Abstract
CP81 is a virulent Campylobacter group III phage whose linear genome comprises 132,454 bp. At the nucleotide level, CP81 differs from other phages. However, a number of its structural and replication/recombination proteins revealed a relationship to the group II Campylobacter phages CP220/CPt10 and to T4-type phages. Unlike the T4-related phages, the CP81 genome does not contain conserved replication and virion modules. Instead, the respective genes are scattered throughout the phage genome. Moreover, most genes for metabolic enzymes of CP220/CPt10 are lacking in CP81. On the other hand, the CP81 genome contains nine similar genes for homing endonucleases which may be involved in the attrition of the conserved gene order for the virion core genes of T4-type phages. The phage apparently possesses an unusual modification of C or G bases. Efficient cleavage of its DNA was only achieved with restriction enzymes recognizing pure A/T sites. Uncommonly, phenol extraction leads to a significant loss of CP81 DNA from the aqueous layer, a property not yet described for other phages belonging to the T4 superfamily.
INTRODUCTION
Campylobacteriosis is a worldwide zoonosis. There are probably more than two million Campylobacter infections in humans annually in the United States. Also, in developing countries, infections with Campylobacter jejuni are very frequent in children, and symptomatic infections early in life are followed later by asymptomatic infections. The bacteria are common commensals of the gastrointestinal tract of various mammals and birds. Consumption of undercooked meats, especially poultry, has been associated with Campylobacter infections (44). The prevalence of Campylobacter-positive chicken is generally high, and transmission of Campylobacter from bird to bird occurs rapidly (25). Intervention strategies mainly focus on biosecurity measures (e.g., improvement of personal hygiene, avoiding of mixed farming, and control of rodents and insects) and postslaughter decontamination of poultry carcasses (19). Since these measures are expensive and not always efficient and since suitable vaccines are not available, bacteriophages have been proposed to reduce the Campylobacter counts on chicken. Indeed, phage administration in the laboratory reduced C. jejuni colonization of the broiler gut and the contamination on chicken skin by several orders of magnitude (3, 6, 16, 20, 29, 50).
Phages intended for therapeutic applications or for the control of pathogens in food production have to fulfill a number of requirements. First and foremost, phages have to be safe in order to avoid any undesired side effect. Therefore, a basic understanding of the biology and genetics of the phage to be used is mandatory. A minimal standard is the acquisition of the genome sequence of the phage to exclude undesired genes. Unfortunately, up to now, genome sequences of Campylobacter phages are scarce. Most Campylobacter phages have been investigated only with respect to their host ranges, morphology, and genome size (3, 4, 11, 16, 22, 26, 30). Almost all Campylobacter phages isolated thus far are members of the family Myoviridae. According to their genome sizes estimated by pulsed-field gel electrophoresis (PFGE), the phages are classified into three groups (group I, 320 kb; group II, 180 to 190 kb; group III, 130 to 140 kb) (40). No group I Campylobacter phage has yet been sequenced. Recently, the nucleotide sequences of two group II phages (CP220 and CPt10) have been published (49). These two phages share a DNA sequence identity of 96% and some protein matches with T4-type phages. Group III Campylobacter phages have been isolated in the United Kingdom, Denmark, and South Korea (22, 26, 30). Perhaps due to chemical modification of bases in the phage DNA (4, 11), Campylobacter phage genomes are highly refractory to genomic analysis. Therefore, no group III phage genome sequences have yet been reported. Here, we present the first genome sequence of a group III Campylobacter phage (CP81) whose gene products share protein sequence identity with proteins of the T4 phage family in a genome that is smaller than any hitherto reported T4-like phage. This observation raises interesting questions with respect to the evolution of the T4 gene set.
MATERIALS AND METHODS
Isolation, propagation, and purification of the phage.
CP81 was isolated from the skin of a retail chicken portion purchased from a supermarket in Bavaria, Germany. To recover phage particles from the skin, 15 to 20 ml of SM buffer (41) was added to the sample, which was then incubated overnight at 45°C on a stirrer. Subsequently, the meat juice was centrifuged at 10,000 × g, followed by filtration of the supernatant through a 0.2-μm-pore-size nitrocellulose membrane (VWR International, Darmstadt, Germany). Phage activity was detected by spotting 10 μl of the CP81 suspension onto a lawn of the C. jejuni reference strain NCTC11168 (18, 21). Group II phage NCTC12684 was obtained from the National Collection of Type Cultures, Health Protection Agency, United Kingdom. This phage has already been characterized (40). The cultivation of bacteria, plaque assays, single plaque isolations, and propagation of the phages were performed as previously described (22, 41). Phages were concentrated and purified by CsCl-step gradients (41).
Examination of phage morphology.
Solutions of purified phages were investigated by transmission electron microscopy using the negative-staining procedure according to Steven et al. (45). Samples were stained with 2% uranyl acetate (pH 4.3), 2% ammonium molybdate (pH 7.0), or 2% sodium phosphotungstate (pH 7.0). Except for uranyl acetate, 0.04% trehalose was added to the staining solution (23). Samples were examined using a Tecnai Spirit (FEI, Hillsboro, OR) equipped with an Eagle 4K charge-coupled device (CCD) camera. Magnifications were calibrated using negatively stained catalase crystals (52). For reproducibility, samples were adjusted to eucentric focus. Measurements were performed using the EMMENU, version 4.0, software (TVIPS, Gauting, Germany).
SDS-PAGE and structural protein analysis by MS.
SDS-PAGE was conducted according to Laemmli (27). To analyze CP81 structural proteins, CsCl-purified phage particles were supplemented with loading buffer (Roth, Karlsruhe, Germany), boiled for 10 min, and separated at 20 mA on a one-dimensional 15% (wt/vol) SDS gel at 15°C. Structural proteins were visualized by Coomassie brilliant blue R-250 (Bio-Rad, Hercules, CA) staining. Protein bands were excised from the gel, washed twice with 50% (vol/vol) acetonitrile in 5 mM ammonium bicarbonate for 60 min, digested with porcine trypsin (Merck, Darmstadt, Germany), and purified as described previously (24). For identification of the proteins, high-pressure liquid chromatography (HPLC) coupled to a mass spectrometer (MS) was used, and automated tandem MS (MS/MS) fragmentation was performed during the HPLC run. For the determination of peptide sequences, MS/MS spectra were obtained using a QStar XL hybrid mass spectrometer (Applied Biosystems, Foster City, California) with a nanoelectrospray source. The obtained data were submitted to the Mascot web server database (Matrix Science, Boston, MA) (35).
Isolation, sequencing, and bioinformatic analysis of CP81 DNA.
To isolate phage DNA, purified CP81 particles were disintegrated by treatment with proteinase K-SDS at 56°C for 2 h. Thereafter, the phage DNA was ethanol precipitated and washed by standard procedures (41). Library generation for the 454 FLX sequencing was carried out according to the manufacturer's standard protocols (Roche/454 Life Sciences, Branford, CT). Briefly, high-molecular-weight phage DNA was randomly sheared by nebulization into fragments ranging in size from 400 bp to 900 bp. The fragments were end polished and the 454 A and B adaptors required for the emulsion PCR and sequencing were added to the ends of the fragments by ligation. The concentration of the resulting fragment library was measured by fluorimetry and sequenced on a 1/16 PicoTiterPlate (PTP) on a Genome Sequencer FLX system using the Roche/454 titanium chemistry. A total of 60,773 sequence reads with an average read length of 350 nucleotides were obtained, yielding 22.3 Mb of sequence data. The sequence reads were assembled using the Roche/454 Newbler software at default settings (release 2.3 [091027_1459]; 454 Life Sciences Corp.). Assembly resulted in a single 132,454-bp contig comprised of 55,196 reads with average sequence coverage of more than 145 per consensus base.
Protein-encoding open reading frames (ORFs) were predicted using the algorithms of ORF-Finder (http://www.ncbi.nlm.nih.gov/) and the Glimmer, version 2.0, microbial gene-finding software combined with manual curation (12). As basic premises for the ORF analysis, the presence of ATG, TTG, or GTG as a potential start codon and a length of at least 25 encoded amino acids were stated. Gene products of the ORFs were compared to the annotated proteins of the nonredundant GenBank database BLASTp (http://www.ncbi.nlm.nih.gov/BLAST/). For gene products with low e-values (>0.001) or no sequence similarity, the PSI-BLAST algorithm was used (1). Identification of putative tRNA genes was performed using tRNAscan-SE (31). Putative Rho-independent transcription terminators were identified using TRANSTERM (5).
The CP81 genome was compared to other T4-like phages using the Georgia Gwinnett College (GGC) T4-like genome website (http://phage.ggc.edu) comprising the following phages: enterobacteria phage T4 (NC000866); Synechococcus cyanophages Syn9 and S-PM2 (NC008296 and NC006820); Prochlorococcus cyanophages P-SSM2 and P-SSM4 (NC006883 and NC006884); Vibrio phage KVP40 (NC005083); enterobacteria phages RB32, RB43, RB49, and RB69 (NC008515, NC007023, NC005066, and NC004928, respectively); Aeromonas phages Aeh1, 25, and 31 (NC005260, NC008208, and NC007022, respectively) (32–34, 36, 39, 46, 51); Shigella phage phiSboM-AG3 (NC013693); and Delftia phage phiW-14 (NC013697).
Restriction endonuclease analysis and in vitro amplification of the CP81 genome.
Restriction endonucleases were supplied by Fermentas (St. Leon-Rot, Germany). Digests were performed according to the manufacturer's recommendations and analyzed by agarose gel electrophoresis using standard procedures (41). The CP81 genome was amplified by use of the REPLI-g whole-genome amplification kit (Qiagen, Hilden, Germany) according to the supplier's recommendations.
Nucleotide sequence accession number.
The complete nucleotide sequence of the CP81 genome was submitted to EMBL under the accession number FR823450.
RESULTS
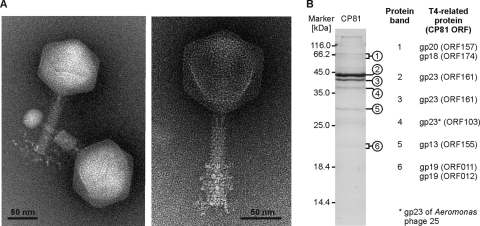
CP81 is the prototype of several very similar group III phages isolated in Germany, Denmark, and the United Kingdom (4, 22). Electron micrographs of negatively stained CP81 particles revealed virions comprised of an isometric head (diameter, 96.4 ± 2.9 nm; n = 40), a contractile tail (97.5 ± 5.47 by 21.2 ± 2.13 nm), and a baseplate with a bundle of presumably six tail fibers, which are about half as long as the tail (Fig. 1A). Single particles with contracted tails and full heads found in the phage band of the CsCl gradient mostly exhibited globular structures at the end of their tails. Binding of phages to cell wall fragments was not observed in the phage preparation. CP81 is a typical myovirus, but it does not share a morphological resemblance to typical T4-like phages. Unlike the T-evens, PseudoT-evens, and SchizoT-evens, the CP81 head is not elongated (prolate). ExoT-even cyanophages, which are more distantly related to T4, possess isometric heads, but the heads of these phages are smaller than the CP81 head while the tails of ExoT-evens are significantly longer (48). CP81 exhibited a rather narrow host range by lysing 60 out of 305 C. jejuni strains (∼20%) belonging to the Campylobacter strain collection of the Bundesinstitut für Risikobewertung (BfR).
Fig. 1.
Morphology and structural proteins of CP81. (A) Electron micrographs of negatively stained virions. The micrographs on the left and on the right show particles stained with phosphotungstate and ammonium molybdate, respectively (see Materials and Methods). The head diameter was measured between the sides parallel and diagonal to the tail. Both axes were identical within the error range. A collar structure as in T4 was not observed. (B) SDS-PAGE of structural CP81 proteins (27). As a marker, the unstained molecular mass marker of Fermentas (St. Leon-Rot, Germany) was used. Visible bands were excised from the gel and analyzed by mass spectrometry (53).
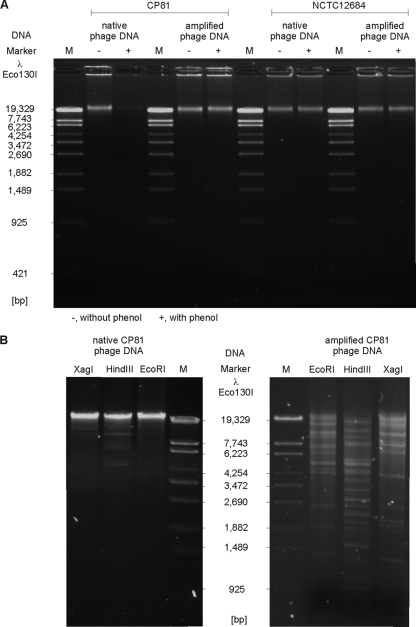
Progress in genome analysis of Campylobacter phages has so far been hampered by the recalcitrant chemical nature of the DNA of Campylobacter phages, particularly those of group III (6, 30, 40). We tested the susceptibility of CP81 DNA to a wide range of restriction enzymes (see Table S1 in the supplemental material). Most of the investigated enzymes did not cut the phage DNA at all. Inefficient cleavage might reflect the very low G+C content of 26.1% determined for CP81 by genome sequencing (see below), which is even lower than the content reported for its host C. jejuni NCTC11168 (30.6%) (38). Indeed, efficient cleavage was achieved using the enzymes DraI (TTTAAA; 455 sites), SmiI (ATTTAAAT; 51 sites), and VspI (ATTAAT; 224 sites) whose recognition sites are exclusively composed of the bases adenine and thymine (Fig. 2). Nevertheless, the CP81 genome also contains numerous theoretical recognition sites for enzymes (e.g., BclI, 32 sites), which were in reality not cleaved. As BclI recognizes the sequence TGATCA, CP81 most likely contains modified C or G bases. We tested a number of restriction endonucleases that cleave methylated DNA (CpG, Dam, Dcm, EcoBI, and EcoKI), but even these enzymes did not give clear fragment patterns (data not shown). The following observation suggests that CP81 (and probably other group III members as well) posses a novel DNA modification that is distinct from those of group II and T4-type phages.
Fig. 2.
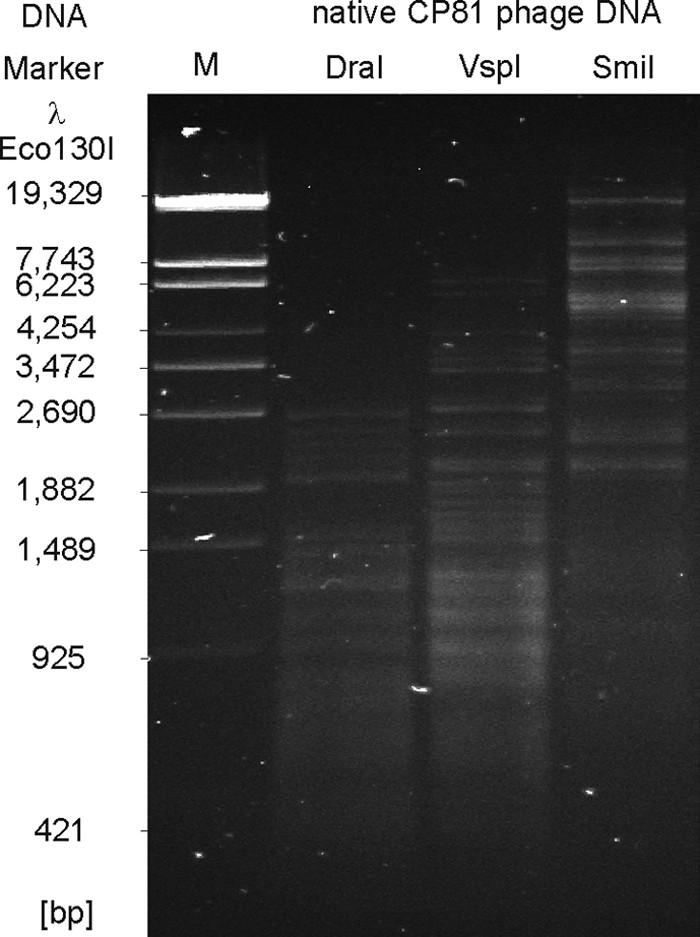
Restriction patterns of native CP81 DNA. The enzymes DraI, VspI, and SmiI recognize pure A/T sequences.
Viral DNA is routinely isolated by disintegration of purified particles with proteinase K and SDS, followed by phenol-chloroform extractions at pH 8.0. Similarly to other phages, CP81 DNA could be recovered in large amounts after treatment of particles with proteinase K-SDS and subsequent precipitation of the released DNA by ethanol. However, in contrast to, e.g., group II Campylobacter phage NCTC12684, phenol extraction of the proteinase K-SDS-treated phage did not result in the partitioning of the phage DNA into the aqueous layer above the phenol phase (Fig. 3A). Ether extractions revealed that CP81 DNA remained associated with the interphase (data not shown). Upon amplification of native CP81 DNA by use of a whole-genome amplification kit (see Materials and Methods), the phage DNA could be phenol extracted without loss (Fig. 3A). Furthermore, the amplified DNA was susceptible to cleavage by restriction enzymes (e.g., EcoRI and HindIII) whose recognition sites comprise C and G bases (Fig. 3B). Molecular cloning and sequencing of those restriction fragments confirmed the amplification of CP81 DNA (data not shown). These results indicate that native CP81 DNA contains a chemical modification of C or G bases that mediates both resistance against cleavage by most restriction endonucleases and distinct partition of the DNA between the phenol and water phase. The treatment with phenol renders the CP81 DNA less hydrophilic, a characteristic thus far not reported for other phages. We screened the CP81 genomic sequence for ORFs that might code for enzymes involved in phage DNA base modification, but the annotation did not give any clues about the nature of this modification.
Fig. 3.
Analysis of native and amplified CP81 DNA. (A) Loss of native CP81 DNA after extraction with phenol. Native DNA of group II phage NCTC12684 and the amplified DNAs of both phages were not lost after phenol extraction. (B) Susceptibility of native and amplified CP81 DNA to cleavage by restriction endonucleases whose recognition sites are composed of A/T and G/C sequences.
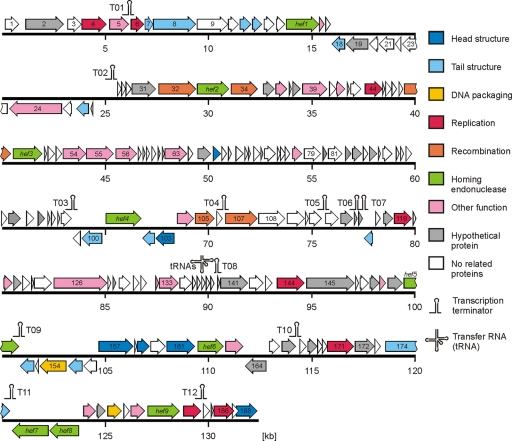
CP81 has a linear genome of 132,454 bp, of which 94% are coding sequences. Using PCR and Bal-31 exonuclease assays, it became evident that the CP81 genome is a circularly permuted molecule (data not shown). We identified 188 putative ORFs (product length of >35 amino acids), 165 on the plus strand and 23 on the minus strand (Fig. 4 and Table 1; see also Table S2 in the supplemental material). At the nucleotide level, CP81 lacks even short regions of sequence identity with T4-type phages and shows only very short regions of DNA sequence identity with the completely sequenced group II Campylobacter phages CP220 and CPt10 (see Fig. S1A in the supplemental material). Likewise, a PROMMER analysis searching for low-level amino acid identities (a match over 10 amino acids giving a point) revealed only a few scattered matches with Campylobacter phages CP220 and CPt10 and practically no matches with a phylogenetically broad range of T4-like phages (Escherichia coli phages T4, RB43, RB49, and RB69; Vibrio phage KVP40; Aeromonas phage Aeh1 and cyanophage S-PM2) (see Fig. S1B).
Fig. 4.
Map of the CP81 genome. Putative genes are colored according to the predicted functions of their products. The positions of putative transcription terminators are indicated.
Table 1.
Characteristics of the CP81 genomic sequence
| Parameter | Value |
|---|---|
| Genome size (bp) | 132,454 |
| Base content (% G+C) | 26.1 |
| Total no. of CDCs (ORFs) | 188 |
| Plus strand | 165 |
| Minus strand | 23 |
| % of genome coding for proteins | 94 |
| No. of start codons | |
| AUG | 184 |
| GUG | 2 |
| UUG | 2 |
| No. of Rho transcription terminators | 12 |
| No. of tRNA genes | 5 |
| No. of related gene products | 106 |
| No. of CP220/CPt10-related proteins (% identity) | 70 (47a [24–99]) |
| No. of T4-type phage-related proteins (% identity) | 29 (5a [23–51]) |
| No. of proteins related to other phages (% identity) | 15a (24–43) |
| No. of proteins related to Campylobacter/Helicobacter (% identity) | 13a (31–98) |
| No. of proteins related to other bacteria (% identity) | 21a (24–61) |
Number of related proteins with the best NCBI BlastP match.
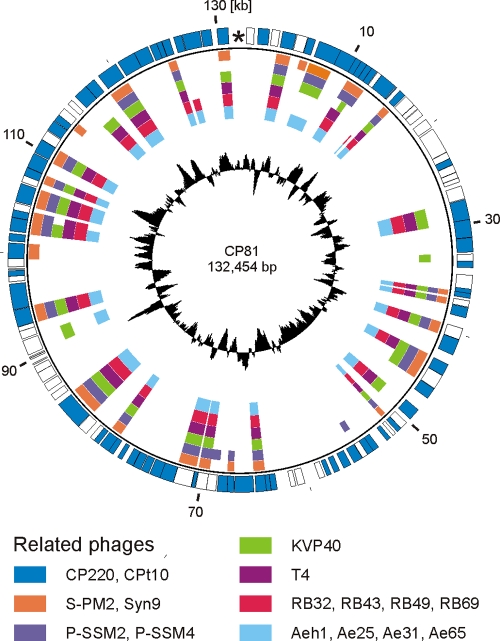
When individual CP81 proteins were investigated by BLAST searches, almost half (92) of the predicted CP81 products exhibited significant homologies (e-value of <0.001) to known proteins: 70 and 29 of the CP81 products were similar to proteins of CP220/CPt10 and T4-type phages, respectively (Fig. 5 and Table 1). With phages from the T4 superfamily, the highest overall levels of protein sequence identity (up to 43%) were found with the ExoT-even cyanophages Syn9/S-PM2 of Synechococcus and P-SSM2/P-SSM4 of Prochlorococcus (9, 32, 46, 51). Twenty-five CP81 gene products (13.3%) are at least 23% identical to T4 proteins (see Table S2 in the supplemental material). Notably, CP81 and CP220/CPt10 are similarly related to T4. The comparison of the Campylobacter phages with each other revealed identity values of T4-like proteins up to 48% while some other proteins of the Campylobacter phages are up to 99% identical (see Table S2).
Fig. 5.
Relationship of CP81 to T4-type phages. Circular illustration of the CP81 genome and similarities of predicted products to proteins of the group II phages CP220/CPt10 and to phages belonging to the T4 superfamily. The inner circle (black) shows a plot of the G+C content deviation along the CP81 genome.
The genomes of the group II Campylobacter phages CP220 (177.5 kb) and CPt10 (175.7 kb) are within the genome size range (164 to 255 kb) of typical T4 phages, while the CP81 genome (132.5 kb) lies well below the smallest described T4 genome. Thus, CP220 and CPt10 have a considerably larger genome than CP81 and encode proteins not encountered in CP81. Most prominent among them are membrane proteins, transposases, and metabolic enzymes including 12 proteins with S-adenosylmethionine (SAM) domains (49). Conversely, the CP81 genome showed an expansion of one protein family, represented by nine ORFs, that is present only in a single member of group II Campylobacter phages. The products of the nine related CP81 ORFs share 39 to 49% amino acid identity with the homing endonuclease function (HEF) protein of the T4-related phage U5 (42) (Fig. 4). Homing endonuclease genes are mobile genetic elements that can promote their own horizontal transfer by inserting into a cognate site not containing the endonuclease gene (43). They are common “molecular parasites” of T4 phage genomes (15). The CP81 HEFs are on average 53% identical to each other and similarly related (45 to 51% identical) to the CP220/CPt10 endonuclease.
In the PROMMER dot plot display, many internal repeats are scattered over the entire CP81 genome (see Fig. S2B in the supplemental material). Internal repeats within the CP81 genome were also observed at the DNA sequence level (Fig. S2A). Perfect nucleotide alignments ranged in length up to 77 bp. The majority of these matches were provided by the hef1 to hef9 genes. All hef genes shared sequence identity of >20 bp. Inverted repeats of >20 bp in length were provided by four predicted phage transcriptional terminators. A 22-bp direct repeat was seen in the putative portal vertex protein ORF157.
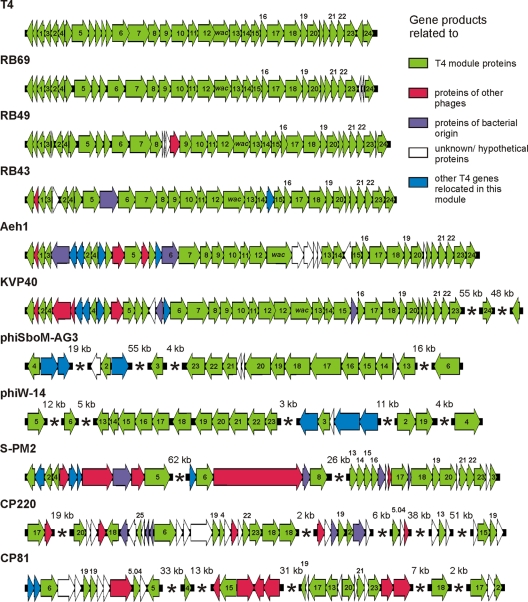
T4-like phages possess colinear genomes consisting of conserved modules (e.g., virion structural module or DNA replication module) interspersed by clusters of variable genes (10, 54). On the CP81 genome, we identified 14 genes whose predicted products share protein sequence identity with structural T4-type proteins (see Table S2 in the supplemental material). However, in strong contrast to the T4-type genome organization, the structural genes of CP81 (some of them were confirmed by mass spectrometry; see below) are not arranged in a module on the CP81 genome (Fig. 4). There is one short cluster of three genes which have an arrangement similar to that in T4-related phages: ORFs 157, 159, and 161 correspond to the portal vertex protein (gp20 of T4), prohead protease (gp21), and major capsid protein (gp23), respectively (Fig. 6). The ORFs 157 and 161 gene products were, indeed, identified by mass spectrometry as coding for virion structural proteins (Fig. 1B).
Fig. 6.
Virion structural modules of T4-type phages. The T-even, PseudoT-even, SchizoT-even, and ExoT-even subgroups are represented by T4/RB69, RB49/RB43, Aeh1/KVP40, and S-PM2, respectively (10, 14). Genes are denoted according to the nomenclature of T4 and colored on the basis of similarities of their products to other proteins.
We observed three cases where more than one CP81 gene product shared sequence relatedness with a T4-type phage protein. The first case relates to the putative major head protein of CP81. In addition to the putative major capsid protein encoded by ORF161, CP81 encodes in ORF103 a second T4 gp23-like protein that is 24% identical to gp23 of Aeromonas phage 25. The ORF103 product was identified as a minor structural protein by peptide mass fingerprinting (Fig. 1B). This situation closely resembles that of T4 phage, where the major head protein gp23 shares sequence identity with the head vertex protein gp24, also a minor structural protein of the virion (7). However, instead of being adjacent as in T4, ORF103 and ORF161 are located on opposite DNA strands. Remarkably, the two CP81 gp23 genes are accompanied by homing endonuclease genes (hef4 and hef6). In both T4 and CP81, the “duplicated” major virion genes share only low-level amino acid identity (CP81, 16%) suggesting that they more likely represent two separate gene import events than an ancient gene duplication event within the CP81 genome.
Even more conspicuous is the case of the putative tail tube protein gene (gp19 in T4), which occurs in three copies in the CP81 genome. ORF11 and ORF12 are located side by side on the plus strand near the 5′ end of the genome far away from other structural genes while ORF152 is situated 90 kb away on the opposite strand and in the vicinity of the major gp23 protein gene (ORF161) (Fig. 4). Comparison of the three gp19 proteins of CP81 showed that they are related but differ in size (ORF11, 181 aa; ORF12, 193 aa; ORF152, 244 aa). The ORF11 and ORF12 gene products are less (27%) identical to each other than to gp19 of T4-type phages (29% and 36% identity, respectively). The ORF152 product shares no significant identity with the ORF11 and ORF12 products but is 23% identical to the tail tube protein of the T4-related Synechococcus phage S-RSM4. Peptides derived from both ORF11 and ORF12 were identified by mass spectrometry in the structural proteins of CP81 virions separated by SDS-PAGE (Fig. 1B). In contrast, an ORF152 product could not be detected, suggesting that it is produced in amounts too small to obtain a band by SDS-PAGE or that ORF152 is a pseudogene. It is again striking that near the tail tube genes, homing endonuclease genes are located (hef1 and hef5).
Finally, two CP81 genes were detected whose products share amino acid identity with the head/DNA packaging T4 gp17 homologue. As the CP81 ORFs 154 and 179 match different regions of the T4 gp17 homologue, the genetic event is a splitting of a gene into two halves and not a gene duplication. Homing endonuclease genes may also play a role in this case. ORF154 and ORF179 are located several kilobases apart on opposite strands and are in the vicinity of hef genes. ORF179 is, in fact, bracketed by hef7 and hef8 on one side and by hef9 on the other side, suggesting a mechanistic role of the hef genes for the splitting of the T4 gp17 homologue in CP81. Splitting of duplicated genes was also observed in T4 phages (7).
We additionally found ORFs which encode homologues of the T4 tail completion proteins gp13 and gp14, the tail sheath stabilizer gp15, and the tail sheath protein gp18. Peptides corresponding to ORF13 and ORF18 were identified by mass spectrometry as structural proteins of CP81 (Fig. 1B). However, these tail genes plus the genes for the tail tube homologues of gp19 do not form a tail module as in T4 phages but are scattered across the CP81 genome.
Again in contrast to T4 phages where DNA replication genes form a conserved gene cluster, CP81 ORFs that encode proteins involved in DNA replication and recombination processes are dispersed over the whole CP81 genome (Fig. 4). We identified putative genes for homologues of the T4 DNA ligase gp30 (ORF171), single-strand binding protein gp32 (ORF183), DNA primase/helicase gp41 (ORF4), DNA polymerase gp43 (ORF126), sliding clamp loader gp44 (ORF44), sliding clamp gp45 (ORF6), DNA primase gp61 (ORF119), RNase H (ORF186), and RNA-DNA helicase UvsW (ORF144). This list of proteins suggests that a T4-like replisome structure is probably also produced by the Campylobacter phage. Moreover, as in T4, recombination and DNA replication might be connected processes because phage CP81 also carries key genes involved in T4-like homologous recombination, namely, ORF32 (topoisomerase II, gp39), ORF49 (RecA-like recombinase, UvsX), ORF105 (recombination endonuclease, gp47), and ORF107 (recombination protein, gp46).
The CP81 genome was screened against a database of undesired genes (DUG) described and validated in a previous safety analysis of bacteriophage genomes (13, 54). No significant hit was obtained with the CP81 sequence. Control runs with E. coli phage lambda yielded such hits, demonstrating that the CP81 sequence was not giving a false-negative result. Thus, phage CP81 is apparently free from critical genes (e.g., virulence factors or antibiotic resistance determinants) that would exclude its possible use for applications within the food chain.
DISCUSSION
The comparative genomics of T4-related bacteriophages has attracted substantial interest because (i) E. coli phage T4 is one of the most intensively investigated systems in molecular biology, (ii) T4-related phages became the focus of intensive genome sequencing efforts, and (iii) T4-related phages were found in very distant bacterial hosts like cyanobacteria, allowing a wide-ranging view into bacteriophage genome evolution (37). Researchers had already identified a set of essential genes for T4 phage multiplication while genome comparisons defined a core set of genes as a unifying feature of all T4-related phages (34). With the inclusion of many T4-like cyanophages into the T4 phage group, the core set for T4-like myoviruses from diverse hosts and environments is now stabilizing at 38 virion structural and DNA replication genes (47).
The observation of structural genes with a conserved gene order (synteny) and detectable protein sequence identity over phylogenetically distantly related bacterial hosts has puzzled virologists and was interpreted as evidence for substantial horizontal gene transfer. In contrast to the situation of lambdoid coliphages, where the structural genes define several individual modules like DNA packaging, head morphogenesis, head-to-tail linker, tail morphogenesis, and tail fiber genes, structural genes of T4-like phages frequently represent only two modules (tail fiber genes versus the rest of the morphogenesis genes). There is apparently a relatively strong resistance to the fragmentation of T4-like structural genes in evolution. The cyanophage S-PM2, for example, shares with T4 coliphage a practically uninterrupted gene cluster of structural genes corresponding to g13 to g23 of T4 phage (Fig. 6). How far the reach of T4-related phages extends is currently an open question. Molecular taxonomists using proteome comparison techniques argued that cyanophages like S-PM2 should not be classified as T4-like phages (28). However, gene content analyses maintain cyanophages in the periphery of the T4-related phage group (47). Recent sequencing work with Campylobacter group II phages (49) and the present report provide interesting new observations for the debate on the reach of T4-related phages. Group II and group III Campylobacter phages contain genes whose products share detectable sequence identity with the most conserved proteins of T4-related phages, namely, the products of structural genes next to the major head gene and DNA replication genes. However, the highly characteristic gene order of this cluster found in all previously defined T4-related phages is not maintained in CP220/CPt10 and CP81. Does this speak for several independent horizontal gene transfer events of T4-type genes into Campylobacter phages? This idea is at odds with current notions about the evolution of T4-like phages, which state that the core genome of T4-related phages is resistant to dispersal in evolution (17). How could genes that are interacting so strongly when building a phage capsid then “travel” independently in evolution? The observation of a “fragmented” virion gene cluster does not necessarily argue against a horizontal gene transfer event in Campylobacter. It may well be possible that the attrition of the T4-type synteny of structural genes occurred in Campylobacter at a time point when the essential set of interacting T4 genes had already arrived en bloc. The gene constellation of g15, g17, g18, (g19), g20, g21, and g23 in Campylobacter phages CP81 and CP220 would thus define a minimal interacting protein set for building a T4-related virion structural skeleton. It should be noted that CP81 does not represent a morphology diagnostic for T4-related phages. However, T4-like cyanophages like S-PM2 do not exhibit the characteristic prolate T4 head either (8). CP81 deviates in another respect from the standard picture of T4-related phages. T4-related phages showed up to now a minimal genome size of about 160 kb while the upper size range was less well defined and could reach up to 250 kb in vibriophages or cyanophages (37). With 132 kb, the CP81 genome size is below any other genome size of a T4-related phage. Might this smaller genome size be a consequence of the attrition of the g13 to g23 cluster in CP81? Two other recently sequenced T4-like phages from proteobacteria, phiW-14 and phiSboM-AG3, also show genome sizes below 160 kb (157.5 kb and 158 kb, respectively), but both still show a complete and perfectly conserved g13 to g23 gene set (Fig. 6). In dot plot analyses, CP81 does not share more sequence relatedness with the small-genome-sized phiW-14 and phiSboM-AG3 phages than with the normal-genome-sized T4-like phages (data not shown). Genome size reduction is thus an unlikely reason for the attrition of the T4 virion gene order. We speculate that the high number of homing endonuclease genes in the CP81 genome may be a more likely reason for the loss of synteny of g13 to g23. The selfish homing endonuclease genes might lead to highly recombinogenic regions in the phage genome by providing short stretches of sequence conservation sufficient for homologous recombination (see Fig. S2 in the supplemental material) as previously suggested for other small repeats in T4 phages (2). At the same time, the nuclease activity of this mobile element might err in sequence recognition, leading to double-stranded DNA breaks. In fact, in cyanophage endonuclease-flanked gene cassettes are located in variable locations on the phage genomes (47). The conspicuous vicinity of homing endonuclease genes next to the dispersed T4-like genes observed in phage CP81 would at least represent a bioinformatic hint to these mobile elements as motors of phage genome evolution.
Our observations challenge still other notions: T4-related phages that infect the more distant phylogenetic relatives exhibited also the highest degree of divergence from each other (37). Why, then, are the T4-like proteins of CP81 not more closely related to T4 coliphages (as the relatedness of their bacterial hosts would indicate) than to T4-like phages from cyanobacteria? This observation is difficult to explain with previously presented evidence for vertical and horizontal evolution in the T4 phage group. Apparently, even for such an intensively investigated group of bacterial viruses like T4 phages, surprises can still be found in close evolutionary relatives of E. coli. Further phage sequencing is thus likely to modify our understanding of phage evolution.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maria-Margarida Vargas for excellent technical assistance and Katharina Bratz for initial work on CP81. We are also indebted to Erich Lanka for helpful discussions and careful reading of the manuscript.
The work of J. A. Hammerl was supported by a grant of the Bundesanstalt für Landwirtschaft und Ernährung (BLE project: CAMPYQUANT).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arbiol C., Comeau A. M., Kutateladze M., Adamia R., Krisch H. M. 2010. Mobile regulatory cassettes mediate modular shuffling in T4-type phage genomes. Genome Biol. Evol. 2:140–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atterbury R. J., Connerton P. L., Dodd C. E., Rees C. E., Connerton I. F. 2003. Application of host-specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6302–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atterbury R. J., Connerton P. L., Dodd C. E., Rees C. E., Connerton I. F. 2003. Isolation and characterization of Campylobacter bacteriophages from retail poultry. Appl. Environ. Microbiol. 69:4511–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown C. M., Dalphin M. E., Stockwell P. A., Tate W. P. 1993. The translational termination signal database. Nucleic Acids Res. 21:3119–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carvalho C. M., et al. 2010. The in vivo efficacy of two administration routes of a phage cocktail to reduce numbers of Campylobacter coli and Campylobacter jejuni in chickens. BMC Microbiol. 10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chibani-Chennoufi S., Canchaya C., Bruttin A., Brüssow H. 2004. Comparative genomics of the T4-Like Escherichia coli phage JS98: implications for the evolution of T4 phages. J. Bacteriol. 186:8276–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clokie M. R., Millard A. D., Mann N. H. 2010. T4 genes in the marine ecosystem: studies of the T4-like cyanophages and their role in marine ecology. Virol. J. 7:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comeau A. M., Arbiol C., Krisch H. M. 2010. Gene network visualization and quantitative synteny analysis of more than 300 marine T4-like phage scaffolds from the GOS metagenome. Mol. Biol. Evol. 27:1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Comeau A. M., Bertrand C., Letarov A., Tétart F., Krisch H. M. 2007. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology 362:384–396 [DOI] [PubMed] [Google Scholar]
- 11. Connerton P. L., et al. 2004. Longitudinal study of Campylobacter jejuni bacteriophages and their hosts from broiler chickens. Appl. Environ. Microbiol. 70:3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Denou E., et al. 2009. T4 phages against Escherichia coli diarrhea: potential and problems. Virology 388:21–30 [DOI] [PubMed] [Google Scholar]
- 14. Desplats C., Krisch H. M. 2003. The diversity and evolution of the T4-type bacteriophages. Res. Microbiol. 154:259–267 [DOI] [PubMed] [Google Scholar]
- 15. Edgell D. R., Gibb E. A., Belfort M. 2010. Mobile DNA elements in T4 and related phages. Virol. J. 7:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. El-Shibiny A., et al. 2009. Application of a group II Campylobacter bacteriophage to reduce strains of Campylobacter jejuni and Campylobacter coli colonizing broiler chickens. J. Food Prot. 72:733–740 [DOI] [PubMed] [Google Scholar]
- 17. Filée J., Bapteste E., Susko E., Krisch H. M. 2006. A selective barrier to horizontal gene transfer in the T4-type bacteriophages that has preserved a core genome with the viral replication and structural genes. Mol. Biol. Evol. 23:1688–1696 [DOI] [PubMed] [Google Scholar]
- 18. Gaynor E. C., et al. 2004. The genome-sequenced variant of Campylobacter jejuni NCTC11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186:503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gibbens J. C., Pascoe S. J., Evans S. J., Davies R. H., Sayers A. R. 2001. A trial of biosecurity as a means to control Campylobacter infection of broiler chickens. Prev. Vet. Med. 48:85–99 [DOI] [PubMed] [Google Scholar]
- 20. Goode D., Allen V. M., Barrow P. A. 2003. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl. Environ. Microbiol. 69:5032–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gundogdu O., et al. 2007. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics 8:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hansen V. M., Rosenquist H., Baggesen D. L., Brown S., Christensen B. B. 2007. Characterization of Campylobacter phages including analysis of host range by selected Campylobacter Penner serotypes. BMC Microbiol. 7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harris J. R., Gebauer W., Markl J. 1995. Keyhole limpet haemocyanin: negative staining in the presence of trehalose. Micron 26:25–33 [Google Scholar]
- 24. Hertwig S., et al. 2003. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 331:605–622 [DOI] [PubMed] [Google Scholar]
- 25. Humphrey T. J., Jorgensen F., Mattick K. L. 2000. Fit to eat? Food scares and safe food production. Microbiol. Today 27:10–12 [Google Scholar]
- 26. Hwang S., et al. 2009. Isolation and characterization of bacteriophages specific for Campylobacter jejuni. Microbiol. Immunol. 53:559–566 [DOI] [PubMed] [Google Scholar]
- 27. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 28. Lavigne R., et al. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loc Carrillo C. M., et al. 2005. Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ. Microbiol. 71:6554–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loc Carrillo C. M., Connerton P. L., Pearson T., Connerton I. F. 2007. Free-range layer chickens as a source of Campylobacter bacteriophage. Antonie Van Leeuwenhoek 92:275–284 [DOI] [PubMed] [Google Scholar]
- 31. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of tRNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mann N. H., et al. 2005. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187:3188–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miller E. S., et al. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller E. S., et al. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567 [DOI] [PubMed] [Google Scholar]
- 36. Petrov V. M., et al. 2006. Plasticity of the gene functions for DNA replication in the T4-like phages. J. Mol. Biol. 361:46–68 [DOI] [PubMed] [Google Scholar]
- 37. Petrov V. M., Ratnayaka S., Nolan J. M., Miller E. S., Karam J. D. 2010. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol. J. 7:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poly F., Threadgill D., Stintzi A. 2004. Identification of Campylobacter jejuni ATCC 43431-specific genes by whole microbial genome comparisons. J. Bacteriol. 186:4781–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pope W. H., et al. 2007. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine Synechococcus. J. Mol. Biol. 368:966–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sails A. D., Wareing D. R., Bolton F. J., Fox A. J., Curry A. 1998. Characterisation of 16 Campylobacter jejuni and C. coli typing bacteriophages. J. Med. Microbiol. 47:123–128 [DOI] [PubMed] [Google Scholar]
- 41. Sambrook J., Russel D. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. Sandegren L., Nord D., Sjöberg B. M. 2005. SegH and Hef: two novel homing endonucleases whose genes replace the mobC and mobE genes in several T4-related phages. Nucleic Acids Res. 33:6203–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sandegren L., Sjöberg B. M. 2004. Distribution, sequence homology, and homing of group I introns among T-even-like bacteriophages: evidence for recent transfer of old introns. J. Biol. Chem. 279:22218–22227 [DOI] [PubMed] [Google Scholar]
- 44. Shane S. M. 2000. Campylobacter infection of commercial poultry. Rev. Sci. Tech. 19:376–395 [DOI] [PubMed] [Google Scholar]
- 45. Steven A. C., et al. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200:351–365 [DOI] [PubMed] [Google Scholar]
- 46. Sullivan M. B., Coleman M. L., Weigele P., Rohwer F., Chisholm S. W. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sullivan M. B., et al. 2010. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ. Microbiol. 12:3035–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tétart F., et al. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Timms A. R., et al. 2010. Evidence for a lineage of virulent bacteriophages that target Campylobacter. BMC Genomics 11:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wagenaar J. A., van Bergen M. A., Mueller M. A., Wassenaar T. M., Carlton R. M. 2005. Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet. Microbiol. 109:275–283 [DOI] [PubMed] [Google Scholar]
- 51. Weigele P. R., et al. 2007. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ. Microbiol. 9:1675–1695 [DOI] [PubMed] [Google Scholar]
- 52. Wrigley N. G. 1968. The lattice spacing of crystalline catalase as an internal standard of length in electron microscopy. J. Ultrastruct. Res. 24:454–464 [DOI] [PubMed] [Google Scholar]
- 53. Zabala B., Hammerl J. A., Espejo R. T., Hertwig S. 2009. The linear plasmid prophage Vp58.5 of Vibrio parahaemolyticus is closely related to the integrating phage VHML and constitutes a new incompatibility group of telomere phages. J. Virol. 83:9313–9320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zuber S., et al. 2007. Genome analysis of phage JS98 defines a fourth major subgroup of T4-like phages in Escherichia coli. J. Bacteriol. 189:8206–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.