Abstract
Replication of the papillomavirus genome is initiated by the assembly of a complex between the viral E1 and E2 proteins at the origin. The E1 helicase is comprised of a C-terminal ATPase/helicase domain, a central domain that binds to the origin, and an N-terminal regulatory region that contains nuclear import and export signals mediating its nucleocytoplasmic shuttling. We previously reported that nuclear accumulation of E1 has a deleterious effect on cellular proliferation which can be prevented by its nuclear export. Here we have shown that nuclear accumulation of E1 from different papillomavirus types blocks cell cycle progression in early S phase and triggers the activation of a DNA damage response (DDR) and of the ATM pathway in a manner that requires both the origin-binding and ATPase activities of E1. Complex formation with E2 reduces the ability of E1 to induce a DDR but does not prevent cell cycle arrest. Transient viral DNA replication still occurs in S-phase-arrested cells but surprisingly is neither affected by nor dependent on induction of a DDR and of the ATM kinase. Finally, we provide evidence that a DDR is also induced in human papillomavirus type 31 (HPV31)-immortalized keratinocytes expressing a mutant E1 protein defective for nuclear export. We propose that nuclear export of E1 prevents cell cycle arrest and the induction of a DDR during the episomal maintenance phase of the viral life cycle and that complex formation with E2 further safeguards undifferentiated cells from undergoing a DDR when E1 is in the nucleus.
INTRODUCTION
Human papillomaviruses (HPVs) are small double-stranded DNA viruses that infect the differentiating epithelium of the skin or mucosa (reviewed in references 10 and 91). About 25 types infect the anogenital tract (6, 19), characterized either as low-risk or high-risk types according to their association with benign or malignant hyperproliferative lesions. Clinically, low-risk HPV types cause benign warts while high-risk types are associated with lesions that can progress to cancer (28, 56, 63, 86).
The HPV life cycle is dependent on the differentiation program that keratinocytes undergo within a stratified epithelium. Viral DNA replication is required during the three distinct phases of the viral life cycle (reviewed in references 30 and 36). Upon infection of cells from the basal layer of the epithelium, the viral genome is established as a nuclear episome and is replicated by up to 50 to 100 copies (reviewed in reference 30). These episomes are then maintained at a constant copy number by low levels of replication in the lower layers of the epithelium. During this maintenance phase, viral DNA replication is thought to occur only once per cell cycle, during S phase, and in synchrony with replication of the host DNA (32). Finally, as the infected keratinocytes reach the uppermost differentiated layers of the epithelium, the copy number of the viral episome is amplified to very high levels (reviewed in reference 30), presumably through multiple rounds of replication in S-phase-arrested cells (23, 32). These episomes drive the expression of the late capsid proteins (27) and become encapsidated to form new virions, which are eventually shed with the top layer of the epithelium.
Two viral proteins, E1 and E2, are required for replication and amplification of the viral episome (reviewed in reference 74). Initiation of viral DNA replication relies on the capacity of E2 to bind specific sequences within the viral origin of replication and to simultaneously interact with the viral helicase E1 (1, 3). Through these interactions, E2 recruits several monomers of E1 to the viral origin (25, 26, 45, 50, 61, 82) and facilitates their assembly into a functional double hexamer that will unwind DNA ahead of the replication fork (64, 65, 77, 80, 81, 85) and serve as a platform for the assembly of host DNA replication factors, such as RPA, polymerase α-primase, and topoisomerase I (12, 13, 29, 44, 48, 58).
The HPV E1 helicase can be divided into three main functional regions. The C-terminal half of E1 comprises an ATPase/helicase domain typical of superfamily III (31, 33), which can self-assemble into hexamers and interact with E2, polymerase α-primase, and topoisomerase I (12, 48, 75, 81, 82, 84). The origin-binding domain (OBD) is located in the center of E1 and is required for dimerization and binding to the viral origin (80). Together, the OBD and C-terminal domain are sufficient to catalyze viral DNA replication in vitro but not in vivo (2, 76; this study), thereby suggesting that the N-terminal region of E1 has a regulatory role during viral DNA replication in vivo. Many elements within the N-terminal region are required for efficient viral DNA replication in vivo (14, 52, 54). Among these, a bipartite nuclear localization signal (NLS), a Crm1-dependent nuclear export signal (NES), a cyclin E/A-binding motif (CBM), and specific Cdk2 phosphorylation sites (S92 and S106 in HPV type 31 (HPV31) and S89, S93, and S107 in HPV11) have been shown to regulate the nucleocytoplasmic shuttling of E1 (18, 23, 46, 88). Recently, we showed that the shuttling of E1 needs to be tightly regulated since overaccumulation of this helicase in the nucleus inhibits cellular proliferation by causing cell cycle arrest in S phase (23). As part of this study, we also demonstrated that the nuclear export of E1 is required for the maintenance of the viral genome in undifferentiated keratinocytes but not for its amplification in differentiated cells. These findings led us to suggest that the Crm1-dependent export of E1 serves to limit its nuclear accumulation during the maintenance phase of the genome in order to prevent cell cycle arrest. Conversely, blocking the nuclear export of E1 in differentiated cells would create a favorable environment for viral DNA amplification by increasing the levels of E1 in the nucleus and blocking cells in S phase.
Cell cycle arrest is often the result of a cellular DNA damage response (DDR) and activation of cell cycle checkpoints (reviewed in references 4 and 5). Interestingly, one of our groups has recently reported that a DDR is activated in keratinocytes immortalized with HPV and is required for the amplification of the viral episome (53). Furthermore, the induction of a DDR caused by the E1- and E2-dependent replication of integrated HPV genomes has been shown to promote genomic instability (35–37). Activation of a DDR can originate from two types of DNA breakage: double-stranded and single-stranded breaks (DSBs and SSBs, respectively). DSBs induce the activation of the ataxia-telangiectasia mutated (ATM)-dependent pathway, which is characterized by the phosphorylation of ATM and its downstream effector kinase, Chk2. SSBs, however, lead to the activation of the ATM and Rad3-related (ATR) pathway, which results in the activation of the ATR kinase and phosphorylation of its cognate downstream effector kinase, Chk1. Upon their activation, both pathways trigger the activation of checkpoints that block cell cycle progression and facilitate DNA repair (reviewed in references 4 and 5).
In this study, we have demonstrated that the ability of E1 to block cell cycle progression and inhibit cellular proliferation is conserved among papillomaviruses and is dependent on the activities of the E1 OBD and ATPase/helicase domain. We have determined that E1-expressing cells are arrested in early S phase and interestingly that this cell cycle block is accompanied by the activation of a DDR and specifically of the ATM pathway. We have shown that high concentrations of E2 can prevent the induction of a DDR by E1 but cannot relieve its inhibitory effect on cell cycle progression. We also have demonstrated that transient viral DNA replication neither requires nor is affected by this DDR. Finally, we have provided evidence, using keratinocytes which express a nuclear export-defective E1 from the viral episome, that overaccumulation of E1 in the nucleus is sufficient to induce a DDR. We suggest that the ability of E1 to cause cell cycle arrest and to induce a DDR is prevented by nuclear export during the episomal maintenance phase of the viral life cycle but not in differentiated cells, where a DDR has been shown to facilitate amplification of the viral genome (53).
MATERIALS AND METHODS
Plasmid construction.
The plasmid used to express HPV31 E2 fused to a 3-Flag (3F) epitope and the plasmids used in the HPV31 transient DNA replication assay have been described previously (24). The red fluorescent protein (RFP)-PCNA expression plasmid was a kind gift of M. Cristina Cardoso (72). Plasmids expressing HPV31 E1 fused to enhanced yellow fluorescent protein (EYFP), as well as E1 from HPV16, HPV11, and bovine papillomavirus type 1 (BPV-1) fused to green fluorescent protein (GFP), were previously described (14, 22). The plasmid expressing HPV31 E1 fused to GFP was constructed by inserting a PCR fragment that contains a modified E1 open reading frame (ORF), bearing a silent mutation that inactivates an internal splicing donor site, between the XhoI and BamHI sites of plasmid pGFP2-C2 (BioSignal Packard-Perkin-Elmer). Plasmids expressing amino acids 1 to 159 and 1 to 332 of HPV31 E1 fused to EYFP were constructed by introducing the appropriate E1 PCR fragment between the XhoI and BamHI sites of plasmid pEYFP-C1 (Clontech). Plasmids expressing E1 fragments that lack the E1 NLS (amino acids 160 to 332, 160 to 629, and 332 to 629) as fusions to EYFP and to the NLS of SV40 large T antigen (LTA) were constructed similarly, with the exception that the PCR fragments were introduced between the BamHI and SalI sites of plasmid AB-2531, which expresses EYFP fused at its C terminus to the SV40 LTA NLS (PKKKRKV). Plasmid AB-2531 was constructed by inserting a PCR fragment encoding the SV40 LTA NLS between the BglII and EcoRI sites of plasmid pEYFP-C1 (Clontech). Amino acid substitutions in the origin binding (K265A/R267A) and ATPase (K463A) domains of HPV31 E1 were introduced by mutagenesis of the EYFP-E1 expression plasmid using the QuikChange kit (Stratagene). All DNA constructs were verified by sequencing. Further details on their construction will be provided upon request.
Cell culture, transfection, and whole-cell extract preparation.
The human cervical carcinoma cell lines C33A and HeLa, ATM-deficient line (primary skin fibroblasts, HDSF, AG04405A; Coriell Institute), as well as the osteosarcoma U2OS line, were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/ml of penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine. Human foreskin keratinocytes (HFKs) that have been immortalized with either the HPV31 wild type (WT) or NES mutant genomes were described previously (23). These cells lines were maintained in KGM (Clonetics) or in E medium in the presence of mitomycin C (Boehringer Mannheim)-treated fibroblast feeders. Transfections of C33A cells were performed using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Whole-cell extracts were prepared 24 h posttransfection by resuspension of the transfected C33A cells in lysis buffer (phosphate-buffered saline [PBS] supplemented with 0.1% Triton X-100, protease and phosphatase inhibitors [10 μg of antipain, 2 μg of leupeptin, 1 μg of pepstatin, and 2 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, and 1 mM orthovanadate {Na3VO4}]) and freezing at −80°C overnight. Cell extracts were thawed and boiled in Laemmli buffer (42) prior to SDS-PAGE.
Confocal fluorescence microscopy.
C33A cells (∼8 × 105) were transfected with the amount of EYFP-31E1 expression plasmid needed and grown on coverslips. When indicated, 3F-31E2 WT and a plasmid containing the viral origin of replication were cotransfected with EYFP-31E1. Twenty-four hours posttransfection, cells were fixed with 4% formaldehyde. For the detection of γH2AX, pATM, pChk2, and pChk1, cells were treated as described in the next section. Otherwise, cells were directly permeabilized with 0.2% Triton X-100, and their DNA was stained with a solution of TO-PRO-3 (1 μM) (Molecular Probes). Cells were mounted using Vectashield mounting medium (Vector Laboratories). Images were acquired using a LSM710 confocal laser coupled to an Axiovert 100 M inverted scanning microscope (Zeiss) and analyzed using the ZEN 2009 LE software program.
Immunofluorescence.
Twenty-four hours posttransfection, cells were permeabilized with 0.5% Triton X-100 for 10 min at 4°C, treated with 50 mM NH4Cl in PBS for 10 min at room temperature, and then blocked with PBS containing 10% bovine serum albumin (BSA) for 3 to 4 h at room temperature. Cells were then incubated with a dilution of the primary antibody (1:500 for detection of γH2AX, pATM, and pChk2; 1:200 for pChk1) overnight at 4°C. After two washes in PBS-BSA, cells were incubated with the secondary antibody (anti-mouse Alexa 633 or anti-rabbit Alexa 633) for 30 min at room temperature. Where indicated, DNA was stained with a solution of 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml) (catalog no. D1306; Invitrogen). Cells were mounted using Vectashield mounting medium (Vector Laboratories). Images were acquired as described above.
Antibodies. (i) Immunofluorescence.
Phospho-H2AX Ser 139 (γH2AX) and phospho-ATM Ser1981 (pATM) were detected using mouse monoclonal antibody from Millipore (clone JBW301, catalog no. 05-636; Upstate) and Rockland (catalog no. 200-301-400), respectively, and then with an Alexa Fluor 633-conjugated goat anti-mouse secondary antibody from Molecular Probes (catalog no. A21050). Similarly, phospho-Chk2 Thr 68 (pChk2) and phospho-Chk1 Ser 345 (pChk1) were detected using rabbit polyclonal antibodies from Cell Signaling (catalog no. 2661 and 2348, respectively) and then with an Alexa Fluor 633-conjugated goat anti-rabbit secondary antibody from Molecular Probes (catalog no. A21070).
(ii) Western blotting.
GFP and EYFP fusion proteins were detected using a mixture of two mouse monoclonal antibodies purchased from Roche (catalog no. 11814460001), while β-tubulin was detected using a mouse monoclonal antibody from Sigma-Aldrich (catalog no. T0426). For Western blot analysis, proteins were transferred onto polyvinylidene difluoride membranes and detected using a horseradish peroxidase-conjugated sheep anti-mouse secondary antibody from GE Healthcare (catalog no. NA931) and an enhanced chemiluminescence detection kit (GE Healthcare).
Transient HPV DNA replication assay.
Transient HPV31 DNA replication was performed as described previously (24) but using EYFP-31E1 instead of Flag-tagged E1 (3F-31E1). Briefly, ∼5 × 104 C33A cells were transfected with four plasmids encoding, respectively, EYFP-31E1, 3F-31E2, the minimal origin of DNA replication together with a firefly luciferase (Fluc) reporter gene (pFLORI31), and Renilla luciferase (Rluc) (pRL). Under standard conditions, 1.25, 2.5, and 5 ng of E1 were used along with a constant quantity of the other plasmids (1×) (10 ng of 3F-31E2, 2.5 ng of pFLORI31, and 0.5 ng of pRL). When indicated, 2×, 4×, and 8× of 3F-31E2 (20, 40, and 80 ng, respectively) and pFLORI31 (5, 10, and 20 ng, respectively) were used. For all conditions, replication of the origin-containing plasmid was quantified 24 or 72 h posttransfection by measuring the levels of firefly and Renilla luciferase activities using the Dual-Glo luciferase assay system (Promega). E1 mutants were analyzed in duplicate in two separate experiments.
Cell cycle analysis.
When several conditions were analyzed for a given transfected population (asynchronous, mimosine block and release in nocodazole), cells were prepared as previously described (23). When a single condition was analyzed (asynchronous), ∼8 × 105 C33A cells were transfected with 1 μg of the indicated plasmid(s) in 6-well plates and directly prepared for cell cycle analysis 24 or 48 h later. Briefly, cells were trypsinized and either fixed and stained with a solution containing 50 μg/ml of propidium iodide (PI) (catalog no. P4170; Sigma-Aldrich) or stained directly using fresh medium supplemented with 6.3 μg/ml of Hoechst (catalog no. B2261; Sigma-Aldrich) and 5 mM verapamil (catalog no. V4629; Sigma-Aldrich) for 30 min at 37°C. The DNA content of EYFP-expressing cells was determined by flow cytometry on a FACScan (PI staining) or FACS BD LSR (Hoechst staining) flow cytometer using the Cellquest Pro software program (BD). Cell cycle distribution was further analyzed and quantified using the FlowJo (v8.1) and ModFit LT software programs, respectively. Specifically, the synchronization wizard within this software was used to construct a model for the cell cycle distribution of C33A cells expressing EYFP alone (i.e., control cells). In this model, the G1 and G2/M peaks are fixed manually and locked for further analysis. Next, the algorithm within Modfit was used to model the S phase (i.e., the region between the G1 and G2/M peaks) to a rectangle model and to separate it in 3 distinct compartments with equal spacing. These compartments were fixed and used to determine the percentages of cells in early, mid-, and late S phase. Once fixed, the same model was used to determine the percentages of cells in G1, early S, mid-S, late S, and G2/M phase for cells expressing EYFP-E1. High-EYFP-expressing cells were excluded from the analysis of cells released in nocodazole since control cells expressing high levels of EYFP and GFP alone were unable to reenter the cell cycle (data not shown).
Quantification of γH2AX and incorporation of bromodeoxyuridine (BrdU). (i) γH2AX analysis.
C33A cells (∼8 × 105) were transfected with 1 to 2 μg of the indicated plasmid(s) in 6-well plates. Twenty-four hours later, cells were trypsinized and fixed in 70% ethanol overnight at −20°C. Fixed cells were permeabilized in PST (PBS supplemented with 4% FBS and 0.1% Triton X-100) for 10 min on ice. Cells were subsequently incubated with the primary antibody (γH2AX) diluted to 1:500 in PST for 2 h at room temperature. Cells were then washed twice with PBS-2% FBS solution and incubated with a 1:400 dilution of the secondary antibody (mouse Alexa Fluor 633) for 1 h at room temperature. Finally, cells were washed and resuspended in PBS-2% FBS solution for fluorescence-activated cell sorter (FACS) analysis. Since the induction of γH2AX is correlated with the levels of expression of E1, the EYFP signal was analyzed separately for low-, medium-, and high-EYFP-expressing cells. Fluorescence intensities ranging from 5 to 20 were considered low, those from 20 to 115 were considered medium, and those from 115 to 1,000 were considered to indicate high-EYFP-expressing cells.
(ii) Incorporation of BrdU.
BrdU incorporation was measured using the allophycocyanin (APC) BrdU flow kit from BD Pharmingen (catalog no. 522598) according to the manufacturer's recommendations. Briefly, 24 h posttransfection, cells were pulsed with 10 μM BrdU for 1 h and then fixed, permeabilized, and treated with DNase to enable access of the APC antibody to the BrdU incorporated in the DNA. Next, cells were stained with anti-BrdU-APC and 7-amino-actinomycin D (7-AAD) and analyzed by FACS.
(iii) Flow cytometry analysis.
The γH2AX, BrdU, and 7-AAD contents of EYFP-expressing cells were determined by flow cytometry on a FACS BD LSR flow cytometer using the Cellquest Pro software program (BD). γH2AX and BrdU were further analyzed and quantified using the FlowJo (v8.1) software program, while cell cycle distribution was analyzed by FlowJo and quantified using ModFit LT software. When indicated, hydroxyurea (HU) (2 mM) or aphidicolin (Aph) (5 μg/ml) was added to the transfected cells 4 h posttransfection.
Colony formation assay.
C33A cells (∼8 × 105) were transfected with 1 μg of the indicated plasmid in a 6-well plate. Twenty-four hours posttransfection, cells were trypsinized and seeded on 100-mm plates in fresh medium supplemented with 500 μg/ml of G418 or 15 μg/ml of bleomycin (Bleocin; EMD Millipore catalog no. 203408). Antibiotic selection was maintained for a period of approximately 3 weeks. Cells were then fixed for 10 min in cold methanol and stained for 2 min with methylene blue (1% [wt/vol] in 60% MeOH-H2O).
RESULTS
The ability to arrest S-phase progression is a conserved feature of papillomavirus E1.
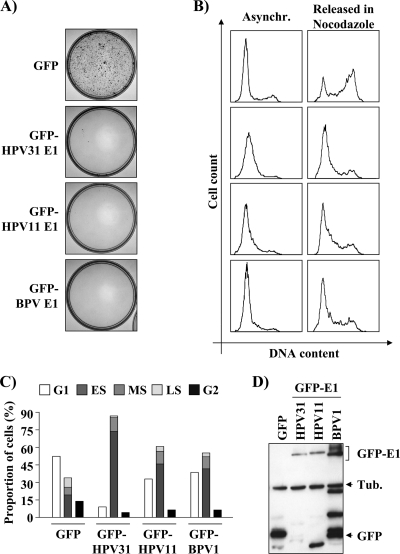
We have recently reported that nuclear accumulation of HPV31 E1 inhibits the proliferation of transfected C33A cells by arresting them in S phase (23). To determine if this activity is conserved among papillomavirus E1 proteins, we tested the ability of two other E1 proteins, from HPV11 and BPV, to inhibit proliferation of C33A cells in a colony formation assay (CFA). These experiments, which were performed with GFP-E1 fusion proteins expressed from a vector containing a Zeocin resistance gene, revealed that the HPV11 and BPV E1 proteins were as good as HPV31 E1 at inhibiting colony formation (Fig. 1A). To determine if this inhibition was due to an arrest in S phase, we analyzed the cell cycle distribution of C33A cells transiently expressing these GFP-E1 proteins (i.e., of GFP-positive cells). As anticipated, we found that HPV11 and BPV E1 also led to an accumulation of cells in S phase (Fig. 1B and C and Table 1), similarly to HPV31 E1 and unlike GFP alone, which was used as a negative control. To further characterize this cell cycle arrest, we tested if E1-expressing cells were able to resume cell cycle progression following a block in G1/S by mimosine. In these experiments, cells were released from the mimosine block into medium containing nocodazole to prevent their progression past the G2/M boundary. Consistent with the results obtained with asynchronous cells, the majority of E1-expressing cells were unable to progress to G2/M, although 73% of the GFP-transfected cells were able to do so (Fig. 1B and Table 2). Western blotting showed that HPV31 and -11 E1 were expressed at comparable levels. As for BPV E1, it was expressed at slightly higher levels but also showed an increase in low-molecular-weight degradation products (Fig. 1D). Collectively, the results presented above suggest that the capacity of E1 to prevent cellular proliferation by blocking S-phase progression is a conserved activity of papillomavirus E1.
Fig. 1.
E1 proteins from different papillomavirus types inhibit cellular proliferation. (A) Colony formation assays (CFA). C33A cells were transfected with the indicated GFP-E1 expression vector (also containing a Zeocin resistance gene). Following 4-weeks' selection in bleomycin, colonies were stained with methylene blue. (B) Cell cycle analysis. C33A cells transiently expressing the indicated GFP-E1 or GFP were trypsinized 48 h posttransfection, and their DNA was stained with Hoechst stain and analyzed by flow cytometry. For each condition, the cells were either grown asynchronously (Asynchr.) or synchronized with mimosine for 24 h and then released in nocodazole for an additional 24 h (Released in Nocodazole). The cell cycle profile of each sample is shown as a histogram and quantified in Tables 1 and 2. (C) Graphical representation of the cell cycle distributions shown in panel B. ES, MS, and LS stand for early, mid-, and late S phase, respectively. (D) Western blot analysis of total protein extracts prepared from transfected C33A cells expressing the GFP-E1 proteins of the various papillomaviruses used in this study. E1 proteins were detected using an anti-GFP antibody, and tubulin (Tub.) was used as a loading control.
Table 1.
Cell cycle distribution of GFP- and EYFP-expressing C33A cells
| Transfected plasmid(s)a | % cells |
|||||
|---|---|---|---|---|---|---|
| G0/G1 | S | G2/M | Intra-S partitioningb |
|||
| E | M | L | ||||
| Mock | 32 | 53 | 15 | 47 | 26 | 26 |
| GFP | 52 | 34 | 14 | 56 | 21 | 24 |
| GFP-HPV31 E1 | 9 | 87 | 4 | 85 | 14 | 1 |
| GFP-HPV11 E1 | 33 | 61 | 7 | 75 | 18 | 7 |
| GFP-BPV1 E1 | 39 | 55 | 6 | 76 | 18 | 5 |
| GFP-HPV16 E1 WT | 16 | 81 | 3 | 78 | 17 | 5 |
| GFP-HPV16 E1 ATPase | 43 | 44 | 14 | 61 | 23 | 16 |
| EYFP | 43 | 43 | 14 | 47 | 30 | 23 |
| EYFP-31E1 1-629 | 24 | 72 | 4 | 75 | 19 | 6 |
| EYFP-31E1 1-159 | 49 | 40 | 11 | 48 | 25 | 28 |
| EYFP-31E1 1-332 | 49 | 40 | 11 | 50 | 30 | 20 |
| EYFP-NLS | 46 | 39 | 15 | 44 | 28 | 28 |
| EYFP-NLS-31E1 160-332 | 45 | 43 | 12 | 47 | 28 | 26 |
| EYFP-NLS-31E1 160-629 | 37 | 59 | 4 | 66 | 22 | 12 |
| EYFP-NLS-31E1 332-629 | 45 | 44 | 11 | 45 | 27 | 27 |
| EYFP | 38 | 48 | 14 | 54 | 25 | 21 |
| EYFP-31E1 WT | 20 | 74 | 6 | 77 | 16 | 7 |
| EYFP-31E1 OBD | 38 | 49 | 13 | 53 | 24 | 22 |
| EYFP-31E1 ATPase | 37 | 55 | 9 | 64 | 22 | 15 |
| EYFP-31E1 NES | 18 | 75 | 7 | 88 | 7 | 5 |
| EYFP-31E1 NES/OBD | 44 | 44 | 12 | 55 | 25 | 20 |
| EYFP-31E1 NES/ATPase | 40 | 44 | 16 | 61 | 25 | 14 |
Five thousand cells positively stained with Hoechst stain or doubly stained with Hoechst stain and EYFP/GFP were selected for mock- and EYFP/GFP-transfected cells, respectively.
The percentage of cells estimated to be in the early (E), mid (M)-, or late (L) S phase according to the Modfit software program.
Table 2.
Cell cycle distribution of GFP- and EYFP-expressing C33A cells released from mimosine blocks
| Transfected plasmid(s)a | % cells: |
|
|---|---|---|
| In G1/early S phase | Out of G1/early S phaseb | |
| Mock | 20 | 80 |
| GFP | 27 | 73 |
| GFP-HPV31 E1 | 74 | 26 |
| GFP-HPV11 E1 | 67 | 33 |
| GFP-BPV1 E1 | 67 | 33 |
| GFP-HPV16 E1 WT | 70 | 30 |
| GFP-HPV16 E1 ATPase | 39 | 61 |
| EYFP | 23 | 77 |
| EYFP-31E1 WT | 58 | 42 |
| EYFP-31E1 OBD | 34 | 66 |
| EYFP-31E1 ATPase | 35 | 65 |
| EYFP-31E1 NES | 64 | 36 |
| EYFP-31E1 NES/OBD | 35 | 65 |
| EYFP-31E1 NES/ATPase | 41 | 59 |
Five thousand cells positively stained with Hoechst or doubly stained with Hoechst and EYFP/GFP were selected for mock- and EYFP/GFP-transfected cells, respectively.
The percentage of cells that resumed cell cycle progression following their release from a G1/S block with mimosine was quantified by counting the numbers of cells in mid- and late S phase and in G2/M phase.
The origin-binding and helicase domains of E1 are necessary and sufficient to inhibit cellular proliferation.
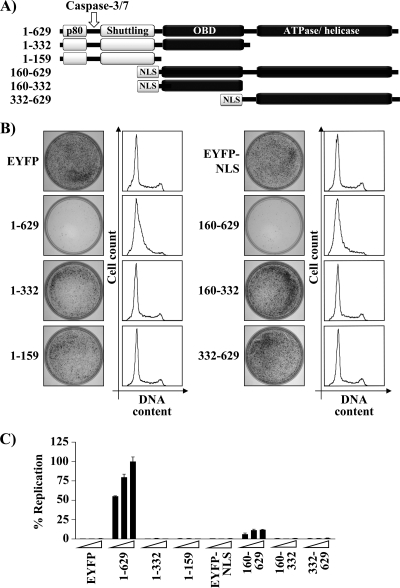
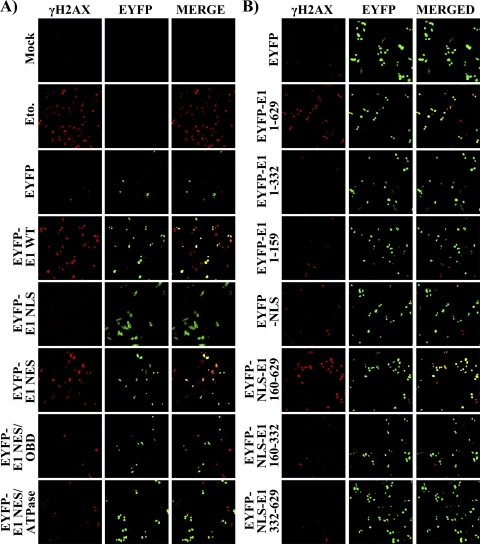
To determine which domains of E1 are required for inhibiting cellular proliferation, we performed colony formation assays with a series of HPV31 E1 truncations spanning the N-terminal region, the OBD, and/or the C-terminal enzymatic domain (Fig. 2A). All truncations were fused to EYFP, and those lacking the N-terminal region of E1 were fused to the SV40 large T antigen NLS. The addition of the NLS was necessary since we previously showed that the antiproliferative effect of E1 is dependent on its nuclear accumulation (23). The results in Fig. 2B show that the two N-terminal fragments of E1 lacking the ATPase/helicase domain (E1 amino acids [aa] 1 to 159 and 1 to 332) were unable to inhibit cellular proliferation. Accordingly, neither fragment had an effect on cell cycle progression (Fig. 2B and Table 1). In contrast, the E1 fragment comprised of the OBD and ATPase/helicase domain (E1 aa 160 to 629) was capable of preventing cellular proliferation and cell cycle progression to an extent similar to that of wild-type (i.e., full-length) E1 (Fig. 2B and Table 1). Neither the OBD nor the ATPase/helicase domain on its own was capable of inhibiting proliferation or cell cycle progression. All truncated proteins were properly localized to the nucleus and expressed at the correct molecular weight, as determined by confocal fluorescence microscopy and Western blotting (data not shown). Interestingly, whereas the E1 fragment spanning the OBD and ATPase/helicase domain (E1 aa 160 to 629) could inhibit cellular proliferation, it could support only very low levels of transient DNA replication (Fig. 2C). From these results, we conclude that the OBD and ATPase/helicase domain of E1 are necessary and sufficient to inhibit cellular proliferation and cell cycle progression and consequently that none of the previously described interactions of the N-terminal domain of E1 with cellular proteins such as p80 (14) and cyclin E/A-Cdk2 (23, 46) are required for this antiproliferative effect (Fig. 2A). Finally, these results show that this effect is also independent of the capacity of E1 to efficiently replicate DNA.
Fig. 2.
The OBD and ATPase/helicase domain of E1 are both required for its antiproliferative activity. (A) Schematic representation of HPV31 E1 truncations used in this analysis, highlighting the location of the p80-binding site (aa 10 to 40), the caspase-3/7 cleavage site, the shuttling module (aa 85 to 125), the origin-binding domain (OBD), and the ATPase/helicase domain. E1 fragments lacking the shuttling module were fused to the SV40 large T antigen nuclear localization signal (NLS). (B) Colony formation assay and cell cycle analysis. CFA and cell cycle analysis were done essentially as described in the legend for Fig. 1 with the exception that cells were selected in G418-containing medium for 3 weeks before staining the colonies with methylene blue. (C) Transient DNA replication activities of E1 truncations. The DNA replication activity of each E1 fragment was determined using 1.25, 2.5, and 5 ng of expression vector 72 h posttransfection. Replication activities are reported as a percentage of the Fluc/Rluc ratio obtained with the 5 ng of the 1–629 E1 expression vector. Error bars represent standard deviations. Cells transfected with EYFP or EYFP-NLS only were used as negative controls.
The origin-binding and ATPase activities of E1 are required for arresting S-phase progression.
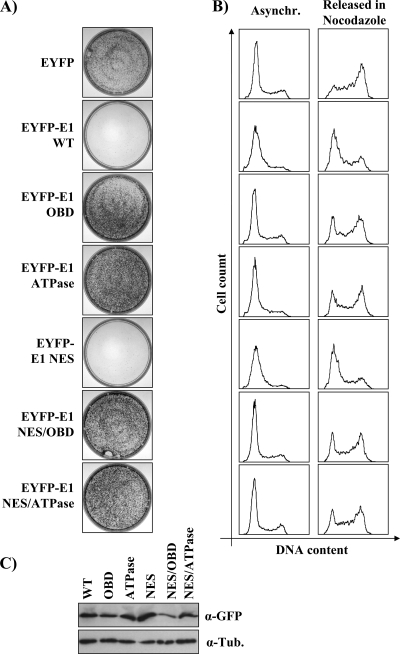
To substantiate the results presented above, we performed colony formation assays with EYFP-31E1 mutant proteins carrying amino acid substitutions that abrogate the origin-binding (OBD mutant; K265A/R267A) (80, 81) or ATPase (ATPase mutant; K463A) activity (81, 84) of E1. The results presented in Fig. 3A show that both E1 mutants failed to inhibit cellular proliferation. However, we observed by fluorescence microcopy that the E1 OBD mutant was not entirely localized to the nucleus, in contrast to the wild type and the ATPase E1 mutant, since it was present in both the nucleus and cytoplasm in approximately 15% of the transfected cells (data not shown). To prevent cytoplasmic accumulation of the E1 OBD mutant, we generated a version lacking the nuclear export signal (NES) and confirmed that it was entirely nuclear (data not shown). For comparison, we also used NES-defective versions of the ATPase mutant and of wild-type E1. When tested in CFAs, both the OBD and ATPase mutant lacking the NES failed to inhibit colony formation, thus confirming that the origin-binding and ATPase activities of E1 are required for preventing cellular proliferation (Fig. 3A). When analyzed by Western blotting, the NES/OBD mutant was reproducibly found to be expressed at lower levels than the wild-type protein (Fig. 3C). To ensure that the inability of this mutant to prevent cellular proliferation was not due to its lower expression, we analyzed the cell cycle distribution of EYFP-E1-expressing cells by flow cytometry, since this technique allows the comparison of cells expressing similar levels of EYFP fluorescence. Figure 3B shows that the E1 OBD and ATPase mutants were unable to prevent S-phase progression, as determined by the ability of cells expressing these proteins to progress to G2/M upon release from a mimosine block (Fig. 3B and Table 1 and 2). All together, these results indicate that the origin-binding and enzymatic activities of E1 are both required for arresting cells in S phase.
Fig. 3.
Amino acid substitutions in the origin-binding and ATPase domains of E1 abolish its antiproliferative activity. (A and B) CFAs (A) or cell cycle analysis (B) of cells transfected with EYFP alone or the indicated EYFP-E1 constructs carrying amino acid substitutions in the OBD, ATPase domain, and NES, as described in the text. For CFAs, cells were selected in G418-containing medium before staining. Cell cycle analyses were performed as described for Fig. 1. (C) Western blot analysis in which E1 proteins were detected using an anti-GFP antibody (α-GFP) and tubulin was used as a loading control (α-Tub.).
To test the generality of these findings, we repeated these experiments with HPV16 E1 fused to GFP. As observed for BPV, HPV11, and HPV31 E1, we found that HPV16 E1 was able to inhibit cellular proliferation by imposing an S-phase arrest (Tables 1 and 2 and data not shown). Furthermore, we determined that these antiproliferative effects of E1 could be abrogated by the mutation of its ATP-binding site (Table 1 and 2 and data not shown) (87). Thus, the enzymatic activity of HPV16 E1 is also required for its growth-inhibitory effects, similar to what was observed for HPV31 E1.
E1 inhibits the proliferation of different cell lines.
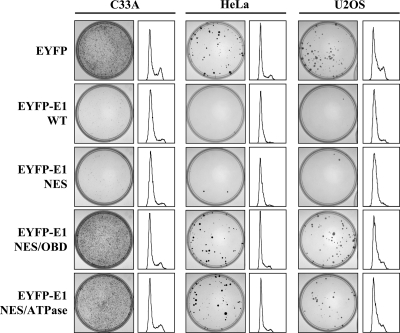
Next, we investigated if E1 could also prevent the proliferation of cell lines other than C33A. We tested its effect in HeLa cells, which express HPV18 E6 and E7, and in U2OS cells, which in contrast to C33A cells express wild-type p53. As shown in Fig. 4, EYFP-31 E1 was able to inhibit the proliferation of HeLa and U2OS cells to an extent similar to that with C33A cells. Importantly, proliferation of all three cell lines was not affected by the E1 NES/OBD and NES/ATPase mutants, thus suggesting that the mechanism by which E1 affects cellular proliferation is the same in each cell line. We then characterized the antiproliferative effect of E1 by flow cytometry and determined that S-phase progression was blocked in all three cell lines expressing the wild type but not the mutant proteins (Fig. 4 and Table 3). Note that in these experiments, DNA content was measured by propidium iodide (PI) staining since Hoechst staining was found to be deleterious to HeLa and U2OS cells (data not shown). Consistent with the ability of E1 to induce a cell cycle arrest in HeLa cells, we found that coexpression of HPV31 E6 and/or E7 in C33A cells did not prevent the deleterious effect of EYFP-31E1 on cell cycle progression (data not shown). Together, the results presented above indicate that the ability of E1 to inhibit S-phase progression and cellular proliferation is independent of the expression of E6 and E7 and that of p53 (68).
Fig. 4.
E1 inhibits cellular proliferation in p53-positive and -negative cell lines. CFAs and cell cycle distributions of EYFP- and EYFP-E1-transfected C33A, HeLa, or U2OS cells. CFA were performed in G418-containing medium. Cell cycle distributions were determined 24 h posttransfection using cells stained with propidium iodide. Results are represented as histograms here and are quantified in Table 3.
Table 3.
Cell cycle distribution of EYFP-expressing C33A, HeLa, and U2OS cells
| Transfected plasmid(s)a | % cells |
|||||
|---|---|---|---|---|---|---|
| G0/G1 | S | G2/M | Intra-S partitioningb |
|||
| E | M | L | ||||
| Mock C33A | 47 | 31 | 22 | 48 | 32 | 19 |
| EYFP | 50 | 29 | 21 | 48 | 34 | 17 |
| EYFP-31E1 WT | 21 | 70 | 9 | 86 | 14 | 0 |
| EYFP-31E1 NES | 25 | 66 | 9 | 88 | 12 | 0 |
| EYFP-31E1 NES/OBD | 64 | 24 | 12 | 50 | 33 | 17 |
| EYFP-31E1 NES/ATPase | 32 | 51 | 17 | 84 | 12 | 4 |
| Mock HeLa | 47 | 43 | 10 | 63 | 21 | 16 |
| EYFP | 57 | 36 | 8 | 64 | 19 | 17 |
| EYFP-31E1 WT | 33 | 65 | 2 | 80 | 18 | 2 |
| EYFP-31E1 NES | 38 | 58 | 4 | 78 | 19 | 3 |
| EYFP-31E1 NES/OBD | 60 | 33 | 7 | 52 | 33 | 15 |
| EYFP-31E1 NES/ATPase | 58 | 37 | 5 | 65 | 27 | 8 |
| Mock U2OS | 37 | 39 | 24 | 51 | 49 | 0 |
| EYFP | 32 | 44 | 24 | 59 | 41 | 0 |
| EYFP-31E1 WT | 35 | 54 | 11 | 83 | 17 | 0 |
| EYFP-31E1 NES | 27 | 65 | 8 | 78 | 22 | 0 |
| EYFP-31E1 NES/OBD | 45 | 43 | 12 | 58 | 37 | 5 |
| EYFP-31E1 NES/ATPase | 42 | 46 | 12 | 59 | 37 | 4 |
Five thousand cells positively stained with propidium iodide (PI) or doubly stained with PI and EYFP were selected for mock- and EYFP-transfected cells, respectively.
The percentage of cells estimated to be in the early (E), mid (M), or late (L) S phase according to the Modfit software program.
E1 arrests cells in early S phase.
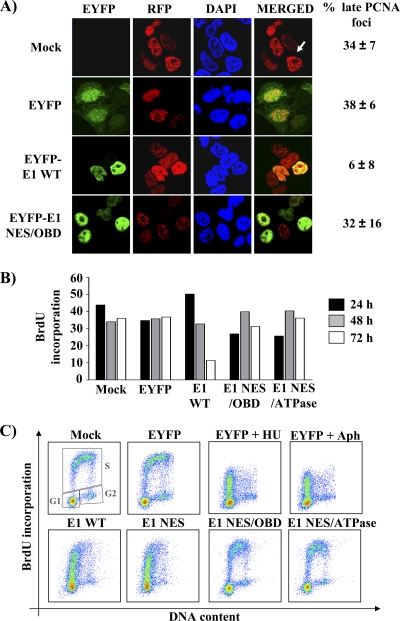
The flow cytometry results presented above suggested that E1-expressing cells accumulate in S phase. To confirm these results by an alternative method, we investigated the effect of E1 on the assembly of PCNA foci, whose size increases from early to late S phase (62). As can be seen in Fig. 5A, large PCNA foci were never detected in cells expressing wild-type EYFP-31E1 but could be readily observed in cells expressing the NES/OBD E1 mutant. These results suggest that cells expressing wild-type E1 fail to reach the end of S phase. To pinpoint the position in S phase in which E1-expressing cells were arrested, we set out to measure their capacity to carry out DNA synthesis (by BrdU incorporation) as a function of their position in the cell cycle (determined by 7-AAD staining). First, we determined by flow cytometry the percentage of E1-expressing cells that could incorporate BrdU at 24, 48, and 72 h posttransfection, following a 1-h pulse-labeling period. As anticipated, BrdU incorporation gradually decreased over time in cells expressing wild-type E1, consistent with the induction of an S-phase arrest, but not in those expressing the E1 NES/OBD and NES/ATPase mutants (Fig. 5B). On the basis of these findings, we decided to measure BrdU incorporation as a function of the cell cycle at 24 h posttransfection. This analysis revealed that cells expressing either a wild-type or NES mutant E1 protein could incorporate BrdU in early S phase only. This was in contrast to the control cells (mock or EYFP transfected) or cells expressing the E1 NES/OBD or NES/ATPase mutant (Fig. 5C). As additional controls, we confirmed that the BrdU incorporation profile of E1-expressing cells was similar to that of EYFP-transfected cells treated with the DNA synthesis inhibitors aphidicolin and hydroxyurea (Fig. 5C) (40, 57). Collectively, the results presented above indicate that E1 arrests cell cycle progression in early S phase.
Fig. 5.
E1 arrests cell cycle progression in early S phase. (A) Cellular localization of RFP-PCNA in transfected cells expressing either the wild type or an origin-binding-defective E1 protein. Twenty-four hours posttransfection, cells were fixed, mounted, and visualized by ×40 fluorescence confocal microscopy. Nuclei (DNA) were stained with DAPI. Cells containing large PCNA foci (such as the one indicated by the white arrow) were quantified and are reported on the right side of the panel as a percentage of the total number of RFP-PCNA-expressing cells. (B) BrdU incorporation. Twenty-four, forty-eight, and seventy-two hours posttransfection, cells expressing EYFP or EYFP-E1 wild-type or NES/OBD or NES/ATPase mutant proteins were pulsed for 1 h with 10 μM BrdU. The percentage of cells that incorporated BrdU was then determined by flow cytometry and is reported in the histogram. (C) BrdU incorporation throughout the cell cycle. Cells expressing EYFP or EYFP-E1 wild type or the indicated mutant proteins were pulsed with BrdU 24 h posttransfection and analyzed as described for panel B. In addition, cells were stained with 7-AAD to measure their DNA content. For each sample, BrdU incorporation is represented as a function of DNA content (cell cycle distribution) in a scatter plot. Boxes indicate the populations of cells in G1, S, and G2. EYFP-transfected cells treated with hydroxyurea (HU) and aphidicolin (Aph) were used as controls.
E1 induces a DNA damage response and activates the ATM pathway.
Since inhibition of cell cycle progression can be induced by a DNA damage checkpoint response, we investigated if E1 can trigger the phosphorylation of histone H2AX on serine 139 (γH2AX), a hallmark of a DNA damage response (DDR) (9, 49, 59, 60). We examined the presence of γH2AX in cells expressing EYFP-E1 by confocal immunofluorescence microscopy. As a positive control, we treated untransfected cells with a 50 μM concentration of the topoisomerase II inhibitor etoposide for 24 h in order to induce a DDR (70). As shown in Fig. 6A, etoposide treatment resulted in a robust induction of γH2AX compared to results for untreated cells (Mock). A similar induction of γH2AX was observed in cells expressing wild-type E1 but not in those expressing either EYFP alone, an E1 mutant that accumulates in the cytoplasm (NLS), or nuclear E1 mutants defective for origin binding or ATPase activity (Fig. 6A). Interestingly, we consistently observed that the E1 NES/ATPase mutant protein weakly activates γH2AX, perhaps because this mutant protein retains the ability to bind DNA through its intact ODB (see Discussion). The induction of γH2AX by HPV31 E1 was also observed in HeLa and U2OS cells, indicating that it is independent of E6, E7, and p53 (data not shown). Finally, evidence linking the induction of a DDR with cell cycle arrest came from the observation that among the different HPV31 E1 fragments characterized in Fig. 2, only those that caused cell cycle arrest were able to induce the phosphorylation of γH2AX (Fig. 6B).
Fig. 6.
Nuclear accumulation of enzymatically active E1 induces a DNA damage response. (A) The indicated EYFP and EYFP-E1 wild-type or mutant proteins were transiently expressed in C33A cells. Twenty-four hours posttransfection, cells were fixed and stained for phospho-serine 139 of γH2AX. Mounted cells were then visualized by ×20 confocal fluorescence microscopy. Mock-transfected cells and cells treated for 24 h with 50 μM etoposide (Eto.) were used as negative and positive controls, respectively. (B) Analysis similar to that for panel A but using the E1 fragments described in the legend for Fig. 2.
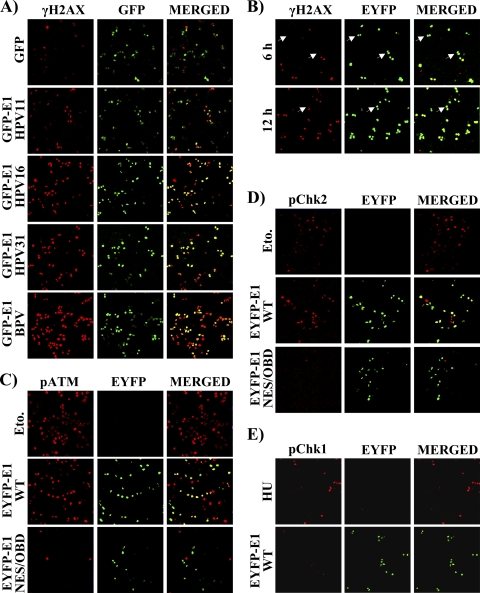
The ability of E1 to induce a DDR appears to be conserved among papillomaviruses, since γH2AX was also detected in cells expressing GFP-E1 from HPV11, HPV16, and BPV (Fig. 7A). In a time course experiment, we found that the expression of HPV31 EYFP-E1 and the induction of γH2AX could be detected as early as 6 and 12 h posttransfection and followed similar kinetics, consistent with γH2AX being triggered soon after expression of E1 (Fig. 7B). Since DNA damage is known to induce the activation of the ATM or ATR checkpoints, depending of the type of DNA lesions created (reviewed in references 38 and 90), we investigated which of these two DDR pathways was induced by E1. Specifically, we monitored the activation of key components of the ATM and ATR pathways by immunofluorescence microscopy. Activation of the ATM pathway was assessed by detection of phosphoserine 1981 of ATM (pATM) and, in separate experiments, phosphothreonine 68 of Chk2 (pChk2). Activation of the ATR pathway was monitored using an antibody against phosphoserine 345 of Chk1 (pChk1). As shown in Fig. 7C and D, the levels of both pATM and pChk2 were increased in cells expressing wild-type E1 to an extent similar to that in cells treated with etoposide, used as a positive control. As anticipated, cells expressing the NES/OBD or NES/ATPase E1 mutant or expressing EYFP alone showed significantly reduced levels of these markers (Fig. 7C and D and data not shown). In contrast, pChk1 was detected in only a few E1-expressing cells, unlike the case with cells treated with etoposide or hydroxyurea, which were used as positive controls (Fig. 7E and data not shown). These results suggest that E1 induces a DNA damage response and concomitant activation of the ATM pathway.
Fig. 7.
The ability of E1 to induce a DDR is a conserved feature of papillomavirus E1 and involves activation of the ATM pathway. (A) The ability of E1 to induce a DDR is conserved in HPV31, -16, and -11 and BPV E1. The activation of γH2AX was analyzed in C33A cells expressing the indicated GFP or GPF-E1 proteins. To analyze γH2AX activation, cells were fixed 24 h posttransfection, stained for phosphoserine 139 of γH2AX, mounted, and visualized by ×20 fluorescence confocal microscopy. (B) Expression of E1 over time correlates with activation of γH2AX. The EYFP-E1 wild type was transiently expressed in C33A cells. Six and twelve hours posttransfection, cells were analyzed as described for panel A. (C to E) E1 activates components of the ATM pathway. The indicated EYFP or EYFP-E1 wild-type or mutant proteins were transiently expressed in C33A cells. Twenty-four hours posttransfection, cells were fixed and stained for phosphoserine 1981 of ATM (pATM) (C), phosphothreonine 68 of Chk2 (pChk2) (D), or phosphoserine 345 of Chk1 (pChk1) (E). Mock-transfected cells were used as negative controls, and cells treated for 24 h with 50 μM etoposide (Eto.) or 2 mM hydroxyurea (HU) were used as positive controls.
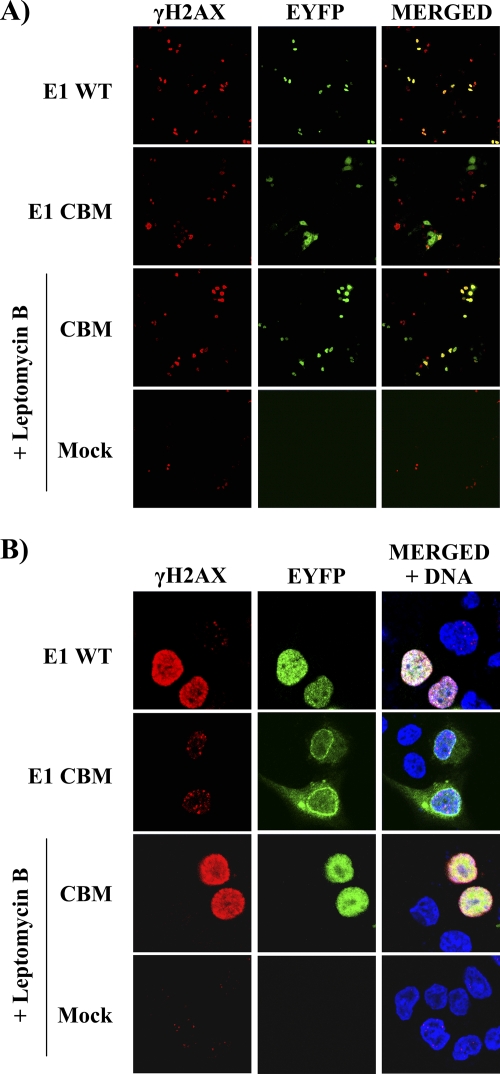
γH2AX foci are induced in cells containing low levels of nuclear E1.
In experiments such as those presented above, we consistently observed that γH2AX was present throughout the nucleus of E1-expressing cells (pan-nuclear staining) rather than being localized into the punctuated nuclear foci that are a hallmark of DNA double-strand breaks (DSBs). We reasoned that this pan-nuclear staining was in part a consequence of the amount of E1 being expressed in these cells and, accordingly, that nuclear foci may become apparent in cells expressing lower levels of E1. To investigate this possibility, we made use of an E1 mutant carrying a defective cyclin-binding motif (CBM). This mutant accumulates only at low levels in the nucleus because it is subject to rapid Crm1-dependent nuclear export (23). As anticipated, we observed by fluorescence microscopy that the activation of γH2AX was barely detectable in E1 CBM-expressing cells, especially when visualized at magnification ×20 (Fig. 8A). At magnification ×63, we were able to detect that the low levels of γH2AX in these cells were localized into discrete nuclear foci (Fig. 8B). This was in contrast to the high levels of γH2AX observed at magnification ×20 and the pan-nuclear staining observed at magnification ×63 in cells expressing wild-type E1 (Fig. 8A and B). Furthermore, we determined that increasing the amounts of the E1 CBM mutant in the nucleus by treatment of the cells with leptomycin B to inhibit its Crm1-dependent nuclear export resulted in pan-nuclear γH2AX staining (Fig. 8A and B). These results indicate that the levels of nuclear E1 affect the pattern of γH2AX staining in cells and suggest that γH2AX first accumulates in discrete foci and later progresses to pan-nuclear staining as the amount of nuclear E1 increases. Furthermore, the accumulation of γH2AX in discrete nuclear foci raises the possibility that E1 induces a DDR by creating DSB in the host genome (see Discussion).
Fig. 8.
γH2AX staining is punctuated in cells that accumulate low levels of E1 in their nucleus. The activation of γH2AX in cells transiently expressing EYFP-E1, either wild type (WT) or a cyclin-binding motif mutant derivative (CBM), was analyzed by immunofluorescence confocal microscopy 24 h posttransfection. Where indicated, the E1 CBM-expressing cells were treated with 7.5 ng/ml of leptomycin B (LMB) for 4 h. Detection of γH2AX was performed at a magnification of ×20 (A) or ×63 (B).
The E1-induced DDR is attenuated by E2.
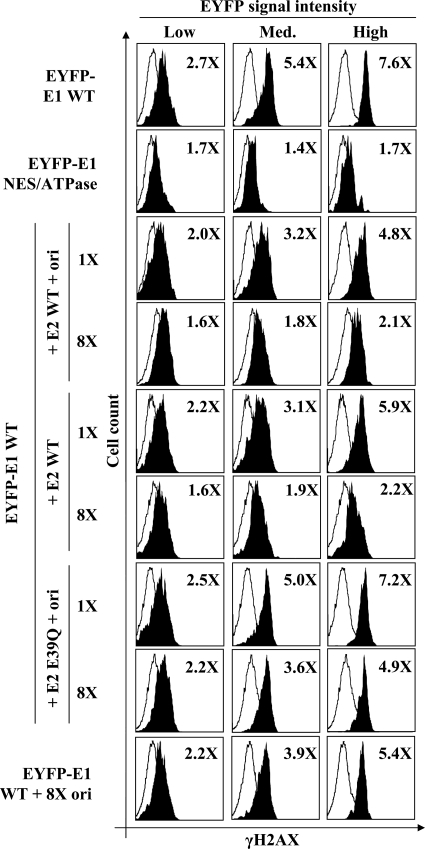
The results presented so far indicate that E1 expression triggers a robust DDR. To test if a similar DDR would be induced under viral DNA replication conditions, we investigated if E2, with or without a plasmid containing the viral origin (ori), had any effect on the levels of γH2AX induced by EYFP-E1. First, we confirmed that the levels of γH2AX could be accurately measured by flow cytometry. As can be seen in Fig. 9 (top row), the amounts of γH2AX were found to be 2.7-fold higher in cells expressing low levels of E1 than in control cells (EYFP-E1-negative cells of the same transfected cell population) and up to 7.6-fold higher in those expressing high levels of E1. These results confirmed that the levels of γH2AX increase as a function of E1 expression. Furthermore, this effect was specific to wild-type E1, since only low levels of γH2AX were detected in cells expressing the E1 NES/ATPase mutant protein (Fig. 9, second row). We then used this method to quantify the effect of E2 and the viral origin on E1-induced γH2AX activation. Specifically, we cotransfected C33A cells with a vector expressing EYFP-E1 with or without a plasmid encoding E2 and one containing the origin, in the same ratios as those used in our standard transient DNA replication assay. We also cotransfected increasing amounts of E2 or E2 plus the origin at levels 2-, 4- and 8-fold higher than those used in the DNA replication assay in order to favor complex formation between E1 and E2. Only the results obtained under standard replication conditions (1×) and those obtained with an 8-fold excess (8×) of E2 in the presence or absence of the origin are presented in Fig. 9. These experiments revealed that the E1-induced activation of γH2AX decreased as a function of the amount of E2 and origin transfected (compare 1× and 8× in Fig. 9). At 8× E2 and ori, the activation of γH2AX was reduced close to background levels, even in cells expressing high levels of EYFP-E1. Results obtained with intermediate amounts of E2 (2× and 4×) indicated that its inhibitory effect on γH2AX induction was dose dependent (data not shown). E1-induced γH2AX levels were decreased by E2 alone in the absence of the origin but not by the origin alone (Fig. 9). This effect of E2 was found to be dependent on its interaction with E1, since it was diminished by the E39Q substitution in E2, which reduces its ability to interact with E1 and to support transient DNA replication by approximately 50 to 60% under our standard assay conditions (24; data not shown). Finally, we confirmed by confocal microscopy that the induction by E1 of pChk2, a downstream effector of the ATM pathway, is also attenuated by E2 (data not shown). Collectively, the aforementioned results indicate that E2 significantly reduces, but does not completely eliminate, the E1-induced DDR.
Fig. 9.
E2 and the viral origin attenuate the E1-induced DDR. γH2AX levels in cells transiently expressing wild-type EYFP-E1 or the NES/ATPase mutant derivative, with or without E2, and containing or lacking the viral origin, are indicated. γH2AX levels were quantified by flow cytometry. For each sample, γH2AX levels were determined as a function of EYFP-E1 expression (low-, medium-, or high-EYFP-E1-expressing cells; shaded histograms), and the non-EYFP-expressing cells of the same cell population were used as an internal control (open histograms). For each condition, the fold increase in γH2AX activation is indicated in the upper right corner and was obtained by dividing the average value of activation measured for the EYFP-E1-positive cells by that of the controls cells.
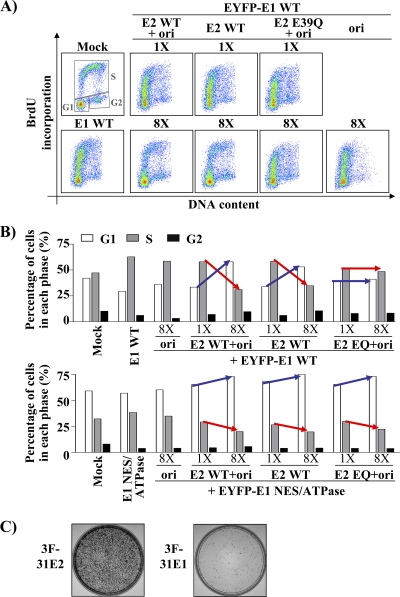
Cells expressing E1 and E2 are arrested in very early S phase.
The finding that E2 attenuates the E1-induced DDR prompted us to investigate if it could also alleviate the S-phase arrest caused by E1. To do so, we performed the same experiments as above with a gradient of E2, with or without the origin, but this time monitored both cell cycle progression and DNA synthesis. Cells were transfected with E1 and increasing amounts of E2 or E2 plus ori and, 24 h later, tested for their ability to carry out DNA synthesis, using a 1-h pulse with BrdU. BrdU incorporation was then plotted as a function of the cell cycle as previously described (Fig. 5C), and the percentages of cells in G1, S, and G2 were quantified using the gates shown for the mock-transfected cells in Fig. 10A. Quantification was performed on cells transfected with 1× and 8× E2 (Fig. 10A), with or without the ori, and numbers obtained are presented in the histogram of Fig. 10B (upper panel). From this analysis, we determined that E2 but not the origin alone leads to a dose-dependent accumulation of E1-expressing cells in G1 (blue arrow in Fig. 10B) and a concomitant decrease of cells in S phase (red arrow). Note that we show in the following section that cells expressing E1 and increasing amounts of E2 can still replicate the ori-containing plasmid. Since HPV DNA replication requires host DNA replication factors, this is consistent with these cells being arrested in S phase rather than in G1. Given this result and the fact that the flow cytometry analysis cannot distinguish between cells in G1 from those in very early S, it is likely that cells expressing E1 and E2 are arrested very early in S phase, close to the G1/S boundary, rather than in G1 per se.
Fig. 10.
E2 and the viral origin modulate the E1-induced cell cycle arrest. BrdU incorporation in EYFP-E1-expressing cells throughout the cell cycle. (A) Scatter plots obtained with cells transfected with a wild-type E1 expression vector, either alone or together with two different amounts (1× and 8×) of wild-type or E39Q E2 expression vector and the origin, as indicated. BrdU incorporation and DNA content were determined by flow cytometry. (B) Proportions of cells in G1, S, and G2 were quantified from the samples shown in panel A and from those transfected with a plasmid expressing the EYFP-E1 NES/ATPase mutant protein as a control (data not shown). Gates showed for the mock-transfected sample were used to quantity the cell cycle distribution of each sample, and the results were reported in a histogram. Results obtained with cells expressing EYFP-E1 are shown in the upper panel, while those obtained with cells expressing the NES/ATPase mutant E1 are shown in the lower panel. The blue and red arrows highlight the concomitant changes in the proportions of cells in G1 and S phase, respectively, brought about by increasing amounts of E2 with or without the origin. (C) CFA performed with 3F-HPV31 E2 or with 3F-HPV31 E1 expression vector. Cells were selected in G418-containing medium before staining.
In similar experiments, we found that the ability of E1 and E2 to cause cell cycle arrest in G1/very early S was slightly enhanced in the presence of the origin and, importantly, that this was not observed with the E39Q E2 mutant (Fig. 10B). This last result suggested that cell cycle arrest in G1/very early S phase is dependent on complex formation between E1 and E2 and by inference that it is not caused by E2 alone. To substantiate the notion that E2 is not sufficient to cause this cell cycle arrest, we determined the effect of wild-type E2 in cells expressing an E1 NES/ATPase mutant that does not interfere with cell cycle progression. As anticipated, only 10% of the cells expressing E2 and the mutant E1 accumulated in G1 (Fig. 10B, lower panel, and data not shown). Importantly, and in contrast to the results obtained with wild-type E1, this weak cell cycle inhibitory effect was also observed with E39Q E2 (Fig. 10B), suggesting that it was independent of complex formation between E1 and E2. From these results, we conclude that E2 has only a modest effect on cell cycle progression, a result supported by the observation that it does not prevent cellular proliferation in a colony formation assay (Fig. 10C). Collectively, the results presented above indicate that complex formation between E1 and E2 causes cells to arrest in G1/very early S. The evidence supporting the hypothesis that E1- and E2-expressing cells are arrested in very early S rather than in G1 is presented below.
Transient HPV DNA replication neither requires nor is affected by a DDR.
The results presented in Fig. 9 and 10 indicated that E2, when expressed in large amounts, reduces the DDR caused by E1 to almost background levels and causes an accumulation of E1-expressing cells in G1/very early S phase. To determine if these effects would impinge on viral DNA replication, we set out to test the effect of increasing amounts of E2 and the origin on E1-catalyzed DNA replication, using our luciferase assay (24).
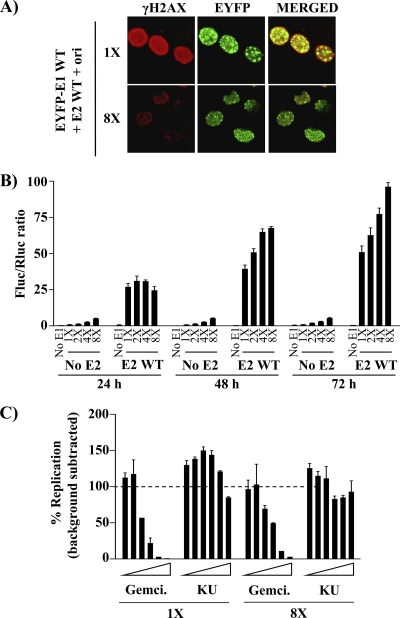
First, we used confocal fluorescence microscopy to examine the intracellular localization of EYFP-E1 under our standard DNA replication assay conditions or in the presence of an 8-fold excess of E2 and the origin. Under standard conditions, we confirmed that E1 is localized primarily into large nuclear foci (Fig. 11A), as previously reported (23, 78). When E2 and the ori were present in excess, EYFP-E1 was still localized into nuclear foci but of a smaller size. Whether these smaller foci form in G1 and become larger during S phase is under investigation. Importantly, we confirmed that γH2AX was strongly induced under standard DNA replication conditions and significantly reduced in the presence of excess E2 and ori, as anticipated from the flow cytometry quantification showed in Fig. 9. Interestingly, we noted that part of the remaining γH2AX signal observed at 8× was localized within these foci (Fig. 11A). Next, we performed the luciferase DNA replication assay in cells transfected with a constant and limiting amount of EYFP-E1 and a 1-, 2-, 4-, or 8-fold excess of E2 and the ori. As controls, the same conditions were used in the absence of E2 to prevent DNA replication. As expected, these control transfections gave rise to increasingly higher Fluc/Rluc ratios proportional to the amount of transfected ori plasmid (which contains the cytomegalovirus (CMV)-Fluc reporter gene) but they did not increased at 48 and 72 h posttransfection, confirming the lack of replication (Fig. 11B, “No E2”). In contrast, the levels of transient DNA replication in cells expressing E1 and E2 gradually increased over time, even in the presence of an excess of E2 and ori (Fig. 11B, E2 WT). Thus, viral DNA replication can take place in E1-expressing cells containing an excess of E2 and ori, indicating that these cells must be in very early S phase rather than late G1 and furthermore that viral DNA replication occurs independently of a robust DDR (which is attenuated by high concentrations of E2). To ascertain the latter suggestion, we tested the effect of an ATM inhibitor (KU-55933) on transient DNA replication. This inhibitor had only a weak effect on HPV DNA replication, at concentrations up to 10 μM, in contrast to the nucleotide analog gemcitabine, which was used as a positive control (Fig. 11C). The inhibitory activity of KU-55933 was confirmed by monitoring its effect on the phosphorylation of Thr68 of Chk2, a well-established ATM phosphorylation site (data not shown). Collectively, these findings indicate that HPV DNA replication not only occurs independently of a robust DDR (at high concentrations of E2 and ori) but remarkably that it remains unabated in the face of a DDR and checkpoint response (at low concentrations of E2 and ori), as demonstrated recently by King et al. (39).
Fig. 11.
Effect of increasing amounts of E2 and the origin on E1-catalyzed DNA replication. (A) Activation of γH2AX in cells transfected with an EYFP-E1 expression plasmid and two different amounts (1× and 8×) of wild-type E2 expression vector plus the origin. Twenty-four hours posttransfection, E1-expressing cells were fixed, stained for γH2AX, mounted, and visualized by ×40 fluorescence confocal microscopy. (B) Transient DNA replication activity of E1 in the presence of increasing amounts of E2 and the origin. DNA replication activity of EYFP-E1 was determined 24, 48, and 72 h posttransfection. Cells were transfected with 5 ng of EYFP-E1 expression plasmid and increasing amounts of wild-type E2 expression vector plus the origin (E2 WT), as indicated (1×, 2×, 4×, and 8×). Background signal was measured in the absence of E2 (5 ng of E1 and increasing amounts of ori-plasmid) (No E2) or in the absence of E1 (No E1). Replication activities are reported as Fluc/Rluc ratios, and error bars represent standard deviations. (C) Effect of the ATM kinase inhibitor KU-55933 on viral DNA replication. Transient HPV DNA replication assays were performed under standard conditions (1×) or in the presence of an excess of E2 and the origin (8×). KU-55933 (KU) was tested at increasing concentrations ranging from 0.156 to 10 μM. Gemcitabine (Gemci., from 1.56 to 100 nM) and 0.1% dimethyl sulfoxide (DMSO) were used as positive and negative controls, respectively. DNA replication levels (Fluc/Rluc ratios) from which background signals were subtracted are reported as percentages of the activity measured in the presence of DMSO, which was set at 100%.
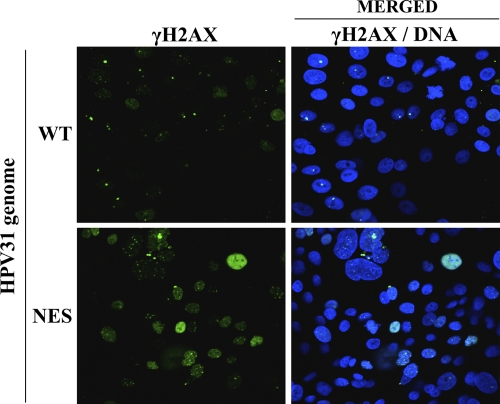
Keratinocytes expressing an NES-defective E1 mutant genome show increased levels of γH2AX.
We previously reported that a mutant HPV31 genome encoding NES-defective E1 is poorly maintained as an episome in immortalized keratinocytes and is gradually lost upon cellular division (i.e., upon several cell passages). This and other results, including those presented herein, led us to suggest that nuclear accumulation/retention of the NES E1 mutant protein is detrimental to cellular proliferation. Since we show here that this antiproliferative effect of E1 is accompanied by a DDR, we set out to determine if a similar DDR was induced in keratinocytes containing the E1 NES mutant genome. To do so, we compared the levels of γH2AX in keratinocytes immortalized with the mutant genome or wild-type HPV31, using immortalized cells at an early passage in which the E1 NES mutant genomes is still maintained as an episome, albeit in smaller amounts (23). As can be seen in Fig. 12, higher levels of γH2AX were indeed induced by the E1 NES mutant as compared to results for the wild type, thus confirming that high levels of E1 in the nucleus are sufficient to induce a DDR. Thus, under physiological conditions, the nuclear export of E1 serves to prevent the induction of a DDR in undifferentiated keratinocytes.
Fig. 12.
γH2AX activation in keratinocytes immortalized with a wild-type or E1 NES mutant genome. Early-passage keratinocytes immortalized with the indicated HPV31 genome were fixed and stained with an antibody against γH2AX. Nuclei (DNA) were stained with DAPI.
DISCUSSION
In this article, we report that the antiproliferative activity of E1 is conserved in HPV11, -31 and -16 and in bovine papillomavirus (BPV) and that it requires the origin-binding and ATPase activities of the protein. We provide evidence that E1 induces a cell cycle arrest independently of the viral oncogenes E6 and E7, since the block was also detected in HeLa cells, and independently of p53, since it was observed in cells that express either wild-type (U2OS) or R275C mutant (C33A) p53 (15, 16). Furthermore, we have shown that the arrest takes place during the early events of S phase, when BrdU is incorporated into DNA and when PCNA has not yet assembled into large foci. Interestingly, this arrest is accompanied by the induction of a DDR and the activation of the ATM pathway, since E1-expressing cells exhibit elevated levels of γH2AX, pATM Ser1981, and pChk2 Thr68. We also show that complex formation between E1 and E2 reduces the levels of γH2AX induced by E1 and leads to a redistribution of E1-expressing cells to an earlier part of S phase, close to the G1/S boundary. Importantly, we present evidence that transient viral DNA replication is neither affected by nor dependent on a DDR. The biological significance of these findings is highlighted by the observation that keratinocytes immortalized with an HPV31 genome encoding an NES mutant E1 show drastically higher levels of γH2AX than cells maintaining a wild-type episome, suggesting that nuclear export of E1 is indeed required to alleviate the activation of a DDR in infected cells.
Induction of a cell cycle arrest and activation of a DDR by E1.
In a recent article (23), we reported that sustained accumulation of E1 in the nucleus is detrimental to cellular proliferation. Here we have shown that E1 induces a cell cycle block in early S phase that is accompanied by the induction of a DDR and that both events require the highly conserved origin-binding and ATPase activities of E1. Accordingly, we found that E1 from low-risk (HPV11) and high-risk (HPV31 and -16) HPV types and from BPV were all able to induce cell cycle arrest, inhibit cellular proliferation, and induce a DDR (albeit to different extents) (Table 4).
Table 4.
Activation of γH2AX by E1 from different papillomaviruses
| Protein expressed | Fold activation of γH2AXa |
|---|---|
| GFP | 2.7 |
| GFP-HPV31 E2 | 2.3 |
| GFP-HPV31 E1 | 6.9 |
| GFP-HPV11 E1 | 4.2 |
| GFP-BPV E1 | 6.2 |
| GFP-HPV16 E1 | 7.0 |
The activation of γH2AX in cells expressing the indicated GFP fusion protein was analyzed by flow cytometry as described in the legend for Fig. 9.
We previously showed that an HPV11 E1 fragment comprising the OBD and C-terminal ATPase domain is sufficient to replicate DNA in vitro, indicating that it can assemble into a functional helicase (2). In this study, the findings that an analogous fragment of HPV31 E1 has antiproliferative activity and supports low levels of transient DNA replication raise the possibility that E1 can assemble onto host DNA and unwind it nonspecifically. This nonspecific unwinding activity may account for the ability of E1 to induce cell cycle arrest and a concomitant DDR. These effects appear to be specific to E1, however, since they were not observed with the related large T-antigen helicase from SV40, whose origin-binding and ATPase domains are structurally and functionally similar to those of E1 (11, 20, 21, 43, 47). Indeed, we observed that large T antigen was unable to interfere with cellular proliferation in a CFA and induced only low levels of DDR markers compared to results for E1 (data not shown). One of the main differences between E1 and large T antigen is that the former has a higher nonspecific DNA-binding activity, at least in vitro (80, 83; A. Fradet-Turcotte and J. Archambault, unpublished results). Accordingly, it has been reported that E1 is capable of replicating a DNA template that does not contain the viral origin (8), indicating that it can efficiently bind and unwind nonviral DNA. These observations further support the possibility that E1 can unwind host DNA nonspecifically, thus accounting for its capacity to interfere with cell cycle progression and to induce a DDR. Consistent with this hypothesis, we found that the formation of a complex between E1 and E2, which has been reported to inhibit the nonspecific DNA-binding activity of E1 (8, 67, 73), greatly reduces its capacity to induce a DDR (Fig. 9). However, the fact that E2 did not prevent E1-induced cell cycle arrest suggests either that E2 does not reduce the magnitude of the DDR below the threshold necessary to induce cell cycle arrest or alternatively that the ability of E1 to induce a DDR and to inhibit cell cycle progression are two mechanistically distinct events. In support of the latter possibility, we observed in CFAs that E1 inhibits the proliferation of ATM-deficient cells (data not shown), indicating that this growth-inhibitory effect is independent of the activation of the ATM kinase. Similarly, we observed that the ability of E1 to block C33A cells in S phase is not prevented by treatment with the ATM inhibitor KU-55933 (data not shown).
Possible mechanisms by which E1 blocks cell cycle progression.
Although the exact mechanism by which E1 interferes with cell cycle progression remains unknown, several hypotheses can be drawn from the existing literature. For instance, the depletion of cellular DNA replication factors or of components of the origin-firing machinery by small interfering RNA (siRNA) has been shown to arrest the progression of the cell cycle in S phase (51, 55, 79, 89). Given that the OBD and ATPase domain of E1, which are required to induce cell cycle arrest, have been reported to interact with the polymerase α-primase, topoisomerase I, and RPA (reviewed in reference 77), it is possible that E1, by competing for these factors, makes them unavailable for host DNA synthesis. Alternatively or in addition, E1 may also interfere with the firing of host origins in early S phase, as previously reported for the Rep78 helicase from adeno-associated virus (AAV) (7). Rep78 has been suggested to interfere with cell cycle progression by interacting with Cdc25A and inhibiting its phosphatase activity, which is required for the firing of origins (7, 66). Interestingly, Rep78 also induces a DDR which, similar to that caused by E1, is not sufficient to block cell cycle progression. However, if Cdc25A was inactivated by E1, we would anticipate that Cdk2, a well-characterized Cdc25A substrate, would also be inactive and thus unable to phosphorylate E1 and prevent its nuclear export. The finding that E1 is nuclear in E1-arrested cells indicates that it has been phosphorylated by Cdk2, and we surmise that Cdc25A must be active in these cells. Therefore, while it is possible that E1 sequesters Cdc25A away from the origin firing complexes, it is unlikely that it inhibits its phosphatase activity. Whether E1 interacts with Cdc25A or other proteins, such as CDC45 and Dbf4/cdc7 (reviewed in reference 66), which are also required for the firing of host origins, remains to be investigated.
Possible mechanisms by which E1 induces a DDR.
We have shown that activation of γH2AX occurs soon after the expression of E1 and can be prevented by high concentrations of E2. Typically, the activation of γH2AX and of the ATM pathway is triggered by double-stranded breaks (DSBs) in the host DNA and by the activation of cell cycle checkpoints (reviewed in reference 4). As our results demonstrate, induction of a DDR is dependent on the origin-binding and ATPase activities of E1 and can be reduced by E2, consistent with the nonspecific unwinding of host DNA by E1 being a source of DSBs. In addition to causing DSBs, it is possible that E1 triggers a DDR by interacting directly with components of the DNA repair machinery, such as the MRN complex, whose Mre11, Rad50, and Nbs1 subunits colocalize with HPV18 and -11 E1 in cells (35; Thomas Broker, personal communication). Indeed, it has recently been shown that the tethering of these proteins to DNA is sufficient to induce a DDR in the absence of DSB (71).
Activation of γH2AX in defined nuclear foci is considered a hallmark of DSBs, whereas its pan-nuclear activation has been associated with apoptosis during S phase (17). Our results with an E1 CBM mutant suggest that both phenomena can happen in E1-expressing cells depending on the nuclear levels of E1. Indeed, we found that in cells expressing the E1 CBM mutant, which accumulates only at low levels in the nucleus and does not arrest cell cycle progression (23), γH2AX is localized in discrete nuclear foci. Furthermore, when the nuclear accumulation of this E1 mutant was increased by inhibiting its nuclear export using leptomycin B (LMB) or by inhibiting its NES, γH2AX staining became pan-nuclear (Fig. 8 and data not shown). On the basis of these observations, we propose that the accumulation of γH2AX in discrete nuclear foci results from the induction of DSB by low levels of E1 in the nucleus, whereas its pan-nuclear accumulation observed at higher E1 concentrations arises from the accumulation of such foci combined with the cells being permanently arrested in S phase and possibly undergoing apoptosis.
Activation of an ATM-dependent DDR is not required for transient HPV DNA replication.
An ATM-dependent DDR is activated by many double-stranded DNA viruses, for different reasons. For example, similarly to HPV, Epstein-Barr virus (EBV) activates the ATM pathway but does not rely on its activation to replicate its genome (41). In contrast, the activation of ATM is required for the phosphorylation of SV40 large T antigen and for the efficient replication of viral DNA (69). Kadaja et al. (35) reported that replication of integrated HPV genomes by E1 and E2 leads to the induction of the ATM pathway and, to a lesser extent, of the ATR pathway and to the recruitment of ATM, Chk2, ATRIP, pChk1, Ku70/80, and the MRN complex at these integration sites. In this study, we found that E1 is sufficient to activate the ATM pathway regardless of the presence or the absence of the genome, suggesting that the induction of the ATM pathway reported by Kadaja et al. (35) may have been caused at least in part by E1. Although E1 activates the ATM pathway, viral DNA replication does not require the induction of this DDR nor is affected by it. These findings are in agreement with those of King et al. (39), who showed that replication of an HPV origin-containing plasmid by E1 and E2 from HPV11 and -16 is not affected by treatment of the cells with 50 μM etoposide during 24 h. Finally, our observation that HPV DNA replication can occur independently of the activation of the ATM pathway is consistent with the previous observation by one of our groups (C.A. Moody and L.A. Laimins) that maintenance of the HPV31 episome in immortalized keratinocytes is unaffected by the ATM inhibitor KU-55933 (53). In that same study, it was found that despite being unnecessary for genome maintenance, the ATM pathway is required for amplification of the viral episome in differentiated keratinocytes, in part to induce the activity of caspase-3/7 and proteolytic cleavage of the N-terminal domain of E1 (52, 53). This study also showed that activation of the ATM-dependent DDR in differentiating keratinocytes immortalized with the HPV31 genome was dependent on E6 and E7, suggesting that viral oncogenes contribute significantly to the activation of this pathway during amplification of the viral genome (53).
Although our first observation that nuclear accumulation of E1 leads to the activation of a DDR was made with transfected cells under conditions where E1 is expressed at high levels, a similar DDR activation was observed in immortalized keratinocytes expressing an NES mutant E1 from the HPV31 episome. As such, these results suggest that the accumulation of E1 in the nucleus of infected cells is sufficient to induce a DDR. Since nuclear accumulation of E1 has been shown to be detrimental to cellular proliferation, our findings are consistent with a model where sustained accumulation of E1 in the nucleus induces an S-phase arrest, which in turn induces apoptosis (pan-nuclear γH2AX). Consistent with this model, Jolly et al. recently reported that leptomycin B, which inhibits the Crm1-exportin pathway, essential for the nuclear export of E1 (23), increases apoptosis in a cell line maintaining the HPV16 episome (34).
Collectively, our study suggests that both the nuclear accumulation and the nonspecific DNA unwinding activity of E1 need to be tightly regulated by the nuclear export of E1 and by its interaction with E2, respectively, in order to avoid the premature activation of the ATM pathway in undifferentiated keratinocytes. Failure to adequately control the nuclear accumulation and activity of E1 in the lower layers of an infected epithelium may lead to the induction of undesired DNA breaks in the host genome that are recombinogenic and could favor integration of the viral genome. This in turn would increase the probability of generating integration events that disrupt the E2 open reading frame and lead to overexpression of the viral E6 and E7 proteins. For lesions containing high-risk HPVs but not for those associated with low-risk types, such integration events represent a critical risk factor for progression to cancer. Thus, failure to properly regulate the nuclear accumulation and activity of E1 from high-risk HPV types may be a more important contributor to HPV-induced carcinogenesis than previously suspected.
ACKNOWLEDGMENTS
We thank Nozomi Sakakibara and Allison McBride for sharing data before publication and all members of the Archambault laboratory for critical reading of the manuscript and for helpful discussions.
This work was supported by grants from the Canadian Institutes for Health Research and from the National Cancer Institute to J.A. and L.A.L., respectively. A.F.-T. and M.L. hold a studentship from the Fonds de la Recherche en Santé du Québec (FRSQ) and from the Canadian Institutes of Health Research (CIHR), respectively. C.A.M. is supported by a K99 Pathway to Independence Award from the National Cancer Institute.
Footnotes
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Abbate E. A., Berger J. M., Botchan M. R. 2004. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 18:1981–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amin A. A., et al. 2000. Identification of domains of the HPV11 E1 protein required for DNA replication in vitro. Virology 272:137–150 [DOI] [PubMed] [Google Scholar]
- 3. Androphy E. J., Lowy D. R., Schiller J. T. 1987. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature 325:70–73 [DOI] [PubMed] [Google Scholar]
- 4. Bartek J., Lukas J. 2001. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13:738–747 [DOI] [PubMed] [Google Scholar]
- 5. Bartek J., Lukas J. 2001. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 490:117–122 [DOI] [PubMed] [Google Scholar]
- 6. Bernard H. U., et al. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berthet C., Raj K., Saudan P., Beard P. 2005. How adeno-associated virus Rep78 protein arrests cells completely in S phase. Proc. Natl. Acad. Sci. U. S. A. 102:13634–13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonne-Andrea C., Tillier F., McShan G. D., Wilson V. G., Clertant P. 1997. Bovine papillomavirus type 1 DNA replication: the transcriptional activator E2 acts in vitro as a specificity factor. J. Virol. 71:6805–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462–42467 [DOI] [PubMed] [Google Scholar]
- 10. Chow L. T., Broker R. 1997. Small DNA tumor viruses, p. 267–301 In Nathanson N. (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, PA [Google Scholar]
- 11. Clertant P., Seif I. 1984. A common function for polyoma virus large-T and papillomavirus E1 proteins? Nature 311:276–279 [DOI] [PubMed] [Google Scholar]
- 12. Clower R. V., Fisk J. C., Melendy T. 2006. Papillomavirus E1 protein binds to and stimulates human topoisomerase I. J. Virol. 80:1584–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conger K. L., Liu J. S., Kuo S. R., Chow L. T., Wang T. S. 1999. Human papillomavirus DNA replication. Interactions between the viral E1 protein and two subunits of human DNA polymerase alpha/primase. J. Biol. Chem. 274:2696–2705 [DOI] [PubMed] [Google Scholar]
- 14. Cote-Martin A., et al. 2008. Human papillomavirus E1 helicase interacts with the WD repeat protein p80 to promote maintenance of the viral genome in keratinocytes. J. Virol. 82:1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crook T., et al. 1992. Clonal p53 mutation in primary cervical cancer: association with human-papillomavirus-negative tumours. Lancet 339:1070–1073 [DOI] [PubMed] [Google Scholar]
- 16. Crook T., Wrede D., Vousden K. H. 1991. p53 point mutation in HPV negative human cervical carcinoma cell lines. Oncogene 6:873–875 [PubMed] [Google Scholar]
- 17. de Feraudy S., Revet I., Bezrookove V., Feeney L., Cleaver J. E. 2010. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc. Natl. Acad. Sci. U. S. A. 107:6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng W., et al. 2004. Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J. Virol. 78:13954–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Villiers E. M., Fauquet C., Broker T. R., Bernard H. U., zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27 [DOI] [PubMed] [Google Scholar]
- 20. Enemark E. J., Chen G., Vaughn D. E., Stenlund A., Joshua-Tor L. 2000. Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol. Cell 6:149–158 [PubMed] [Google Scholar]
- 21. Enemark E. J., Joshua-Tor L. 2006. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270–275 [DOI] [PubMed] [Google Scholar]
- 22. Fradet-Turcotte A., Brault K., Titolo S., Howley P. M., Archambault J. 2009. Characterization of papillomavirus E1 helicase mutants defective for interaction with the SUMO-conjugating enzyme Ubc9. Virology 395:190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fradet-Turcotte A., Moody C., Laimins L. A., Archambault J. 2010. Nuclear export of human papillomavirus type 31 E1 is regulated by Cdk2 phosphorylation and required for viral genome maintenance. J. Virol. 84:11747–11760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fradet-Turcotte A., Morin G., Lehoux M., Bullock P. A., Archambault J. 2010. Development of quantitative and high-throughput assays of polyomavirus and papillomavirus DNA replication. Virology 399:65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frattini M. G., Laimins L. A. 1994. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc. Natl. Acad. Sci. U. S. A. 91:12398–12402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frattini M. G., Laimins L. A. 1994. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology 204:799–804 [DOI] [PubMed] [Google Scholar]
- 27. Frattini M. G., Lim H. B., Laimins L. A. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. U. S. A. 93:3062–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gissmann L., et al. 1983. Human papillomavirus types 6 and 11 DNA sequences in genital and laryngeal papillomas and in some cervical cancers. Proc. Natl. Acad. Sci. U. S. A. 80:560–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han Y., Loo Y. M., Militello K. T., Melendy T. 1999. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 73:4899–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hebner C. M., Laimins L. A. 2006. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev. Med. Virol. 16:83–97 [DOI] [PubMed] [Google Scholar]
- 31. Hickman A. B., Dyda F. 2005. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15:77–85 [DOI] [PubMed] [Google Scholar]
- 32. Hoffmann R., Hirt B., Bechtold V., Beard P., Raj K. 2006. Different modes of human papillomavirus DNA replication during maintenance. J. Virol. 80:4431–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. James J. A., et al. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11:1025–1035 [DOI] [PubMed] [Google Scholar]
- 34. Jolly C. E., Gray L. J., Parish J. L., Lain S., Herrington C. S. 2009. Leptomycin B induces apoptosis in cells containing the whole HPV 16 genome. Int. J. Oncol. 35:649–656 [DOI] [PubMed] [Google Scholar]
- 35. Kadaja M., Isok-Paas H., Laos T., Ustav E., Ustav M. 2009. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 5:e1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kadaja M., Silla T., Ustav E., Ustav M. 2009. Papillomavirus DNA replication—from initiation to genomic instability. Virology 384:360–368 [DOI] [PubMed] [Google Scholar]
- 37. Kadaja M., et al. 2007. Genomic instability of the host cell induced by the human papillomavirus replication machinery. EMBO J. 26:2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kastan M. B., Bartek J. 2004. Cell-cycle checkpoints and cancer. Nature 432:316–323 [DOI] [PubMed] [Google Scholar]
- 39. King L. E., et al. 2010. Human papillomavirus E1 and E2 mediated DNA replication is not arrested by DNA damage signalling. Virology 406:95–102 [DOI] [PubMed] [Google Scholar]
- 40. Krek W., DeCaprio J. A. 1995. Cell synchronization. Methods Enzymol. 254:114–124 [DOI] [PubMed] [Google Scholar]
- 41. Kudoh A., et al. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156–8163 [DOI] [PubMed] [Google Scholar]
- 42. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 43. Li D., et al. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512–518 [DOI] [PubMed] [Google Scholar]
- 44. Loo Y. M., Melendy T. 2004. Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J. Virol. 78:1605–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lusky M., Hurwitz J., Seo Y. S. 1994. The bovine papillomavirus E2 protein modulates the assembly of but is not stably maintained in a replication-competent multimeric E1-replication origin complex. Proc. Natl. Acad. Sci. U. S. A. 91:8895–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma T., Zou N., Lin B. Y., Chow L. T., Harper J. W. 1999. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. U. S. A. 96:382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mansky K. C., Batiza A., Lambert P. F. 1997. Bovine papillomavirus type 1 E1 and simian virus 40 large T antigen share regions of sequence similarity required for multiple functions. J. Virol. 71:7600–7608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Masterson P. J., Stanley M. A., Lewis A. P., Romanos M. A. 1998. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J. Virol. 72:7407–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Modesti M., Kanaar R. 2001. DNA repair: spot(light)s on chromatin. Curr. Biol. 11:R229–R232 [DOI] [PubMed] [Google Scholar]
- 50. Mohr I. J., et al. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694–1699 [DOI] [PubMed] [Google Scholar]
- 51. Montagnoli A., et al. 2004. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 64:7110–7116 [DOI] [PubMed] [Google Scholar]
- 52. Moody C. A., Fradet-Turcotte A., Archambault J., Laimins L. A. 2007. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc. Natl. Acad. Sci. U. S. A. 104:19541–19546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moody C. A., Laimins L. A. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 5:e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morin G., et al. 2011. A conserved amphipathic helix in the N-terminal regulatory region of the papillomavirus E1 helicase is required for efficient viral DNA replication. J. Virol. 85:5287–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moyer S. E., Lewis P. W., Botchan M. R. 2006. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U. S. A. 103:10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Munger K., et al. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 78:11451–11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Oguro M., Suzuki-Hori C., Nagano H., Mano Y., Ikegami S. 1979. The mode of inhibitory action by aphidicolin on eukaryotic DNA polymerase alpha. Eur. J. Biochem. 97:603–607 [DOI] [PubMed] [Google Scholar]
- 58. Park P., et al. 1994. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc. Natl. Acad. Sci. U. S. A. 91:8700–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rogakou E. P., Boon C., Redon C., Bonner W. M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 [DOI] [PubMed] [Google Scholar]
- 61. Sanders C. M., Stenlund A. 2001. Mechanism and requirements for bovine papillomavirus, type 1, E1 initiator complex assembly promoted by the E2 transcription factor bound to distal sites. J. Biol. Chem. 276:23689–23699 [DOI] [PubMed] [Google Scholar]
- 62. Schermelleh L., et al. 2007. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 35:4301–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schiffman M., Castle P. E., Jeronimo J., Rodriguez A. C., Wacholder S. 2007. Human papillomavirus and cervical cancer. Lancet 370:890–907 [DOI] [PubMed] [Google Scholar]
- 64. Schuck S., Stenlund A. 2005. Assembly of a double hexameric helicase. Mol. Cell 20:377–389 [DOI] [PubMed] [Google Scholar]
- 65. Schuck S., Stenlund A. 2005. Role of papillomavirus E1 initiator dimerization in DNA replication. J. Virol. 79:8661–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sclafani R. A., Holzen T. M. 2007. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 41:237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sedman J., Stenlund A. 1995. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 14:6218–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sengupta S., Harris C. C. 2005. p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 6:44–55 [DOI] [PubMed] [Google Scholar]
- 69. Shi Y., Dodson G. E., Shaikh S., Rundell K., Tibbetts R. S. 2005. Ataxia-telangiectasia-mutated (ATM) is a T-antigen kinase that controls SV40 viral replication in vivo. J. Biol. Chem. 280:40195–40200 [DOI] [PubMed] [Google Scholar]
- 70. Smart D. J., et al. 2008. Assessment of DNA double-strand breaks and gammaH2AX induced by the topoisomerase II poisons etoposide and mitoxantrone. Mutat. Res. 641:43–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Soutoglou E., Misteli T. 2008. Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320:1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sporbert A., Domaing P., Leonhardt H., Cardoso M. C. 2005. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 33:3521–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stenlund A. 2003. E1 initiator DNA binding specificity is unmasked by selective inhibition of non-specific DNA binding. EMBO J. 22:954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stenlund A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 4:777–785 [DOI] [PubMed] [Google Scholar]
- 75. Sun Y., Han H., McCance D. J. 1998. Active domains of human papillomavirus type 11 E1 protein for origin replication. J. Gen. Virol. 79(Pt. 7):1651–1658 [DOI] [PubMed] [Google Scholar]
- 76. Sun Y. N., Lu J. Z., McCance D. J. 1996. Mapping of HPV-11 E1 binding site and determination of other important cis elements for replication of the origin. Virology 216:219–222 [DOI] [PubMed] [Google Scholar]
- 77. Sverdrup F., Myers G. 1997. The E1 proteins, p. 37–53 In Myers G., Baker C., Münger K., Sverdruo F., McBride A., Bernard H.-U. (ed.), Human papillomavirus. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 78. Swindle C. S., et al. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Taricani L., Shanahan F., Parry D. 2009. Replication stress activates DNA polymerase alpha-associated Chk1. Cell Cycle 8:482–489 [DOI] [PubMed] [Google Scholar]
- 80. Titolo S., Brault K., Majewski J., White P. W., Archambault J. 2003. Characterization of the minimal DNA binding domain of the human papillomavirus e1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein. J. Virol. 77:5178–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Titolo S., et al. 2000. Identification of domains of the human papillomavirus type 11 E1 helicase involved in oligomerization and binding to the viral origin. J. Virol. 74:7349–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Titolo S., et al. 1999. Role of the ATP-binding domain of the human papillomavirus type 11 E1 helicase in E2-dependent binding to the origin. J. Virol. 73:5282–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Titolo S., Welchner E., White P. W., Archambault J. 2003. Characterization of the DNA-binding properties of the origin-binding domain of simian virus 40 large T antigen by fluorescence anisotropy. J. Virol. 77:5512–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. White P. W., et al. 2001. Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J. Biol. Chem. 276:22426–22438 [DOI] [PubMed] [Google Scholar]
- 85. Wilson V. G., West M., Woytek K., Rangasamy D. 2002. Papillomavirus E1 proteins: form, function, and features. Virus Genes 24:275–290 [DOI] [PubMed] [Google Scholar]
- 86. Woodman C. B., Collins S. I., Young L. S. 2007. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer. 7:11–22 [DOI] [PubMed] [Google Scholar]
- 87. Yasugi T., Vidal M., Sakai H., Howley P. M., Benson J. D. 1997. Two classes of human papillomavirus type 16 E1 mutants suggest pleiotropic conformational constraints affecting E1 multimerization, E2 interaction, and interaction with cellular proteins. J. Virol. 71:5942–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yu J. H., Lin B. Y., Deng W., Broker T. R., Chow L. T. 2007. Mitogen-activated protein kinases activate the nuclear localization sequence of human papillomavirus type 11 E1 DNA helicase to promote efficient nuclear import. J. Virol. 81:5066–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zegerman P., Diffley J. F. 2010. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467:474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhou B. B., Elledge S. J. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433–439 [DOI] [PubMed] [Google Scholar]
- 91. zur Hausen H., de Villiers E. M. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427–447 [DOI] [PubMed] [Google Scholar]