunoblotting using streptavidin-conjugated AP, l-AHA was clearly incorporated into both cellular and viral proteins and did not appear to substantially impact viral protein translation (Fig. 5A). We did note that this technique resulted in low levels of background staining of the membrane even in the absence of l-AHA addition to cells (Fig. 5A, lane 1). Because there was no l-AHA incorporated into proteins in this lane, we suspect this background staining is a result of nonspecific binding of streptavidin-AP to proteins present in the membrane. As the specific staining of l-AHA proteins was easily visualized above this background level, we concluded that this background staining would not interfere with measuring cellular and viral protein synthesis in subsequent experiments. Importantly, when virus titers determined from l-AHA-treated infected cells were compared with titers from infected cells in which no l-AHA was present, there was no impact on the MRV titer (Fig. 5B). These data suggested that l-AHA labeling did not interfere with viral protein function necessary for MRV replication and that this technology was suitable for examining MRV translation.
Fig. 5.
l-AHA labeling does not interfere with MRV replication. L929 cells were infected with T3D with and without the addition of l-AHA, and at 0, 6, 12, 18, and 24 h, cells were harvested and processed. (A) Following lysis, samples were separated on SDS-PAGE, transferred to nitrocellulose, and then probed with streptavidin-conjugated AP or σNS polyclonal antibodies, followed by AP-conjugated α-rabbit IgG. Bound AP conjugates were detected by chemiluminescence. The positions of MRV proteins in the blot are indicated. (B) Lysed samples were subjected to standard MRV plaque assay on L929 cells. The plaques were counted, and the relative increases in virus titer from time zero were calculated. The means and standard deviations of two experimental replicates are shown.
MRV and cellular translation are inhibited when SGs are present.
To examine the impact of SGs on viral translation, we sought to create an environment in which MRV mRNAs were being translated to experimentally detectable levels but where the virus was not able to prevent drug-induced SG formation. Our previous data suggested that there is a major decrease in virus-induced SGs after 6 h p.i (36). We found that we could detect substantial levels of viral protein synthesis at 6 h p.i., and if we treated infected cells with SG-inducing drugs at this time, the virus was not yet capable of interfering with SG formation induced by drugs (data not shown and Fig. 6B and C).
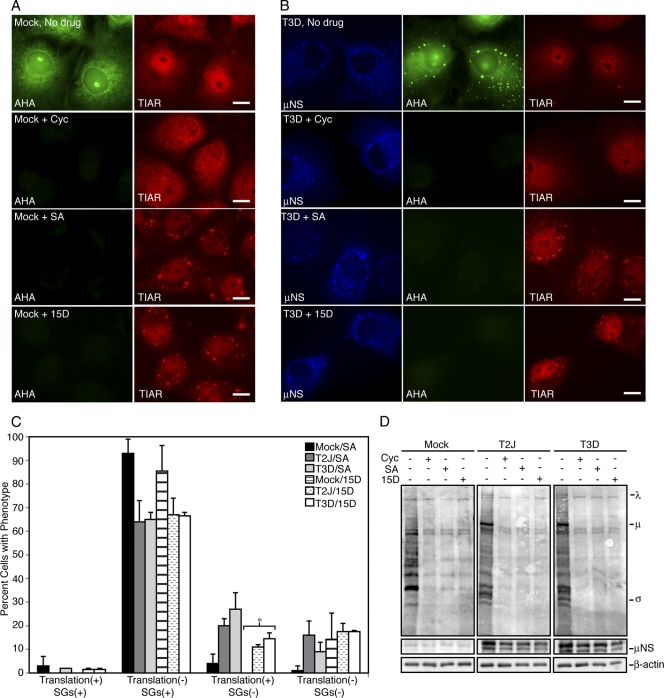
Fig. 6.
Viral and cellular translation are inhibited when SGs are present. (A and B) Cos-7 cells were mock infected (A) or infected with T3D (B), and at 6 h p.i., the cells were left untreated (No drug; top row) or treated with cycloheximide (Cyc; second row), SA (third row), or 15D-PGJ2 (15D; bottom row) for 45 min and then labeled with l-AHA for 30 min in the presence of drugs. The cells were fixed and permeabilized, labeled with biotin, and then stained with Alexa 488-conjugated streptavidin (AHA) (left column in panel A; middle column in panel B), rabbit α-μNS polyclonal antiserum (μNS) (left column in panel B), and goat α-TIAR polyclonal antibodies (TIAR) (right columns), followed by Alexa 350-conjugated donkey α-rabbit IgG or Alexa 594-conjugated donkey α-goat IgG. Bars = 10 μm. (C) Following treatment as in panel A, cells were counted based on their translation and SG phenotype. The percentage of cells containing each phenotype out of the total number of cells counted was calculated, and the means and standard deviations of two experimental replicates are shown. Infected groups that were statistically different from mock-infected cells are indicated by an asterisk (P < 0.005). (D) Cos-7 cells were mock infected or infected with T2J or T3D and were treated with drugs as in panel A for 60 min, at which point l-AHA was added in the presence of drugs for an additional 60 min. Proteins were labeled with biotin and then separated on SDS-PAGE and transferred to nitrocellulose by electroblotting. l-AHA-labeled proteins were detected by incubation of blots with AP-conjugated streptavidin. As protein loading and infection controls, identical sample volumes were examined in parallel using rabbit anti-β-actin polyclonal antibodies or rabbit α-μNS polyclonal antibodies followed by AP-conjugated goat α-rabbit IgG. The positions of MRV proteins on the AHA blot are indicated.
To visualize protein synthesis on an individual cell basis, Cos-7 cells were infected at 5 CIU/cell with MRV T2J or T3D, and at 6 h.p.i. the cells were left untreated or were treated with cycloheximide, SA, or 15D-PGJ2 for 45 min, at which point l-AHA was added for 30 min. The cells were then fixed, permeabilized, and subjected to a biotin-alkyne click reaction. Labeled cells were stained with virus- and SG-specific antibodies and Alexa 488-conjugated streptavidin and then examined by immunofluorescence microscopy for MRV infection, SG formation, and new protein synthesis. Both uninfected and infected, untreated cells synthesized new proteins during the labeling period (Fig. 6A and B, top rows, and data not shown [T2J]). Cycloheximide inhibited translation of new proteins without inducing SGs in both infected and uninfected cells (Fig. 6A and B, second rows, and data not shown [T2J]). SA and 15D-PGJ2 induced SGs in both uninfected and infected cells, and neither was able to support new protein synthesis (Fig. 6A and B, third and fourth rows). Quantification of these results showed that at early times p.i., MRV does not substantially inhibit SG formation and that the presence of SGs strongly correlates with inhibition of protein translation in uninfected and infected cells whether the SGs are induced by an eIF2α phosphorylation-dependent (SA) or -independent (15D-PGJ2) mechanism (Fig. 6C).
To examine the entire cell population, these experiments were repeated; however, after l-AHA incorporation, proteins were harvested and labeled with biotin, separated on SDS-PAGE, transferred to nitrocellulose, and then blotted with AP-conjugated streptavidin to visualize new protein synthesis. Similar to what was seen on an individual cell basis, new cellular and viral protein synthesis was detected in both uninfected and infected, untreated cells, but cycloheximide, SA, and 15D-PGJ2 inhibited both cellular and viral translation (Fig. 6D). These results confirm that both eIF2α phosphorylation-dependent and -independent induction of SGs correlates with the inhibition of cellular and viral translation in MRV-infected cells.
Viral mRNAs escape translational shutoff when SGs are disrupted.
SGs were not observed at late times in MRV-infected cells irrespective of eIF2α phosphorylation induced by the virus or by SA or following eIF2α phosphorylation-independent translation inhibition induced by 15D-PGJ2 (Fig. 2 to 4). To determine whether viral or cellular proteins are synthesized when SGs are disrupted by MRV, Cos-7 cells were infected with MRV T2J or T3D, and then, at 24 h p.i., the cells were left untreated or were treated with cycloheximide, SA, or 15D-PGJ2. l-AHA was incorporated into proteins that were translated in the absence and presence of drugs, and then the cells were fixed, permeabilized, and examined by immunofluorescence microscopy for MRV infection, SG formation, and new protein synthesis. In the absence of drugs, infected cells were found to synthesize new proteins during the labeling period (Fig. 7A, top row, and data not shown [T2J]). Cycloheximide inhibited translation of new proteins in infected cells (Fig. 7A, second row, and data not shown [T2J]). In contrast to mock-infected cells, which contained SGs and did not support new protein synthesis in the presence of SA or 15D-PGJ2 (Fig. 6A, third and bottom rows, and 7B), the majority of MRV-infected cells did not contain SGs and were capable of supporting new protein synthesis (Fig. 7A, third and bottom rows). Quantification of these results showed that there is a significant difference between uninfected and MRV-infected cells following drug treatment with regard to translation activity and SG presence, with the majority of mock-infected cells containing SGs and being translationally inactive and the majority of MRV-infected cells lacking SGs and being translationally active (Fig. 7B).
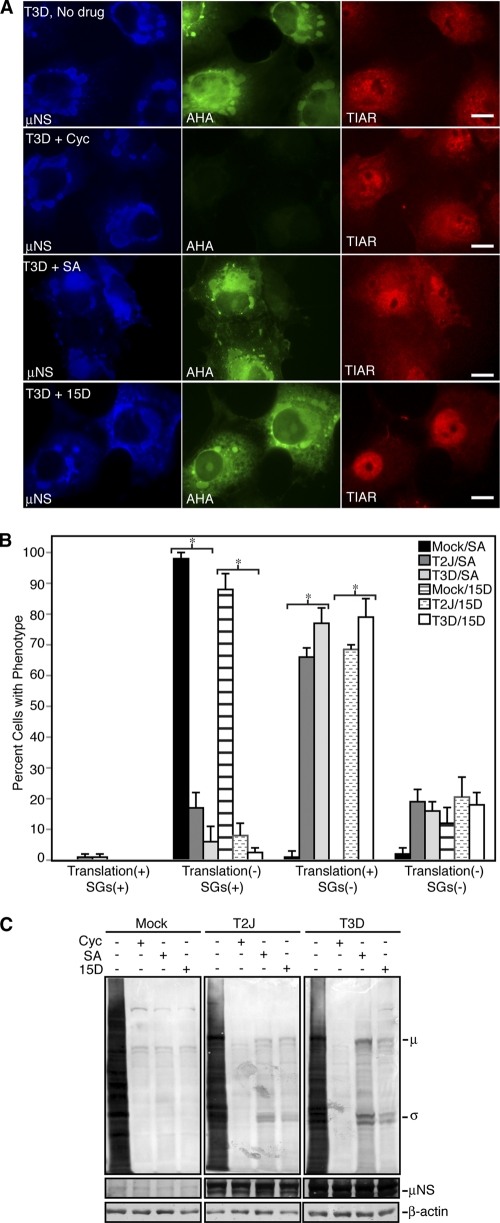
Fig. 7.
MRV mRNAs escape translational shutoff when SGs are disrupted. (A) Cos-7 cells were infected with T3D, and at 24 h p.i., the cells were left untreated (No drug; top row) or treated with cycloheximide (Cyc; second row), SA (third row), or 15D-PGJ2 (15D; bottom row) for 45 min and then labeled with l-AHA for 30 min in the presence of drugs. The cells were fixed, permeabilized, labeled with biotin, and then stained with Alexa 488-conjugated streptavidin (AHA; middle column), rabbit α-μNS polyclonal antiserum (μNS; left column), and goat anti-TIAR polyclonal antibodies (TIAR; right column), followed by Alexa 350-conjugated donkey α-rabbit IgG or Alexa 594-conjugated donkey α-goat IgG. Bars = 10 μm. (B) Following treatment as in panel A, cells were counted based on their translation and SG phenotype. The percentage of cells containing each phenotype out of the total number of cells counted was calculated, and the means and standard deviations of three experimental replicates are shown. Infected groups that were statistically different from mock-infected cells are indicated by an asterisk (P < 0.005). (C) Cos-7 cells were mock infected or infected with T2J or T3D and were treated with drugs as in panel A for 60 min, at which point l-AHA was added in the presence of drugs for an additional 60 min. Proteins were labeled with biotin and then separated on SDS-PAGE and transferred to nitrocellulose by electroblotting. l-AHA-labeled proteins were detected by incubation of blots with AP-conjugated streptavidin. As protein-loading and infection controls, identical sample volumes were examined in parallel using rabbit anti-β-actin polyclonal antibodies or rabbit α-μNS polyclonal antibodies followed by AP-conjugated goat α-rabbit IgG. The positions of MRV proteins on the AHA blot are indicated.
While this result showed that protein synthesis is active in MRV-infected cells where SGs were absent, it did not determine if MRV interference with SGs correlates with release of all protein translation inhibition or if only viral mRNA is translated under these conditions. To answer this question, we repeated these experiments; however, instead of visualizing proteins by fluorescence microscopy, we separated labeled proteins on SDS-PAGE, transferred them to nitrocellulose, and blotted them with AP-conjugated streptavidin. Examination of the proteins synthesized in the absence and presence of drugs confirmed that cycloheximide, SA, and 15D-PGJ2 induced nearly complete shutoff of cellular protein synthesis in the absence of MRV infection (Fig. 7C). Cycloheximide also induced shutoff of protein synthesis in MRV-infected cells. Remarkably, proteins that migrated at the predicted size of MRV proteins were the only obvious translation products in the presence of SA and 15D-PGJ2 in MRV-infected cells (Fig. 7C). We confirmed that these proteins were viral by immunoblotting the same membrane with virus protein-specific antibodies (data not shown). Taken together with data obtained when MRV could not prevent SG formation (Fig. 6), these data suggest that MRV translation is inhibited in the presence of phosphorylated eIF2α (SA) or sequestered eIF4A (15D-PGJ2) when SGs are present but that at late times in infection, even though the drugs are still present, disruption of SGs by MRV correlates with escape of viral mRNA from host cell translational shutoff.
MRV escape from host translation inhibition is independent of PKR.
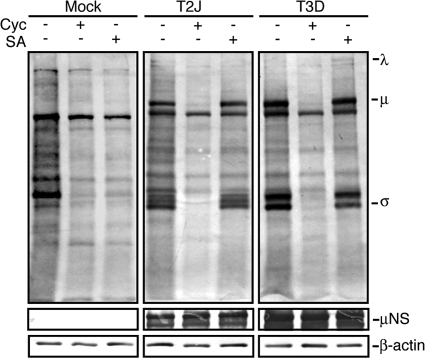
In some MRV strains, virus dsRNA binding protein σ3 is thought to inhibit PKR phosphorylation of eIF2α and prevent host cell shutoff (11, 22, 41). However, in strains in which translational shutoff is not inhibited, it remains unclear if σ3 inhibition of PKR plays a role in virus mRNA escape from cellular translational shutoff. Because we found that MRV translation was able to escape SA-induced translational shutoff, which occurs through HRI kinase phosphorylation of eIF2α and not through PKR (24, 27), we suspected that MRV escape from host translational shutoff may occur independently of σ3 inhibition of PKR. To determine if PKR inhibition was necessary for MRV translational escape from SA-induced eIF2α phosphorylation, we infected PKR−/− cells with MRV T2J and T3D, and at 24 h p.i., the cells were left untreated or were treated with cycloheximide or SA and labeled with l-AHA. Similar to what has been previously reported (27), SA induced high levels of eIF2α phosphorylation in PKR−/− cells even in the presence of MRV infection (data not shown), suggesting that translation initiation should be inhibited in the presence of SA in these experiments. In the absence of PKR, untreated uninfected and infected cells were translationally active (Fig. 8). In cycloheximide-treated uninfected and infected cells, both viral and cellular translation were inhibited. In SA-treated cells, cellular translation was inhibited, suggesting that eIF2α was indeed phosphorylated to levels sufficient to interfere with translation initiation in PKR−/− cells. However, viral translation was still active under these conditions (Fig. 8), suggesting that MRV mRNA can escape phospho-eIF2α-induced host cell translational shutoff mediated by SA activation of HRI. This finding suggests that the translation of MRV mRNAs when eIF2α is phosphorylated and cellular translation is inhibited does not depend on σ3 inhibition of PKR.
Fig. 8.
SA does not inhibit MRV translation in PKR−/− cells. PKR−/− cells were mock infected or infected with T2J or T3D, and at 24 h p.i., the cells were left untreated or treated with cycloheximide (Cyc) or SA for 60 min, at which point l-AHA was added and the cells were incubated an additional 60 min. Proteins were labeled with biotin, separated on SDS-PAGE, and transferred to nitrocellulose by electroblotting. l-AHA-labeled proteins were detected by incubation of blots with AP-conjugated streptavidin. As protein-loading and infection controls, identical sample volumes were examined in parallel using rabbit α-β-actin polyclonal antibodies or rabbit α-μNS polyclonal antibodies followed by AP-conjugated goat α-rabbit IgG. The positions of MRV proteins on the AHA blot are indicated.
DISCUSSION
MRV disrupts SGs in infected and drug-treated cells independently of eIF2α phosphorylation.
We recently reported that cells form SGs in response to MRV infection at early times p.i. in an eIF2α phosphorylation-dependent manner and that SGs are disrupted as viral infection proceeds in a manner that is dependent on viral protein synthesis (36). In this work, we show that in addition to disrupting SGs over time in infected cells, MRV also prevents SG formation following treatment with the SG-inducing drugs SA, 15D-PGJ2, and NSC119893. MRV disruption of SGs in infected and drug-treated cells occurred both in the presence of phosphorylated eIF2α (Fig. 2, 3, and 4A) and independently of phosphorylated eIF2α (Fig. 4C), suggesting that MRV acts downstream of this cellular stress signal to interfere with SG formation. These data support a hypothesis in which MRV encodes a protein or nucleoprotein complex that disrupts or interferes with SG formation, perhaps by direct or indirect interference with other signals involved in SG formation or SG effecter protein aggregation. Other viruses have been shown to encode viral factors that actively disrupt SGs by interfering with SG effecter proteins (6, 49). The identification of the mechanism behind MRV interference with SG formation is under way.
SG disruption correlates with MRV escape from host translational shutoff.
Sequestration of translation initiation factors in SGs likely contributes to the inhibition of translation that occurs following infection with many viruses. It therefore is not surprising that viruses have developed mechanisms to disperse SGs to release translation initiation factors that are necessary to carry out viral protein synthesis. Although many viruses have been shown to disrupt SGs, most studies have not yet delineated the importance of SG disruption in successful viral infection. It has been shown that some viruses, such as vesicular stomatitis virus (VSV), Sindbis virus, and herpes simplex virus (HSV), replicate better in TIA-1−/− cells, which are deficient in the ability to form SGs (21), suggesting that the function of SGs in translation inhibition may be detrimental to successful virus infection. In the case of poliovirus, it was found that the ability of the virus protease to disrupt SGs via G3BP cleavage was important for successful virus infection, as expression of a noncleavable G3BP inhibited poliovirus replication (49). Additionally, the binding of TIAR/1 to West Nile virus RNA disrupts SGs and also appears to promote viral RNA replication (21), though it has yet to be shown that SG disruption per se plays an important role in this process. In this work, we provide evidence that in the case of MRV, SG disruption may be an important step in virus circumvention of the host immune response by demonstrating that MRV translation is inhibited when SGs are present but is not inhibited when SGs are disrupted by the virus. Correlation between the release of MRV mRNAs from translation inhibition and SG disruption occurred in the presence of phosphorylated eIF2α and when eIF4A was inhibited, bolstering the argument that SGs themselves impact viral translation.
Although our data support a hypothesis that SGs play a role in regulation of MRV translation and that SG disruption appears to coincide with viral mRNA escape from cellular translational inhibition, it is not likely that SG disruption is sufficient for viral mRNA translation in the stressed environment. An obvious question that arises is how MRV mRNAs are able to compete for the limited ternary complex that is available in the cell when eIF2α is phosphorylated following infection or SA treatment. One possibility is that MRV translation is refractory to a high concentration of phosphorylated eIF2α as a result of an unidentified alternative translation initiation pathway. Many viral and cellular mRNAs contain specialized structures or elements within their sequences that allow them to be preferentially translated in an environment of high levels of phosphorylated eIF2α. These can include certain internal ribosome entry sites (IRESs), such as that found in hepatitis C virus RNA, which recruits eIF2α and tRNAMet independently of ternary complex (46); upstream open reading frames (uORFs), which induce ribosome stalling that leads to preferential translation of some stress-related mRNAs, such as ATF4 (48); and mRNAs that can recruit tRNAMet independently of eIF2α, such as Sindbis virus 26S RNA (38). A recent study showed that another member of the family Reoviridae, rotavirus, can also translate viral proteins in the presence of high levels of phosphorylated eIF2α (31). MRV and other members of the family Reoviridae may have developed a noncanonical pathway for translation initiation. Early studies suggested that MRV synthesizes capped mRNAs at early times in infection and switches to synthesis of uncapped messages to escape host cell translational shutoff at later times in infection (19, 42, 50). Although our data showing inhibition of MRV translation in the presence of SA at early, but not late, times might be interpreted as supporting a scenario where MRV mRNAs are differentially capped throughout infection, there have been many studies that suggest that MRV mRNAs are capped throughout the viral life cycle (2, 4). Moreover, the finding that MRV protein synthesis was also inhibited by 15D-PGJ2 in a manner that correlates with SG presence and absence in infection (Fig. 6 and 7) argues that it is in fact SG presence and not eIF2α phosphorylation that prevents MRV translation. These data also suggest that MRV mRNA translation occurs via a pathway that is independent of eIF4A. A detailed examination of MRV translation, including identification of the cellular translation initiation factors that are required, identification of the viral proteins that are involved, and examination of viral RNA to identify important sequences or structures, is warranted in the future.
MRV translation: role of σ3 inhibition of PKR.
Previous data implicated the MRV σ3 dsRNA binding protein in prevention of cellular translational shutoff following infection with some MRV strains (22, 40, 41). The MRV σ3 protein has also been shown to functionally replace other viral PKR inhibitors when expressed in cells infected with these viruses (1, 22). For these reasons, the PKR-inhibitory activity of σ3 was suspected to be involved in the ability of MRV mRNAs to escape translational shutoff in strains that could not prevent this cellular response to infection. Several lines of evidence from this study suggest that this may not be the case. First, strains of MRV that differ in their abilities to prevent host cell translational shutoff (T3D, T1L, and T2J) were able to continue translating mRNA in the presence of SA at late times in infection (Fig. 7 and data not shown), suggesting that the ability to prevent host cell translational shutoff and the ability to escape host cell translational shutoff likely occur through independent mechanisms. Second, because SA induces eIF2α phosphorylation via the HRI kinase (24, 27) and not PKR, σ3-mediated PKR inhibition would not impact eIF2α phosphorylation under these conditions. Moreover, SA treatment of infected cells induced levels of eIF2α phosphorylation similar to those in mock-infected cells (Fig. 4), suggesting that even if PKR were involved in SA induction of eIF2α phosphorylation, σ3 did not significantly inhibit PKR under these circumstances. Finally, MRV mRNA translation is not inhibited in the presence of SA, which induces eIF2α phosphorylation via the HRI kinase in PKR−/− cells (Fig. 8), strongly suggesting that σ3 inhibition of PKR is not necessary for MRV escape from host translational shutoff.
A model for the role of SGs in MRV infection.
Based on previous data (36) and this study, we have developed a working model for the roles of SGs and SG disruption in MRV infection. In this model, when any strain of MRV infects cells, the cells attempt to inhibit viral replication by activation of PKR (or other stress kinases), phosphorylation of eIF2α, inhibition of ternary complex, and formation of SGs. The presence of SGs inhibits viral translation, as is artificially shown by treatment of infected cells at this early time with SA or 15D-PGJ2. However, there must be a delicate balance between SG formation and viral translation, as ultimately, the virus is able to disperse SGs as infection proceeds, and this ability is dependent on the accumulation of viral proteins in the cell (36). In MRV strains where σ3 binds dsRNA so that PKR and eIF2α phosphorylation are inhibited as infection proceeds, no SGs form, and neither virus nor cellular translation is inhibited. In MRV strains where σ3 dsRNA binding activity cannot prevent PKR activation, eIF2α is phosphorylated as infection proceeds and ternary complex is limited. Cellular, but not viral, translation is inhibited in these infected cells. Virus mRNA escape from translational inhibition appears to correlate with SG disruption, as is shown artificially by the ability of MRV, but not cellular mRNAs, to be translated in cells following treatment with SA or 15D-PGJ2 only when the virus is capable of disrupting SGs. SG disruption is likely not sufficient for MRV translation in the presence of phosphorylated eIF2α and limited ternary complex, but our data suggest that it may be a necessary step toward virus escape from cellular translational shutoff.
ACKNOWLEDGMENTS
We thank Scott Kimball for PKR−/− cell lines, Bryan Bellaire for assistance with confocal microscopy, and Jason Buehler for technical assistance.
This work was supported by NIH NIAID grant K22 AIO65496 and the Roy J. Carver Charitable Trust grant 08-3181 to C.L.M. Other assistance to C.L.M. was provided by the College of Veterinary Medicine Office of the Dean, the Office of Biotechnology, and the Office of the Provost, Iowa State University.
Footnotes
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Beattie E., et al. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 69:499–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brendler T., Godefroy-Colburn T., Yu S., Thach R. E. 1981. The role of mRNA competition in regulating translation. III. Comparison of in vitro and in vivo results. J. Biol. Chem. 256:11755–11761 [PubMed] [Google Scholar]
- 3. Dang Y., et al. 2006. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870–32878 [DOI] [PubMed] [Google Scholar]
- 4. Detjen B. M., Walden W. E., Thach R. E. 1982. Translational specificity in reovirus-infected mouse fibroblasts. J. Biol. Chem. 257:9855–9860 [PubMed] [Google Scholar]
- 5. Dieterich D. C., Link A. J., Graumann J., Tirrell D. A., Schuman E. M. 2006. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U. S. A. 103:9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Emara M. M., Brinton M. A. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furlong D. B., Nibert M. L., Fields B. N. 1988. σ1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gilks N., et al. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hahon N. 1965. Assay of variola virus by the fluorescent cell-counting technique. Appl. Microbiol. 13:865–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huismans H. 1971. Host cell protein synthesis after infection with bluetongue virus and reovirus. Virology 46:500–503 [DOI] [PubMed] [Google Scholar]
- 11. Huismans H., Joklik W. K. 1976. Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology 70:411–424 [DOI] [PubMed] [Google Scholar]
- 12. Imani F., Jacobs B. L. 1988. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 σ3 protein. Proc. Natl. Acad. Sci. U. S. A. 85:7887–7891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kedersha N., Anderson P. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969 [DOI] [PubMed] [Google Scholar]
- 14. Kedersha N., et al. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF2α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim W. J., Kim J. H., Jang S. K. 2007. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J. 26:5020–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kimball S. R., Horetsky R. L., Ron D., Jefferson L. S., Harding H. P. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284:C273–C284 [DOI] [PubMed] [Google Scholar]
- 18. Krishnamoorthy T., Pavitt G. D., Zhang F., Dever T. E., Hinnebusch A. G. 2001. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 21:5018–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lemieux R., Zarbl H., Millward S. 1984. mRNA discrimination in extracts from uninfected and reovirus-infected L-cells. J. Virol. 51:215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leroux A., London I. M. 1982. Regulation of protein synthesis by phosphorylation of eukaryotic initiation factor 2α in intact reticulocytes and reticulocyte lysates. Proc. Natl. Acad. Sci. U. S. A. 79:2147–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li W., et al. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989–12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lloyd R. M., Shatkin A. J. 1992. Translational stimulation by reovirus polypeptide σ3: substitution for VAI RNA and inhibition of phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 66:6878–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu J., O'Hara E. B., Trieselmann B. A., Romano P. R., Dever T. E. 1999. The interferon-induced double-stranded RNA-activated protein kinase PKR will phosphorylate serine, threonine, or tyrosine at residue 51 in eukaryotic initiation factor 2α. J. Biol. Chem. 274:32198–32203 [DOI] [PubMed] [Google Scholar]
- 24. Lu L., Han A. P., Chen J. J. 2001. Translation initiation control by heme-regulated eukaryotic initiation factor 2α kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 21:7971–7980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matts R. L., Levin D. H., London I. M. 1983. Effect of phosphorylation of the alpha-subunit of eukaryotic initiation factor 2 on the function of reversing factor in the initiation of protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 80:2559–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mazroui R., et al. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. Mol. Biol. Cell 17:4212–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McEwen E., et al. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925–16933 [DOI] [PubMed] [Google Scholar]
- 28. McInerney G. M., Kedersha N. L., Kaufman R. J., Anderson P., Liljestrom P. 2005. Importance of eIF2α phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mendez I. I., Hermann L. L., Hazelton P. R., Coombs K. M. 2000. A comparative analysis of freon substitutes in the purification of reovirus and calicivirus. J. Virol. Methods 90:59–67 [DOI] [PubMed] [Google Scholar]
- 30. Mokas S., et al. 2009. Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell 20:2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montero H., Rojas M., Arias C. F., Lopez S. 2008. Rotavirus infection induces the phosphorylation of eIF2α but prevents the formation of stress granules. J. Virol. 82:1496–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nilsen T. W., Maroney P. A., Baglioni C. 1982. Inhibition of protein synthesis in reovirus-infected HeLa cells with elevated levels of interferon-induced protein kinase activity. J. Biol. Chem. 257:14593–14596 [PubMed] [Google Scholar]
- 33. Pain V. M. 1996. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 236:747–771 [DOI] [PubMed] [Google Scholar]
- 34. Piron M., Vende P., Cohen J., Poncet D. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17:5811–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Preiss T., Hentze M. 2003. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 25:1201–1211 [DOI] [PubMed] [Google Scholar]
- 36. Qin Q., Hastings C., Miller C. L. 2009. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J. Virol. 83:11090–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Samuel C. E., Duncan R., Knutson G. S., Hershey J. W. 1984. Mechanism of interferon action. Increased phosphorylation of protein synthesis initiation factor eIF2α in interferon-treated, reovirus-infected mouse L929 fibroblasts in vitro and in vivo. J. Biol. Chem. 259:13451–13457 [PubMed] [Google Scholar]
- 38. Sanz M. A., Castello A., Ventoso I., Berlanga J. J., Carrasco L. 2009. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS One 4:e4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schieble J. H., Kase A., Lennette E. H. 1967. Fluorescent cell counting as an assay method for respiratory syncytial virus. J. Virol. 1:494–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmechel S., Chute M., Skinner P., Anderson R., Schiff L. 1997. Preferential translation of reovirus mRNA by a σ3-dependent mechanism. Virology 232:62–73 [DOI] [PubMed] [Google Scholar]
- 41. Sharpe A. H., Fields B. N. 1982. Reovirus inhibition of cellular RNA and protein synthesis: role of the S4 gene. Virology 122:381–391 [DOI] [PubMed] [Google Scholar]
- 42. Skup D., Millward S. 1980. mRNA capping enzymes are masked in reovirus progeny subviral particles. J. Virol. 34:490–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Skup D., Millward S. 1980. Reovirus-induced modification of cap-dependent translation in infected L cells. Proc. Natl. Acad. Sci. U. S. A. 77:152–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith J. A., et al. 2006. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 80:2019–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith J. A., Schmechel S. C., Williams B. R., Silverman R. H., Schiff L. A. 2005. Involvement of the interferon-regulated antiviral proteins PKR and RNase L in reovirus-induced shutoff of cellular translation. J. Virol. 79:2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terenin I. M., Dmitriev S. E., Andreev D. E., Shatsky I. N. 2008. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 15:836–841 [DOI] [PubMed] [Google Scholar]
- 47. Tourrière H., et al. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Vattem K. M., Wek R. C. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 101:11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. White J. P., Cardenas A. M., Marissen W. E., Lloyd R. E. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305 [DOI] [PubMed] [Google Scholar]
- 50. Zarbl H., Skup D., Millward S. 1980. Reovirus progeny subviral particles synthesize uncapped mRNA. J. Virol. 34:497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang F., et al. 2001. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J. Biol. Chem. 276:24946–24958 [DOI] [PubMed] [Google Scholar]
- 52. Zweerink H. J., Joklik W. K. 1970. Studies on the intracellular synthesis of reovirus-specified proteins. Virology 41:501–518 [DOI] [PubMed] [Google Scholar]