Abstract
The papillomavirus E1 and E2 proteins are essential for viral genome replication. E1 is a helicase that unwinds the viral origin and recruits host cellular factors to replicate the viral genome. E2 is a transcriptional regulator that helps recruit the E1 helicase to the origin and also plays a role in genome partitioning. We find that when coexpressed, the E1 and E2 proteins from several papillomavirus types localize to defined nuclear foci and result in growth suppression of the host cells. Growth suppression was due primarily to E1 protein function, and nuclear expression of E1 was accompanied by activation of a DNA damage response, resulting in phosphorylation of ATM, Chk2, and H2AX. Growth suppression and ATM activation required the ATPase and origin-specific binding functions of the E1 protein and resulted in active DNA repair, as evidenced by incorporation of nucleotide analogs and detection of free DNA ends. In the presence of the E2 protein, these activities became localized to nuclear foci. We postulate that these foci represent viral replication factories and that a cellular DNA damage response is activated to facilitate replication of viral DNA.
INTRODUCTION
Papillomaviruses (PVs) are double-stranded DNA viruses that infect cutaneous and mucosal epithelial cells of animals. Over 200 papillomavirus types have been reported to date, and phylogenetic classification indicates that there are at least 29 genera (1). Papillomavirus genomes consist of approximately 8 kb of double-stranded DNA, which typically contains seven to eight genes. Two of these genes encode replication proteins, E1 and E2. The E1 protein is a helicase that has been shown by structural and biochemical studies to be essential for initiation of viral DNA replication. The E1 protein by itself has low affinity for the viral origin of replication, which contains specific, palindromic E1 binding sites. However, the multifunctional E2 protein binds to specific sites adjacent to the E1 binding sites and helps recruit E1 in a cooperative manner. When loaded onto the viral replication origin, the E1 and E2 proteins recruit host replication factors such as RPA, topoisomerase I, and Polα/primase to initiate viral DNA replication (reviewed in reference 36). The E2 protein can function both as a transcriptional transactivator and repressor of viral early genes and for some papillomavirus types has also been shown to tether the viral genome to host chromatin to maintain and partition the extrachromosomal genomes.
Eukaryotic cells have many different strategies to combat viral infection. Many viruses induce a cellular DNA damage response (DDR), either indirectly by virtue of viral DNA replicative intermediates that resemble damaged DNA or directly by viral protein function (reviewed in reference 51). The host cell induces the DNA damage response in an attempt to arrest cell growth and allow repair of genomic DNA damage, thus maintaining genomic stability. Two of the major regulators of the DNA damage response are the ATM (ataxia telangiectasia mutated) and the ATR (ATM and Rad3-related) kinases, which belong to the phosphoinositide-3-kinase-related protein kinases (PIKKs) (reviewed in reference 7). These kinases initiate a signal transduction cascade that activates many pathways and proteins to maintain genome integrity. While there is much overlap in the substrates of the ATM and ATR pathways, the ATR pathway is generally induced by single-stranded DNA at stalled replication forks whereas the ATM pathway is activated by double-strand DNA breaks (DSB) resulting from collapsed replication forks or ionizing irradiation (reviewed in reference 51). Among many of the substrates that are phosphorylated by activated ATM are H2AX and Chk2, while phosphorylation of Chk1 is associated with an induced ATR pathway.
In the context of a viral infection, the host DNA damage response can be disadvantageous to viral production, and thus viruses have evolved mechanisms to overcome host cellular defenses and “reprogram” these responses for their own benefit (reviewed in references 4 and 51). For instance, in HIV infection, the Vpr accessory protein arrests cells in G2/M phases of the cell cycle and induces the ATR pathway by binding to chromatin and inducing Chk1 phosphorylation and the formation of phosphorylated H2AX (γH2AX) and 53BP1 nuclear foci (33). This G2 arrest results in increased viral expression and production (17). In another example, detection of newly synthesized viral DNA during lytic replication of Epstein-Barr virus (EBV) results in an ATM DNA damage response (30). However, the ATM signaling cascade is modified by the viral BGLF4 kinase, resulting in promotion of an S-phase-like environment for viral DNA replication, inhibition of cellular DNA replication (29), and promotion of viral DNA circularization by homologous recombinational repair (31). Other small DNA viruses, such as polyomavirus and simian virus 40 (SV40), are similar to papillomaviruses and rely on cellular replication factors for DNA synthesis. SV40 and polyomaviruses reprogram and exploit the cellular DNA damage response for self-propagation (8, 19). SV40 large T antigen can induce DNA damage in the absence of the viral replication origin, thus activating the ATM, ATR, and Fanconi anemia (FA) homologous recombination pathways, which are required for efficient replication of SV40 (2). Recently, it was shown that “high-risk” oncogenic papillomaviruses also require the ATM pathway to efficiently amplify viral genomes in differentiated cells (39). However, the exact mechanism for the induction of DDR in cells harboring human papillomavirus (HPV) remains unclear.
In this study we have shown that coexpression of the papillomavirus E1 and E2 proteins from various papillomavirus types resulted in cell growth suppression. The viral replication helicase, E1, was found to be the primary cause of the growth arrest, and we show that this was due at least in part to an induction of the ATM DNA damage response pathway. Furthermore, E1 protein expression resulted in cellular DNA synthesis, and this required both the specific DNA binding and ATPase functions of E1. Furthermore, coexpression of the E1 and E2 proteins resulted in the formation of nuclear foci that recruited proteins associated with the ATM response. We propose that recruitment of the cellular replication and repair machinery to these nuclear foci is advantageous for papillomavirus replication.
MATERIALS AND METHODS
Plasmids.
All E2 proteins were tagged with an N-terminal Flag epitope and were expressed from the metallothionein-inducible promoter in the pMEP4 plasmid, as described previously (40). The corresponding E1 genes were cloned into a similar plasmid, pMEP9, in which the hygromycin resistance gene was replaced with the neomycin resistance gene. The E1 proteins were also expressed from the inducible methallothionein promoter, and each E1 protein was tagged with an N-terminal Glu-Glu (EE) epitope. To increase expression, regions at the 5′ ends of some of the E1 and E2 genes were resynthesized using optimal human codons, and the resulting fragments were cloned into the E1 and E2 genes using convenient restriction sites, as described previously (40). The recoded amino acid residues were as follows: bovine papillomavirus type 1 (BPV1) E1 (1 to 274), BPV E2 (none), HPV8 E1 (1 to 239), HPV8 E2 (none), HPV11 E1 (1 to 649), HPV11 E2 (1 to 148), HPV16 E1 (1 to 151), HPV16 E2 (1 to 227), HPV31 E1 (1 to 402), and HPV31 E2 (1 to 372). To generate plasmids encoding the mutated HPV16 K230A S231A and K483A E1 proteins, the SpeI and NsiI fragments from the pTZ18U:HPV16W12 K230A S231A and K483A plasmids (44) were cloned into the pMEP9 EE-E1 vector, using standard procedures. The Stratagene QuikChange site-directed mutagenesis kit (Agilent) was used to generate mutated HPV16 E2 proteins using the pTZ:Flag-HPV16 E2 recoded plasmid as a template. The primers used were as follows: E2 E39A, 5′GGAAGCACATGAGACTGGCGTGCGCCATCTACTACAAGGCC-3′ and 5′GGCCTTGTAGTAGATGGCGCACGCCAGTCTCATGTGCTTCC-3′; E2 R37A and I73A, 5′CCACATCGACTACTGGAAGCACATGGCACTGGAGTGCGCCATCTAC-3′, 5′GTAGATGGCGCACTCCAGTGCCATGTGCTTCCAGTAGTCGATGTGG-3′, 5′GTAAGAACAAGGCCCTGCAGGCCGCCGAGCTCCAGCTGACCCTGGAGACC-3′, and 5′GTAAGAACAAGGCCCTGCAGGCCGCCGAGCTCCAGCTGACCCTGGAGACC-3′; E2 R302A/R304A, 5′GCTAATACTTTAAAATGTTTAAAATATAAATTTAAAAAGCATTGTACATTG-3′ and 5′CAATGTACAATGCTTTTTAAATTTATATTTTAAACATTTTAAAGTATTAGC-3′. The mutated genes were subcloned into the pMEP4 vector with BamHI and HindIII digestion. The HPV16 genomes used for cotransfection studies were either the “prototype” genome, p1203 (42), or the W12-derived genome (14). The plasmid containing the HPV16 origin (nucleotides 7838 to 130) has been described previously (9). The pTZ19U control DNA is a commercially available plasmid.
Inhibitors.
Ten micromolar KU-55933 (Calbiochem) and 2.5 mM caffeine (C8960; Sigma) were added to cell culture medium, as indicated.
Cell culture and transfection.
The monkey kidney epithelial cell line, CV-1, and the cervical carcinoma-derived cell line, C33-A, were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Human foreskin keratinocytes (HFK) were grown in F medium (3:1 [vol/vol] F-12 [Ham]-DMEM, 5% FBS, 0.4 μg/ml hydrocortisone, 5 μg/ml insulin, 8.4 ng/ml cholera toxin, 10 ng/ml epidermal growth factor (EGF), 24 μg/ml adenine, 100 U/ml penicillin, and 100 μg/ml streptomycin) and 10 μM Y-27632, where indicated, in the presence of irradiated 3T3-J2 feeder cells, as described previously (39). HFKs immortalized by and harboring HPV genomes were grown in F-medium in coculture with irradiated 3T3 feeders. Transfections were performed using FuGENE 6 (Roche) with 6 μg of each DNA (12 μg total) for a 100-mm plate and 0.4 μg (each) DNA (0.8 μg total) for a 12-well plate using a DNA/FuGENE ratio of 1:3 (μg/μl) according to the protocol provided by the manufacturer.
Cell growth suppression assay.
CV-1 cells (0.5 × 106) were seeded onto a 100-mm plate and transfected with 6 μg of E1 and/or E2 expression plasmids balanced with the respective empty vectors, pMEP4 or pMEP9 (total DNA, 12 μg) and 36 μl of FuGENE 6 (Roche) according to the protocol provided by the manufacturer. Twenty-four hours later, the cells were trypsinized and transferred to 100-mm plates at 1:9 or 1:27 ratios, with or without 0.1 or 1 μM CdSO4, in medium containing 200 μg/ml hygromycin (Roche) and 400 μg/ml G418 (Gibco; Invitrogen). The cell growth suppression assays shown in the figures are from the experiments conducted in the absence of CdSO4. The medium was changed every other day until the pMEP4/9 vector control plate was 60 to 80% confluent. The cells were fixed in 3.7% formaldehyde for 10 min and stained overnight with 0.14% methylene blue.
Western blotting.
CV-1 cells were transfected with different combinations of pMEP9-E1, pMEP4-E2, or the empty pMEP4 or pMEP9 plasmid using FuGENE 6 (Roche), as described above. Protein expression was induced by treatment with 3 μM CdSO4 for 4 h prior to harvesting at 48 h posttransfection. Cells were lysed in ice-cold RIPA buffer (20 mM HEPES [pH 7.3], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS) containing protease inhibitor Complete and PhosSTOP tablets (Roche). Equal amounts of total protein (between 6 to 15 μg per lane) were separated in NuPAGE Bis-Tris gels (Invitrogen), as recommended by the manufacturer. The proteins were transferred to an Immobilon P membrane (Millipore), and the membranes were treated with SuperSignal Western blot enhancer (Thermo Scientific). Proteins were probed with mouse monoclonal anti-EE for E1 (1:4 dilution) and M2 anti-Flag (1:10,000 dilution; Sigma) for E2 and further detected with the appropriate secondary antibodies. Protein bands were identified using the SuperSignal Dura substrate (Thermo Scientific), and images were captured by a Kodak Image Station system and by autoradiography.
Immunofluorescence.
CV-1 cells cultured on Superfrost plus slides were transfected with HPV16 E1 and/or E2 expression plasmids or the corresponding empty vectors, as described above. For HFK, cells were seeded on glass coverslips and cultured with irradiated J2 feeders in F medium containing Y-27632 (3). The cells on slides or coverslips were treated with 3 μM CdSO4 induction for 4 h prior to fixation with 4% paraformaldehyde (PFA) at either 24 to 28 or 48 h posttransfection for HFK or CV-1, respectively. The cells were permeabilized in 0.1% Triton X-100 for 15 min and blocked in 0.25% bovine serum albumin (BSA)-gelatin in phosphate-buffered saline (PBS). The cells were stained with primary antibody (see “Antibodies,” below) for 1 h at 37°C. After washing, the slides were stained with DyLight secondary antibodies (Jackson ImmunoResearch) for 30 min at 37°C. The slides were washed and mounted with 4′,6-diamidino-2-phenylindole (DAPI) containing Prolong Gold (Invitrogen). Images were collected on a Leica TCS-NT/SP5 confocal microscope (Leica Microsystems, Exton, PA) using a 63× or 40× oil immersion objective with a numerical aperture (NA) of 1.32 at zoom 1, 3, or 6. Images were processed using the Leica AS Lite (version 2.1.1, build 4443) and Adobe Photoshop CS3 (Adobe Systems) software programs. For quantitative analysis, the IMARIS software program, version 7.2 (Bitplane), was used to measure the levels of E1, E2, and phosphorylated Chk2 in each cell. Briefly, individual nuclei (excluding the cytoplasm) were analyzed for the mean signal intensity from four different color channels. Ten or more fields containing 30 to 150 nuclei were counted for each sample (totaling 600 to 1,000 nuclei per sample). The background intensity was subtracted from each signal. Nuclei that were expressing E1 or E2 above subjectively determined threshold levels were scored as positive, whereas cells with signals lower than the threshold were scored negative.
Antibodies.
The following antibodies and dilutions were used for immunoblotting: mouse monoclonal anti-EE (hybridoma cells expressing antibodies against the EE epitope were kindly provided by Gernot Walter [18]; 1:4 dilution of cell culture supernatant) and M2 anti-Flag (Sigma; 1:10,000 dilution) for E2. The following antibodies and dilutions were used for immunofluorescence: chicken anti-EE (Bethyl Laboratories, Inc., A190-109A; 1:100), M2 mouse and rabbit anti-Flag (Sigma F3165 and F7425; 1:500 and 1:200, respectively), rabbit anti-phosphorylated Thr68 Chk2 (Cell Signaling 2661; 1:100), mouse anti-phosphorylated Ser1981 ATM (Abcam ab81292; 1:100), mouse anti-phosphorylated Ser15 p53 (Cell Signaling 9286; 1:100), rabbit anti-phosphorylated Ser139 H2AX (Cell Signaling 2577; 1:100), rabbit anti-phosphorylated Ser345 pChk1 (Cell Signaling 2349; 1:100), rabbit anti-NBS1 (Novus NB100-143; 1:200), and rabbit anti-caspase-3 cleaved at Asp175 (Cell Signaling 9661; 1:200).
Nucleotide incorporation assay.
The Click-iT EdU cell proliferation assay (Invitrogen) was used to detect incorporation of nucleotide analogue EdU in cells expressing E1, E2, E1-E2, or pMEP vectors. HFKs grown in Y-27632 with irradiated J2 feeders were seeded on coverslips and transfected with appropriate E1, E2, or vector plasmids using FuGENE 6 as described above. Cells were treated with 3 μM CdSO4 for 4 h and with 50 μM EdU for 30 min prior to fixation with 4% PFA. The protocol provided by the manufacturer was used to detect EdU, followed by immunostaining of E1 and E2, as described above (see “Immunofluorescence”).
TUNEL assay.
To detect DNA breaks in E1- or E2-expressing cells, a terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (Roche) was used. HFKs grown in Y-27632 with irradiated J2 feeders were seeded on coverslips and transfected with appropriate E1, E2, or vector plasmids using FuGENE 6, as described above. Cells were treated with 3 μM CdSO4 for 4 h prior to fixation at 24 to 28 h posttransfection. Cells on coverslips were fixed in 4% PFA and permeabilized in 0.1% Triton X-100 followed by the TUNEL reaction, as directed by the manufacturer (Roche). The coverslips were washed in PBS and stained with chicken anti-EE and M2 anti-Flag, as described above (see “Immunofluorescence”).
RESULTS
Papillomavirus E1 proteins arrest cell growth.
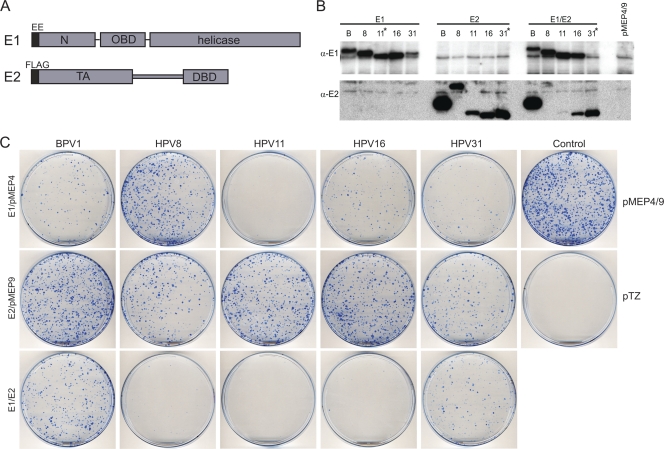
We have previously shown that all papillomavirus E2 proteins can interact with host mitotic chromosomes, and this is thought to be important for viral genome partitioning. However, E2 proteins encoded by the alphapapillomaviruses are only observed bound to chromosomes under certain fixation conditions (40), and it is not known whether these conditions unmask or stabilize this association. In an effort to determine whether other viral proteins could stabilize the association of alpha-PV E2 proteins with mitotic chromatin, E1 proteins from HPV11, -16, and -31 were coexpressed with the corresponding E2 proteins. The E1 proteins were found to be poorly expressed, as had been observed previously for the alphapapillomavirus E2 proteins, and so an N-terminal portion of each E1 open reading frame (ORF) was recoded to increase protein expression levels. As shown in Fig. 1A, the E2 proteins contained an N-terminal Flag epitope and the E1 proteins contained an N-terminal EE epitope. Both epitope tags have been shown previously to have no effect on protein function (13, 37). In each case, the recoding process substantially increased E1 expression to detectable levels (Fig. 1B and data not shown). Plasmids encoding either the E1 or E2 proteins, along with respective neomycin and hygromycin resistance markers, were transfected into either CV-1 or C33A cells. However, this combination proved to be growth inhibitory to both cell types, and it was almost impossible to obtain cell lines expressing both proteins. Since we routinely establish stable E2 expression plasmids in these cell lines, it appeared that the E1 protein was likely responsible for the observed growth arrest.
Fig. 1.
Growth-inhibitory effect of papillomavirus E1 expression on CV-1 cells. (A) Map of the E1 and E2 proteins. The E1 protein is tagged with a Glu-Glu (EE) epitope and is expressed from the pMEP9 vector. The E2 protein is tagged with a Flag epitope and is expressed from the pMEP4 vector. E1 has three major domains: the N-terminal domain (N), the origin binding domain (OBD), and a C-terminal helicase domain. The E2 protein has two domains: the transactivation (TA) and DNA binding (DBD) domains. In most cases, a part of or the entire gene was codon optimized for increased protein expression. Recoded amino acid residues were BPV1 E1 (1 to 274), BPVE2 (none), HPV8 E1 (1 to 239), HPV8 E2 (none), HPV11 E1 (1 to 649), HPV11E2 (1 to 148), HPV16 E1 (1 to 151), HPV16 E2 (1 to 227), HPV31 E1 (1 to 402), and HPV31 E2 (1 to 372). (B) Western blot analysis of the E1 and E2 proteins expressed in transiently transfected CV-1 cells. Cells were transfected with combinations of pMEP9-E1, pMEP4-E2, or the empty pMEP4 or pMEP9 plasmid. Cells were treated with 3 μM CdSO4 for the last 4 h prior to extraction at 48 h posttransfection. The blot was probed with anti-EE for E1 and anti-Flag for E2. The HPV11 E1 and HPV31 E2 and E1-E2 protein lysates were diluted 5-fold with lysate from nontransfected cells to reduce the visual interference of overexposure from these samples (marked with asterisks). A total of 6 μg of cell lysate was loaded in each lane. (C) Cell growth suppression assay in CV-1 cells. CV-1 cells were transfected with papillomavirus E1 and E2 expression plasmids balanced with the respective empty vectors, pMEP4 and pMEP9. Cell growth was compared to that of the positive control or pMEP4 or -9 vector and the negative-control vector pTZ18R. After 24 h, cells were split 1:9 into 100-mm-diameter plates. After approximately 2 weeks of selection in 200 μg/ml hygromycin- and 400 μg/ml G418-containing medium, cells were fixed with formalin and the resulting colonies were stained with methylene blue. Similar results were obtained with and without CdSO4 in the medium.
To analyze this further, we tested the growth-inhibitory properties of the E1 and E2 proteins in a cell growth suppression assay. To determine whether growth arrest was specific to the alpha genus of papillomaviruses, we also analyzed pairs of E1 and E2 proteins from the beta (HPV8) and delta (BPV1) genera of papillomaviruses. As described above for the alpha-E1 proteins, it was also necessary to recode N-terminal portions of these E1 proteins to increase protein expression. However, the BPV1 and HPV8 E2 proteins were expressed well and did not require codon optimization (40). As shown in Fig. 1C, all five E1 proteins suppressed CV-1 cell growth. The HPV11 E1 protein appeared to be most growth inhibitory, but as shown in the Western blots obtained from transiently transfected cells in Fig. 1B, the proteins were expressed at different levels and so it is difficult to conclude that they have inherently different activities. The HPV8 E1 protein had the least effect on cell growth, and this may be because it was found localized mostly in the cytoplasm of the cell (see below). Fradet-Turcotte et al. recently demonstrated that nuclear but not cytoplasmic E1 was growth inhibitory (16). For the most part, the E2 proteins were not growth inhibitory to CV-1 cells except for HPV31 E2. This protein was completely recoded and is therefore expressed at much higher levels than the other E2 proteins, and this gave rise to a mild growth inhibition. The HPV8 and HPV16 E2 proteins enhanced the growth suppression properties of their respective E1 proteins. It should be noted that E1 and E2 are expressed from the inducible metallothionein promoter; however, cell growth suppression is observed even in the absence of promoter induction (Fig. 1C). The metallothionein promoter is somewhat “leaky,” but the resulting low levels of E1 and E2 are sufficient for the observed cell growth inhibition, demonstrating that this is not an artifact of overexpression.
Expression of papillomavirus E1 protein induces a DNA damage response.
Although it was very difficult to obtain cell lines expressing the E1 or E1 and E2 proteins, cells expressing these proteins could be detected by immunofluorescence after transient transfection of CV-1 cells. However, no mitotic cells expressing either the E1 or the E1-E2 proteins could be observed. This led us to conclude that these cells were blocked in the cell cycle. Since cellular growth arrest often occurs in response to cellular checkpoint activation, we wanted to see if a DNA damage response was activated in cells that are transiently expressing E1 proteins.
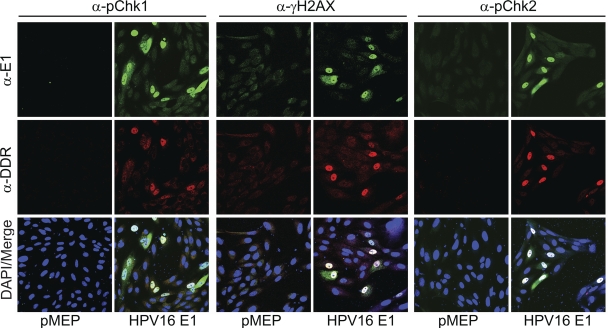
To test this, CV-1 cells were transfected with the HPV16 E1 expression vectors and analyzed for markers of the ATM and ATR DNA damage response. In addition to staining for the E1 protein, cells were costained with antibodies against phosphorylated Chk1 (pChk1), Chk2 (pChk2), and H2AX (γH2AX). As shown in Fig. 2, all cells expressing the E1 protein induced an ATM DNA damage response as indicated by upregulation of phosphorylated Chk2 and γH2AX. As described previously, phosphorylated Chk1 was also detected in a subset of E1-expressing cells, indicating that the ATR pathway was also induced (25, 39). The fact that pChk1 was detected in only a subset of E1-expressing cells suggested that it was activated in a cell cycle-dependent fashion, consistent with the finding that the ATR pathway is activated in S and G2 phases of the cell cycle (23). A combination of the ATM/ATR inhibitors (KU-55933 and caffeine) eliminated the majority of the signal from all of the activated DNA damage response markers tested, confirming that these pathways were required to activate Chk1, Chk2, and H2AX (data not shown). Because the correlation between E1 expression and the observed induction of pChk2 was relatively higher than that for other markers, subsequent studies focused on the ATM response.
Fig. 2.
HPV16 E1 activates the pChk1, pChk2, and γH2AX proteins. CV-1 cells were transfected with an HPV16 E1 expression plasmid or empty vectors (pMEP9) on coverslips. Cells were stained and with chicken anti-EE (α-E1) (in green) and antibodies to various DNA damage response (DDR) proteins, pChk1 (Ser345), pChk2 (Thr68), and histone γH2AX (Ser139) (in red).
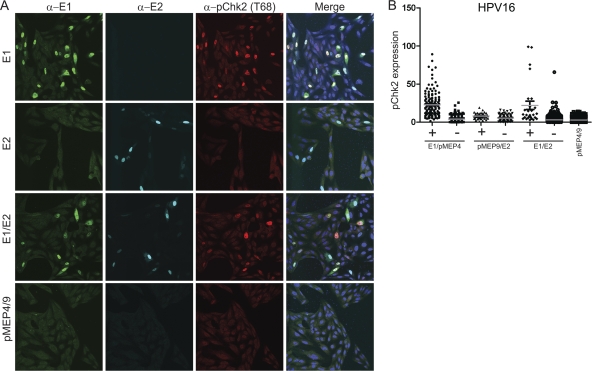
To determine whether the ATM pathway was activated by E1 proteins from other papillomaviruses, we assessed the phosphorylation of Chk2 at position Thr68 (α-pChk2), which has been shown to phosphorylate p53, which in turn activates the G1 checkpoint (5, 20). Since the levels of protein expression were different in each cell, imaging software was used to analyze the level of pChk2 in E1- and E2-expressing cells on a cell-by-cell basis. As can be seen in Fig. 3A, pChk2 was activated primarily in CV-1 cells that are expressing HPV16 E1 or E1 and E2 together. HPV16 E2 expression by itself does not affect pChk2 levels, consistent with its inability to affect cell growth. The HPV16 E1 protein is located in both the cytoplasm and the nucleus; however, only nuclear E1 activated pChk2 (see below). To quantitate the levels of nuclear E1, E2, and phosphorylated Chk2 on a cell-by-cell basis, the imaging analysis software Imaris (Bitplane) was used to measure the signal intensity from each cell within multiple fields of cells. Only a subset of cells expressed the viral proteins, and so within each cell line the cells were divided into two categories: those that expressed detectable levels of the viral proteins in the nucleus and those that did not. The level of pChk2 in each of these categories is shown in Fig. 3B and in the right panel of Fig. 3C.
Fig. 3.
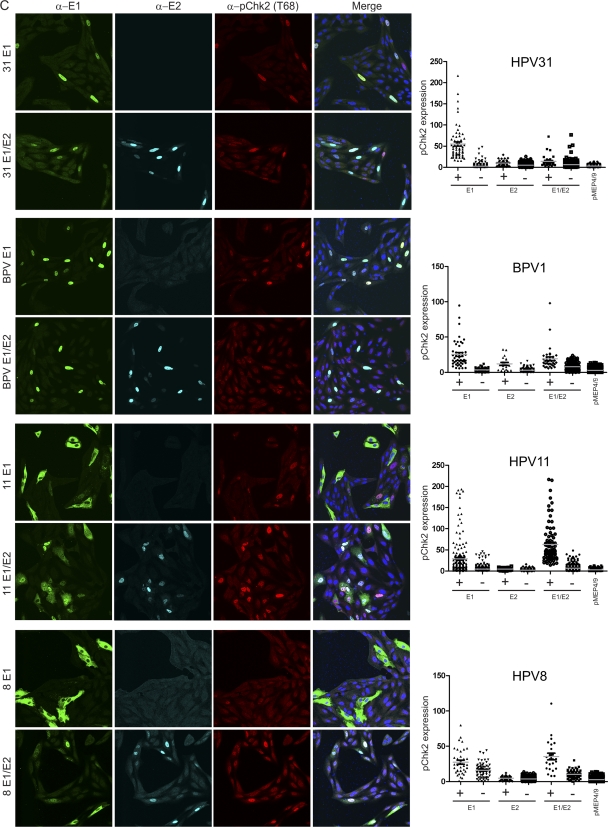
Nuclear expression of papillomavirus E1 proteins induces phosphorylated Chk2. (A) Increased levels of phosphorylated Chk2 are detected in HPV16 E1-expressing cells. CV-1 cells were transfected with HPV16 E1 and E2 expression vectors or the corresponding empty vectors. Cells were stained with chicken anti-EE (shown in green), M2 or rabbit anti-Flag (in cyan), and anti-phosphorylated Chk2 (Thr68) (in red.) The imaging procedure is as described in the legend for Fig. 2A except that a 40× oil immersion objective with zoom 1 was used instead of the 63× objective lens. (B) Quantitative representation of the level of phosphorylated Chk2 signal in the presence (+) or absence (−) of detectable nuclear E1 or E2 or E1-E2 together. IMARIS version 7.2 was used to analyze the level of phosphorylated Chk2 in each cell. Briefly, each individual nucleus was analyzed for the intensity of signals from four different color channels using a surface tool. Approximately 10 or more fields of panels containing 30 to 150 nuclei from each sample were counted (totaling 600 to 1,000 nuclei per sample). A background threshold was subtracted from each signal. Nuclei that were expressing E1 or E2 above threshold levels were scored as positive, while cells that had expression lower than the threshold were scored negative. (C) The E1 protein from several different papillomavirus types induces pChk2. CV-1 cells were transfected with HPV11, -31, or -8 or BPV1 E1 or E1-E2 expression vectors as described above for panel A. The images of E2 expression alone are not shown. Quantitative analyses as described for panel B are shown to the right of the panel of confocal images.
To determine whether the E1 proteins from HPV8, -11, and -31 and BPV1 could also upregulate the ATM pathway, we analyzed the pChk2 levels in CV-1 cells transfected with HPV8, -11, and -31 and BPV1 E1 and E2 expression vectors. As shown in Fig. 3C, when expressed alone, E1 proteins from HPV31 and BPV1 are localized mostly in the nucleus, while E1 proteins from HPV8 and HPV11 were localized in the cytoplasm as well as in the nucleus. Nonetheless, for each virus, cells with nuclear E1 contained phosphorylated Chk2 at levels much higher than those of cells not expressing detectable E1 or control cells. As had been observed for HPV16, expression of the E2 protein alone had no effect on Chk2 phosphorylation (Fig. 3C, right panels). When HPV8 and HPV11 E1 and E2 were expressed together, Chk2 phosphorylation increased, and we surmise that this is due to increased nuclear import of the E1 protein by the E2 protein. This is consistent with a recent report that only the nuclear HPV31 E1 protein has growth-inhibitory properties (16). However, there was no significant increase, and perhaps a decrease, in pChk2 when the E2 protein of BPV1 and HPV31 E2 was coexpressed with its corresponding E1 protein. It is possible that at some ratios E2 can inhibit the growth-inhibitory properties of E1, perhaps by interfering with the formation of the E1 hexameric helicase. However, it is difficult for us to compare and contrast the effects observed with different viruses because of the various levels and ratios of E1 and E2 expressed from our vectors (Fig. 1B).
To corroborate the observed induction of pChk2, similar experiments were carried out in CV-1 cells transfected with plasmids expressing either E1, E2, or both proteins from all five papillomaviruses. Cells were stained with antibodies against pChk1 or γH2AX. Again, a very strong correlation was found between cells expressing the nuclear E1 protein and activation of these DNA damage response markers (data not shown).
Human keratinocytes containing replicating HPV genomes show activated pChk2.
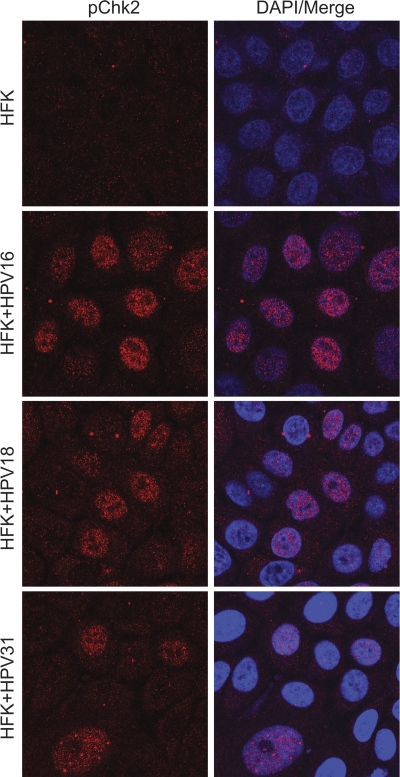
Low levels of the E1 and E2 proteins are produced in proliferating cells that maintain the viral genomes as extrachromosomal elements. High levels of E1 and E2 are thought to be expressed in differentiated cells that are vegetatively producing virus. For example, high levels of the E2 protein have been observed in cells that are amplifying the viral genome in BPV1 fibropapillomas (41). HPV31 has been shown to induce an ATM response in both differentiated and undifferentiated keratinocytes. To determine whether the ATM response is also induced in cells containing other viral types, we stained human keratinocytes that maintained the HPV16, -18, or -31 genome as extrachromosomal elements with antibodies against phosphorylated Chk2. As shown in Fig. 4, all three HPV-containing cell lines had induced levels of phosphorylated Chk2 compared to that of control keratinocytes. Thus, pChk2 induction is observed in keratinocytes containing replicating HPV genomes, and the observations made for HPV31 by Moody and Laimins (39) can be extended to other viral types.
Fig. 4.
HFKs containing replicating HPV genomes show upregulation of phosphorylated Chk2. Primary HFKs and HFKs immortalized by and containing replicating HPV16, -18, and -31 genomes were grown on coverslips and fixed in 4% PFA. The cells were stained with antibodies against phosphorylated Chk2 (Thr68) and processed as described in Materials and Methods.
Papillomavirus E1 and E2 proteins localize to nuclear foci when coexpressed.
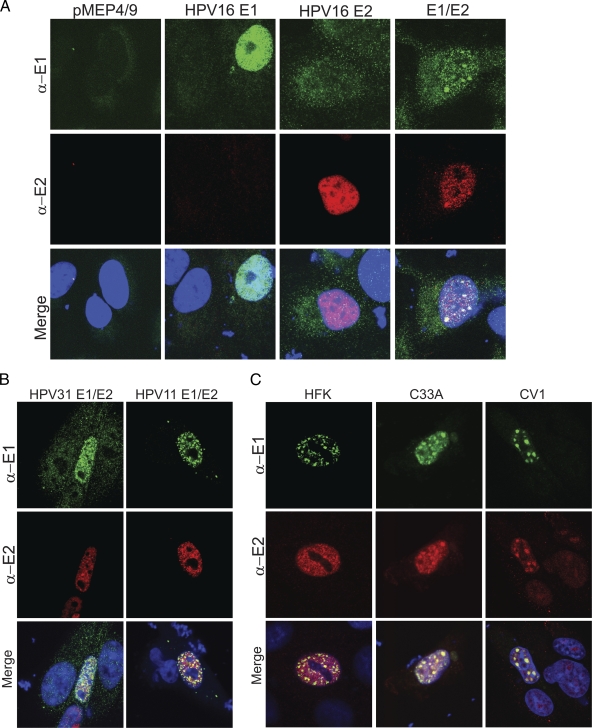
When expressed individually, the HPV16 E1 and E2 proteins were observed in fine granular or diffuse nuclear patterns. However, when they were expressed together, large punctate foci containing both proteins could be observed in the interphase nuclei of a subset of cells (Fig. 5A). Similar foci have been observed previously and, based on colocalization with RPA and sites of BrdU incorporation, were thought to be replication foci (47). Similar foci were also observed upon expression of the HPV11 and HPV31 E1 and E2 proteins (Fig. 5B). As shown in Fig. 5C, these foci were also observed in C33A cells and in human foreskin keratinocytes, which closely resemble the natural target cells of papillomavirus infection, with all three pairs of alpha-HPV E1 and E2 proteins.
Fig. 5.
The alphapapillomavirus E1 and E2 proteins colocalize in defined nuclear foci. (A) HPV16 E1 and E2 nuclear foci. CV-1 cells cultured on slides were transfected with HPV16 E1 and E2 expression plasmids or the corresponding empty vectors. The E1 and E2 proteins were detected with chicken anti-EE (α-E1) and mouse M2 anti-Flag (α-E2) antibodies, respectively, and DyLight secondary antibodies. Cellular DNA was counterstained with DAPI (blue). The E1 protein is detected as green and E2 as red. (B) HPV11 and -31 E1 and E2 nuclear foci. CV-1 cells were transfected with HPV11 or -31 E1 and E2 expression vectors as described for panel A. The characteristic foci seen with E1 and E2 coexpression are not observed when the proteins are expressed individually, similar to the observation with HPV16 (data not shown). (C) HPV16 E1 and E2 coexpressed in C33A and HFK localize to defined foci. HFK and C33A cells were transiently transfected with HPV16 E1 and E2 expression plasmids. For HFK, the cells were fixed with 4% PFA 24 h after transfection with 3 μM CdSO4 induction 4 h prior to fixation. The staining and imaging procedures are described in Materials and Methods.
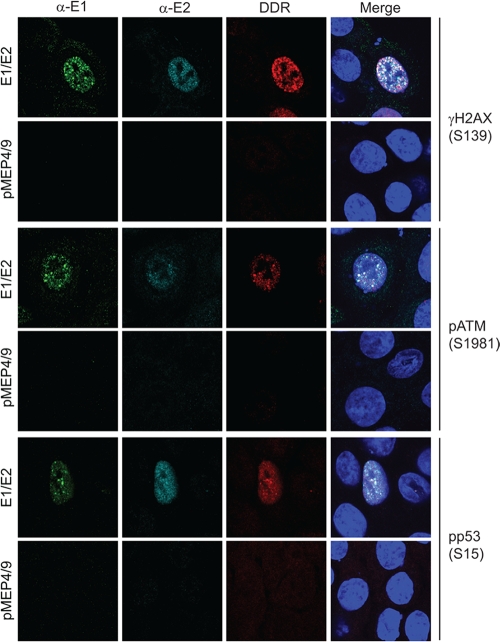
The observation that the HPV E1 proteins suppressed cell growth and activated a DNA damage response suggested to us that the foci might be DNA repair foci. Many viruses evoke a DNA damage response, and often they take advantage of this response to replicate their own genomes (51). Furthermore, the papillomaviruses have been shown to evoke an ATM DNA damage response in differentiated cells in the productive stage of the viral life cycle (39). To see if the E1-E2 foci recruited DNA damage response proteins, human keratinocytes transiently expressing HPV16 E1 and E2 were analyzed for markers of the ATM and ATR DNA damage response pathways. In addition to staining for the E1 and/or E2 proteins, cells were costained with antibodies against H2AX phosphorylated on serine 139 (γH2AX), ATM phosphorylated on serine 1981 (pATM), and p53 phosphorylated on serine 15 (pp53). As shown in Fig. 6, HFKs expressing the E1 and E2 proteins induced an ATM DNA damage response as indicated by upregulation of γH2AX, pATM, and pp53 and partial recruitment of these proteins to the E1-E2 foci.
Fig. 6.
HPV16 E1 and E2 foci partially colocalize with DDR proteins from the ATM pathway. HFKs conditionally immortalized by culture in Y-27632 were transfected with HPV16 E1 and E2 expression plasmids or empty vectors (pMEP4/9) on coverslips. Cells were stained and processed as described in Materials and Methods, with chicken anti-EE (α-E1) (in green), M2 or rabbit anti-Flag (α-E2) (in cyan), and various DNA damage response (DDR) proteins, anti-phosphorylated ATM (Ser1981), p53 (Ser15), and histone γH2AX (Ser139) (in red).
E1 and E2 foci recruit DNA repair response proteins by inducing DNA breaks.
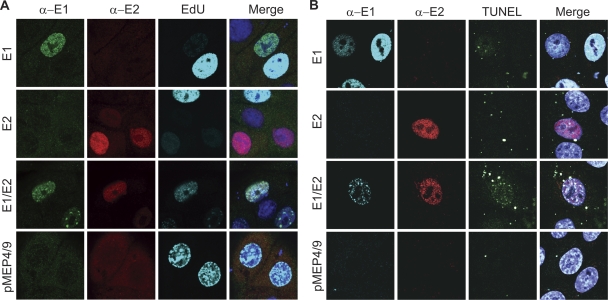
HPV11 E1-E2 nuclear foci have been noted previously, in both the presence and absence of viral origin-containing DNA, and were thought to be replication centers for the virus since they recruited RPA and incorporated nucleotide analogs (47). Since we had observed that E1-E2 expression resulted in cell growth suppression and likely cell cycle arrest, we wanted to examine this further. To further determine whether E1- or E1-E2-expressing cells were blocked in S phase, the cells were pulse-labeled with EdU to detect incorporation into newly replicated DNA. It was initially hoped that this would confirm or determine that cells were in S phase and pinpoint whether they were blocked at early or late stages. Cells in early, mid, or late S phase exhibit different nuclear replication patterns: early-replicating euchromatic DNA forms a multitude of tiny foci throughout the nucleus; fewer, larger foci are apparent in mid-S-phase; and large foci are observed around the periphery of the nucleus and nucleolus as heterochromatin replicates in late S phase. As shown in Fig. 7A, E1- and E1-E2-expressing HFK did incorporate EdU; however, the amount incorporated seemed much lower than that for the majority of labeled cells. This led us to question whether these cells were in fact actually in S phase or actively replicating host DNA or were incorporating EdU as part of a repair, and not replication, process. Furthermore, in the presence of the E1 and E2 proteins, EdU was found concentrated within the E1-E2 nuclear foci in 100% of cells.
Fig. 7.
The E1-E2 foci recruit DDR proteins by causing cellular genomic instability. (A) EdU incorporation in E1-expressing cells. HFKs conditionally immortalized by culture in Y-27632 were transfected with HPV16 E1, E2 (total DNA amount balanced with empty vectors), E1-E2, and pMEP4/9 expression vectors. Cells were treated with 3 μM CdSO4 for 4 h prior to fixation and 50 μM EdU for 30 min prior to fixation. The procedure provided by the Click-iT EdU cell proliferation assay was used for the detection of EdU (shown in cyan) followed by immunostaining of E1 (α-E1; in green) and E2 (α-E2; in red) as described in Materials and Methods. (B) Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) signal incorporation in E1-expressing cells. HFKs conditionally immortalized by culture in Y-27632 were transfected with HPV16 E1, E2 (total DNA amount balanced with empty vectors), E1-E2, and pMEP4/9 vectors. Cells were treated with 3 μM CdSO4 4 h prior to fixation at 24 h posttransfection. Cells on coverslips were fixed in 4% PFA, permeabilized in 0.1% Triton X-100, and subjected to the TUNEL reaction. The cells were subsequently stained with anti-EE (α-E1) and anti-Flag (α-E2) antibodies, as described in Materials and Methods. The observed nonnuclear speckles in the TUNEL channel are due to transfected DNA.
The ATM response is usually induced in response to double-stranded DNA breaks, and γH2AX and the MRN (MRE11-Rad50-NBS1) complex usually mark the broken ends of DNA to recruit repair proteins. Orf 36 of herpesvirus 68 and BGLF4 of EBV can induce the ATM response by directly phosphorylating H2AX rather than inducing actual DNA damage (49). However, large T antigen of SV40 (an origin binding helicase analogous to E1) can induce an ATM response by directly damaging host DNA (2). To test whether E1 was directly damaging host cellular DNA, we employed the comet assay, which uses electrophoresis to directly detect broken DNA in the nucleus. This technique indicated that cells transiently transfected with E1 did show more DNA damage than control cells, but because only a low percentage of cells expressed the viral proteins, it was difficult to conclude that these cells specifically harbored DNA breaks. Therefore, we employed the in situ TUNEL assay to determine whether cells specifically expressing E1 also contained DNA breaks. As shown in Fig. 7B, a proportion of cells expressing the E1 protein (∼5 to 20% in three independent experiments) showed evidence of DNA damage as evidenced by incorporation of dUTP to free ends of DNA. This could indicate that E1-mediated DNA damage occurs only in cells at specific stages of the cell cycle. Control cells or cells expressing the E2 protein showed no TUNEL signal. However, when E1 and E2 were coexpressed, the TUNEL signal was specifically localized to the E1-E2 foci in the majority (∼80%) of cells. It is possible that E1 alone or E1-E2 expression might cause stalling of cells at different stages in the cell cycle or the foci may concentrate the sites of DNA damage, allowing more-sensitive visualization by TUNEL. The TUNEL assay is often used to detect cells undergoing apoptosis. However, we were unable to detect cleaved caspase-3 in cells expressing the viral proteins, confirming that we are detecting DNA damage and not apoptotic nuclear breakdown (data not shown).
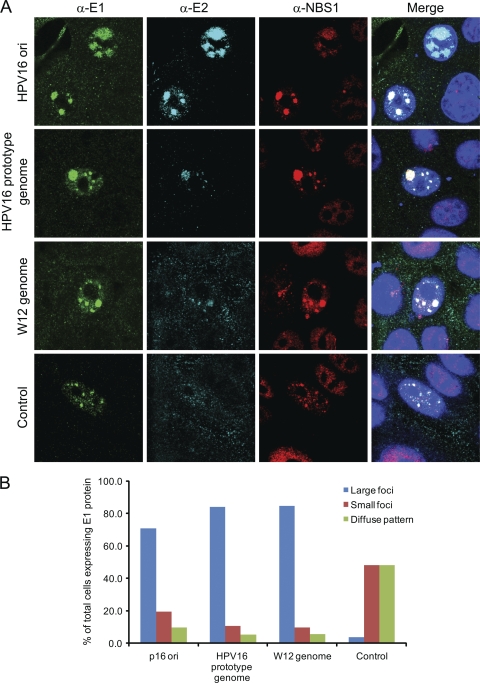
The experiments above demonstrated that the E1-E2 foci can form in the absence of viral origin-containing DNA. To determine whether origin-containing DNA would change the appearance or formation of the foci, HPV16 E1 and E2 expression vectors were cotransfected along with HPV16 viral genomic DNA or viral origin-containing DNA. The resulting E1-E2 DNA damage response foci were notably larger than those in cells cotransfected with control, non-origin-containing DNA (Fig. 8). The viral origin DNA would be predicted to stabilize and enhance the E1-E2 interaction within foci.
Fig. 8.
(A) HFKs conditionally immortalized by culture in Y-27632 were transfected with HPV16 E1 and E2 expression plasmids along with either an HPV16 origin-containing plasmid or HPV16 genomes (either prototype or W12) or a nonspecific plasmid, pTZ19U (control). Cells were stained with chicken anti-EE (α-E1; in green), rabbit anti-Flag (α-E2; in cyan), and anti-NBS1 (in red). (B) For each group of transfected cells, those displaying E1-E2 foci were divided into the three categories shown, according to the size of the foci. Between 38 and 54 cells were counted in each category. The percentage of cells in each category is shown.
Furthermore, in this experiment we showed that all E1-E2 foci (irrespective of the presence of origin DNA) contained large amounts of the NBS1 protein, further confirming recruitment of another marker of the DNA damage response.
Analysis of E1 and E2 functions required for growth suppression, induction of the ATM pathway, and formation of E1-E2 nuclear foci.
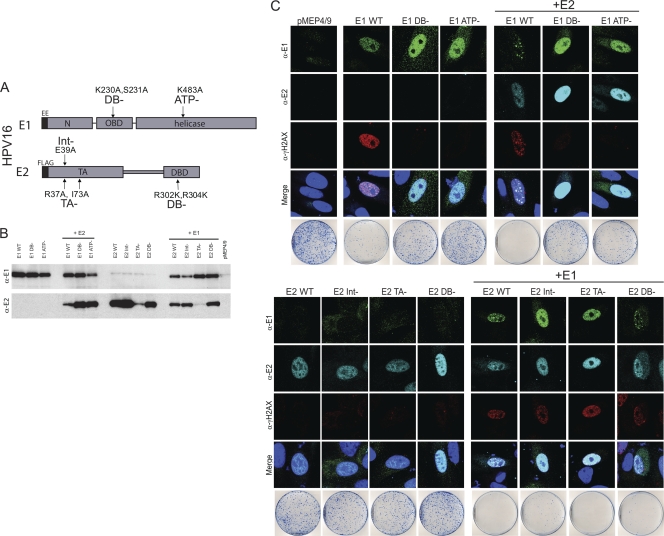
We have shown that the papillomavirus E1 proteins can induce an ATM DNA repair response resulting in growth suppression. Expression of the E2 protein, in addition to E1, results in the formation of E1-E2 nuclear foci that recruit pATM, pp53, and γH2AX (Fig. 6). To determine what functions of the E1 and E2 proteins are required for these activities, we generated point mutations in the HPV16 E1 and E2 genes that specifically inactivated well-characterized functions of the proteins. The E2 protein was mutated in the DNA binding function (R302K/R304K), in the transcriptional regulatory function (R37A/I73A), and in a residue known to disrupt the E1-E2 interaction (E39A). In E1, the origin-specific DNA binding (K230A/S231A) and ATPase (K483A) functions were specifically targeted (Fig. 9A and B). Immunoblot analysis showed that the levels of the mutated E1 and E2 proteins were somewhat variable in extracts of transiently transfected CV-1 cell extracts (Fig. 9B), but immunofluorescence was assayed in cells expressing relatively similar amounts of each protein.
Fig. 9.
Nuclear focus formation requires the specific DNA binding and ATPase functions of E1 and the transactivation and E1 interaction functions of the E2 protein. (A) Diagram of the specific mutations generated in the HPV16 E1 and E2 proteins. The following mutations were made: E1 mutations that eliminate specific DNA binding (DB-) and ATP binding (ATP-) activities; mutations in E2 that interfere with interaction with the E1 protein (Int-), specific DNA binding (DB-), and the transcriptional regulation function of E2 (TA-). (B) Western blotting of mutated E1 and E2 proteins transiently expressed in CV-1 cells. CV-1 cells were transfected with E1 or E2 expression plasmids and corresponding empty vectors. Cells were treated with 3 μM CdSO4 4 h prior to harvesting at 48 h posttransfection. Proteins were detected with anti-EE for E1 (α-E1) and anti-Flag for E2 (α-E2). (C) The characteristic foci observed with wild-type E1 and E2 require the ATPase and DNA binding functions of E1 and the transactivation and E1 interaction functions of E2. CV-1 cells were transfected with wild-type and mutated E1 or E2 expression vectors as described for panel A, and cells were stained with antibodies against the EE epitope (α-E1), Flag (α-E2), and γH2AX (α-γH2AX). The staining and imaging procedures and cell growth suppression assay are as described in Materials and Methods.
The mutated HPV16 E1 and E2 proteins were tested for their ability to activate an ATM response as measured by H2AX, Chk2, and Chk1 phosphorylation in CV-1 cells (Fig. 9 and data not shown) and in human foreskin keratinocytes (data not shown). As shown in Fig. 9C, 100% of cells expressing the wild-type E1 protein contained γH2AX. E1 proteins defective in the DNA binding or ATPase functions, respectively, induced a low level of γH2AX response in 17% and 9% of cells, respectively. Between 6 and 12% of cells transfected with control vectors or expressing either wild-type or mutated E2 proteins had low levels of γH2AX, suggesting that this was a background basal level of γH2AX-positive cells. Thus, a functional E1 protein is required for significant induction of the DNA damage response. Coexpression of the wild-type E2 protein resulted in the formation of E1-E2 nuclear foci, and γH2AX was recruited to these foci. The DNA binding and ATPase-defective E1 proteins did not upregulate γH2AX and, when coexpressed with the E2 protein, did not form the characteristic E1-E2 nuclear foci. In CV-1 cells but not in HFKs, these proteins were often detected in a different punctate nuclear pattern in the presence of E2. However, these foci were quite different from the ATM-related foci and instead colocalized with the pericentromeric region of the host chromosomes (data not shown). Therefore, the DNA binding and ATPase activities of E1 are required for activation of the ATM pathway and for the formation of nuclear DDR foci with E2.
When expressed alone, the E2 proteins did not form nuclear foci and did not result in H2AX phosphorylation. However, when expressed with E1, wild-type E2 colocalized with E1 in the characteristic nuclear DDR foci. The DNA binding-defective E2 protein (R302K/R304K) was also able to form E1-E2 foci that recruited γH2AX. The E2 proteins defective in the E1 interaction or in the transcriptional regulatory function were not able to form nuclear foci with E1 but also had no effect on E1-mediated upregulation of γH2AX.
The experiments described above demonstrated that expression of E1 alone (in the absence of E2) was sufficient for cell growth inhibition and for induction of the ATM response. Thus, we can conclude that formation of nuclear DDR foci of E1 was not essential for these phenomena. However, in some cases coexpression of the E2 protein modulated E1-mediated growth inhibition (Fig. 1C), and this could potentially be mediated by formation of the E1-E2 nuclear foci. Therefore, the mutated HPV16 E1 and E2 proteins were tested for their ability to suppress cellular growth of CV-1 cells. As shown in Fig. 8, the DNA binding and ATPase functions of E1 were essential for cell growth inhibition in CV-1 cells. Coexpression of the wild-type E2 protein did not rescue the ability of these mutated proteins to inhibit CV-1 growth. When expressed alone, neither wild-type nor mutated E2 proteins affected CV-1 cell growth. As was shown above in Fig. 1C, the wild-type E2 protein could enhance the growth-inhibitory effect of E1. This enhancement of growth suppression by the E2 protein required the ability to interact with E1 but not the transcriptional regulatory or DNA binding functions of E2.
A summary of the phenotypes of the mutated E1 and E2 proteins is shown in Table 1. Expression of E1 alone was sufficient for activation of an ATM response and for growth suppression, and this required the DNA binding and ATPase functions of E1. The E2 protein could concentrate the E1 protein and the DDR response into punctate foci within the nucleus and could increase E1-mediated growth suppression. However, the R37A/I73A E2 protein (defective in transcriptional regulation) was unable to form DDR foci with E1 but could still enhance cellular growth suppression. Therefore, we conclude that the E2 protein likely enhances growth suppression in these experiments by promoting nuclear localization of the E1 protein. However, localization of both the E1 and E2 proteins to specific regions of the nucleus could recruit and concentrate cellular replication proteins to facilitate replication of the viral genome.
Table 1.
Activities of the E1 and E2 proteins required for ATM DNA damage response, growth suppression, and nuclear focus formation
| Phenotype | Protein expresseda |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| None | E1 wt | E1 DB− | E1 ATP− | E1 wt + E2 wt | E1 DB− + E2 wt | E1 ATP− + E2 wt | E1 wt + E2 Int− | E1 wt + E2 TA− | E1 wt + E2 DB− | |
| ATM induction | − | +++ | − | − | +++ | − | − | +++ | +++ | +++ |
| Growth suppression | − | ++ | − | − | +++ | − | − | +++ | +++ | +++ |
| Formation of E1/E2 DDR foci | − | − | − | − | +++ | − | − | − | − | +++ |
wt, wild type. Mutation effects are described as follows: DB− and ATP−, elimination of specific DNA binding and ATP binding activities, respectively; Int− and TA−, interference with interaction with the E1 protein and the transcriptional regulation function of E2, respectively. The relative magnitude of each activity is indicated as follows: +++, high; ++, medium; −, none.
DISCUSSION
In this study we have shown that the E1 helicase from various papillomavirus types can arrest cell growth. This is due, at least in part, to activation of the ATM DNA damage response and requires the DNA binding and ATPase functions of E1. This E1-mediated ATM activation results in recruitment of the cellular DNA repair machinery and results in actual DNA replication and/or repair, as evidenced by incorporation of labeled nucleotides and labeling of free DNA ends.
The replicative process of many viruses induces a similar DNA repair response, and many viruses use this response to their advantage since it allows recruitment of cellular components that can synthesize DNA. In normal cells, the ATM response results in cell cycle arrest until DNA damage is repaired. This arrest inhibits cellular DNA replication and permits only repair of DNA damage. Use of the repair machinery to replicate viral DNA would be advantageous since it would allow the virus to synthesize its DNA without competition from that of the host (6). The replication machinery induced by the ATM/ATR pathway could also be more efficient for the virus, since break-induced repair pathways do not require origin licensing proteins to reinitiate DNA replication (34). In fact, it has been shown that papillomaviruses amplify their DNA in the G2 phase of the cell cycle in cells that have already completed S phase (6) and that efficient amplification of the viral genome requires an activated ATM response in differentiated cells (39). The DNA damage response does not seem to interfere with HPV DNA replication, since a recent study showed that transient viral replication is not inhibited by DNA damage-inducing agents, indicating that E1 and E2 can replicate the viral genome in the presence of a DNA damage response (28).
The mechanism by which the E1 protein destabilizes the host genome has yet to be determined. It is possible that E1 unwinds chromosomal DNA, resulting in aberrant replicative intermediates and stalled replication forks and activation of both ATM and ATR pathways. High concentrations of the E1 protein have been shown to promote frequent loading of the helicase at the viral origin, resulting in “onion-skin” replication intermediates (35). In cells with integrated genomes, E1 expression induces amplification of integrated origins, leading to genomic instability and recruitment of the MRN complex, pATM, and pChk2 and phosphorylation of H2AX (25, 26). We propose that in the absence of either integrated or extrachromosomal viral genomes, the active E1 helicase can nonspecifically bind and unwind cellular DNA, resulting in a DNA damage response. This explains why the DNA binding and ATPase activities of E1 are required to induce this response. In the presence of E2, this activity becomes localized to specific foci, perhaps because of the ability of the E2 protein to bind to specific regions of host chromatin. In the studies presented here, the E1-mediated growth-suppressive effect was observed at uninduced, and therefore very low, levels of the E1 protein. Thus, it is unlikely to be due to an artifact of overexpression. Notably, expression of SV40 Large T antigen has also been shown to cause overt DNA damage and induce a DNA damage response in the absence of viral DNA (2).
Although the primary cause of growth arrest was found to be the E1 protein, in some cases coexpression of E2 could enhance this effect. This seemed to be due primarily to increasing the nuclear localization of the E1 protein. Nuclear expression of the E1 protein is detrimental to cellular proliferation (16). The E1 proteins from several papillomaviruses have been shown to shuttle between the nucleus and the cytoplasm using a nuclear localization signal (NLS) and a nuclear export sequence (NES) located in the N-terminal domain (11, 16, 21). Nucleocytoplasmic shuttling is regulated by phosphorylation of the E1 protein and by complex formation with the E2 protein. In the dividing cells of a papilloma, E1 is encoded in a very low abundance message (10), and it must be exported to the cytoplasm to allow cells to proliferate and to promote long-term maintenance of the viral genome (16). Thus, the virus might have evolved to maintain the expression of the E1 protein at a minimal level in the basal layer. Upon differentiation, the induction of a DNA damage response (by increased expression of E1) would not have serious consequences because these cells are destined to terminally differentiate and be shed from the epithelium.
Coexpression of the E1 and E2 proteins results in the formation of nuclear foci containing E1 and E2 in several cell types (CV-1, C33a, and HFK). The formation of defined HPV11 E1-E2 foci in the absence of the viral genome or origin has been observed previously, and the foci were suggested to be viral replication foci because of their colocalization with RPA and BrdU (47). Because E1 and E2 expression was accompanied by cell growth arrest in our experiments, we further investigated the possibility that the observed E1-E2 foci were DNA repair foci. These foci partially colocalized with phosphorylated ATM, Chk2, and H2AX and incorporated nucleotide analogs such as EdU. Furthermore, we noted that the EdU signals in the foci were much less intense than the signals observed in cells undergoing active DNA replication in S phase. This decreased signal suggested that these may be sites of DNA repair. To further analyze the condition of DNA at these sites, we employed the TUNEL assay. The TUNEL assay is most often used to detect free 3′OH ends of DNA resulting from fragmentation due to apoptosis. However, the TUNEL assay can also detect sites of DNA repair in nuclear foci (27, 48, 50).
In our experiments, the E1 and E2 proteins were expressed in various cell types in the absence of other viral gene products. We think it likely that the resulting observations represent a model for the vegetative stage of viral genome amplification, since damage to the cellular DNA in differentiated cells is of no consequence to the host. We observed similar findings for the “low-risk” HPV11 and BPV1 E1 proteins, and so the observed damage to host DNA does not correlate with host genetic instability and carcinogenesis. In dividing cells, expression and nuclear localization of the E1 protein are likely to be tightly regulated to prevent damage to host DNA and to ensure that the virus does not induce cell growth inhibition or apoptosis (16). In “high-risk” viruses, suppression of cellular checkpoints by other viral gene products, such as E6 and E7, could lead to propagation of E1-mediated DNA damage to the host DNA and genomic instability.
As shown in Fig. 4 and as previously shown by Moody and Laimins (39), the ATM pathway is activated in dividing keratinocytes that are replicating the viral genome as persistent extrachromosomal elements. However, under these circumstances, induction of the ATM response does not result in growth inhibition, suggesting that the virus has suppressed the checkpoint. HPV oncogenes promote an S-phase environment by relaxing the G1/S checkpoint, but this also results in replication stress and a subsequent DNA damage response (12, 46). However, DNA damage-mediated growth arrest is abrogated in cells expressing the HPV E6 and E7 oncoproteins (15, 24). HPV16 E7 can attenuate the DNA damage checkpoint by accelerating the proteolytic turnover of claspin, a regulator of the ATR DNA damage checkpoint in the G2 phase of the cell cycle (45), and E7 has been shown to bind to phosphorylated ATM (39). We predict that similar mechanisms suppress the checkpoints induced by the E1 protein in HPV-infected dividing cells.
The formation of foci is a strategy often used by viruses and cells to increase the local protein concentration and promote the interaction of proteins. It has been shown that the E2 protein is essential for loading E1 onto the origin in a low-E1-abundance environment (43). E2 also helps recruit cellular factors that are necessary for viral genome amplification. Thus, we propose that the observed E1-E2 foci are replication factories for the viral genome and that cellular DNA damage response proteins are recruited, at least in part, by local DNA damage caused by the E1 protein. Several functions of the E1 and E2 proteins are required for the formation of these foci. Not surprisingly, a mutation in E2 that disrupts E1-E2 binding abrogates focus formation. E2-specific DNA binding is not required for focus formation, but a mutation in the N-terminal domain that is important for transcriptional activation and repression is required. This mutation, R37A/I73A, also abrogates the chromatin association function of the E2 protein from many papillomavirus types (32, 37, 38). The E2 proteins have been shown to bind to specific regions of host chromatin (22; M.-K. Jang and A. McBride, unpublished), and future studies will determine whether these sites are important for the formation of the viral replication factories. The E1 ATP binding and specific DNA binding functions are also required for focus formation, suggesting that cellular proteins involved in the DNA damage/DNA repair response might also be involved in the formation of the foci.
ACKNOWLEDGMENTS
We thank Amelie Fradet-Turcotte and Jacques Archambault for sharing data prior to publication and Sandra Chapman for generation of the HPV-containing HFK lines. We are also grateful to members of the McBride laboratory for critical reading of the manuscript.
This work was funded by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Bernard H. U., et al. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boichuk S., Hu L., Hein J., Gjoerup O. V. 2010. Multiple DNA damage signaling and repair pathways deregulated by simian virus 40 large T antigen. J. Virol. 84:8007–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chapman S., Liu X., Meyers C., Schlegel R., McBride A. A. 2010. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Invest. 120:2619–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chaurushiya M. S., Weitzman M. D. 2009. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst.). 8:1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chehab N. H., Malikzay A., Appel M., Halazonetis T. D. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 14:278–288 [PMC free article] [PubMed] [Google Scholar]
- 6. Chow L. T., Duffy A. A., Wang H. K., Broker T. R. 2009. A highly efficient system to produce infectious human papillomavirus: elucidation of natural virus-host interactions. Cell Cycle 8:1319–1323 [DOI] [PubMed] [Google Scholar]
- 7. Cimprich K. A., Cortez D. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dahl J., You J., Benjamin T. L. 2005. Induction and utilization of an ATM signaling pathway by polyomavirus. J. Virol. 79:13007–13017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Del Vecchio A. M., Romanczuk H., Howley P. M., Baker C. C. 1992. Transient replication of human papillomavirus DNAs. J. Virol. 66:5949–5958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng W., et al. 2003. mRNA splicing regulates human papillomavirus type 11 E1 protein production and DNA replication. J. Virol. 77:10213–10226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng W., et al. 2004. Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J. Virol. 78:13954–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duensing S., Munger K. 2003. Centrosome abnormalities and genomic instability induced by human papillomavirus oncoproteins. Prog. Cell Cycle Res. 5:383–391 [PubMed] [Google Scholar]
- 13. Ferran M. C., McBride A. A. 1998. Transient viral DNA replication and repression of viral transcription are supported by the C-terminal domain of the bovine papillomavirus type 1 E1 protein. J. Virol. 72:796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flores E. R., Allen-Hoffmann B. L., Lee D., Sattler C. A., Lambert P. F. 1999. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology 262:344–354 [DOI] [PubMed] [Google Scholar]
- 15. Foster S. A., Demers G. W., Etscheid B. G., Galloway D. A. 1994. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J. Virol. 68:5698–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fradet-Turcotte A., Moody C., Laimins L. A., Archambault J. 2010. Nuclear export of human papillomavirus type 31 E1 is regulated by Cdk2 phosphorylation and required for viral genome maintenance. J. Virol. 84:11747–11760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goh W. C., et al. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 4:65–71 [DOI] [PubMed] [Google Scholar]
- 18. Grussenmeyer T., Scheidtmann K. H., Hutchinson M. A., Eckart W., Walter G. 1985. Complexes of polyoma virus medium T antigen and cellular proteins. Proc. Natl. Acad. Sci. U. S. A. 82:7952–7954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hein J., et al. 2009. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J. Virol. 83:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirao A., et al. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824–1827 [DOI] [PubMed] [Google Scholar]
- 21. Hsu C. Y., Mechali F., Bonne-Andrea C. 2007. Nucleocytoplasmic shuttling of bovine papillomavirus E1 helicase downregulates viral DNA replication in S phase. J. Virol. 81:384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jang M. K., Kwon D., McBride A. A. 2009. Papillomavirus E2 proteins and the host BRD4 protein associate with transcriptionally active cellular chromatin. J. Virol. 83:2592–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jazayeri A., et al. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8:37–45 [DOI] [PubMed] [Google Scholar]
- 24. Jones D. L., Munger K. 1997. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J. Virol. 71:2905–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kadaja M., Isok-Paas H., Laos T., Ustav E., Ustav M. 2009. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 5:e1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kadaja M., et al. 2007. Genomic instability of the host cell induced by the human papillomavirus replication machinery. EMBO J. 26:2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanoh M., et al. 1999. Significance of myocytes with positive DNA in situ nick end-labeling (TUNEL) in hearts with dilated cardiomyopathy: not apoptosis but DNA repair. Circulation 99:2757–2764 [DOI] [PubMed] [Google Scholar]
- 28. King L. E., et al. 2010. Human papillomavirus E1 and E2 mediated DNA replication is not arrested by DNA damage signalling. Virology 406:95–102 [DOI] [PubMed] [Google Scholar]
- 29. Kudoh A., et al. 2006. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4-MCM6-MCM7 complex during Epstein-Barr virus productive replication. J. Virol. 80:10064–10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kudoh A., et al. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156–8163 [DOI] [PubMed] [Google Scholar]
- 31. Kudoh A., et al. 2009. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J. Virol. 83:6641–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kurg R., Sild K., Ilves A., Sepp M., Ustav M. 2005. Association of bovine papillomavirus E2 protein with nuclear structures in vivo. J. Virol. 79:10528–10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lai M., Zimmerman E. S., Planelles V., Chen J. 2005. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J. Virol. 79:15443–15451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lydeard J. R., et al. 2010. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 24:1133–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mannik A., Runkorg K., Jaanson N., Ustav M., Ustav E. 2002. Induction of the bovine papillomavirus origin “onion skin”-type DNA replication at high E1 protein concentrations in vivo. J. Virol. 76:5835–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McBride A. A. 2008. Replication and partitioning of papillomavirus genomes. Adv. Virus Res. 72:155–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McPhillips M. G., Oliveira J. G., Spindler J. E., Mitra R., McBride A. A. 2006. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 80:9530–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McPhillips M. G., Ozato K., McBride A. A. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 79:8920–8932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moody C. A., Laimins L. A. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 5:e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oliveira J. G., Colf L. A., McBride A. A. 2006. Variations in the association of papillomavirus E2 proteins with mitotic chromosomes. Proc. Natl. Acad. Sci. U. S. A. 103:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Penrose K. J., Garcia-Alai M., Prat-Gay G., McBride A. A. 2004. CK2 phosphorylation-induced conformational switch triggers degradation of the papillomavirus E2 protein. J. Biol. Chem. 279:22430–22439 [DOI] [PubMed] [Google Scholar]
- 42. Romanczuk H., Howley P. M. 1992. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc. Natl. Acad. Sci. U. S. A. 89:3159–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sedman T., Sedman J., Stenlund A. 1997. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J. Virol. 71:2887–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Soeda E., Ferran M. C., Baker C. C., McBride A. A. 2006. Repression of HPV16 early region transcription by the E2 protein. Virology 351:29–41 [DOI] [PubMed] [Google Scholar]
- 45. Spardy N., et al. 2009. Human papillomavirus 16 E7 oncoprotein attenuates DNA damage checkpoint control by increasing the proteolytic turnover of claspin. Cancer Res. 69:7022–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spardy N., et al. 2007. The human papillomavirus type 16 E7 oncoprotein activates the Fanconi anemia (FA) pathway and causes accelerated chromosomal instability in FA cells. J. Virol. 81:13265–13270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Swindle C. S., et al. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Taatjes D. J., Sobel B. E., Budd R. C. 2008. Morphological and cytochemical determination of cell death by apoptosis. Histochem. Cell Biol. 129:33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tarakanova V. L., et al. 2007. Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host. Microbe 1:275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vafa O., et al. 2002. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol. Cell 9:1031–1044 [DOI] [PubMed] [Google Scholar]
- 51. Weitzman M. D., Lilley C. E., Chaurushiya M. S. 2010. Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64:61–81 [DOI] [PubMed] [Google Scholar]