Abstract
Adenosine deaminase acting on RNA 1 (ADAR1) is a double-stranded RNA binding protein and RNA-editing enzyme that modifies cellular and viral RNAs, including coding and noncoding RNAs. This interferon (IFN)-induced protein was expected to have an antiviral role, but recent studies have demonstrated that it promotes the replication of many RNA viruses. The data from these experiments show that ADAR1 directly enhances replication of hepatitis delta virus, human immunodeficiency virus type 1, vesicular stomatitis virus, and measles virus. The proviral activity of ADAR1 occurs through two mechanisms: RNA editing and inhibition of RNA-activated protein kinase (PKR). While these pathways have been found independently, the two mechanisms can act in concert to increase viral replication and contribute to viral pathogenesis. This novel type of proviral regulation by an IFN-induced protein, combined with some antiviral effects of hyperediting, sheds new light on the importance of ADAR1 during viral infection and transforms our overall understanding of the innate immune response.
INTRODUCTION
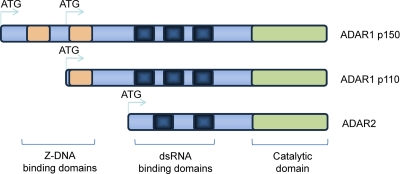
The adenosine deaminases acting on RNA (ADARs) are double-stranded RNA (dsRNA) binding enzymes that catalyze RNA editing of cellular and viral dsRNAs by deamination, which converts adenosines into inosines (6, 22, 54). Inosine is recognized as a guanosine, and thereby deamination alters the sequence- or structure-specific recognition of RNAs, their translation, and, consequently, the amino acid sequences of several proteins. This process also affects noncoding RNA, and the modification of microRNA (miRNA) sequences is very important in the RNA interference (RNAi) pathway that regulates posttranscriptional gene expression (35, 53, 54). In vertebrate cells, there are three genes that code for the ADAR1, ADAR2, and ADAR3 proteins. The mammalian Adar1 gene encodes two forms of the ADAR1 protein: the interferon (IFN)-inducible ∼150-kDa form (p150) found in both the cytoplasm and the nucleus and the constitutively expressed ∼110-kDa form (p110) found only in the nucleus (40, 90). These two forms are generated through alternative promoters (one of which is IFN inducible) and alternative splicing of exon I (27). Both forms are active deaminases with a C-terminal catalytic deaminase domain, three centrally located dsRNA binding domains (dsRBDs), and either one (p110) or two (p150) Z-DNA binding domains (Z-DBDs) at the N terminus (33, 34) (Fig. 1). ADAR2 has two dsRBDs and a deaminase domain, which mediates the RNA-editing activity. ADAR3 has a similar structure and is expressed specifically in brain cells, but its deaminase activity has not been demonstrated (54).
Fig. 1.
Schematic representation of ADAR1 p150, ADAR1 p110, and ADAR2. ADAR1 p150 has two Z-DBDs, three dsRNA binding domains, and a catalytic domain. ADAR1 p110 is generated through an alternative promoter and alternative splicing of exon I and is lacking the first Z-DBD. ADAR2 has two dsRNA binding domains and a catalytic domain. Blue arrows indicate the translation initiation sites.
Modifications by ADAR enzymes in coding and noncoding dsRNAs have the potential to affect several biological processes (19, 35, 46). ADAR-edited transcripts are produced mainly in the central nervous systems of mammals, Drosophila, and squids, where they increase the diversity of proteins produced from a single gene (59, 61, 72). A large, ever-growing number of introns, untranslated regions, and miRNAs have been shown to be edited by ADARs, which suggests a wide role in posttranscriptional processes (35, 53, 54). ADAR1 is essential to embryogenesis, as shown by the early lethality of embryos with a defect in hematopoiesis in the fetal liver and stress-induced apoptosis (82, 83). Furthermore, ADAR1 is an essential regulator of hematopoietic stem cell maintenance and a suppressor of IFN signaling via downregulation of IFN-inducible transcripts. This role likely protects organisms from the excessive IFN activation associated with autoimmune disorders, cancer, and chronic inflammation (30).
RNA-editing patterns characteristic of ADAR enzymes have also been observed in several viral RNAs, including those of hepatitis delta virus (HDV) (10, 45), human immunodeficiency virus type 1 (HIV-1) (17, 62), human respiratory syncytial virus (48), influenza virus (74, 78), Epstein-Barr virus (EBV) (36), Rift Valley fever virus (75), measles virus (MV) (2, 12, 13, 75), lymphocytic choriomeningitis virus (LCMV) (92), polyomavirus (42), and Drosophila sigma virus (9). In addition, ADAR-type sequence changes have been observed in hepatitis C virus (HCV) (76), human herpesvirus 8 (HHV8) (23), mammalian parainfluenza virus 3 (50), avian leukosis virus (28), and avian Rous-associated virus type 1 (20), providing indirect evidence that ADARs likely influence the replication of these viruses by RNA editing (Table 1).
Table 1.
Viruses affected or likely affected by ADAR activity
| Virus | Type | Host(s) | ADAR activitya | ADAR effectb | Reference(s) |
|---|---|---|---|---|---|
| Avian leukosis virus | RNA | Birds | RNA editing, possibly due to ADAR | ND | 28 |
| Epstein-Barr virus | DNA | Humans | RNA editing | Proviral via editing of viral miRNAs | 36 |
| Hepatitis C virus (replicon) | RNA | Humans | RNA editing, possibly due to ADAR | Antiviral | 76 |
| Hepatitis delta virus | Satellite RNA | Humans | RNA editing | Optimalc | 10, 45 |
| Human herpesvirus 8 | DNA | Humans | RNA editing | Proviral via editing of viral miRNAs | 23 |
| Human immunodeficiency virus type 1 | RNA | Humans | RNA editing; PKR inhibition | Proviral | 14, 17, 18, 62 |
| Human respiratory syncytial virus | RNA | Humans | RNA editing, possibly due to ADAR | Proviral through antibody escape | 48, 67 |
| Influenza virus | RNA | Mammals, birds | RNA editing; NS1-ADAR1 binding | ND | 51, 74, 78 |
| Lymphocytic choriomeningitis virus | RNA | Mammals | RNA editing, possibly due to ADAR | Possibly antiviral via loss of protein function in heterologous system | 92 |
| Measles virus | RNA | Humans | PKR inhibition; RNA editing | Proviral; possibly antiviral via hyperediting and persistence in brain | 12, 13, 60, 75, 80 |
| Parainfluenza virus 3 | RNA | Mammals | RNA editing, possibly due to ADAR | ND | 50 |
| Polyomavirus | DNA | Mammals | Antisense RNA editing, possibly due to ADAR | Possibly antiviral via RNA nuclear retention | 42 |
| Rift Valley fever virus | RNA | Mammals | RNA editing | ND | 75 |
| Rous-associated virus type 1 | RNA | Birds | RNA editing, possibly due to ADAR | None on replication; increased transforming activity | 20 |
| Sigma virus | RNA | Drosophila | RNA editing, possibly due to ADAR | None detected | 9 |
| Vesicular stomatitis virus | RNA | Mammals | PKR inhibition | Proviral | 43, 52 |
NS1, nonstructural protein 1.
ND, not determined.
Both antiviral and proviral effects of ADAR result in optimal replication.
As an IFN-induced protein, ADAR1 p150 is expected to be part of the cell response against viral infection and to block viral replication (29, 64, 70). While this antiviral activity seems likely with the HCV replicon, LCMV, and polyomavirus, evidence of wide involvement of ADAR1 as part of the innate immune response against virus infections is lacking. In contrast, ADAR1 interferes with DNA sensors, allowing downregulation of innate immune responses that in excess could be harmful to the host (85). Furthermore, an increasing number of studies have demonstrated that ADAR1 enhances the replication of several RNA viruses (Table 1). Such effects are seen as part of the HDV replication cycle for optimal production of the satellite viral particles (38, 39) and were recently observed with HIV-1 (14, 17, 62), MV (80), and vesicular stomatitis virus (VSV) (43, 52). In the examples described below, experiments clearly demonstrate that ADAR1 is responsible for the enhancement of viral replication via two mechanisms: RNA editing and inhibition of the dsRNA-activated protein kinase (PKR).
ADAR1 ENHANCES THE REPLICATION OF RNA VIRUSES VIA RNA EDITING
RNA editing, possibly due to ADARs, is found in a number of viruses but has been well analyzed for only a few of them. Several lines of evidence clearly show that ADAR1 has direct proviral effects via RNA editing: ADAR1 is required for optimal HDV replication (38, 39), while ADAR1 and ADAR2 promote the production of HIV-1 virions (17, 18, 62).
Hepatitis delta virus.
HDV is a subviral satellite of hepatitis B virus (HBV) that increases the severity of HBV-related disease (66). The RNA genome of HDV is a single-stranded (ss) circular molecule that can resemble an imperfect dsRNA with short (∼15-bp) double-stranded regions interspersed with numerous mismatches, bulges, and internal loops. The hepatitis delta virus antigen (HDAg) is the sole HDV protein. HDV produces two forms of HDAg (HDAg-S and HDAg-L) that have distinct roles in the viral replication cycle. The HDV replication cycle begins with the production of HDAg-S, which is encoded by the infectious viral genome and supports viral RNA replication (77). The large form is subsequently produced by RNA editing and the specific deamination of adenosine 1012 in the antigenome. The edited adenosine is within the amber stop codon (UAG) that terminates HDAg-S synthesis. After replication and transcription, deamination of this adenosine to inosine leads to the production of an mRNA in which the UAG amber termination codon has been changed to UGG (tryptophan [W]), thereby extending the open reading frame to encode HDAg-L (11, 45). Because of the codon change produced by this editing event, the editing site is called the amber/W site.
HDV-editing activity is accomplished by ADAR1 p110 in the nuclei of HuH-7 hepatoma cells, and the secondary structure of HDV RNA determines the level of editing (10, 44, 63, 88). Experiments using knockdown or overexpression of ADAR1 were conducted to determine to what extent the editing activity is essential for viral RNA replication. Short interfering RNA (siRNA)-mediated knockdown of ADAR1 expression led to decreased HDV amber/W editing and HDAg-L expression, as well as to reduced virus production (39). When ADAR1 or ADAR2 was overexpressed, the amber/W site hyperediting induced excessive production of HDAg-L and extensive non-amber/W site editing with a concomitant production of dominant negative HDAg variants (38). In addition, IFN treatment of HuH-7 cells induced the production of ADAR1 p150, which was able to edit the amber/W site and further increase HDAg-L production (31). These changes were detrimental to viral RNA replication, indicating that ADAR1 plays an essential role in optimal HDV production via RNA editing.
Human immunodeficiency virus type 1.
HIV-1 is a retrovirus that causes AIDS and harbors extensive sequence variations, primarily due to recombination and to the lack of fidelity of its reverse transcriptase (49, 55, 81). The observation that ADAR1 mRNA and protein expression increases during HIV infection of primary lymphocytes or lymphocytic cell lines suggests a possible function of ADAR1 during HIV-1 replication (14, 62). Two studies have investigated the role of the editing function of ADAR1 in HIV-1 expression and replication (17, 62). In both studies, overexpression of ADAR1 caused increased virion production of up to 6-fold in HEK 293T cells (17) and between 2- to 10-fold in COS7 cells (62), as measured by HIV-1 p24 in the cell culture supernatants. In addition, siRNAs that reduce the endogenous levels of ADAR1 in HEK 293T cells were also shown to reduce the level of viral protein production (14, 62), further supporting the proviral activity of ADAR1. To determine which role the editing function of ADAR1 plays in this enhancement, the two studies used catalytically inactive mutants but reached different conclusions. In one study, both overexpressed ADAR1, and ADAR1-editing-negative mutant E955A upregulated HIV-1 protein production and increased virion production in HEK 293T cells (17), while in the second study, an editing-negative mutant with mutations in positions 910 to 912 did not enhance virion production in COS7 cells (62). Both mutants were confirmed to lack editing activity, raising the possibility that the discrepancy may be due to differences in cell type.
Edited adenosines in the HIV-1 RNA were found in the 5′ untranslated region and the Tat and Rev sequences and in the vicinity of Rev responsive element (RRE) RNA within the Env gene (17, 62). To evaluate whether ADAR-mediated editing could affect viral replication, an HIV-1 mutant with a 3-nucleotide edited sequence downstream of the RRE was constructed to mimic the naturally edited RNA. This mutant showed enhanced expression compared to that of the wild-type, confirming that the altered sequence increases viral replication (62). In terms of viral infectivity, both virion release and infectivity increased significantly with overexpression of ADAR1, while the catalytic domain mutant did not show this increase, indicating that these properties are regulated by the RNA-editing ability of ADAR1 (17). Interestingly, a recent study showed that ADAR2 also edits HIV-1 RNA and increases viral production and virion release, but in contrast to ADAR1, ADAR2 does not increase the infectious potential of the virus (18). Overall, ADAR1 and ADAR2 appear to enhance HIV-1 replication through an RNA-editing mechanism. In addition, the differences in results suggest that ADAR1 may use an additional mechanism to promote HIV-1 replication.
ADAR1 ENHANCES THE REPLICATION OF RNA VIRUSES VIA PKR INHIBITION
PKR, an IFN-inducible kinase, plays an important role in antiviral immunity, apoptosis, cell proliferation, and stress signaling (24, 25, 68). It is composed of two N-terminal dsRBDs and a C-terminal catalytic kinase domain. Binding of 30- to 50-bp or longer dsRNA or structured ssRNA, such as viral RNA produced during replication, can mediate PKR autophosphorylation and activation. PKR then catalyzes the phosphorylation of the protein synthesis initiation factor eIF2α, leading to inhibition of translation. Recent data show that ADAR1 exerts a proviral activity by binding to PKR via its first dsRBD, followed by inhibition of PKR and eIF2α phosphorylation (14, 52, 84). This effect was demonstrated on MV, VSV, and HIV-1 (14, 17, 43, 52, 80). The mechanism occurs through ADAR1 binding to PKR.
Vesicular stomatitis virus.
VSV is a member of the Rhabdoviridae family with a negative-sense RNA genome that replicates in the cytoplasm of infected cells. VSV is essentially asymptomatic for humans; however, cattle, horses, and pigs show lesions in the mucous membranes of the mouth and nose, while the virus is neuropathic in mice (87). VSV can exploit defects in the translational regulation of cancer cells, allowing its use as an oncolytic virus (5).
VSV infection is lethal for mice lacking PKR (PKR−/−), and PKR−/− murine embryo fibroblasts (MEFs) are more permissive to VSV infection than wild-type fibroblasts (3, 73). The effect of ADAR1 on this virus was assayed in murine NIH 3T3, GP+E86, and wild-type MEF cells, all expressing PKR (52). These cells were more susceptible to VSV infection when they stably expressed ADAR1 p150, as shown by 11-, 32- and 66-fold-higher viral titers in NIH 3T3, GP+E86, and MEF cells, respectively (52). VSV susceptibility was 90-fold higher for MEF PKR−/− cells than for their PKR+/+ counterparts, but this increased susceptibility was not increased further by ADAR1, strongly suggesting that this effect is dependent upon PKR expression (52). Variants and mutants of ADAR1 assayed in MEF cells showed that variants missing the Z-DBDs and/or a dsRBD did not increase the level of VSV infection relative to the increase shown with the wild-type ADAR1, whereas a C-terminally-truncated protein in the deaminase domain stimulated VSV infection 60-fold, indicating that this proviral function is independent of RNA editing (52). In contrast, when ADAR1 was stably overexpressed in human HEK 293 cells, VSV growth did not change significantly, suggesting host differences in susceptibility (43). To study the effect of decreased ADAR1 expression on VSV titer, two types of studies were performed. First, transient silencing of endogenous ADAR1 with siRNA in VSV-infected HEK 293T cells reduced the viral titer 8-fold compared to that for the control, making these cells more resistant to VSV infection (52). Second, HeLa cell lines in which ADAR1 expression was stably knocked down by RNAi (ADAR1kd) were constructed (80) and used to measure VSV titer (43). ADAR1kd cells expressed less than 15% of the amount of ADAR1 p110 and less than 10% of the amount of ADAR1 p150 expressed by the parental cell lines (43, 80). When these cells were treated with beta interferon (IFN-β), the knockdown of ADAR1 decreased the VSV titer about 10-fold more than IFN inhibition but had no effect on nontreated cells (43). While the knockdown of ADAR1 had no effect on the total levels of PKR, it increased PKR and eIF2α phosphorylation in cells treated with IFN (43). Overall, both studies showed an enhancement of VSV replication mediated by ADAR1 that was independent of its RNA-editing activity and dependent on its inhibitory effect on PKR activation.
Measles virus.
MV is a negative-stranded RNA virus that is a member of the Paramyxoviridae family. It replicates in the cytoplasm of the host cell and causes an acute respiratory illness in humans (65, 89). A persistent infection of the central nervous system can also occur in rare cases, leading to a fatal neurological disease known as subacute sclerosing panencephalitis (SSPE), which is correlated with an ADAR-mediated hyperediting activity in MV sequences (2, 13, 37). The MV vaccine (MVvac) strains are devoid of SSPE consequences and show selectivity against tumor cells, which indicates their therapeutic potential as oncolytic viruses (21).
To study the influence of ADAR1 on the growth of MV, HeLa ADAR1kd cells were used (80). These cells and ADAR1-sufficient HeLa cells were infected with a modified MVvac carrying the green fluorescent protein (GFP) gene. While there was a high level of GFP reporter signals present in the parental ADAR1-sufficient cell line, the GFP signal from an MV virus deficient for the C gene, a virulence factor, was 30-fold lower in HeLa ADAR1kd cells, suggesting that ADAR1 enhances the replication of the virus (80). MV is a cytolytic virus that induces cell death by apoptosis mediated by PKR activation (79). In HeLa ADAR1kd cells, apoptosis induced by the wild-type virus and MV deficient for the V gene, another virulence factor, was enhanced by 30% and 40%, respectively. Consistent with these observations, PKR phosphorylation was increased after MV infection with the three MV variants. In contrast, very low PKR activation levels were detected following MV infection of HeLa ADAR1/PKR-sufficient cells (80). Activation of PKR in infected HeLa ADAR1kd cells also correlated with increased phosphorylation of eIF2α and interferon regulatory factor 3 (IRF-3). These observations point out an antiapoptotic and proviral role for ADAR1 in MV infection through the suppression of dsRNA-dependent pathways such as PKR and IRF-3.
HIV-1.
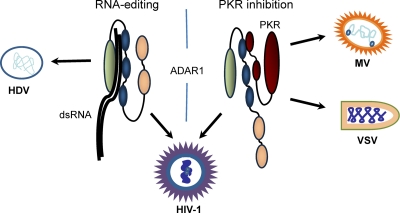
In addition to the effect of ADAR1 on HIV-1 replication via RNA editing, an effect via PKR inhibition was also characterized. In lymphocytic Jurkat T cells infected with HIV-1, PKR was transiently activated, but this activation was reversed when virus production was high enough to be visible by immunoblotting against the capsid protein (14). This reduction in PKR phosphorylation also correlated with an increase in the ADAR1 p150 and p110 forms in the cells. In addition, PKR-ADAR1 interactions dramatically increased at the peak of HIV-1 production compared to those in noninfected cells (14). The overexpression of ADAR1 in HEK 293T cells increased HIV-1 production 2- to 8-fold in the absence of exogenous PKR and completely rescued PKR-induced inhibition of HIV-1 production. This increased viral production correlated with decreased phosphorylation of PKR and eIF2α, and ADAR1 counteracted the inhibitory effect of PKR (14, 17). A similar, but less pronounced, effect on eIF2α phosphorylation was also observed with ADAR2 (18). ADAR1 proteins mutated or deleted in the catalytic deaminase domain were still able to increase HIV-1 production, whereas the absence of the dsRBDs was deleterious for this effect, showing that this activity was independent of RNA editing (14, 17). Furthermore, in astrocytic cells that inadequately support HIV-1 replication due to a heightened PKR response, ADAR1 increased HIV-1 production in a manner similar to that of another PKR inhibitor, the TAR RNA binding protein (TRBP) (14, 56). These results strongly suggest that ADAR1 proviral activity on HIV-1 is also mediated in large part by PKR inhibition (Fig. 2).
Fig. 2.
ADAR1 enhancement of the replication of RNA viruses via RNA editing and inhibition of PKR. (Left) ADAR1 enhances the replication of HDV and HIV-1 via RNA editing. (Right) ADAR1 enhances the replication of MV, VSV, and HIV-1 via the inhibition of PKR.
PKR inhibition by ADAR1 in the context of these viruses raised a question about the mechanism of interaction between these two proteins. Immunoprecipitation assays with ADAR1 and PKR in the presence of RNase A and V1 showed a direct protein-protein interaction only when the first dsRBD of ADAR1 was present (52). Furthermore, a two-hybrid screen using PKR as bait isolated ADAR1 domains, all of which had the first dsRBD, thereby confirming the interaction and a likely inhibition through this domain (14).
CONCLUSIONS
ADAR1 has a new role in enhancing viral replication.
Because ADAR1 is synthesized from an IFN-stimulated gene, it would appear obvious that the protein should contribute to the cell innate immune pathway that would react against virus replication (29, 64, 70). In addition, the similarity in function with APOBEC3G, a DNA cytidine deaminase acting against HIV-1 in the absence of the Vif protein, would further suggest an antiviral role (1, 74). Although this hypothesis may be true for some viruses, in the case of the RNA viruses described here, HDV, HIV-1, VSV, and MV, it has now been clearly shown that ADAR1 enhances viral replication via RNA editing and inhibition of PKR (Table 1 and Fig. 2). For HIV-1, both mechanisms act in concert to increase viral replication (14, 17, 62). Because of the biological significance of ADARs in editing cellular miRNAs and by inhibiting RNAi function independently of editing, ADARs could also have an impact on viral replication by editing and modifying the function of viral miRNAs (32, 35, 53, 54, 71, 91). Indeed, this function has been shown to enhance the replication of Epstein-Barr virus and human herpesvirus 8, two herpesviruses, but has not been demonstrated thus far for RNA viruses (23, 36). Considering that HIV-1 carries viral siRNAs (7, 41, 58) and that VSV replication is downregulated by cellular miRNAs (57), further research may reveal if ADAR1 can edit them and modify their function.
ADAR1 enhances or prevents viral replication in various cellular contexts.
Because every biological function is the result of a balance among different activities, the above findings can be complemented by observations that for some viruses, ADAR1 could also have dual roles, both proviral and antiviral (69). Indeed, viral RNA editing by ADARs increases sequence diversity that might be favorably selected in a specific cellular context but might also be deleterious when the sequences are hyperedited. HDV, for example, is typical of a virus that evolved with the optimal amount of ADAR1, which becomes deleterious for the virus when increased or decreased (38, 39). The ADAR1 editing activity in MV also reflects this complexity by exhibiting various results in different cells. The rare cases of human SSPE have been correlated with hyperediting of several MV genes in human brain cells, which leads to a persistent, less fitted but more pathogenic virus, suggesting an antiviral role for ADAR1 (2, 12, 13, 60, 75). Increased MV replication has been observed in murine embryonic cells with a deletion of ADAR1 p150 expression (86); however, this effect was not correlated to a decrease in editing compared to that of the control. These cells are derived from nonviable embryos, and the increased replication could be due to other disturbed mechanisms. In contrast, ADAR1 knockdown in human cells induced a decrease in MV replication attributed to an absence of PKR inhibition, indicating an enhancement of viral replication mediated by ADAR1 (80).
ADAR1 has a new function in modulating the function of another IFN-induced protein.
For VSV, HIV-1, and MV, ADAR1 inhibits PKR-induced apoptosis mediated by cellular stresses and viruses and therefore contributes to a high level of virus replication (14, 16, 17, 43, 52, 80). While it seems unexpected that an IFN-induced protein would counteract the IFN response by specific inhibition, this finding correlates with and explains part of the observed general downregulation of the innate immune pathway mediated by ADAR1 (30, 85). The similarity with another autoregulatory loop between miRNA-103/107 and Dicer, an endonuclease which is itself involved in the maturation of miRNAs, emphasizes that multiregulation of cellular processes is necessary to maintain cell function (47). The autoregulation between two IFN-induced proteins during viral infection emphasizes that the cell response is not intended to counteract viral replication but is the result of the concomitant evolution of viruses with that of their hosts. While PKR is regulated by a large number of cellular and viral RNAs and proteins, only a few factors have been shown to regulate ADAR1 activity (15, 25, 26, 29, 64). This novel type of proviral regulation by an IFN-induced protein changes and complements our understanding of the innate immune response. This response may not always be detrimental to the virus because cells need to reach an equilibrium to avoid death, due to either viral replication or to cell apoptosis induced by a hyperactive antiviral response. In the case of HIV-1, the manipulation of the host response by the virus seems to lead to both virus replication and cell survival, but this equilibrium is unstable and ultimately leads to chronic inflammation of lymphoid cells, which contributes to disease progression (8, 15). For many lytic viral infections, the extent of cell death via viral replication and a hyperactive host immune response could determine the extent of cell survival and disease remission and could also be exploited for increased oncolysis of tumor cells (4, 21, 24–26).
ACKNOWLEDGMENTS
We are grateful to Robert Scarborough for helpful discussions and comments on the manuscript.
The work done in our laboratory was supported by the Canadian Institutes for Health Research and by the Canadian Foundation for AIDS Research.
Biographies

Jean-François Gélinas (M.Sc., 2010) obtained his B.Sc. and M.Sc. from McGill University, Montréal, Canada. His M.Sc. work was on the role of ADAR1 and PKR on HIV-1 expression. He now works as a Research Assistant at the Centre de Recherche du Centre Hospitalier de l'Université de Montréal.

Guerline Clerzius (Ph.D., 2010) obtained her B.Sc. and M.Sc. from the Université de Montréal, Montréal, Canada. She obtained her Ph.D. from McGill University, Montréal, Canada, for her work on the multiple levels of PKR regulation during HIV-1 replication. She is now a Clinical Research Associate at the Radiation Oncology Department, Jewish General Hospital, Montréal.

Eileen Shaw (B.Sc., 2010) obtained her B.Sc. from the University of British Columbia, Vancouver, Canada. She is currently an M.Sc. student at McGill University, Montréal, Canada, working on the regulation of PKR activation by cellular factors during HIV-1 replication.

Anne Gatignol (Ph.D., 1988) obtained her Ph.D. from the Université de Toulouse, France. She did her postdoctoral training at the George Washington University, Washington, DC, and the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, working on the regulation of HIV-1 expression by viral and cellular factors. She started as an independent researcher of the Institut National de la Santé et de la Recherche Médicale at the Institut Cochin, Paris, France. She is now a Group Leader at the Lady Davis Institute for Medical Research and an Associate Professor at McGill University, Montréal, Québec, Canada. She works on virus-cell interactions in the regulation of HIV-1 replication. Her projects relate to understanding gene expression and PKR regulation by TRBP, ADAR1, and PACT during HIV replication and the interplay between HIV and the RNA interference pathway, as well as using ribozymes and RNA interference against the virus.
Footnotes
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Albin J. S., Harris R. S. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med. 12:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baczko K., et al. 1993. Clonal expansion of hypermutated measles virus in a SSPE brain. Virology 197:188–195 [DOI] [PubMed] [Google Scholar]
- 3. Balachandran S., et al. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129–141 [DOI] [PubMed] [Google Scholar]
- 4. Barber G. N. 2004. Vesicular stomatitis virus as an oncolytic vector. Viral Immunol. 17:516–527 [DOI] [PubMed] [Google Scholar]
- 5. Barber G. N. 2005. VSV-tumor selective replication and protein translation. Oncogene 24:7710–7719 [DOI] [PubMed] [Google Scholar]
- 6. Bass B. L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71:817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bennasser Y., Le S. Y., Benkirane M., Jeang K. T. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607–619 [DOI] [PubMed] [Google Scholar]
- 8. Brenchley J. M., Douek D. C. 2008. The mucosal barrier and immune activation in HIV pathogenesis. Curr. Opin. HIV AIDS 3:356–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carpenter J. A., Keegan L. P., Wilfert L., O'Connell M. A., Jiggins F. M. 2009. Evidence for ADAR-induced hypermutation of the Drosophila sigma virus (Rhabdoviridae). BMC Genet. 10:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Casey J. L. 2006. RNA editing in hepatitis delta virus. Curr. Top. Microbiol. Immunol. 307:67–89 [DOI] [PubMed] [Google Scholar]
- 11. Casey J. L., Bergmann K. F., Brown T. L., Gerin J. L. 1992. Structural requirements for RNA editing in hepatitis delta virus: evidence for a uridine-to-cytidine editing mechanism. Proc. Natl. Acad. Sci. U. S. A. 89:7149–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cattaneo R., Billeter M. A. 1992. Mutations and A/I hypermutations in measles virus persistent infections. Curr. Top. Microbiol. Immunol. 176:63–74 [DOI] [PubMed] [Google Scholar]
- 13. Cattaneo R., et al. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55:255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clerzius G., et al. 2009. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J. Virol. 83:10119–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clerzius G., Gélinas J. F., Gatignol A. 2011. Multiple levels of PKR inhibition during HIV-1 replication. Rev. Med. Virol. 21:42–53 [DOI] [PubMed] [Google Scholar]
- 16. Der S. D., Yang Y. L., Weissmann C., Williams B. R. 1997. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc. Natl. Acad. Sci. U. S. A. 94:3279–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doria M., Neri F., Gallo A., Farace M. G., Michienzi A. 2009. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 37:5848–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doria M., et al. 2011. ADAR2 editing enzyme is a novel human immunodeficiency virus-1 proviral factor. J. Gen. Virol. 92:1228–1232 [DOI] [PubMed] [Google Scholar]
- 19. Farajollahi S., Maas S. 2010. Molecular diversity through RNA editing: a balancing act. Trends Genet. 26:221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Felder M. P., et al. 1994. Functional and biological properties of an avian variant long terminal repeat containing multiple A to G conversions in the U3 sequence. J. Virol. 68:4759–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galanis E. 2010. Therapeutic potential of oncolytic measles virus: promises and challenges. Clin. Pharmacol. Ther. 88:620–625 [DOI] [PubMed] [Google Scholar]
- 22. Gallo A., Galardi S. 2008. A-to-I RNA editing and cancer: from pathology to basic science. RNA Biol. 5:135–139 [DOI] [PubMed] [Google Scholar]
- 23. Gandy S. Z., et al. 2007. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication. J. Virol. 81:13544–13551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. García M. A., et al. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. García M. A., Meurs E. F., Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811 [DOI] [PubMed] [Google Scholar]
- 26. George C. X., et al. 2009. Tipping the balance: antagonism of PKR kinase and ADAR1 deaminase functions by virus gene products. J. Interferon Cytokine Res. 29:477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. George C. X., Samuel C. E. 1999. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. U. S. A. 96:4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hajjar A. M., Linial M. L. 1995. Modification of retroviral RNA by double-stranded RNA adenosine deaminase. J. Virol. 69:5878–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haller O., Kochs G., Weber F. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hartner J. C., Walkley C. R., Lu J., Orkin S. H. 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 10:109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hartwig D., et al. 2006. The large form of ADAR 1 is responsible for enhanced hepatitis delta virus RNA editing in interferon-alpha-stimulated host cells. J. Viral Hepat. 13:150–157 [DOI] [PubMed] [Google Scholar]
- 32. Heale B. S., et al. 2009. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 28:3145–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herbert A., et al. 1997. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. U. S. A. 94:8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Herbert A., Rich A. 2001. The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc. Natl. Acad. Sci. U. S. A. 98:12132–12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hundley H. A., Bass B. L. 2010. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem. Sci. 35:377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iizasa H., et al. 2010. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem. 285:33358–33370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jabbour J. T., Duenas D. A., Sever J. L., Krebs H. M., Horta-Barbosa L. 1972. Epidemiology of subacute sclerosing panencephalitis (SSPE). A report of the SSPE registry. JAMA 220:959–962 [PubMed] [Google Scholar]
- 38. Jayan G. C., Casey J. L. 2002. Increased RNA editing and inhibition of hepatitis delta virus replication by high-level expression of ADAR1 and ADAR2. J. Virol. 76:3819–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jayan G. C., Casey J. L. 2002. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J. Virol. 76:12399–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim U., Wang Y., Sanford T., Zeng Y., Nishikura K. 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. U. S. A. 91:11457–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klase Z., et al. 2007. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumar M., Carmichael G. G. 1997. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. U. S. A. 94:3542–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Z., Wolff K. C., Samuel C. E. 2010. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology 396:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Linnstaedt S. D., Kasprzak W. K., Shapiro B. A., Casey J. L. 2009. The fraction of RNA that folds into the correct branched secondary structure determines hepatitis delta virus type 3 RNA editing levels. RNA 15:1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luo G. X., et al. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maas S., Kawahara Y., Tamburro K. M., Nishikura K. 2006. A-to-I RNA editing and human disease. RNA Biol. 3:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martello G., et al. 2010. A microRNA targeting dicer for metastasis control. Cell 141:1195–1207 [DOI] [PubMed] [Google Scholar]
- 48. Martínez I., Melero J. A. 2002. A model for the generation of multiple A to G transitions in the human respiratory syncytial virus genome: predicted RNA secondary structures as substrates for adenosine deaminases that act on RNA. J. Gen. Virol. 83:1445–1455 [DOI] [PubMed] [Google Scholar]
- 49. Menéndez-Arias L. 2002. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog. Nucleic Acid Res. Mol. Biol. 71:91–147 [DOI] [PubMed] [Google Scholar]
- 50. Murphy D. G., Dimock K., Kang C. Y. 1991. Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology 181:760–763 [DOI] [PubMed] [Google Scholar]
- 51. Ngamurulert S., Limjindaporn T., Auewaraku P. 2009. Identification of cellular partners of influenza A virus (H5N1) non-structural protein NS1 by yeast two-hybrid system. Acta Virol. 53:153–159 [DOI] [PubMed] [Google Scholar]
- 52. Nie Y., Hammond G. L., Yang J. H. 2007. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J. Virol. 81:917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nishikura K. 2006. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol. 7:919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79:321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Onafuwa-Nuga A., Telesnitsky A. 2009. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 73:451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ong C. L., et al. 2005. Low TRBP levels support an innate human immunodeficiency virus type 1 resistance in astrocytes by enhancing the PKR antiviral response. J. Virol. 79:12763–12772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Otsuka M., et al. 2007. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27:123–134 [DOI] [PubMed] [Google Scholar]
- 58. Ouellet D. L., et al. 2008. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 36:2353–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palladino M. J., Keegan L. P., O'Connell M. A., Reenan R. A. 2000. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 102:437–449 [DOI] [PubMed] [Google Scholar]
- 60. Patterson J. B., et al. 2001. Evidence that the hypermutated M protein of a subacute sclerosing panencephalitis measles virus actively contributes to the chronic progressive CNS disease. Virology 291:215–225 [DOI] [PubMed] [Google Scholar]
- 61. Patton D. E., Silva T., Bezanilla F. 1997. RNA editing generates a diverse array of transcripts encoding squid Kv2 K+ channels with altered functional properties. Neuron 19:711–722 [DOI] [PubMed] [Google Scholar]
- 62. Phuphuakrat A., et al. 2008. Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J. Virol. 82:10864–10872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Polson A. G., Bass B. L., Casey J. L. 1996. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380:454–456 [DOI] [PubMed] [Google Scholar]
- 64. Randall R. E., Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 65. Rima B. K., Duprex W. P. 2009. The measles virus replication cycle. Curr. Top. Microbiol. Immunol. 329:77–102 [DOI] [PubMed] [Google Scholar]
- 66. Rizzetto M. 2009. Hepatitis D: thirty years after. J. Hepatol. 50:1043–1050 [DOI] [PubMed] [Google Scholar]
- 67. Rueda P., Garcia-Barreno B., Melero J. A. 1994. Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A-G substitutions (hypermutations). Virology 198:653–662 [DOI] [PubMed] [Google Scholar]
- 68. Sadler A. J., Williams B. R. 2007. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 316:253–292 [DOI] [PubMed] [Google Scholar]
- 69. Samuel C. E. 2011. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 411:180–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Samuel C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Scadden A. D., Smith C. W. 2001. RNAi is antagonized by A→I hyper-editing. EMBO Rep. 2:1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Seeburg P. H. 1996. The role of RNA editing in controlling glutamate receptor channel properties. J. Neurochem. 66:1–5 [DOI] [PubMed] [Google Scholar]
- 73. Stojdl D. F., et al. 2000. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J. Virol. 74:9580–9585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Suspène R., et al. 2011. Double-stranded RNA adenosine deaminase ADAR-1-induced hypermutated genomes among inactivated seasonal influenza and live attenuated measles virus vaccines. J. Virol. 85:2458–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Suspène R., et al. 2008. Inversing the natural hydrogen bonding rule to selectively amplify GC-rich ADAR-edited RNAs. Nucleic Acids Res. 36:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Taylor D. R., Puig M., Darnell M. E., Mihalik K., Feinstone S. M. 2005. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J. Virol. 79:6291–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Taylor J. M. 2009. Replication of the hepatitis delta virus RNA genome. Adv. Virus Res. 74:103–121 [DOI] [PubMed] [Google Scholar]
- 78. Tenoever B. R., et al. 2007. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science 315:1274–1278 [DOI] [PubMed] [Google Scholar]
- 79. Toth A. M., Devaux P., Cattaneo R., Samuel C. E. 2009. Protein kinase PKR mediates the apoptosis induction and growth restriction phenotypes of C protein-deficient measles virus. J. Virol. 83:961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Toth A. M., Li Z., Cattaneo R., Samuel C. E. 2009. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J. Biol. Chem. 284:29350–29356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wainberg M. A., Jeang K. T. 2008. 25 years of HIV-1 research—progress and perspectives. BMC Med. 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang Q., Khillan J., Gadue P., Nishikura K. 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290:1765–1768 [DOI] [PubMed] [Google Scholar]
- 83. Wang Q., et al. 2004. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 279:4952–4961 [DOI] [PubMed] [Google Scholar]
- 84. Wang Y., Samuel C. E. 2009. Adenosine deaminase ADAR1 increases gene expression at the translational level by decreasing protein kinase PKR-dependent eIF-2alpha phosphorylation. J. Mol. Biol. 393:777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang Z., et al. 2008. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. U. S. A. 105:5477–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ward S. V., et al. 2011. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc. Natl. Acad. Sci. U. S. A. 108:331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Whelan S. P. 2008. Vesicular stomatitis virus, p. 291–299 In Mahy B. W., van Regenmortel M. H. (ed.), Encyclopedia of Virology, 3rd ed., vol. T-2 Academic Press, Oxford, United Kingdom [Google Scholar]
- 88. Wong S. K., Lazinski D. W. 2002. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. U. S. A. 99:15118–15123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yanagi Y., Takeda M., Ohno S. 2006. Measles virus: cellular receptors, tropism and pathogenesis. J. Gen. Virol. 87:2767–2779 [DOI] [PubMed] [Google Scholar]
- 90. Yang J. H., et al. 2003. Intracellular localization of differentially regulated RNA-specific adenosine deaminase isoforms in inflammation. J. Biol. Chem. 278:45833–45842 [DOI] [PubMed] [Google Scholar]
- 91. Yang W., et al. 2005. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J. Biol. Chem. 280:3946–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zahn R. C., Schelp I., Utermohlen O., von Laer D. 2007. A-to-G hypermutation in the genome of lymphocytic choriomeningitis virus. J. Virol. 81:457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]