Abstract
As part of an ongoing study of early human immunodeficiency virus type 1 (HIV-1) infection in sub-Saharan African countries, we have identified 134 seroconverters (SCs) with distinct acute-phase (peak) and early chronic-phase (set-point) viremias. SCs with class I human leukocyte antigen (HLA) variants B*44 and B*57 had much lower peak viral loads (VLs) than SCs without these variants (adjusted linear regression beta values of −1.08 ± 0.26 log10 [mean ± standard error] and −0.83 ± 0.27 log10, respectively; P < 0.005 for both), after accounting for several nongenetic factors, including gender, age at estimated date of infection, duration of infection, and country of origin. These findings were confirmed by alternative models in which major viral subtypes (A1, C, and others) in the same SCs replaced country of origin as a covariate (P ≤ 0.03). Both B*44 and B*57 were also highly favorable (P ≤ 0.03) in analyses of set-point VLs. Moreover, B*44 was associated with relatively high CD4+ T-cell counts during early chronic infection (P = 0.02). Thus, at least two common HLA-B variants showed strong influences on acute-phase as well as early chronic-phase VL, regardless of the infecting viral subtype. If confirmed, the identification of B*44 as another favorable marker in primary HIV-1 infection should help dissect mechanisms of early immune protection against HIV-1 infection.
INTRODUCTION
HIV-1 viral load (VL or viremia), in the typical form of viral RNA concentration in plasma, has both epidemiological and clinical implications because of its dual impact on transmission (18, 30, 71) and on the rate of disease progression (58, 59). In most studies, a quantitative measure of VL is used without reference to estimated date of infection (EDI), under the assumption that patients are seldom observed during acute-phase (peak) infection and that the early chronic-phase (set-point) VL is usually stable for years in patients with no apparent manifestations of immunodeficiency. Factors known or suspected to influence VL range from viral mutations (changes in replication fitness or switches in coreceptor tropism) (15, 28, 39, 72) to host genes that govern innate and adaptive immune responses (54, 75, 81, 83, 84, 86).
Within the human nuclear genome, human leukocyte antigen (HLA) class I genes are the most convincing (and universally applicable) quantitative trait loci for HIV-1 viremia (14, 16, 17, 66). However, the individual HLA alleles, haplotypes, and supertypes with reported impacts on HIV-1 VL are not always clear because their distribution and patterns of linkage disequilibrium often differ from one population to another (7, 53, 63). Consensus findings have been limited to a few variants like B*27 and B*57 (9, 80), although more recent work based on African cohorts has yielded consensus results for additional variants, including A*74, B*13, and B*81, that are often favorable (49, 81). The HLA class I heavy chains encoded by these alleles differentially present highly conserved viral epitopes for cytotoxic T-lymphocyte (CTL) responses (5, 29, 39). Such HLA-restricted immune mechanisms provide clues for designing better vaccine antigens that can drive immune responses toward desirable epitopes, i.e., those are highly relevant to viral fitness (13, 37, 38, 65, 91).
We and others have reported previously that both host and viral factors can independently impact HIV-1 VL during the early and chronic phases of infection (54, 81, 84, 85). Host factors ranged from coreceptor gene (e.g., CCR5) polymorphisms to HLA variants (24, 50, 83, 84). The importance of viral characteristics is reflected by a correlation between donor and recipient VL after controlling for the effects of age and gender, as well as HLA (85). Supporting evidence from several recent studies suggests that the magnitude of the influence of the transmitted virus on the recipient's VL can be highly variable (26, 27). While heterogeneity in viral subtypes and other characteristics might raise some doubts about the relative contribution of host factors, our analyses of recent African seroconverters (SCs) with acute- and early chronic-phase VL measurements indicate that HLA class I factors can influence VL during primary HIV-1 infection with at least two major viral subtypes (A1 and C).
(Part of this work was presented as a poster at the 18th Conference on Retroviruses and Opportunistic Infections, Boston, MA, 27 February to 2 March 2011 [86a].)
MATERIALS AND METHODS
Study population.
Between 2006 and 2010, over 370 HIV-1 seroconverters (SCs) were enrolled from Kenya, Rwanda, Uganda, South Africa, and Zambia in studies sponsored by the International AIDS Vaccine Initiative (IAVI). The procedures for written informed consent and all other research protocols were approved by institutional review boards at all sponsoring organizations, with further compliance to human experimentation guidelines set forth by the U.S. Department of Health and Human Services. Clinical and laboratory tests have been described in detail elsewhere (35). For this study, we focused on a subset of 134 SCs observed in two distinct phases of primary HIV-1 infection (PHI). They were selected (Fig. 1) based on (i) the availability of more than 20 SCs from a single study site (grouped by country) with biological specimens for HLA class I genotyping, (ii) a narrow interval (<4 months) between the last seronegative and first seropositive visits to allow reliable calculation of the estimated date of infection (EDI), i.e., either the midpoint between the last seronegative and first seropositive visit or 2 weeks prior to the detection of HIV-1 p24 antigen in plasma, (iii) adequate follow-up for measuring VL during acute phase (<3 months) and early chronic phase (3 to 12 months) without antiretroviral therapy, (iv) the availability of CD4+ T-cell (CD4) counts for at least one of the two targeted infection intervals, (v) no apparent liver malfunction, i.e., serum alanine transferase concentration of <183 IU/liter (3 times the upper range in healthy adult Africans) (34), and (vi) no apparent kidney malfunction, i.e., serum creatinine concentration of <327 μM (3 times the upper range in healthy adult Africans) (34). The remaining SCs (n = 241), excluded from this work, all had insufficient data or uncertain EDI (Fig. 1; also see Table S1 in the supplemental material).
Fig. 1.
Flow chart for selecting and analyzing informative HIV-1 seroconverters (SCs), with a focus on two distinct phases of primary HIV-1 infection. (a) Selection was based on availability and adequacy of clinical data, including acute-phase viral load (VL), early set-point VL, and set-point CD4+ T-cell counts (missing in some subjects). (b) Streamlined association analyses rely on univariate and multivariable models.
HIV-1 VL as primary and CD4 count as secondary outcome.
Plasma VL (RNA copies/ml) was measured using the Amplicor monitor assay, version 1.5 (Roche Applied Science, Indianapolis, IN) (70). For log10 transformation, a VL below the lower limit of detection (50 RNA copies/ml) was assigned a value of 0.849 (half of log1050). CD4 counts were based on T-cell immunophenotyping, with assays done at individual clinics using the FACScount System (Beckman Coulter Ltd., London, United Kingdom). For this study, CD4 counts during the early chronic phase (corresponding to earliest VL set-point) were not collected for 5 of 134 SCs available for analyses.
Identification of HIV-1 subtypes by viral sequencing.
HIV-1 pol sequencing was performed as a routine procedure for monitoring rates of drug resistance mutation and for providing an indication of infecting viral subtypes (70). Briefly, a 1.7-kb amplicon encompassing the pol region was sequenced using five primers and the ABI BigDye terminator kit (version 3.1, Applied Biosystems, Foster City, CA). Sequence identities were established with the REGA HIV-1 Subtyping tool and the Stanford HIV RT and Protease Sequence database (http://hivdb.stanford.edu/). The pol sequences can capture four of five major recombinant forms (87). Samples that could not be assigned a specific subtype or recombinant form were subjected to phylogenetic analysis using CLUSTAL X, version 2.0 (46), MEGA, version 4 (79), and reference sequences from the Los Alamos HIV database (http://www.hiv.lanl.gov). Selective sequencing of the env region was done occasionally to resolve residual ambiguity with pol sequences (70).
HLA genotyping.
Allelic variants at three HLA class I loci (HLA-A, HLA-B, and HLA-C) were resolved to 4-digit specificities using a combination of PCR-based techniques (81, 82). Reference to fully resolved alleles followed the revised nomenclature effective in April 2010 (55). Because of the limited sample size, HLA specificities were analyzed at the 2-digit level unless there was prior evidence for different outcomes associated with HLA alleles at high resolution (4 digits).
Descriptive statistics and correlation analyses.
Using software packages in SAS, version 9.2 (SAS Institute, Cary, NC), SCs were first stratified by country of origin and then compared for demographic data, laboratory findings (VL, viral subtypes, and CD4 counts) (Table 1), and HLA variants of interest (see the supplemental material). Differences between countries and viral subtypes were assessed primarily by (i) analysis of variance (ANOVA) and the t test for quantitative variables with a normal distribution, (ii) nonparametric (e.g., Kruskal-Wallis) test for VL data before log10 transformation, or (iii) χ2 and Fisher exact tests for categorical variables. Several outcome measures were also tested for linearity and strength of correlation, as reflected by Pearson r and Spearman rho (ρ) values, respectively. Individual plots were generated using GraphPad Prism (GraphPad Software, Inc.).
Table 1.
Characteristics of 134 seroconvertersa enrolled from four African countries and suitable for studying primary HIV-1 infection
| Overall characteristicsb | Kenya | Rwanda | Uganda | Zambia |
|---|---|---|---|---|
| No. of subjects | 27 | 35 | 27 | 45 |
| Sex ratio (M/F) | 3.50 (21/6) | 1.06 (18/17) | 1.25 (15/12) | 1.50 (27/18) |
| Age (mean yr ± SD) | 27.4 ± 5.0 | 37.8 ± 9.7 | 34.7 ± 10.2 | 34.6 ± 6.8 |
| Age ≥ 40 [no. (%) of subjects] | 1 (3.7) | 14 (40.0) | 8 (29.6) | 12 (26.7) |
| HIV-1 subtype(s) [no. (%) of subjects] | ||||
| A1 | 17 (63.0) | 29 (82.9) | 8 (29.6) | 0 (0.0) |
| C | 5 (18.5) | 2 (5.7) | 1 (3.7) | 43 (95.6) |
| Others (D, B, recombinants, or unknown) | 5 (18.5) | 4 (11.4) | 18 (66.7) | 2 (4.4) |
| Estimated date of infection (mo/day/yr) | ||||
| Earliest | 04/21/2006 | 02/21/2006 | 04/04/2006 | 06/26/2006 |
| Latest | 11/26/2008 | 09/06/2008 | 03/09/2009 | 05/26/2008 |
| Duration of infection (mo) | ||||
| At acute-phase VL visit [median (IQR)] | 1.8 (1.3–2.2) | 1.5 (0.7–1.9) | 1.7 (1.5–2.5) | 1.9 (1.5–2.1) |
| At set-point VL visit [median (IQR)] | 8.6 (5.5–11.0) | 8.4 (8.2–11.0) | 8.6 (8.1–11.0) | 8.2 (5.5–11.0) |
| VL measures taken within 3 mo [median (IQR)] | 2 (2–3) | 3 (3–4) | 2 (2–3) | 2 (2–3) |
| Acute-phase VL (mean log10 ± SD) | 5.02 ± 1.00 | 5.22 ± 0.79 | 4.78 ± 1.06 | 5.01 ± 0.88 |
| Set-point VL (mean log10 ± SD) | 3.97 ± 1.06 | 3.53 ± 1.20 | 3.76 ± 1.39 | 4.17 ± 0.93 |
| CD4+ T cell count (mean ± SD) | ||||
| At acute-phase VL visit | 536 ± 185 | 566 ± 277 | 643 ± 298 | 497 ± 183 |
| At set-point VL visit | 555 ± 246 | 573 ± 293 | 649 ± 282 | 472 ± 168 |
All seroconverters have defined HLA class I genotypes.
M, male; F, female; IQR, interquartile range, from 25th to 75th percentile.
Hypothesis and statistical models.
Based on recent evidence from a large African cohort (81), we aimed to test a primary hypothesis that favorable HLA variants generally confer a strong impact on early VL, before the virus acquires mutations to facilitate immune escape. Among all the HLA variants that are potentially favorable in one way or another, 11 are highly relevant to outcomes after HIV-1 infection among native Africans; these include A*29, A*74, B*13, B*44, B*57, B*58:01 (often in contrast to B*58:02), B*81, C*18, B*39-C*10 (haplotype), B*42-C*17 (haplotype), and A*30+C*03 (combination), as reported for three sub-Saharan African cohorts (41, 49, 81). Analytical approaches, including generalized linear models (GLMs) and mixed models (“Proc Mixed” in SAS), followed established strategies for testing independent associations (77, 81, 82). Statistical significance was accepted at the level of a P value of ≤0.05, provided that internal consistency was established and that multivariable models could rule out potential confounding by nongenetic factors (Fig. 1b). False discovery probabilities (q values) from screening tests were assessed using the “Proc Multtest” procedure in SAS.
RESULTS
Characteristics of HIV-1 seroconverters available for primary analysis.
Our selection process yielded 134 informative SCs from four countries, including 45 from Zambia (Lusaka and Copper Belt), 35 from Rwanda (Kigali), 27 from Kenya (Kilifi and Nairobi), and 27 from Uganda (Entebbe and Masaka) (Table 1). The estimated dates of infection (EDI) ranged from February 2006 to March 2009. The duration of infection was highly comparable across study sites at the visit corresponding to the acute phase (median, 1.5 to 1.9 months) and the visit corresponding to the early set-point (median, 8.2 to 8.6 months). Based on viral sequencing (successful in 95.5% of SCs), HIV-1 subtypes A1 and C were the most common, being found in 54 (40%) and 51 (38%) SCs, respectively. Additional subtypes and recombinant forms were relatively uncommon (2 for subtype B, 18 for subtype D, and 3 for recombinants); these and six SCs with missing information (VL too low to facilitate viral sequencing) were grouped together (Table 1).
Kenyan SCs differed from others in their lower age (27.4 ± 5.0 years), high male-to-female ratio (3.5), and infection with mixed HIV-1 subtypes (A1, C, and others) (18.5 to 63.0%). Rwandan SCs had relatively high acute-phase VLs (5.22 ± 0.79 log10) accompanied by relatively low set-point VLs (3.53 ± 1.20 log10). Zambians were characterized by the predominance of HIV-1 subtype C infection (95.6%) and relatively low CD4 counts in both the acute phase (497 ± 183 cells/μl) and early set-point (472 ± 168 cells/μl). Ugandans had the highest frequency (66.7%) of viral subtypes other than A1 and C.
Dynamics of HIV-1 viremia during acute and early chronic phases of infection.
Peak (highest) VL during acute-phase infection was readily discernible in 106 SCs because multiple measures within the first 3 months of infection were available. The other 28 SCs each had one single VL measurement taken near the end of the acute phase (Fig. 2a) but still well before set-point VL was established in the early chronic phase (3 to 12 months after EDI) (Fig. 2b).
Fig. 2.
Distribution of acute-phase and set-point viral load (VL) in 134 seroconverters (SCs). The panels display overall correlation between duration of infection and VL (a and b, drawn to different scales), as well as correlation between acute-phase and set-point VL measurements (c). Open and filled circles (a) correspond to two subgroups (see text). Arrow points to two subjects (from two countries) who share almost identical VL results.
Overall, the acute-phase VLs ranged from 2.55 log10 to 7.03 log10, showing no correlation with duration of infection (DOI) (Fig. 2a). The early set-point VLs ranged from below detection (in seven SCs) to 5.69 log10 (Fig. 2b). VLs in these two phases showed a strong linear correlation (Pearson r = 0.61, P < 0.0001) (Fig. 2c); a strong nonlinear correlation was also apparent for VLs before log10 transformation (Spearman ρ = 0.66, P < 0.0001).
Absence of correlation between HIV-1 subtype and viremia.
In 128 of 134 SCs with successful viral sequencing, neither acute-phase nor set-point VL differed by HIV-1 subtype (P ≥ 0.131 by one-way ANOVA) (Fig. 3). The set-point VLs in three SCs with subtype A1 viruses were below the lower limit of detection. Transformation of VLs to log10 did not alter the results, as nonparametric tests (by VL ranking) led to the same conclusions (P ≥ 0.309 by Kruskal-Wallis test). Direct comparison between subtype A1 and subtype C alone confirmed similarities between these groups (P ≥ 0.114 by t test) (Fig. 3).
Fig. 3.
HIV-1 viral load (VL) in 128 HIV-1 seroconverters (SCs) stratified by viral subtype. Acute-phase VL (a) and set-point VL (b) are compared. For each stratum, horizontal bars connected by a vertical line correspond to mean and standard deviation. Five individuals in the third group (filled circles) have subtype B (n = 2) or recombinant forms (n = 3), while the rest have subtype D (open circles). Six subjects with undetermined viral subtypes are excluded here.
Distribution of select HLA variants by patient groups and country of origin.
Selection of SCs for analysis did not lead to obvious bias in either the national origin (see Table S1 in the supplemental material) or distribution of the 11 HLA variants of interest (see Table S2 in the supplemental material). Stratification by country of origin did not show overall genetic heterogeneity (see Table S3 in the supplemental material), but 3 of 11 candidate variants, i.e., B*13, B*39-C*12, and the A*30+C*03 combination, were too rare to facilitate association analyses (see Table S2 in the supplemental material). For the eight remaining variants, the frequency was highest for A*74 (20 subjects) and lowest for A*29, B*81, and C*18 (10 subjects each).
HLA candidates screened and then dismissed by mixed models.
In analyses adjusted for gender, DOI, age at EDI, and country of origin (Table 2), five HLA candidates (A*29, A*74, B*58:01, B*81, and C*18) failed to show appreciable impact on VL or CD4 count (P ≥ 0.051 and q ≥ 0.102). Indeed, mean beta estimates for VL were positive (unfavorable) for A*29 and A*74, which were represented by A*29:02 and A*74:01, respectively. On the other hand, B*42-C*17 lacked internal consistency (Table 2), being somewhat unfavorable for VL (P = 0.042 and q = 0.102) while highly favorable for CD4 count (P < 0.001 and q = 0.004). As expected, B*42:01 and C*17:01 were the individual alleles accounting for the B*42-C*17 haplotype.
Table 2.
Association analyses for eight HLA variants: impact on repeated measures of HIV-1 viral load and CD4+ T-cell count in 134 seroconverters
| HLA varianta | Analysis of VLb |
Analysis of CD4 countb |
||||||
|---|---|---|---|---|---|---|---|---|
| No. of subjects | β ± SE | P | qc | No. of subjects | β ± SE | P | qc | |
| A*29 | 10 | 0.18 ± 0.17 | 0.300 | 0.384 | 10 | −47 ± 39 | 0.231 | 0.582 |
| A*74 | 20 | 0.12 ± 0.13 | 0.336 | 0.384 | 20 | 12 ± 29 | 0.674 | 0.839 |
| B*42-C*17 | 18 | 0.28 ± 0.14 | 0.042 | 0.102 | 18 | 108 ± 31 | <0.001 | 0.004 |
| B*44 | 12 | −0.80 ± 0.17 | <0.001 | 0.004 | 12 | 213 ± 37 | <0.001 | 0.004 |
| B*57 | 11 | −1.06 ± 0.16 | <0.001 | 0.004 | 11 | 30 ± 37 | 0.415 | 0.664 |
| B*58:01 | 14 | −0.02 ± 0.15 | 0.881 | 0.881 | 14 | −7 ± 34 | 0.839 | 0.839 |
| B*81 | 10 | −0.18 ± 0.18 | 0.325 | 0.384 | 10 | 42 ± 40 | 0.291 | 0.582 |
| C*18 | 10 | −0.34 ± 0.17 | 0.051 | 0.102 | 10 | −12 ± 38 | 0.759 | 0.839 |
Of 11 candidates, 3 (B*13, B*39-C*12, and A*30+C*03) are too rare (≤5 subjects each) to allow informative analysis.
Mixed models test virological and immunological outcomes in the first 12 months of infection (see text). For consistency, age, sex, country of origin, and duration of infection are retained as covariates in each test.
False discovery rates (q values) are shown for the multiple hypotheses.
HLA variants persistently associated with acute-phase and set-point viremia.
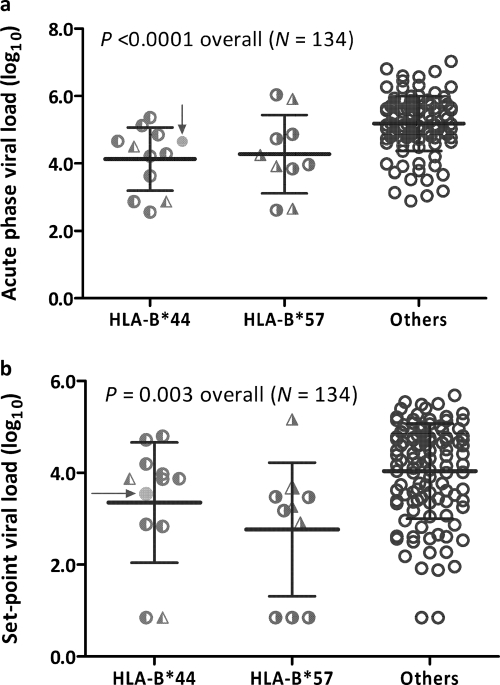
Mixed models consistently identified B*44 and B*57 as markers of relatively low viremia at two intervals of PHI (P < 0.01 and q <0.01 for all tests) (Table 2; also see Table S4 in the supplemental material). SCs with HLA-B*44 (n = 12) and B*57 (n = 11) had significantly lower peak VLs than carriers of other alleles (regression beta values of −1.08 ± 0.26 log10 and −0.83 ± 0.27 log10, respectively) (P ≤ 0.003) (Fig. 4a). These relationships were similar when viral subtype replaced country of origin as a nongenetic covariate in the analytic models (data available upon request). The acute-phase mean VL was also somewhat lower in Ugandans than in Rwandans (adjusted beta value of −0.30 ± 0.21 log10, P > 0.15).
Fig. 4.
Acute-phase (a) and set-point (b) viral load in 134 HIV-1 seroconverters after stratification by HLA variant (B*44, B*57, and others). For each stratum, horizontal bars connected by a vertical line correspond to mean and standard deviation. One subject with both B*44:03 and B*57:02 (solid circle indicated by arrow) is grouped with others with B*44 alone. Half-filled circles represent subjects with B*44:03 and B*57:03, while half-filled triangles represent individuals with B*44:15 and B*57:02.
Both B*44 and B*57 were independently associated with lower set-point VLs in the 134 SCs (Fig. 4b), with adjusted VL differences of −0.75 ± 0.33 log10 for B*44 (P = 0.026) and −1.13 ± 0.34 log10 for B*57 (P = 0.001). Of note, five of seven (or 71%) SCs with undetectable set-point VLs had either B*44 or B*57. Among nongenetic factors, being female or Rwandan was associated with lower VLs (−0.42 ± 0.20 log10, P = 0.037, and −0.53 ± 0.25 log10, P = 0.034, respectively). Age and DOI had no appreciable impact on set-point VL (adjusted P value, >0.16 for all).
Within this study population, B*44 was represented by two 4-digit alleles, namely, B*44:03 (n = 10) and B*44:15 (n = 2) (Fig. 4). Similarly, B*57 was represented by two 4-digit alleles, B*57:03 (n = 6) and B*57:02 (n = 5) (Fig. 4). No inference could be made about possible joint effects of B*44 and B*57 from a single individual who carried both.
Factors associated with CD4 count during early chronic phase of HIV-1 infection.
In analyses similar to those performed for set-point VL, B*44 was associated with higher CD4 counts (167 ± 72, P = 0.022) (Table 3). The trend toward association with higher CD4 counts in B*57-positive SCs (106 ± 74) was not statistically significant (P = 0.16). Higher CD4 counts in females than males (158 ± 44, P < 0.001) and lower CD4 counts in Ugandans than Zambians (Δ = 184 ± 60, P = 0.003) mirrored their respective associations with set-point VL. On the other hand, neither age nor DOI had any appreciable effect on CD4 count (adjusted P value, >0.10). In an alternative model where HIV-1 subtype replaced country of origin as a covariate, HIV-1 subtype C was associated with lower CD4 counts than subtype A1 (Δ = −139 ± 46, P = 0.003), despite the lack of difference in set-point VLs between the two major subtypes (Fig. 3). Other subtypes, however, did not differ from subtype A1 in terms of CD4 count. For gender, HLA-B*44, and B*57, there were negligible differences for comparisons of their effects on CD4 count and VL.
Table 3.
Two multivariable models for testing major correlates of CD4+ T-cell counts in 128 HIV-1 seroconverters at the visit corresponding to the earliest set-point viral load
| Variable in model | Change in CD4 counta |
|||
|---|---|---|---|---|
| Model 1 |
Model 2 |
|||
| β ± SE | P | β ± SE | P | |
| Female | 158 ± 44 | 0.0004 | 171 ± 43 | 0.0001 |
| HLA-B*44 | 167 ± 72 | 0.022 | 159 ± 72 | 0.029 |
| HLA-B*57 | 106 ± 74 | 0.158 | 113 ± 74 | 0.129 |
| HIV-1 subtype Cb | NA | NA | −139 ± 46 | 0.003 |
| Other HIV-1 subtypesb | NA | NA | −27 ± 56 | 0.629 |
For consistency, age and duration of infection are retained as covariates in all tests although they are not associated with the outcome. Country of origin and HIV-1 subtype are analyzed separately (model 1 versus model 2) because of concerns with colinearity. The beta estimates (β) and standard errors have been adjusted for all factors in each model (NA, not applicable).
HIV-1 subtype A1 (the most frequent subtype) served as the reference group.
DISCUSSION
Despite a modest sample size and limited statistical power, our analysis here demonstrated that HLA-B*44 and B*57 can substantially influence the level of HIV-1 viremia in sub-Saharan Africans with primary HIV-1 infection (PHI). The impact of B*44 and B*57 on viremia well exceeded the threshold value (0.30 log10) commonly used to ascribe biological and epidemiological significance (61, 74). In addition, nongenetic host factors, such as age, sex, duration of infection, country of origin and viral subtype, did not obscure the effects attributable to B*44 and B*57—statistical adjustments for those potential confounders did not meaningfully alter the estimates of effect size (i.e., mean regression beta and standard error of the mean). Other HLA candidates had either conflicting (inconsistent) or null associations, so their specific roles in HIV-1 infection might not be generalizable. Larger cohorts may allow further evaluation of these and other variants for country- or virus-specific relationships.
During acute-phase and early chronic infection, a strong association between HLA-B*57 and low viremia was expected, as B*57 variants (mostly B*57:03 and B*57:02 in Africans) have been recognized early and widely as highly favorable (11, 36, 60). The importance of B*57 to clinical and immunological responses during PHI has also been documented (2, 3). Moreover, B*57 in infected partners has been associated with delayed transmission of HIV-1 to their uninfected cohabiting partners in Zambia (82) and more recently in the United States (21). Three dominant, B*57-restricted CTL epitopes have been mapped to the HIV-1 Gag region. Mutations that alter HIV-1 Gag sequences appear to “cripple” viral replication, as suggested by persistently lower VLs upon transmission to new hosts (1, 23). Viruses from B*57-positive individuals can accumulate immune escape mutations, which ultimately lead to functional compensation and pathogenetic consequences (12). In this study, the SCs with B*57 did have relatively high CD4 counts during early chronic infection compared with those of other SCs, but the small sample size limited statistically meaningful inference. An intact CD4 profile in early infection might further translate to delayed disease progression (58, 59).
Relatively little attention has been paid to HLA-B*44, because it has not been definitively associated with virologic, immunologic, or clinical outcomes before, although one study has identified B*44:03 as a favorable allele in the context of HIV-1 subtype C infection in South Africa (49). In our study population, the association of B*44 with relatively low viremia was accompanied by a corroborative association with high CD4 count. Of the two B*44 alleles (B*44:03 and B*44:15) found in this mixed study population, B*44:03 (previously B*4403) is a common allele that has been studied extensively. According to the HIV Molecular Immunology Database (52), nearly a dozen HIV-1-specific CTL epitopes have been attributed to B*44:03 and its related alleles. The most consistent epitopes targeted by B*44:03 are AW11 derived from Gag/P24 (31, 56) and QW11 from Pol/integrase (39, 73). In South Africans, a single CTL escape mutation (at the QW11 epitope) is associated with low viral load (73). In other populations, B*44:02 is a major allele that is often found on the B*44-C*05 haplotype (53). Understanding the early immune responses facilitated by various B*44 alleles should help with the dissection of protective immunity to HIV-1 infection, especially in the context of subtypes A1 and C in sub-Saharan Africa (22).
Viral subtypes are highly relevant to the design of vaccines (42, 57), and multiple reports have indicated that disease progression may differ somewhat by viral subtype (32, 33, 40, 62, 90). In the cohort under study, the levels of viremia during PHI did not differ by the two major subtypes (A1 and C), but CD4 counts did show clear differences between subtypes A1 and C. Dissociation between virologic and immunologic outcomes has been noted earlier for viral subtypes A and D (4). The distribution of HIV-1 subtypes A1 and C in our cohort correlated closely with the country of origin (Table 1), precluding comparison of different viral subtypes within a homogeneous subpopulation (e.g., Ugandans alone or Zambians alone). The accumulation of enough SCs with different viral subtypes in a subpopulation (e.g., Kenyans) will likely permit such comparison.
The HIV-1 VL presumably reflects the equilibrium between viral replication and immunologically mediated viral clearance. In the context of PHI, the acute phase does not involve intense antibody responses (19, 45, 67). Set-point viremia is typically reached within the first few months after infection (6), well before the debut of high-affinity neutralizing antibodies that require extensive somatic mutation (45, 67). The underlying immune control during acute and early chronic infection appears to depend on cellular immune pathways (8, 43), including CTL and natural killer (NK) cell activities mediated by HLA class I allelic products. Thus, B*44 and B*57 most likely mediate the level of viremia through these cellular mechanisms to influence immune control of PHI. Our ability to analyze acute-phase viremia in a subset of carefully selected SCs was critical to demonstrating the early impacts conferred independently by B*44 and B*57.
The acute phase of PHI can have multiple long-term implications. First, early events of host-virus interactions are most devastating to mucosal CD4 cells (the “leaky gut” effect) (6, 10), often followed by a multitude of cytokine responses (78). Second, individuals with acute infection are highly contagious (93)—in a study of HIV-1-discordant couples from Uganda, 43.5% of incident cases of heterosexual HIV-1 transmission involved donor/source partners with acute infection (68). Third, acute infection sets the stage for other pathophysiological events, including immune dominance (3, 25), viral genetic drift or recombination (20, 94), and immune escape (39, 64, 69, 76, 89). It is conceivable that relatively effective control of the initial “viral burst,” as seen frequently in SCs with B*44 and B*57 during acute infection, can limit viral reservoirs and alter other pathways of HIV-1 pathogenesis. Sequencing of viral and proviral DNA during early infection should facilitate further elucidation of early immune pressure.
Overall, the association of HLA class I variants with different levels of viremia in PHI reiterates the importance of HLA-related immune responses to control of HIV-1 infection. For both B*44 and B*57, the magnitude of the impact on viral load reduction as observed here (∼1.0 log10) is higher than earlier estimates (rarely exceeding 0.3 log10) based on more chronically infected seroprevalent subjects (81). In the latest (and largest) study that focuses on HIV-1 controllers (VL of <2,000 copies/ml for ≥12 months), B*57 remains as the most prominent factor in subjects of African and European ancestries (88). However, our results here do not support the attribution of all major favorable effects of the HLA-B molecule to potential peptide-binding properties of three amino acid residues along the peptide-binding groove (88). Indeed, three residues at positions 67, 70, and 97 (around the C and F pockets) all differ between the B*44 and B*57 alleles found in our study population, with Ser, Asn, and Arg for B*44 and Met, Ser, and Val for B*57. One plausible explanation is that other variants in linkage disequilibrium with B*44 might contribute to the observed relationships. Alternatively, at the early phase of HIV-1 infection, the multifaceted HLA-B molecule has deployed its natural killer cell ligand property in addition to its antigen presentation function. In other words, similarity or difference in peptide-binding preferences alone may not fully capture the spectrum of concerted and evolving immune function that is essential to durable containment of HIV-1 infection (44). Either way, ongoing efforts to expand the sub-Saharan African cohorts of seroconverters should allow detailed analysis of HLA sequence motifs and other related properties (e.g., supertypes and frequencies) (47, 48).
Finally, our ability to analyze acute-phase viremia has also produced direct evidence that highly effective viral control, as reflected by undetectable viral load, is not entirely due to defective founder viruses (a plausible alternative hypothesis). For example, seven SCs in our study population had viremia levels of less than 50 RNA copies/ml at the earliest set-point (Fig. 2c), but they all had detectable viremia during acute infection. Indeed, three of them had acute-phase viremia levels of ≥40,000 copies/ml. Since the majority (5/7) of these SCs have B*44 or B*57 (Fig. 4b), frequent testing of HIV-1-seronegative subjects with these alleles may help identify additional SCs whose course of viremia can offer critical insights about mechanisms of early immune control (25, 51, 92). By our calculation (see Table S3 in the supplemental material), about one in six Africans in Kenya, Rwanda, Uganda, and Zambia carry either B*44 or B*57.
Supplementary Material
ACKNOWLEDGMENTS
The bulk of the research described in this study was conducted by the IAVI African HIV Research Network. We thank staff and study participants within the research network for their valuable contributions. We are also grateful to Kui Zhang for advice on statistics, to Dongning He for technical assistance, to Yun Joo Yoo for templates of streamlined statistical programs, and to Josephine Cox for critical reading of an earlier draft of the manuscript. The draft was further reviewed by two scientists designated by IAVI.
This work was made possible in part by generous support from (i) IAVI, (ii) United States National Institute of Allergy and Infectious Diseases (NIAID) through an R01 grant (AI071906 to R.A.K.) and a K02 grant (AI076123 to J.T.), (iii) Fogarty AIDS International Training and Research Program (AITRP) (grant FIC 2D43 TW001042 to S.L.), and (iv) the American people through the United States Agency for International Development (USAID).
The contents are the responsibility of the study authors and do not necessarily reflect the views of USAID or the United States Government.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Allen T. M., Altfeld M. 2008. Crippling HIV one mutation at a time. J. Exp. Med. 205:1003–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altfeld M., et al. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581–2591 [DOI] [PubMed] [Google Scholar]
- 3. Altfeld M., et al. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8+ T cell response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baeten J. M., et al. 2007. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J. Infect. Dis. 195:1177–1180 [DOI] [PubMed] [Google Scholar]
- 5. Bansal A., et al. 2007. Immunological control of chronic HIV-1 infection: HLA-mediated immune function and viral evolution in adolescents. AIDS 21:2387–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brenchley J. M., Price D. A., Douek D. C. 2006. HIV disease: fallout from a mucosal catastrophe? Nat. Immunol. 7:235–239 [DOI] [PubMed] [Google Scholar]
- 7. Cano P., et al. 2007. Common and well-documented HLA alleles: report of the ad-hoc committee of the American Society for Histocompatibility and Immunogenetics. Hum. Immunol. 68:392–417 [DOI] [PubMed] [Google Scholar]
- 8. Cao J., McNevin J., Malhotra U., McElrath M. J. 2003. Evolution of CD8+ T cell immunity and viral escape following acute HIV-1 infection. J. Immunol. 171:3837–3846 [DOI] [PubMed] [Google Scholar]
- 9. Carrington M., O'Brien S. J. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535–551 [DOI] [PubMed] [Google Scholar]
- 10. Centlivre M., Sala M., Wain-Hobson S., Berkhout B. 2007. In HIV-1 pathogenesis the die is cast during primary infection. AIDS 21:1–11 [DOI] [PubMed] [Google Scholar]
- 11. Costello C., et al. 1999. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS 13:1990–1991 [DOI] [PubMed] [Google Scholar]
- 12. Crawford H., et al. 2009. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206:909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Currier J. R., et al. 2002. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J. Immunol. Methods 260:157–172 [DOI] [PubMed] [Google Scholar]
- 14. Dalmasso C., et al. 2008. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PLoS One 3:e3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Derdeyn C. A., et al. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022 [DOI] [PubMed] [Google Scholar]
- 16. Fellay J., et al. 2009. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 5:e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fellay J., et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fideli U. S., et al. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 (HIV-1) in Africa. AIDS Res. Hum. Retroviruses 17:901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fiebig E. W., et al. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879 [DOI] [PubMed] [Google Scholar]
- 20. Fischer W., et al. 2010. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One 5:e12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao X., et al. 2010. HLA-B alleles associate consistently with HIV heterosexual transmission, viral load, and progression to AIDS, but not susceptibility to infection. AIDS 24:1835–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gill D. K., et al. 2010. Equivalence of ELISpot assays demonstrated between major HIV network laboratories. PLoS One 5:e14330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goepfert P. A., et al. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 205:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gonzalez E., et al. 1999. Race-specific HIV-1 disease modifying effects associated with CCR5 haplotypes. Proc. Natl. Acad. Sci. U. S. A. 96:12004–12009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goonetilleke N., et al. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hecht F. M., et al. 2010. HIV RNA level in early infection is predicted by viral load in the transmission source. AIDS 24:941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hollingsworth T. D., et al. 2010. HIV-1 transmitting couples have similar viral load set-points in Rakai, Uganda. PLoS Pathog. 6:e1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Honeyborne I., et al. 2010. HLA-Cw*03-restricted CD8+ T-cell responses targeting the HIV-1 Gag major homology region drive virus immune escape and fitness constraints compensated by intra-codon variation. J. Virol. 84:11279–11288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Honeyborne I., et al. 2007. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J. Virol. 81:3667–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. John G. C., et al. 2001. Correlates of mother-to-child human immunodeficiency virus type 1 (HIV-1) transmission: association with maternal plasma HIV-1 RNA load, genital HIV-1 DNA shedding, and breast infections. J. Infect. Dis. 183:206–212 [DOI] [PubMed] [Google Scholar]
- 31. Jones N. A., et al. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 200:1243–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaleebu P., et al. 2002. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J. Infect. Dis. 185:1244–1250 [DOI] [PubMed] [Google Scholar]
- 33. Kanki P. J., et al. 1999. Human immunodeficiency virus type 1 subtypes differ in disease progression. J. Infect. Dis. 179:68–73 [DOI] [PubMed] [Google Scholar]
- 34. Karita E., et al. 2009. CLSI-derived hematology and biochemistry reference intervals for healthy adults in eastern and southern Africa. PLoS One 4:e4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karita E., et al. 2007. Investigating the utility of the HIV-1 BED capture enzyme immunoassay using cross-sectional and longitudinal seroconverter specimens from Africa. AIDS 21:403–408 [DOI] [PubMed] [Google Scholar]
- 36. Kaslow R. A., et al. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405–411 [DOI] [PubMed] [Google Scholar]
- 37. Kaslow R. A., et al. 2001. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J. Virol. 75:8681–8689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kebba A., et al. 2007. Recent HIV-1 infection in a high-risk Ugandan cohort: implications for phase IIB test-of-concept HIV vaccine trials. Pharmacogenomics 8:409–414 [DOI] [PubMed] [Google Scholar]
- 39. Kiepiela P., et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53 [DOI] [PubMed] [Google Scholar]
- 40. Kiwanuka N., et al. 2008. Effect of human immunodeficiency virus Type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J. Infect. Dis. 197:707–713 [DOI] [PubMed] [Google Scholar]
- 41. Koehler R. N., et al. 2010. Class I HLA-A*7401 is associated with protection from HIV-1 acquisition and disease progression in Mbeya, Tanzania. J. Infect. Dis. 202:1562–1566 [DOI] [PubMed] [Google Scholar]
- 42. Korber B. T., Letvin N. L., Haynes B. F. 2009. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. J. Virol. 83:8300–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koup R. A. 1994. Virus escape from CTL recognition. J. Exp. Med. 180:779–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koup R. A., Graham B. S., Douek D. C. 2011. The quest for a T cell-based immune correlate of protection against HIV: a story of trials and errors. Nat. Rev. Immunol. 11:65–70 [DOI] [PubMed] [Google Scholar]
- 45. Kulkarni S., et al. 2008. Neutralizing antibody responses in recent seroconverters with HIV-1 subtype C infections in India. AIDS Res. Hum. Retroviruses 24:1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 47. Lazaryan A., et al. 2010. Human leukocyte antigen class I supertypes and HIV-1 control in African Americans. J. Virol. 84:2610–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lazaryan A., et al. 2011. The influence of human leukocyte antigen class I alleles and their population frequencies on human immunodeficiency virus type 1 control among African Americans. Hum. Immunol. 72:312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leslie A., et al. 2010. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J. Virol. 84:9879–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu R., et al. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–377 [DOI] [PubMed] [Google Scholar]
- 51. Liu Y., et al. 2006. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J. Virol. 80:9519–9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Los Alamos National Laboratory 2010. HIV Molecular Immunology Database. http://www.hiv.lanl.gov/content/immunology/index.html Last accessed April 2011
- 53. Maiers M., Gragert L., Klitz W. 2007. High-resolution HLA alleles and haplotypes in the United States population. Hum. Immunol. 68:779–788 [DOI] [PubMed] [Google Scholar]
- 54. Mann D. L., et al. 1998. Major histocompatibility complex genotype is associated with disease progression and virus load levels in a cohort of human immunodeficiency virus type 1-infected Caucasians and African Americans. J. Infect. Dis. 178:1799–1802 [DOI] [PubMed] [Google Scholar]
- 55. Marsh S. G., et al. 2010. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75:291–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Matthews P. C., et al. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 82:8548–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McMichael A., Mwau M., Hanke T. 2002. HIV T cell vaccines, the importance of clades. Vaccine 20:1918–1921 [DOI] [PubMed] [Google Scholar]
- 58. Mellors J. W., et al. 1995. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann. Intern. Med. 122:573–579 [DOI] [PubMed] [Google Scholar]
- 59. Mellors J. W., et al. 2007. Prognostic value of HIV-1 RNA, CD4 cell count, and CD4 cell count slope for progression to AIDS and death in untreated HIV-1 infection. JAMA 297:2349–2350 [DOI] [PubMed] [Google Scholar]
- 60. Migueles S. A., et al. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Modjarrad K., Chamot E., Vermund S. H. 2008. Impact of small reductions in plasma HIV RNA levels on the risk of heterosexual transmission and disease progression. AIDS 22:2179–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neilson J. R., et al. 1999. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J. Virol. 73:4393–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Novitsky V., et al. 2001. Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum. Immunol. 62:146–156 [DOI] [PubMed] [Google Scholar]
- 64. Novitsky V., et al. 2009. Timing constraints of in vivo Gag mutations during primary HIV-1 subtype C infection. PLoS One 4:e7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Paris R., et al. 2004. HLA class I serotypes and cytotoxic T-lymphocyte responses among human immunodeficiency virus-1-uninfected Thai volunteers immunized with ALVAC-HIV in combination with monomeric gp120 or oligomeric gp160 protein boosting. Tissue Antigens 64:251–256 [DOI] [PubMed] [Google Scholar]
- 66. Pelak K., et al. 2010. Host determinants of HIV-1 control in African Americans. J. Infect. Dis. 201:1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pellegrin I., et al. 1996. Kinetics of appearance of neutralizing antibodies in 12 patients with primary or recent HIV-1 infection and relationship with plasma and cellular viral loads. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 11:438–447 [DOI] [PubMed] [Google Scholar]
- 68. Pinkerton S. D. 2007. How many sexually-acquired HIV infections in the U. S. A. are due to acute-phase HIV transmission? AIDS 21:1625–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Prado J. G., et al. 2010. Replicative capacity of human immunodeficiency virus type 1 transmitted from mother to child is associated with pediatric disease progression rate. J. Virol. 84:492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Price M. A., et al. 2011. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res. Hum. Retroviruses 27:5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Quinn T. C., et al. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 342:921–929 [DOI] [PubMed] [Google Scholar]
- 72. Rolland M., et al. 2010. Amino-acid co-variation in HIV-1 Gag subtype C: HLA-mediated selection pressure and compensatory dynamics. PLoS One 5:e12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rousseau C. M., et al. 2008. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype C proteome: immune escape and viral load. J. Virol. 82:6434–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Saag M. S., et al. 1996. HIV viral load markers in clinical practice. Nat. Med. 2:625–629 [DOI] [PubMed] [Google Scholar]
- 75. Saah A. J., et al. 1998. Association of HLA profiles with early plasma viral load, CD4+ cell count and rate of progression to AIDS following acute HIV-1 infection. Multicenter AIDS Cohort Study. AIDS 12:2107–2113 [DOI] [PubMed] [Google Scholar]
- 76. Salazar-Gonzalez J. F., et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shrestha S., et al. 2009. Host genetics and HIV-1 viral load set-point in African-Americans. AIDS 23:673–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stacey A. R., et al. 2009. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 83:3719–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 80. Tang J., Kaslow R. A. 2003. The impact of host genetics on HIV infection and disease progression in the era of highly active antiretroviral therapy. AIDS 17:S51–S60 [DOI] [PubMed] [Google Scholar]
- 81. Tang J., et al. 2010. Human leukocyte antigens and HIV type 1 viral load in early and chronic infection: predominance of evolving relationships. PLoS One 5:e9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tang J., et al. 2008. Human leukocyte antigen class I genotypes in relation to heterosexual HIV type 1 transmission within discordant couples. J. Immunol. 181:2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tang J., et al. 2002. Distribution of chemokine receptor CCR2 and CCR5 genotypes and their relative contribution to human immunodeficiency virus type 1 (HIV-1) seroconversion, early HIV-1 RNA concentration in plasma, and later disease progression. J. Virol. 76:662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tang J., et al. 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276–8284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tang J., et al. 2004. HLA allele sharing and HIV type 1 viremia in seroconverting Zambians with known transmitting partners. AIDS Res. Hum. Retroviruses 20:19–26 [DOI] [PubMed] [Google Scholar]
- 86. Tang J., et al. 2002. Host genetic profiles predict virological and immunological control of HIV-1 infection in adolescents. AIDS 16:2275–2284 [DOI] [PubMed] [Google Scholar]
- 86a. Tang J., et al. 2011. HIV-1 viral load during acute infection in sub-Saharan Africans: deciphering the impact of host and viral factors, poster abstr. 510. Abstr. 18th Conf. Retrovir. Opportun. Infect., Boston, MA, 27 February to 2 March 2011 [Google Scholar]
- 87. Tebit D. M., Arts E. J. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 11:45–56 [DOI] [PubMed] [Google Scholar]
- 88. The International HIV Controllers Study 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Treurnicht F. K., et al. 2010. Adaptive changes in HIV-1 subtype C proteins during early infection are driven by changes in HLA-associated immune pressure. Virology 396:213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Vasan A., et al. 2006. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin. Infect. Dis. 42:843–852 [DOI] [PubMed] [Google Scholar]
- 91. Walker L. E., et al. 2009. Design and preclinical development of a recombinant protein and DNA plasmid mixed format vaccine to deliver HIV-derived T-lymphocyte epitopes. Vaccine 27:7087–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang Y. E., et al. 2009. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J. Virol. 83:1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wawer M. J., et al. 2005. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J. Infect. Dis. 191:1403–1409 [DOI] [PubMed] [Google Scholar]
- 94. Zhang M., et al. 2010. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.