Abstract
Singapore grouper iridovirus (SGIV), a major pathogen of concern for grouper aquaculture, has a double-stranded DNA (dsDNA) genome with 162 predicted open reading frames, for which a total of 62 SGIV proteins have been identified. One of these, ORF158L, bears no sequence homology to any other known protein. Knockdown of orf158L using antisense morpholino oligonucleotides resulted in a significant decrease in virus yield in grouper embryonic cells. ORF158L was observed in nuclei and virus assembly centers of virus-infected cells. This observation led us to study the structure and function of ORF158L. The crystal structure determined at 2.2-Å resolution reveals that ORF158L partially exhibits a structural resemblance to the histone binding region of antisilencing factor 1 (Asf1), a histone H3/H4 chaperon, despite the fact that there is no significant sequence identity between the two proteins. Interactions of ORF158L with the histone H3/H4 complex and H3 were demonstrated by isothermal titration calorimetry (ITC) experiments. Subsequently, the results of ITC studies on structure-based mutants of ORF158L suggested Arg67 and Ala93 were key residues for histone H3 interactions. Moreover, a combination of approaches of ORF158L knockdown and isobaric tags/mass spectrometry for relative and absolute quantifications (iTRAQ) revealed that ORF158L may be involved in both the regulation and the expression of histone H3 and H3 methylation. Our present studies suggest that ORF158L may function as a histone H3 chaperon, enabling it to control host cellular gene expression and to facilitate viral replication.
INTRODUCTION
Iridoviruses are large double-stranded DNA (dsDNA) viruses exhibiting icosahedral symmetry. Members of the genus Iridovirus infect either invertebrates or poikilothermic vertebrates (fish, amphibians, and reptiles). The family Iridoviridae contains 5 genera: Megalocytivirus, Iridovirus, Chloriridovirus, Ranavirus, and Lymphocystivirus (33). Members of the genus Ranavirus are a significant cause of disease in ectothermic animals (8). Singapore grouper iridovirus (SGIV), a member of the genus Ranavirus, was isolated from brown-spotted grouper in 1998 after it had caused severe economic losses in 1994 at marine net cage farms in Singapore (9, 27). Subsequently, the whole genome of SGIV, with 140,131 bp, was sequenced with a prediction of 162 open reading frames (ORFs) (29). Proteomic studies identified the existence of 62 SGIV proteins; 25 proteins were identified by matrix-assisted laser desorption ionization–time of flight (MALDI TOF)-TOF MS/MS (29) and 26 by one-dimensional gel electrophoresis MALDI and liquid chromatography MALDI (28), and the other 11 proteins were discovered using the isobaric tags/mass spectrometry for relative and absolute quantifications (iTRAQ) method (4). One hundred twenty-seven ORFs have been characterized at the transcriptional level and classified into three groups: immediate-early, early, and late genes (5). orf158L encodes an early novel protein (5). Sequence analysis of ORF158L did not identify any functional domains or close similarities to other proteins with known functions.
In this investigation, knockdown studies of orf158L using antisense morpholino (asMO) oligonucleotides resulted in significant decrease of virus yields in grouper embryonic cells as measured by 50% tissue culture infective dose (TCID50) assay. These results revealed the importance of ORF158L and prompted us to study its structure and functions.
The crystal structure of ORF158L was determined at 2.2-Å resolution. Although it has a novel architecture, it bears a resemblance to the histone binding region of antisilencing factor 1 (Asf1), with approximately 12% sequence identity observed between the N-terminal regions of the two proteins. Asf1 acts as a histone H3/H4 chaperon, playing roles in nucleosome assembly/disassembly in response to DNA damage and in the regulation of gene expression. In addition, Asf1 is involved in the regulation of histone H3/H4 acetylation (2) and H3 methylation (17, 21, 22).
In general, histone methylation plays critical roles in regulating transcription, maintaining genomic integrity, and contributing to epigenetic memory (2, 25). During a viral infection, virus proteins, such as herpes simplex virus (HSV) VP16, varicella-zoster virus (VZV) IE62, and HIV-1 Tat, can interact with host proteins to regulate histone modifications to initiate virus infection and viral genome replication (13, 23, 31). Studying viral control of histone modifications not only provides insights into the complexities of the virus-host interaction, but also may lead to new strategies for therapeutic intervention (19).
Immunofluorescence and protein-binding studies had confirmed that ORF158L is a histone H3 binding protein. The present structural and functional studies identified a putative histone H3 binding site on ORF158L. Subsequently, structure-based mutational studies showed that mutant Arg67Ala significantly affected histone H3 binding. In addition, the effects of orf158L knockdown studies on histone H3 expression and methylation were investigated using iTRAQ and Western blotting. Our results suggested a key role for ORF158L in histone H3 modulation.
MATERIALS AND METHODS
Cell culture.
Grouper embryonic cells (GECs) from the brown-spotted grouper Epinephelus tauvina (7) were cultured in Eagle's minimum essential medium containing a final concentration of 10% fetal bovine serum, 162 mM NaCl, 100 IU of penicillin G per ml, 0.1 mg streptomycin sulfate per ml, and 5 mM HEPES. The medium was adjusted to pH 7.4 with NaHCO3.
Morpholino antisense knockdown and titration of virus yields.
Both asMO against orf158L (asMO158; 5′-AGTTTGCTACGATGGCCCACCCCAT-3′) and control antisense morpholino (asMOControl; 5′-CCTCTTACCTCAGTTACAATTTATA-3′) were designed with online software (Oligo Design). AsMOs were delivered into GECs by electroporation using a Nucleofector apparatus (Amaxa) and the program T-027. This program is designed by the manufacturer as a general program, independent of any particular cell type. Four groups of GECs were prepared and treated differently in parallel. They were mock infected (GECs not transfected with asMO and not infected by SGIV), directly infected (GECs not transfected with asMO but infected by SGIV), and asMOControl infected and asMO158 infected (GECs first transfected with asMOControl and asMO158, respectively, followed by SGIV infection after incubation at 26°C for 40 h). SGIV infection was carried out using two different doses, a low multiplicity of infection (MOI) of 0.3 and a high MOI of 3. The cells were washed twice with phosphate-buffered saline (PBS) to remove unabsorbed virions at 2 h postinfection (p.i.) and maintained in fresh medium. Both cells and media were collected at three different times (24, 48, and 72 h p.i.) and preserved at −80°C until subsequent titration. After freezing and thawing, the cells were lysed by passing them through a 20-gauge syringe needle 5 to 10 times. Supernatants were collected after brief spins at 2,000 rpm for 1 min and serially diluted from 10−2 to 10−10 for TCID50 assay in GECs. Viral titers were calculated using the Spearman-Karber method (12). Differences between TCID50 values of asMOControl- and asMO158-infected groups or between different time points within each group were analyzed using analysis of variance (ANOVA) with a significance level where P was equal to <0.05 (SPSS version 11.5; SPSS Inc., Chicago, IL).
Immunofluorescence.
After transfection with asMOs, the cells were cultured in a Lab-Tek II CC2 Chamber Slide (Nunc). The cells were then infected by SGIV at an MOI of 3 and incubated at 26°C for 48 h prior to immunofluorescence staining. In brief, the cells were washed with PBS, fixed in 3% (wt/vol) paraformaldehyde, permeabilized with precooled 100% methanol, and subsequently washed with PBS. Samples were blocked with 1% bovine serum albumin (BSA) for 1 h and initially labeled at 37°C with the primary antibodies rabbit anti-histone H3 (ab1791; abcam) and mouse anti-ORF158L. The cells were then washed with PBS, followed by incubation with anti-rabbit Alexa Fluor 594-conjugated secondary antibody (red) and anti-mouse Alexa Fluor 488-conjugated secondary antibody (green) (Invitrogen) for 1 h. Subsequently, the samples were counterstained with Hoechst 33342 (blue) (H21492; Invitrogen) and then washed with PBS. The chambers were removed, and the respective samples were protected with the Fluorsave reagent (CalBiochem) and covered with a cover slide. Finally, the samples were observed using an Olympus Fluoview FV500.
Gene cloning, site-directed mutagenesis, protein expression, and antibody production.
The orf158L gene was PCR amplified and cloned into pET-15b (Novagen). Recombinant protein was overexpressed in Escherichia coli strain BL21(DE3) (Novagen) at 18°C overnight with 100 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The recombinant protein was first purified using an Ni-nitrilotriacetic acid (NTA) column (Qiagen) under native conditions and then by gel filtration chromatography using a Superdex 26/75 column (Amersham). Purified ORF158L was used for generation of monoclonal antibodies in mouse, crystallization, and protein-binding assays. The H3/H4 histone complex was purified from vector pET11a-H3H4 as described by English et al. (11). Histones H3 (M2507S) and H4 (M2504S) were purchased from New England BioLabs (NEB). Site-directed mutations to produce the ORF158L R67A and A93R mutants were carried out by using a protocol from Finnzymes. pET-15b plasmids containing these mutants were amplified in 25 cycles using Phusion High Fidelity DNA polymerase (Finnzymes) with designed primers (R67A 5′ primer, TTCAACGCAACCGGAGCGTACATTACTCATACG; R67A 3′ primer, CGTATGAGTAATGTACGCTCCGGTTGCGTTGAA; A93R 5′ primer. GTACCAAAAGACGCCCGCTACGCGAGAATACAC; and A93R 3′ primer, GTGTATTCTCGCGTAGCGGGCGTCTTTTGGTAC). Mutant proteins were expressed and purified using the same protocol as for the wild type.
Crystallization and structure determination.
Purified native ORF158L was concentrated to 8 mg/ml in a buffer consisting of 20 mM Tris-HCl, pH 8.5, 150 mM NaCl, 1 mM MgCl2, and 1 mM dithiothreitol (DTT). Initial crystallization screening was done under Hampton Research Screen I and II conditions (15). The protein was crystallized at 25°C using the hanging drop vapor diffusion method. The best crystals were obtained when a drop of 1 μl protein solution was mixed with 1 μl of reservoir solution containing 3% glycerol, 18% polyethylene glycol (PEG) 3350, 0.1 M Tris-HCl, pH 8.5, and 0.3 M sodium acetate (NaOAc). Similarly, the recombinant selenium methionine (SelMet) ORF158L was concentrated to 8 mg/ml in the same buffer as the native protein. Crystals of this SelMet protein were grown under essentially the same conditions as for the native ORF158L. Macroseeding was employed to obtain bigger SelMet crystals. The crystals grew to a final size of approximately 0.2 by 0.2 by 0.05 mm. Prior to data collection, both native and SelMet protein crystals were soaked in the reservoir solutions with stepwise increases in the concentrations of the cryoprotectant glycerol from 5% to 25%. The crystals were picked up in a nylon loop and frozen at 100 K in a stream of nitrogen gas. X-ray diffraction data sets were collected at the synchrotron beam line X29, National Synchrotron Light Source, Brookhaven National Laboratory. Diffraction data were processed using the program HKL2000 (26). For phase determination, the resolution range from 50 to 2.5 Å was chosen. Two out of three expected Se sites of 1 asymmetric unit were identified using the program BNP (32). BNP was further used to automatically build ∼90% of the total model. The remaining parts of the model were built manually using the program O (16). Further cycles of model building, alternating with refinement using the program CNS (1), resulted in the final model with an Rwork of 0.197 (Rfree = 0.275). The crystallographic statistics are provided in Table 1.
Table 1.
Crystallographic statistics
| Statistic | Value |
|---|---|
| Data collection statistics | |
| Space group | C2 |
| Unit cell dimensions (Å, degrees) | a = 59.98, b = 41.01, c = 52.33, α = 90, β = 95.32, γ = 90.00 |
| Data set | SAD |
| Resolution range (Å) | 50–2.2 |
| Wavelength (Å) | 0.979 |
| Observed hkl | 47,568 |
| Unique hkl | 12,640 |
| Completeness (%)d | 100 (100) |
| Redundancy | 3.8 (3.8) |
| Overall (I/σI) | 16.1 |
| Rsyma (%) | 5.3 (10.4) |
| Refinement and quality | |
| Resolution range | 50–2.2 |
| Rworkb | 0.197 |
| Rfreec | 0.275 |
| RMSD bond lengths (Å) | 0.011 |
| RMSD bond angles (degrees) | 1.386 |
| Avg B factors | |
| Main-chain atoms (no. of atoms) | 19.63 (399) |
| Side chain atoms (no. of atoms) | 21.50 (677) |
| Waters (no. of atoms) | 29.64 (69) |
| Ramachandran plot | |
| Most-favored regions (%) | 87.6 |
| Additional allowed regions (%) | 12.4 |
| Generously allowed regions (%) | 0 |
| Disallowed regions (%) | 0 |
Rsym = Σ|Ii − <I>|/Σ|Ii|, where Ii is the intensity of the ith measurement and <I> is the mean intensity for that reflection.
Rwork = Σ|Fobs − Fcalc|/Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree, as for Rwork, but for 5% of the total reflections chosen at random and omitted from refinement.
High-resolution bin (2.28 to 2.2 Å); details in parentheses.
ITC.
Isothermal titration calorimetry (ITC) experiments were performed on a VP-ITC instrument (MicroCal). The buffer consisted of 10 mM HEPES, pH 7.4, 150 mM NaCl, and 10% glycerol. The protein solutions and buffers were degassed for 20 min before titration; 300 μl of 175 μM ORF158L (wild type or the R67A and A93R mutants) or ORF102L was loaded into the syringe to titrate 2 ml of 6.7 μM histone H3 or H3/H4 in the stirred (at 310 rpm) cells. Thirty injections, 10 μl each, of ORF158L or ORF102L protein were delivered at 300-s intervals. The collected data were analyzed according to the single-binding-site model using software developed in our laboratory and implemented in Origin 7.0 (OriginLab). Heats of dilution (background heats after protein is saturated with ligand) were taken into account by introducing an additional adjustable parameter. In parallel, the binding of ORF158L to histone H4 was examined under identical conditions.
iTRAQ and Western blotting.
Four groups of cells, namely, mock-infected, directly infected, asMOcontrol-infected, and asMO158-infected cells, were prepared in parallel as mentioned above. The cells were infected at a high MOI of 5 and harvested at 48 h p.i. by centrifugation for 5 min at 500 × g and 4°C and used for iTRAQ analysis and Western blotting according to methods described previously (4). After the protein extraction, four samples of mock-infected, directly infected, and asMOcontrol- and asMO158-infected cells were labeled with four unique iTRAQ reagents, namely, 114, 115, 116, and 117, respectively (provided in the iTRAQ reagent multiplex kit; Applied Biosystems). Protein extracts were also examined by Western blotting using mouse anti-ORF158L, rabbit anti-histone H3 (ab1791; abcam), and rabbit anti-histone H3K79 (ab2886; abcam) antibody monomethylation.
Protein structure accession number.
The X-ray structure Protein Data Bank (PDB) coordinates of ORF158L were deposited in the PDB under number 3RJ2.
RESULTS
Knockdown of orf158L and its effect on virus replication.
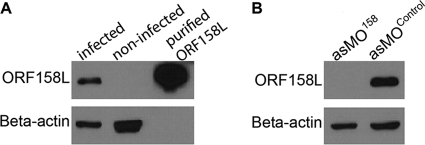
In order to determine if the predicted ORF158L is actually an encoded protein in vivo, we generated a monoclonal antibody against ORF158L using the purified recombinant ORF158L. A single band of the same size as the recombinant ORF158L was detected with the infected cell lysate, while it was not detected in the mock-infected cells (Fig. 1A). To assess the effectiveness of morpholino knockdown on ORF158L expression, both asMO158- and asMOcontrol-transfected cells were infected with SGIV and incubated for 48 h. Cells were then harvested, and the lysate was analyzed by Western blotting using the anti-ORF158L antibody. A clear band, of the same size as ORF158L, was shown in the asMOcontrol knockdown sample, but not in the asMO158 knockdown sample (Fig. 1B).
Fig. 1.
Effect of asMO158 knockdown on ORF158L expression in virus-infected cells. (A) Western blot showing the anti-ORF158L antibody is specific and can detect SGIV ORF158L protein in virus-infected cells and purified recombinant ORF158L. (B) ORF158L protein is detected in asMOControl-transfected cells but not in asMO158 knockdown.
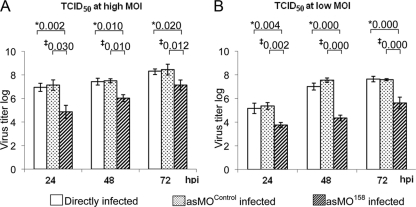
A TCID50 assay was performed for directly infected cells and asMOcontrol- and asMO158-treated cells. Although the cells were infected at a high MOI of 3 or a low MOI of 0.3, the viral titers produced in asMO158-treated cells were at least 10-fold lower than those produced in asMOcontrol-treated cells at all time points (Fig. 2). The reduction is significant (P < 0.05 by one-way ANOVA). Differences in the viral titers of directly infected and asMOcontrol-infected cells were not significant at any time point. For all SGIV-infected grouper cells, the viral titers increased with time courses of 24, 48, and 72 h p.i. For TCID50 data from the asMO158-infected sample, one-way ANOVA showed that increases in viral titers from 24 to 72 h p.i. (P = 0.020) and 48 to 72 h p.i. (P = 0.046) were significant after infection at a high MOI. In parallel, P values were 0.001 and 0.009, respectively, after infection at the low MOI. These results showed that asMO158 knockdown significantly suppressed viral replication but did not abolish viral replication completely.
Fig. 2.
Titration of virus yields after SGIV infection using the TCID50 method. There were three groups of cells: directly infected grouper embryonic cells and asMOControl-infected and asMO158-infected cells. The cells were infected with two different doses. *, the P values shown above the bars represent significant differences between directly infected and asMO158-infected cells; ‡, the P values shown above the bars represent significant differences between asMOControl-infected and asMO158-infected cells. The P values for mean comparisons in the asMO158-infected group at 24 h p.i. and 72 h p.i. and at 48 h p.i. and 72 h p.i. are 0.020 and 0.046 at a high MOI (A) and 0.001 and 0.009, respectively, at a low MOI (B). The data were taken as means of three independent experiments analyzed using one-way ANOVA (SPSS) to show significant differences (P < 0.05).The error bars indicate means ± standard deviations.
Localization of ORF158L and histone H3 proteins.
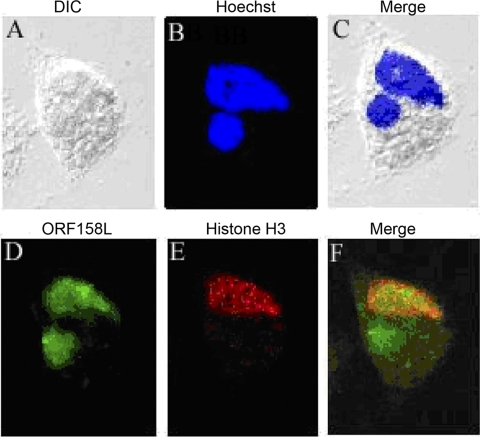
A GEC infected by SGIV under differential interference contrast (DIC) is shown in Fig. 3A. Histone H3 was detected only in the nucleus (Fig. 3E). However, ORF158L protein was localized in both the nucleus and a particular cytoplasmic area (Fig. 3D) where, interestingly, DNA could also be detected using Hoechst staining (Fig. 3B and C). This specific cytoplasmic region containing double-stranded DNA is believed to be the viral assembly site of SGIV (33). Taking the data together, ORF158L was colocalized with the host histone H3 in the nucleus (Fig. 3F). In addition, it was shown to be accumulated in the viral assembly center in the cytoplasm. These studies revealed the importance of ORF158L and thus prompted us to study its structure and function.
Fig. 3.
Colocalization of ORF158 with histone H3. (A) A cell in a DIC image. (B) DNA staining with Hoechst (blue) in the nucleus and the virus assembly center (round shape). (C) Merged image of DIC and Hoechst staining. (D) ORF158L is labeled with Alexa Fluor 488 (green). (E) Histone H3 is labeled with Alexa Fluor 594 (red). (F) Colocalization of ORF158L and histone H3 (yellow). ORF158L is accumulated in the host nucleus and viral assembly center (green area in the cytoplasm). Histone H3 and ORF158L proteins are colocalized in the nucleus.
Overall structure.
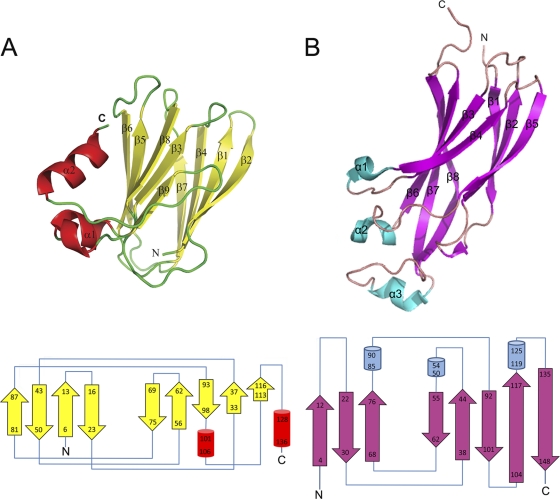
The structure of recombinant SelMet ORF158L was solved by single-wavelength anomalous dispersion (SAD) using the synchrotron data set and refined to a final R factor of 0.197 (Rfree = 0.275) at 2.2-Å resolution. The statistics for the Ramachandran plot from the PROCHECK analysis showed over 87% of nonglycine residues in the most favored region and no residues in the disallowed region (Table 1). The asymmetric unit consists of an ORF158L molecule comprising 133 residues from Ile5 to Ala137. Four residues at the N terminus and 1 residue at the C terminus were disordered and were not included in the model. The organization of ORF158L molecules in the crystal is consistent with the finding that it exists as a monomer in solution studies, such as gel filtration (data not shown). Figure 4A shows a ribbon representation of the structure of ORF158L and its topology diagram. The ORF158L molecule mainly consists of two β-sheets. The first β-sheet consists of four antiparallel β-strands, while the second consists of five antiparallel β-strands. Together, they form an elongated β-sandwich domain. In addition, there are two short α-helices on one surface of the molecule with several loops between the secondary structures.
Fig. 4.
Structure of ORF158L and comparison with Asf1. (A) (Top) Ribbon diagram of ORF158L crystal structure. β-Strands and α-helices are depicted in yellow and red, respectively. (Bottom) Topology diagram of ORF158L. β-Strands, α-helices, and connecting loops are represented by yellow arrows, red cylinders, and blue lines, respectively. (B) (Top) Ribbon representation of Asf1 structure (PDB code 1TEY). β-Strands and α-helices are depicted in magenta and cyan, respectively. (Bottom) Topology diagram of Asf1. β-Strands, α-helices, and connecting loops are represented by magenta arrows, cyan cylinders, and blue lines, respectively. In both ribbon diagrams, the N and C termini and the secondary structures, such as β-strands and α-helices, are labeled. The structure-related figures in this article were prepared using the program PYMOL (10).
Sequence and structural homology.
A BLAST search revealed that ORF158L shows no significant sequence homology with any proteins with known functions. However, it does exhibit some identity (approximately 18% maximum) with two hypothetical proteins from Ostreococcus lucimarinus and Laccaria bicolor. A search for topologically similar proteins in DALI (14), as well as in BioXGEM (6), revealed no structural homologs for the full-length ORF158L. However, several proteins, including Asf1, show partial homology with ORF158L. Based on the colocalization experiment, we found that ORF158L was colocalized with histone H3 protein in nuclei, which provided a clue to its histone binding property. The histone binding property of ORF158L might be required to regulate the host histone H3 during SGIV replication. Asf1 is a histone binding protein, and its complex structure with H3/H4 histone proteins has been reported (23). The superposition of Asf1 (PDB code 1TEY) onto ORF158L yielded a root mean square deviation (RMSD) of 3.1 Å for 72 Cα atoms (or 54%), and the two proteins share 12% sequence identity. Overall, the presence of an architecture with two β-sheets is the most conserved feature between ORF158L and Asf1 (Fig. 4; see Fig. S1 in the supplemental material). Previously, it had been shown that amino acid positions 54 and 94 of Asf1 are critical for interaction with histone 3 (23). A closer one-to-one structural comparison between Asf1 and ORF158L led us to speculate that amino acid position 67 (Arg67 of ORF158L) is the possible equivalent of amino acid position 54 of Asf1 (Asp54 of Asf1). Similarly, position 93 (Ala93 of ORF158L) is equivalent to position 94 of Asf1 (Val94 of Asf1) (Fig. 5; see Fig. S1, S2, and S3 in the supplemental material). These observations suggest that there may be an evolutionarily conserved structural and functional relationship among these proteins, while the amino acid sequences of the proteins have diverged. Further, these observations also led us to the mutagenesis and ITC experiments for the functional characterization of ORF158L.
Fig. 5.
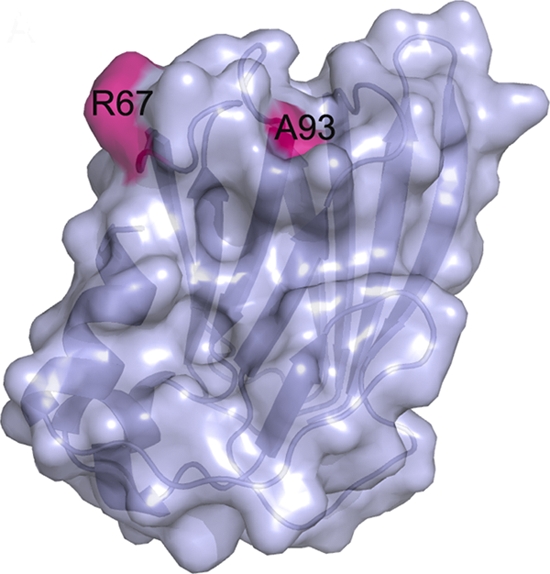
Surface representation of the ORF158L molecule. The locations of H3-interacting residues R67 and A93 are highlighted.
Interactions between ORF158L and histone H3.
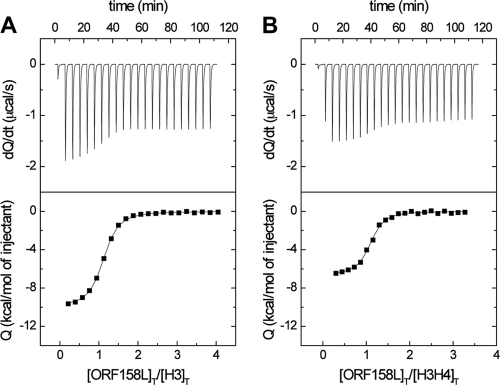
The interaction of ORF158L with histone H3 was examined by ITC. The analysis showed that binding occurred between ORF158L and histone H3 (Fig. 6) with a dissociation constant (Kd) of 0.29 μM and a large favorable change in enthalpy (ΔH) of −10.0 kcal/mol accompanied by a small unfavorable change in entropy (−TΔS) of +1.2 kcal/mol (Table 2). The experimental stoichiometry is 1:1 (n = 1.05). On the other hand, histone H4 in the stirred cells titrated against ORF158L protein showed no interaction between them (data not shown). This indicates that ORF158L binds only with histone H3.
Fig. 6.
Binding affinity measurements with isothermal titration calorimetry. (Top) Calorimetric measurements at 22°C with 30 injections of 10 μl ORF158L into the cell containing histone H3. (Bottom) Energy (kcal/mol) released during each injection. (A) Wild-type ORF158L and H3. (B) Wild-type ORF158L and H3/H4.
Table 2.
Thermodynamic parameters for ORF158L interactionsd
| Proteina | Kab (106 M−1) | Kd (μM) | ΔH (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|---|
| WT ORF158L + H3 | 3.5 ± 0.2 | 0.29 ± 0.02 | −10.0 ± 0.4 | +1.2 ± 0.4 |
| WT ORF158L + H3H4 | 5.2 ± 0.5 | 0.19 ± 0.02 | −6.7 ± 0.4 | −2.3 ± 0.4 |
| R67A ORF158L + H3 | 0.089 ± 0.02 | 11 ± 2 | −0.9 ± 0.3 | −5.8 ± 0.3 |
| A93R ORF158L + H3 | 1.2 ± 0.2 | 0.82 ± 0.1 | −8.2 ± 0.4 | −4.9 ± 0.4 |
| ORF102L + H3 | NDc |
WT, wild type.
Ka, association equilibrium constant.
ND, no interaction was detected.
Values are means ± standard deviations.
In addition, interactions of the ORF158L mutants R67A and A93R with histone H3 were investigated using ITC under the same conditions as for the wild type. These mutations were selected to change a large side chain into a small one and vice versa and to swap these side chain positions. According to the ITC experiments, the R67A mutant exhibited a weak interaction with H3, with a dissociation constant of 11 μM (a 40-fold reduction in binding affinity). However, the mutation A93R showed a mild effect on the interaction with H3 (a 3-fold reduction in affinity) (see Fig. S4 in the supplemental material). This study supports our prediction of a histone H3 binding site on ORF158L and suggests a critical role for R67 in the ORF158L-H3 interaction.
As a negative control, an ITC experiment was conducted using the SGIV ubiquitin-like protein ORF102L instead of ORF158L. No interaction was detected when it was titrated with H3 (see Fig. S4 in the supplemental material).
Knockdown of orf158L and its effects on histone H3 modulation.
iTRAQ, a quantitative proteomics experiment, was carried out to investigate the protein profile after orf158L knockdown by comparing the ratios of the same peptide sequences from different samples that were labeled by different iTRAQ reporter ions. This analysis identified significant changes of two peptides representing histone H3. Variable modifications were not included in the database search for histone proteins due to the limitations of the software. Hence, the identified peptides do not bear modifications, such as methyl and acetyl groups, according to the peptide masses (Table 3). The iTRAQ result illustrated only changes in histone H3 represented by nonmodified peptides. In this data set, variations of histone H3 quantity between directly infected cells and mock-infected cells and orf158L knockdown and control knockdown were indicated by ratios of mass ions 115/114 and 117/116, respectively. The value of the 115/114 ratio was 0.525, suggesting downregulation of histone H3 in the directly infected cells. In contrast, the average ratio of 117/116 was 1.75, indicating that histone H3 was upregulated when orf158L had been knocked down.
Table 3.
Histone H3 is differentially regulated in virus-infected cells after orf158L knockdown
| Protein name | Accession no. | Protein MW | Protein pI | Peptide count | Peptide sequence | Total ion score | Total ion score CIa (%) | Avg iTRAQ ratiob |
|
|---|---|---|---|---|---|---|---|---|---|
| 115/114 | 117/116 | ||||||||
| Histone H3 | Gi|119690002 | 15074.91 | 11.04 | 1 | EIAQBFKTDLR | 73.3 | 99.96657 | 0.524929 | 1.751760 |
| Histone H3 | gi|73761832 | 13666.76 | 11.02 | 1 | EIAQDFKTDXR | 73.3 | 99.96657 | 0.524929 | 1.751760 |
CI, confidence interval.
Protein samples from mock-infected, directly infected, and asMOcontrol- and asMO158-infected cells were labeled with iTRAQ reagents 114, 115, 116, and 117, respectively.
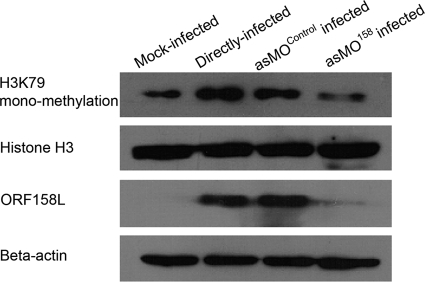
To gain insight into the posttranslational modifications, i.e., the methylation of K79 on histone H3, Western blotting was performed using a monoclonal antibody against this specific modification and revealed an increase of the methylated histone H3 in SGIV-infected cells and a decrease after asMO158 knockdown. However, the overall histone H3 level was unchanged (Fig. 7).
Fig. 7.
Effects of asMO158 knockdown on the expression of host histone H3 and K79 monomethylation of histone H3. The Western blotting result shows that K79 methylation of H3 protein is upregulated after SGIV infection and downregulated in asMO158 knockdown. The total histone H3 is unchanged after infection and knockdown.
DISCUSSION
The predicted orf158L is transcribed in the early phase of SGIV infection (5), and in this study, the protein encoded by this gene was detected for the first time in infected GECs by Western blotting. Knockdown of the orf158L translation during SGIV infection caused a significant decrease in viral replication in GECs. ORF158L was not detected in mature viral particles (28, 29), and thus, it cannot be a viral structural protein. However, the protein accumulated in large amounts at the virus assembly center in infected cells. These observations imply that ORF158L is a transient protein. It may play an important role at an early stage of viral infection, the process of virus assembly, and may also participate in the regulation of the host system to promote virus replication.
ORF158L was shown to colocalize in the nucleus with histone H3 using confocal immunofluorescence microscopy. The detection of histone H3 in the nucleus but not in the viral assembly site suggests that the host histone H3 may not be involved in the virus assembly process. However, since ORF158L was found in both the nucleus and the viral assembly site, it further suggests that ORF158L is not a virus structural protein but that it could shuffle in and out of the nucleus and that it is involved in the virus assembly process.
ORF158L has no sequence homology of any significance with any known protein. Structural comparisons suggest that ORF158L represents an unknown fold that contains a histone binding region. The pairwise structural comparison with the histone H3/H4-interacting N-terminal domain of the protein Asf1, i.e., the histone binding region of Asf1 with ORF158L, shows marked structural homology. Although the two proteins share little or no significant sequence homology, their overall histone binding regions are comparable. It has been shown that for Asf1, its mutations V94R and D54R completely abolished and reduced its histone binding property (23). Similarly, mutations on structurally equivalent residues of ORF158L, such as R67 and A93, significantly affected the binding of ORF158L with histone H3. In particular, the R67A mutant nearly abolished (a 40-fold reduction in binding affinity) the interaction with histone H3. However, the A93R mutant had reduced interactions (a 3-fold reduction in binding) with histone H3. This study indicates that R67 and A93 are key residues that mediate interactions between ORF158L and the host histone H3.
A protein-binding study confirmed that ORF158L binds to the histone complex H3/H4 and histone H3 alone with similar dissociation constants. The binding of ORF158L and histone H3 was illustrated to fit one binding site model. ITC verified that no binding exists between ORF158L and histone H4. This suggests that the binding of ORF158L with the histone H3/H4 complex was mainly due the interaction of ORF158L and histone H3. It is important to mention that, based on nuclear magnetic resonance (NMR) titration and mutational analysis of Asf1, the interaction of Asf1 with histone H3/H4 is mediated mainly by histone H3 (23). However, the X-ray structure of the Asf1-H3/H4 complex revealed that Asf1 also directly binds with histone H4 and that the binding is more extensive at the last 10 residues of H4 (11). Based on this analysis, we speculate that ORF158L may bind with histone H4 only in the presence of histone H3.
The iTRAQ experiment was performed to gain insight into how histone H3 was regulated during SGIV infection. The expression of histone H3, as identified by the non-posttranslationally modified peptide, was downregulated in SGIV-infected GECs but was found to be upregulated when orf158L was knocked down (Table 3). However, posttranslational modifications were not included in our iTRAQ database search. Histone H3 can be acetylated, phosphorylated, or methylated. Methylation mainly occurs at K4, K9, K27, K36, and K79 residues to affect gene replication, transcription, activation, and repression (19, 20). The downregulation of H3K79 methylation was detected in asMO158 knockdown cells. In contrast, the total abundance of H3 remained unchanged (Fig. 7). It is likely that the reduction of posttranslationally modified histone H3 represented by H3K79 methylation and the increase of unmodified histone H3 identified by iTRAQ could keep the total abundance of histone H3 at a constant level in asMO158 knockdown cells. To establish an infection, one of the primary tasks of viruses is to control the gene expression of host cells by regulation of the posttranslational modification of histone (3). Several viral proteins are known to participate in the regulation of cellular histone H3 methylation to initiate viral gene expression. The HSV VP16 and VZV IE62 proteins are able to interact with the host cell factor 1 to repress H3K9 methylation and to activate H3K4 trimethylation, which are pivotal for the replication of these viruses (3, 13, 18, 24). In this study, we showed that the reduction of ORF158L in SGIV-infected cells reduced H3K79 methylation. However, the mechanism by which ORF158L regulates H3 modification remains to be established.
In summary, knockdown by antisense morpholino oligonucleotides showed that ORF158L is crucial for SGIV replication. Structural studies indicate ORF158L has a novel architecture at the conserved histone binding region. Moreover, binding studies demonstrated that ORF158L interacts with histone H3. iTRAQ and Western blotting analyses suggest that ORF158L may be involved in regulation of the expression and posttranslational modification of histone H3. Our results show that ORF158L could be a viral gene-encoded histone chaperon protein. Future experiments will be directed at exploring how ORF158L interacts with histone H3 and regulates the host gene expression to facilitate SGIV replication.
Supplementary Material
ACKNOWLEDGMENTS
We thank Li Zhengjun for molecular biological advice, Lin Qingsong and Lim Teck Kwang for iTRAQ techniques and discussion, and Shashikant Joshi for suggestions.
J.S and his team were supported by the Academic Research Fund, NUS (R154000438112). This work was supported financially by Academic Research Fund Functional Genomic Studies of Singapore Grouper Iridovirus grants (R-154-000-387-112 and R-714-014-007-271) from MOE and the Mechanobiology Institute, Singapore, to C.L.H.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Brünger A. T., et al. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905–921 [DOI] [PubMed] [Google Scholar]
- 2. Campos E. I., Reinberg D. 2009. Histones: annotating chromatin. Annu. Rev. Genet. 43:559–599 [DOI] [PubMed] [Google Scholar]
- 3. Caron C., Col E., Khochbin S. 2003. The viral control of cellular acetylation signaling. BioEssays 25:58–65 [DOI] [PubMed] [Google Scholar]
- 4. Chen L. M., et al. 2008. iTRAQ analysis of Singapore grouper iridovirus infection in a grouper embryonic cell line. J. Gen. Virol. 89:2869–2876 [DOI] [PubMed] [Google Scholar]
- 5. Chen L. M., Wang F., Song W., Hew C. L. 2006. Temporal and differential gene expression of Singapore grouper iridovirus. J. Gen. Virol. 87:2907–2915 [DOI] [PubMed] [Google Scholar]
- 6. Chen Y. C., Lo Y. S., Hsu W. C., Yang J. M. 2007. 3D-partner: a web server to infer interacting partners and binding models. Nucleic Acids Res. 35:W561-W-567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chew-Lim M., et al. 1994. Grouper cell line for propagating grouper viruses. Singap. J. Prim. Ind. 22:113–116 [Google Scholar]
- 8. Chinchar V. G. 2002. Ranaviruses (family Iridoviridae): emerging cold-blooded killers. Arch. Virol. 147:447–470 [DOI] [PubMed] [Google Scholar]
- 9. Chua F. H. C., Ng M. L., Ng K. L., Loo J. J., Wee J. Y. 1994. Investigation of out-breaks of a novel disease, ‘sleepy grouper disease’, affecting the brown-spotted grouper, Epinephelus tauvina Forskal. J. Fish Dis. 17:417–427 [Google Scholar]
- 10. DeLano W. L. 2008. The PyMOL molecular graphic system. Scientific LLC, Palo Alto, CA [Google Scholar]
- 11. English C. M., Adkins M. W., Carson J. J., Churchill M. E., Tyler J. K. 2006. Structural basis for the histone chaperone activity of Asf1. Cell 127:495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hamilton M. A., Russo R. C., Thurston R. V. 1977. Trimmed Spearman-Karber method for estimating median lethal concentrations in toxicity bioassays. Environ. Sci. Technol. 11:714–719 [Google Scholar]
- 13. Hancock M. H., Cliffe A. R., Knipe D. M., Smiley J. R. 2010. Herpes simplex virus VP16, but not ICP0, is required to reduce histone occupancy and enhance histone acetylation on viral genomes in U2OS osteosarcoma cells. J. Virol. 84:1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holm L., Sander C. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20:478–480 [DOI] [PubMed] [Google Scholar]
- 15. Jancarik J., Kim S. H. 1991. Sparse matrix sampling: a screening method for crystallisation of proteins. J. Appl. Crystallogr. 244:409–411 [Google Scholar]
- 16. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110–119 [DOI] [PubMed] [Google Scholar]
- 17. Kim H. J., Seol J. H., Cho E. J. 2009. Potential role of the histone chaperone, CAF-1, in transcription. BMB Rep. 42:227–231 [DOI] [PubMed] [Google Scholar]
- 18. Kristie T. M., Liang Y., Vogel J. L. 2010. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim. Biophys. Acta 1799:257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lachner M., O'Sullivan R. J., Jenuwein T. 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116:2117–2124 [DOI] [PubMed] [Google Scholar]
- 20. Li B., Carey M., Workman J. L. 2007. The role of chromatin during transcription. Cell 128:707–719 [DOI] [PubMed] [Google Scholar]
- 21. Lin L. J., Minard L. V., Johnston G. C., Singer R. A., Schultz M. C. 2010. Asf1 can promote trimethylation of H3 K36 by Set2. Mol. Cell. Biol. 30:1116–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moshkin Y. M., et al. 2009. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol. Cell 35:782–793 [DOI] [PubMed] [Google Scholar]
- 23. Mousson F., et al. 2005. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc. Natl. Acad. Sci. U. S. A. 102:5975–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Narayanan A., Ruyechan W. T., Kristie T. M. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 104:10835–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ng S. S., Yue W. W., Oppermann U., Klose R. J. 2009. Dynamic protein methylation in chromatin biology. Cell. Mol. Life Sci. 66:407–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Emzymol, 276:307–326 [DOI] [PubMed] [Google Scholar]
- 27. Qin Q. W., et al. 2001. Electron microscopic observations of a marine fish iridovirus isolated from brown-spotted grouper, Epinephelus tauvina. J. Virol. Methods 98:17–24 [DOI] [PubMed] [Google Scholar]
- 28. Song W., Lin Q., Joshi S. B., Lim T. K., Hew C. L. 2006. Proteomic studies of the Singapore grouper iridovirus. Mol. Cell Proteomics 5:256–264 [DOI] [PubMed] [Google Scholar]
- 29. Song W. J., et al. 2004. Functional genomics analysis of Singapore grouper iridovirus: complete sequence determination and proteomic analysis. J. Virol. 78:12576–12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. St Germain C., O'Brien A., Dimitroulakos J. 2010. Activating Transcription Factor 3 regulates in part the enhanced tumour cell cytotoxicity of the histone deacetylase inhibitor M344 and cisplatin in combination. Cancer Cell Int. 10:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Duyne R., et al. 2008. Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weeks C. M., Miller R. 1999. Optimizing Shake-and-Bake for proteins. Acta Crystallogr. D 55:492–500 [DOI] [PubMed] [Google Scholar]
- 33. Williams T., Barbosa-Solomieu V., Chinchar V. G. 2005. A decade of advances in iridovirus research. Adv. Virus Res. 65:173–248 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.