Abstract
Physiological functions of sucrose (Suc) transporters (SUTs) localized to the tonoplast in higher plants are poorly understood. We here report the isolation and characterization of a mutation in the rice (Oryza sativa) OsSUT2 gene. Expression of OsSUT2-green fluorescent protein in rice revealed that OsSUT2 localizes to the tonoplast. Analysis of the OsSUT2 promoter::β-glucuronidase transgenic rice indicated that this gene is highly expressed in leaf mesophyll cells, emerging lateral roots, pedicels of fertilized spikelets, and cross cell layers of seed coats. Results of Suc transport assays in yeast were consistent with a H+-Suc symport mechanism, suggesting that OsSUT2 functions in Suc uptake from the vacuole. The ossut2 mutant exhibited a growth retardation phenotype with a significant reduction in tiller number, plant height, 1,000-grain weight, and root dry weight compared with the controls, the wild type, and complemented transgenic lines. Analysis of primary carbon metabolites revealed that ossut2 accumulated more Suc, glucose, and fructose in the leaves than the controls. Further sugar export analysis of detached leaves indicated that ossut2 had a significantly decreased sugar export ability compared with the controls. These results suggest that OsSUT2 is involved in Suc transport across the tonoplast from the vacuole lumen to the cytosol in rice, playing an essential role in sugar export from the source leaves to sink organs.
The disaccharide Suc is a major product of photosynthesis produced in source leaves and is the main carbohydrate transported in the vascular tissue of many plants. The orchestrated production, transport, and storage of Suc, therefore, is essential for normal plant growth and development (Lalonde et al., 2004; Lim et al., 2006; Kühn and Grof, 2010). Suc transport occurs at both the whole plant and intracellular levels, and this sugar is loaded over a short distance to the phloem either apoplastically by the plasma membrane sucrose transporters (SUTs) or symplastically through plasmodesmata, depending on the plant species (Rennie and Turgeon, 2009). Subsequent to phloem loading, Suc is transported over a long distance to sink tissues (ap Rees and Hill, 1994; Williams et al., 2000; Lalonde et al., 2004; Lim et al., 2006; Sauer, 2007). Within cells, Suc is partitioned into organelles, especially vacuoles, for short-term or long-term storage.
SUTs are encoded by small gene families in plants that have been divided into five major clades, SUT1 to SUT5 (Kühn and Grof, 2010). The SUT1 clade represents dicot SUTs, which have the highest substrate affinities. Members of SUT1 and monocot-specific SUT3 clades are localized at the plasma membrane and are responsible for phloem loading or Suc import into sink tissues (Riesmeier et al., 1994; Gottwald et al., 2000; Slewinski et al., 2009). The SUT2 clade members, localized also at the plasma membrane, possess an unusually long N terminus and central loop sequence. Recently, some SUT4 clade members such as Arabidopsis (Arabidopsis thaliana) AtSUT4, barley (Hordeum vulgare) HvSUT2, Lotus japonicus LjSUT4, and poplar (Populus tremula × Populus alba) PtaSUT4 have been assigned to the vacuolar membrane, tonoplast, through proteomic and/or GFP fusion analyses (Endler et al., 2006; Reinders et al., 2008; Payyavula et al., 2011). The SUT5 clade is a largely uncharacterized group of monocot Suc transporters, of which OsSUT5 has a higher affinity for Suc, and its transport activity is less sensitive to pH than transporters in the SUT3 clade (Sun et al., 2010).
The organ- and tissue-specific expression of SUTs suggests that they have distinct functions. In the SUT1 clade, potato (Solanum tuberosum) StSUT1 (Riesmeier et al., 1994), tobacco (Nicotiana tabacum) NtSUT1 (Bürkle et al., 1998), and Arabidopsis AtSUC2 (Gottwald et al., 2000) are expressed in leaf vascular tissues and are essential for phloem loading. Similarly, members of the SUT3 clade, such as rice (Oryza sativa) OsSUT1 and maize (Zea mays) ZmSUT1, are also thought to be responsible for phloem loading (Scofield et al., 2007; Slewinski et al., 2009). For both clades, the expression of a number of members is also observed in sink tissues, suggesting additional functions in relation to Suc transport into sink organs. For instance, AtSUC1 (Stadler et al., 1999; Sivitz et al., 2008), NtSUT3 (Lemoine et al., 1999), and Plantago major SUC1 (PmSUC1; Lauterbach et al., 2007) are expressed in pollen tubes, suggesting their function in growing pollen tubes. From the SUT3 clade, OsSUT1 is expressed in pollen and in nucellar projections and aleurone tissues of immature seeds and thus likely functions in apoplastic Suc transport into these sink organs (Furbank et al., 2001; Hirose et al., 2010). HvSUT1 is also mainly expressed in maternal nucellar projections and in the filial transfer cells of barley (Weschke et al., 2000), suggesting that it has similar functions.
Among the SUT4 clade members that include tonoplast-localized SUTs, HvSUT2 is expressed in leaves, roots, and pericarps (Weschke et al., 2000) and was further found to be expressed in leaf mesophyll cells (Endler et al., 2006). AtSUT4, which functions as a H+-Suc symporter (Weise et al., 2000), is expressed in the companion cells (Schulze et al., 2003) and mesophyll cells of leaves (Endler et al., 2006). Thus, HvSUT2 and AtSUT4 have been suggested to function in the transport and vacuolar storage of photosynthetically derived Suc (Endler et al., 2006). LjSUT4 was demonstrated to be a tonoplast-localized H+-Suc symporter and thus hypothesized to function in Suc transport across the tonoplast from the vacuole lumen to the cytosol (Reinders et al., 2008). Recently, RNA interference (RNAi) transgenic poplar plants with reduced PtaSUT4 expression showed an increased ratio of leaf to stem biomass, indicating a link between vacuolar transport of Suc and biomass partitioning (Payyavula et al., 2011). However, physiological evidence for the function of tonoplast SUTs has not yet been reported in plant mutants.
It has been previously shown that a number of cereals including rice store relatively high ratios of Suc to transitory starch in their leaves, which differs from other plant species, including Arabidopsis, which primarily store starch (Nakano et al., 1995; Winder et al., 1998; Murchie et al., 2002; Trevanion, 2002; Lee et al., 2008). Considering that Suc is temporarily stored in the vacuoles of photosynthetic assimilatory tissues (Riens et al., 1991; Winter et al., 1993; Martinoia et al., 2007; Neuhaus, 2007; Linka and Weber, 2010), this raises the question of the involvement and function of tonoplast-localized SUTs in plant growth and development in cereals. The rice genome contains five SUTs, OsSUT1 to OsSUT5 (Aoki et al., 2003). To date, however, only OsSUT1 has been characterized in detail (Scofield et al., 2007; Hirose et al., 2010; Sun et al., 2010). We aimed to increase our understanding of Suc storage and transport in rice, an important worldwide crop. In this regard, characterization of the remaining OsSUTs is necessary, as these proteins underpin the process of Suc transport in rice. In this study, OsSUT2, a tonoplast-localized SUT, was analyzed by subcellular localization using a GFP fusion protein, via its Suc transport ability in yeast and through its gene expression properties using promoter::GUS transgenic rice. Furthermore, an OsSUT2 mutant allele, ossut2, was characterized in terms of morphological phenotype, primary metabolite levels, and sugar export ability. Finally, a possible physiological role of OsSUT2 is proposed.
RESULTS
Identification of the Rice Tonoplast Suc Transporter OsSUT2
To compare the amino acid similarity between the five reported OsSUTs (Aoki et al., 2003) and SUTs from other plant species, we constructed an unrooted phylogenetic tree using the neighbor-joining method (Supplemental Fig. S1). This analysis revealed that OsSUT2 (LOC_Os12g44380) belongs to the SUT4 clade of tonoplast-localized SUTs including AtSUT4, HvSUT2, LjSUT4, and PtaSUT4 (Endler et al., 2006; Reinders et al., 2008; Payyavula et al., 2011) and the plasma membrane-localized StSUT4 (Chincinska et al., 2008). For the other members of the SUT4 clade, their localization remains to be determined.
To characterize OsSUT2, its full-length cDNA was cloned by reverse transcription (RT)-PCR. This cloned OsSUT2 gene (GenBank accession no. HQ540307) was from the japonica cv Dongjin and found to encode a protein of 501 amino acids with six amino acid differences compared with OsSUT2 from the japonica cv Nipponbare (AB091672; Aoki et al., 2003) and a single amino acid difference compared with OsSUT2 from the indica cv Minghui86 (DQ072592; Sun et al., 2008). OsSUT2 consists of 12 predicted transmembrane spans, short cytoplasmic N- and C-terminal ends, and a short central cytoplasmic loop between transmembrane spans 6 and 7, which are common characteristics of members of the SUT4 clade (Supplemental Fig. S2). This substantial structural similarity to AtSUT4, HvSUT2, LjSUT4, and PtaSUT4 further suggests that OsSUT2 is a strong candidate as a tonoplast-localized SUT.
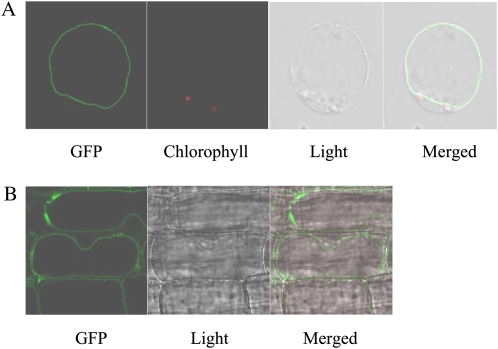
To next determine the subcellular localization of OsSUT2, we generated transgenic rice plants expressing an OsSUT2-GFP fusion construct under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The tonoplast localization of OsSUT2 was demonstrated in protoplasts isolated from the mesophyll cells of OsSUT2-GFP transgenic rice. The GFP signal was clearly evident at the tonoplast, which is on the inside of and is indented by chloroplasts (indicated by chlorophyll autofluorescence; Fig. 1A). When using a regenerated albino plant expressing OsSUT2-GFP, the GFP signal was evident on the inside of the nucleus, thereby confirming that it is on the tonoplast, not the plasma membrane (Fig. 1B). In addition, the leaf mesophyll protoplasts of OsSUT2-GFP transgenic rice were stained with FM4-64, which primarily stains the plasma membrane (Uemura et al., 2004). The GFP signals did not overlap with those of FM4-64 (Supplemental Fig. S3), further indicating that OsSUT2 does not localize to the plasma membrane. These results clearly demonstrate that OsSUT2 resides on the tonoplast.
Figure 1.
Subcellular localization of the OsSUT2-GFP fusion protein. A, Localization of the OsSUT2-GFP fusion protein in leaf mesophyll protoplasts isolated from transgenic rice plants expressing OsSUT2-GFP. B, Localization of the OsSUT2-GFP fusion protein in an albino leaf of transgenic rice. Chlorophyll autofluorescence was used as a chloroplast marker in A. GFP signal and chlorophyll autofluorescence are indicated in green and red, respectively.
Expression Pattern of OsSUT2
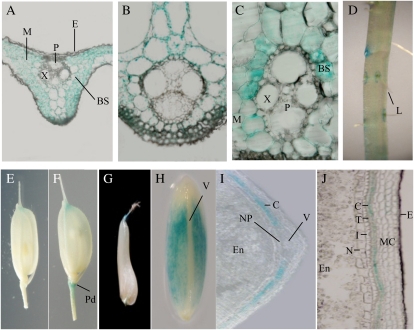
To examine the spatiotemporal expression pattern of OsSUT2 in rice, we generated transgenic rice plants expressing the OsSUT2 promoter::GUS construct (pOsSUT2::GUS) and carried out histochemical GUS analysis. There was no significant variation found in GUS expression among independent transgenic rice plants expressing pOsSUT2::GUS. A representative line with the highest GUS intensity was selected for subsequent analysis. In this line, GUS activity was detectable in all examined rice tissues, including leaves, stems, roots, flowers (spikelets), and immature seeds (Fig. 2). In leaf blades of the pOsSUT2::GUS line, GUS activities were high in mesophyll and bundle sheath cells but no activity was found in the vascular bundles, including the phloem sieve elements and companion cells (Fig. 2, A and B). In epidermal cells, GUS expression was barely detectable. The GUS expression pattern in the leaf sheaths was similar to that of leaf blades, but higher levels of GUS activity were observed in the bundle sheath cells (Fig. 2C). In roots, GUS expression was only detectable in the regions around the emerging lateral roots (Fig. 2D). In spikelets before fertilization, weak GUS activity was detectable at the edges of palea and lemma but not in other floral organs, including the stamens (Fig. 2E). In contrast, following fertilization, the pedicel regions of the spikelets exhibited high GUS activity (Fig. 2F).
Figure 2.
Histochemical localization of GUS expression in leaves, roots, flowers, and seeds of transgenic rice carrying the pOsSUT2::GUS construct. A and B, Cross-sections of leaf blades. C, Cross-sections of leaf sheaths. D, Roots. E, Spikelet before fertilization. F, Spikelet after fertili-zation. G, Immature seed at 3 to 4 DAF. H, Immature seed at 7 to 8 DAF. I, Transverse section of 7- to 8-DAF immature seed. J, Longitudinal section of 7- to 8-DAF immature seed. BS, Bundle sheath cell; C, cross cell layer; E, epidermis; En, endosperm; I, integument; L, lateral root; M, mesophyll cell; MC, mesocarp; N, nucellar epidermis; NP, nucellar projection; P, phloem cell; Pd, pedicel; T, tube cell layer; V, main vascular bundle; X, metaxylem.
Seeds are elongated longitudinally with the cellularization and proliferation of endosperm cells during the prestorage phase (1–6 d after fertilization [DAF]) and thickened during the starch-filling milky phase (7–15 DAF) in rice (Hirose et al., 2002; Lim et al., 2006). In developing immature seeds of transgenic pOsSUT2::GUS lines, high GUS activity was observed in the 7- to 8-DAF seed coat at the early starch-filling stage, while only low activity was observed at 3 to 4 DAF in the prestorage phase seed (Fig. 2, G and H). To further examine OsSUT2 expression in immature seeds, stained seeds were longitudinally or transversely sectioned. In the 7- to 8-DAF immature seeds, OsSUT2 was highly expressed in the cross cell layers but not in the main vascular tissues, nucellar projection, or endosperm cells (Fig. 2, I and J). We also evaluated OsSUT2 expression in immature seeds by real-time PCR. The results confirmed that OsSUT2 is expressed preferentially in the seed coat and not in the endosperm (Supplemental Fig. S4). In addition, this expression pattern is consistent with previously reported RT-PCR results showing that the Minghui86 OsSUT2 is highly expressed in leaf blades and in developing caryopses (Sun et al., 2008).
Suc Transport Activity of OsSUT2 in Yeast
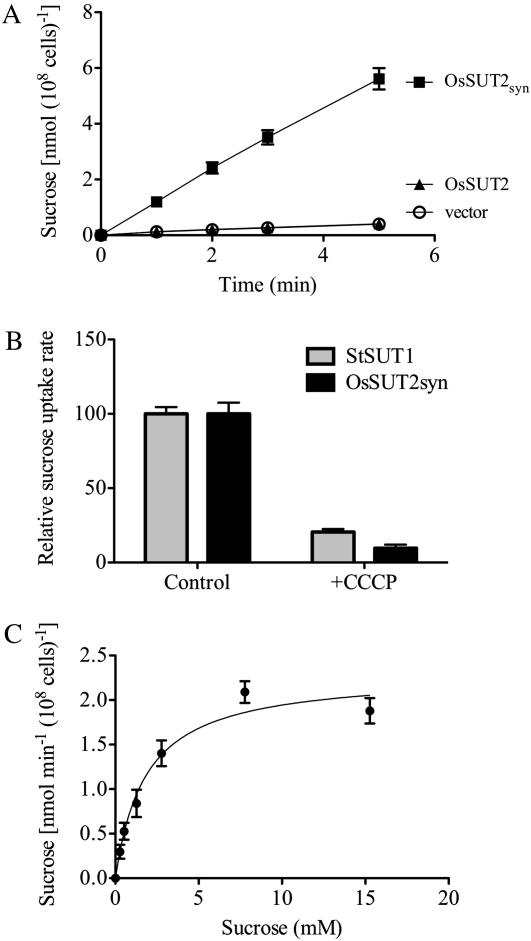
To test whether OsSUT2 has Suc transport activity, we performed assays with yeast cells expressing the rice OsSUT2 cDNA using radiolabeled [14C]Suc at pH 4.0. In this experiment, an expression-optimized synthesized open reading frame of OsSUT2, referred to as OsSUT2syn, was also tested (Sun et al., 2010). Suc transport into yeast cells expressing the expression-optimized OsSUT2syn was found to be linear between 1 and 5 min and approximately 14-fold higher than the uptake levels into yeast cells transformed with the empty vector after 5 min (Fig. 3A). In contrast, the native OsSUT2 rice cDNA was found not to be functional in yeast, even though it encodes an identical protein to the synthetic, expression-optimized clone (Fig. 3A). Changes in codon usage are thought to improve translation efficiency in heterologous systems by relieving translation-limiting regulation or by modifying mRNA structure (Gustafsson et al., 2004).
Figure 3.
Suc transport assay in yeast. A, Time course of Suc uptake in yeast. SEY6210 cells transformed with OsSUT2, OsSUT2syn, or the empty vector were incubated with 14C-labeled Suc at a final concentration of 1 mm at pH 4.0. Results shown are means ± sd for three independent yeast transformants. B, H+ coupling of Suc transport by OsSUT2syn. Yeast cells expressing either OsSUT2syn (black bars) or StSUT1 (gray bars) were incubated in 1 mm [14C]Suc with (+CCCP) or without (Control) the protonophore CCCP (10 μm final concentration). Background uptake rates of cells transformed with the empty vector were subtracted, and the average uptake rate in the absence of CCCP for each transporter was set to 100%. The absolute uptake rates were 51.39 ± 2.35 nmol Suc min−1 (108 cells)−1 for StSUT1, 0.96 ± 0.068 nmol Suc min−1 (108 cells)−1 for OsSUT2, and 0.059 ± 0.002 nmol Suc min−1 (108 cells)−1 for the empty vector. Results shown are means ± sd for three independent yeast transformants. C, Suc uptake kinetics of OsSUT2. Cells were incubated with [14C]Suc at pH 4.0, and uptake rates corrected for the uptake rates of cells transformed with the empty vector were determined. The Km was 1.86 ± 0.38 mm, calculated by a nonlinear regression fit of the Michaelis-Menten equation. The Vmax was 2.30 ± 0.06 nmol Suc min−1 (108 cells)−1. Results shown are based on experiments with three independent yeast transformants. OsSUT2syn, Synthetic cDNA; OsSUT2, native cDNA.
Suc transport by OsSUT2 was strongly inhibited by the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP; Fig. 3B). This indicates that OsSUT2 functions as a H+-Suc symporter. Suc transport by the potato H+-Suc symporter StSUT1 (Schulze et al., 2000) was similarly sensitive to CCCP (Fig. 3B). The kinetics of [14C]Suc uptake into yeast cells expressing OsSUT2 were analyzed; the Km of OsSUT2 was determined to be 1.86 ± 0.38 mm, and the Vmax was 2.30 ± 0.06 nmol Suc min−1 (108 cells)−1 at pH 4.0 (Fig. 3C).
Plant Growth Phenotype of the OsSUT2 Mutant
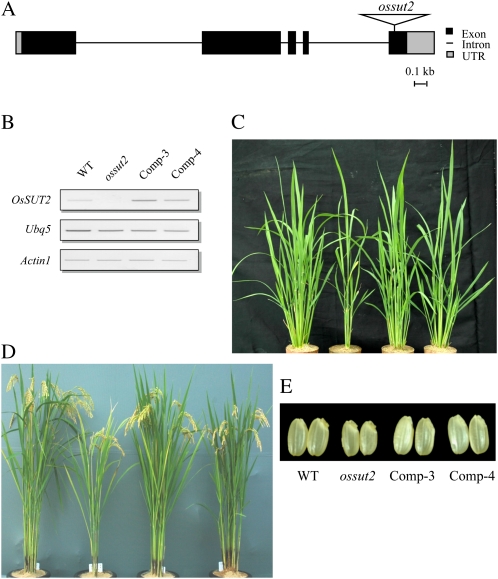
To examine the in planta function of OsSUT2, we isolated a null mutant of this gene from the rice T-DNA mutant population (An et al., 2005a, 2005b; Jeong et al., 2006; http://www.postech.ac.kr/life/pfg/risd/index.html). The isolated mutant allele, ossut2, harbors a T-DNA insertion in the fifth exon of OsSUT2 (Fig. 4A). Using OsSUT2-specific and T-DNA-specific primers, mutants that were homozygous for the insertion were isolated from the segregating progeny. RT-PCR analysis indicated that the endogenous OsSUT2 transcript is absent in ossut2, confirming that it is a null mutant (Fig. 4B).
Figure 4.
Isolation and characterization of the ossut2 mutant. A, Schematic diagram of the rice OsSUT2 gene and the insertion position of the T-DNA. In ossut2, T-DNA is inserted into the fifth exon. UTR, Untranslated region. B, RT-PCR analysis of ossut2 and the complemented lines, Comp-3 and Comp-4. OsSUT2 transcripts are not detectable in the homozygous mutant, while the two complemented lines express OsSUT2 at higher levels than the wild type. UBQ5 and ACTIN1 are PCR controls. C, Four-week-old plants grown in the greenhouse. The leftmost plant is the wild type (WT). The ossut2 mutant and two complemented lines are shown left to right. D, Mature flowering plants of the wild type, ossut2, and complemented lines (left to right) grown in a rice paddy field. E, Husked grains of the same plants shown in D.
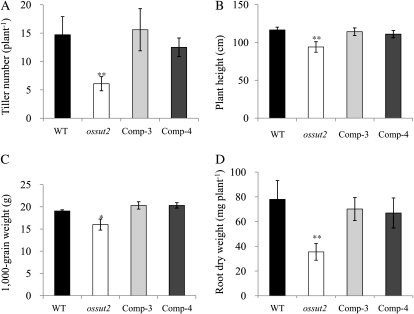
The ossut2 mutant displayed strong plant growth reduction under greenhouse conditions at 4 weeks after sowing (Fig. 4C). The same effect was observed in mature plants grown in rice paddy fields (Fig. 4D). The growth of ossut2 was compared with its wild-type segregant control in terms of tiller number, plant height, 1,000-grain weight, and root weight (Fig. 5). The average tiller number of the ossut2 plants was 6.1, 58.8% lower than that of the wild-type plants (14.8; Fig. 5A). Similarly, the average height was 112.8 cm in the control plants compared with only 91.4 cm in ossut2, a decrease of about 19% (Fig. 5B). The average weight of 1,000 grains was 16 and 19.1 g in ossut2 and control wild-type plants, respectively, a reduction of about 16% in ossut2 (Fig. 5C). Consistently, ossut2 mutant grains were visibly smaller than seeds from control plants (Fig. 4E). In addition, the root dry weight was decreased by 54.5% in ossut2 (35.5 mg per plant) compared with the controls (78.0 mg per plant; Fig. 5D).
Figure 5.
Growth phenotype of mature ossut2 plants grown in a rice paddy field. A, Number of tillers. B, Plant height. C, Weight of 1,000 grains. D, Root dry weight. Each data point represents the mean ± sd from 10 different plants. Black bars, The wild type (WT); white bars, ossut2; light gray bars, Comp-3; dark gray bars, Comp-4. The statistical significance of correlation coefficients is indicated by asterisks: * P < 0.05, ** P < 0.01.
To determine whether this plant growth reduction is indeed due to the ossut2 mutation, we transformed the mutant line with wild-type OsSUT2 cDNA under the control of the maize Ubiquitin1 (Ubi1) promoter. More than 10 independent transgenic rice plants were obtained, among which two lines, designated Comp-3 and Comp-4, showing high expression of OsSUT2 were selected and analyzed in subsequent experiments. RT-PCR analysis indicated that these two lines expressed OsSUT2 at higher levels than the wild-type control (Fig. 4B). In phenotype analysis of Comp-3 and Comp-4, the tiller number, plant height, 1,000-grain weight, grain size, and root dry weight were all found to be restored to levels similar to the control plants (Figs. 4, C–E, and 5). This genetic complementation experiment clearly demonstrated that the ossut2 mutation is responsible for the growth defect phenotype and strongly suggested that the function of OsSUT2 is essential for the normal growth and development of rice plants.
Primary Carbon Metabolites in the ossut2 Mutant
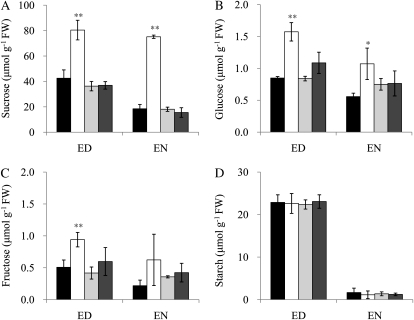
To assess whether the levels of Suc, as well as of other soluble sugars, Glc and Fru, and of starch in the source leaves of ossut2 were altered by the loss of the tonoplast SUT, OsSUT2, we analyzed these metabolites in 4-week-old wild-type and mutant plants. Suc was significantly increased in ossut2 leaves compared with the controls, segregant wild-type and complemented lines (Fig. 6A). In the ossut2 mutant, the Suc content was about 2-fold higher at the end of the day and about 4-fold higher at the end of the night compared with the controls. Similarly, the levels of Glc and Fru were increased in the ossut2 mutant compared with the controls (Fig. 6, B and C). In contrast, the transitory starch levels in ossut2 mutant leaves were not different from the controls (Fig. 6D). The complemented lines all showed similar soluble sugar contents to those of the segregant wild-type plants.
Figure 6.
Leaf soluble sugar and starch contents in ossut2 mutant plants grown for 4 weeks in the greenhouse. The levels of Suc (A), Glc (B), Fru (C), and starch (D) are shown. Each data point represents the mean ± sd from five different plants. ED, End of the day; EN, end of the night; FW, fresh weight. Black bars, The wild type (WT); white bars, ossut2; light gray bars, Comp-3; dark gray bars, Comp-4. The statistical significance of correlation coefficients is indicated by asterisks: * P < 0.05, ** P < 0.01.
Sugar Export Activity of the ossut2 Mutant
To examine whether any change in photosynthetic capacity accompanied the phenotypic manifestations in ossut2, we evaluated the rates of net photosynthesis in this mutant by measuring the CO2 consumption from the fully expanded leaves of 8-week-old rice plants. The ossut2 mutant did not show a significant reduction of net photosynthetic activity compared with wild-type plants at a sequentially changed photon flux density (Supplemental Fig. S5).
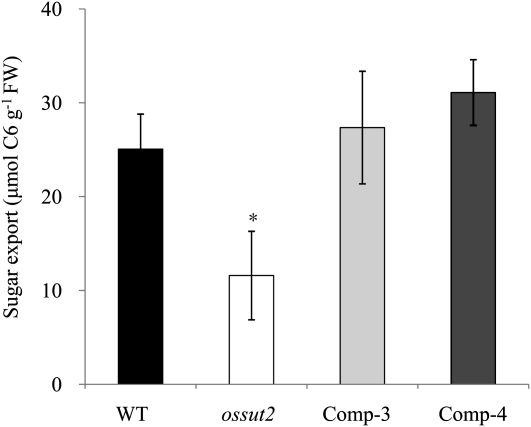
We speculated whether the accumulation of sugars in the source leaves may lead to a decreased transfer of Suc to the sink organs, since grain size and weight and root weight, as well as plant height and tiller number, were reduced in ossut2. To quantify the sugar export ability in this mutant, we detached mature leaf blades of 4-week-old plants at the end of day, incubated these for 16 h under dark conditions, and determined the soluble sugar levels (total of Suc, Glc, and Fru). During this dark phase, detached wild-type leaves exported 25.1 μmol C6 units g−1 fresh weight, whereas ossut2 mutant leaves showed about a 54% reduction (11.6 μmol C6 units g−1 fresh weight) in sugar export. Both complemented lines exhibited a similar sugar export activity level (27.4 and 31.1 μmol C6 units g−1 fresh weight of Comp-3 and Comp-4, respectively) to that of the wild type (Fig. 7).
Figure 7.
Quantification of sugar export activity of the ossut2 mutant. Histograms show the amount of soluble sugars (total of Suc, Glc, and Fru) detected in the mature leaf blades of 4-week-old plants detached at the end of day and incubated in 15 mm EDTA solution for 16 h under dark conditions. FW, Fresh weight. Black bars, The wild type (WT); white bars, ossut2; light gray bars, Comp-3; dark gray bars, Comp-4. The indicated data are means ± sd from three separate experiments. The statistical significance of correlation coefficients is indicated by the asterisk (P < 0.05).
DISCUSSION
OsSUT2 Encodes a Tonoplast H+-Suc Symporter in Rice
This study provides several lines of evidence that OsSUT2 is a functional tonoplast Suc transporter. First, OsSUT2 belongs to the SUT4 clade that includes the previously identified tonoplast-localized SUTs, AtSUT4, HvSUT2, LjSUT4, and PtaSUT4 (Endler et al., 2006; Reinders et al., 2008; Payyavula et al., 2011; Supplemental Fig. S1). Second, OsSUT2-GFP localizes to the tonoplast in rice mesophyll protoplasts and plants (Fig. 1). Third, Suc transport by OsSUT2 expressed in yeast was inhibited by CCCP, a protonophore (Fig. 3). These findings indicate that OsSUT2 functions as a tonoplast H+-Suc symporter. Fourth, we found that the loss of function of OsSUT2 in rice resulted in higher accumulation of Suc in leaves compared with the controls (Fig. 6) and led to a significantly reduced sugar export ability (Fig. 7). This is most likely due to reduced Suc transport from the vacuole to the cytosol, resulting in higher Suc accumulation in the vacuole lumen. Our results are consistent with the observation that PtaSUT4 RNAi transgenic poplar had elevated Suc content in source leaves (Payyavula et al., 2011). Considering that OsSUT2 functions as a H+-Suc symporter in yeast as well as the normal pH gradient across the tonoplast, our data indicate that OsSUT2 functions in Suc transport across the tonoplast from the vacuole lumen to the cytosol.
Suc transport in AtSUT4-expressing yeast cells was previously found to be stimulated by Glc but inhibited by addition of the protonophore CCCP (Weise et al., 2000). Suc transport into HvSUT2-expressing yeast cells was also previously reported to be inhibited by treatments with the protonophore CCCP and by the Suc transport inhibitor p-chloromercuribenzoylsulfonic acid (Weschke et al., 2000). Similarly, Suc transport into Xenopus oocytes by LjSUT4 was found to be inhibited by CCCP, and the extracellular application of Suc induced membrane depolarization in LjSUT4-expressing Xenopus oocytes (Reinders et al., 2008). Hence, our results here are consistent with the contention that all currently identified tonoplast Suc transporters function as H+-Suc symporters rather than antiporters and support the previous hypothesis that they function in Suc transport from the vacuole to the cytosol (Neuhaus, 2007; Reinders et al., 2008). It is likely that the Suc transport activity of OsSUT2 in yeast is due to mistargeting of the overexpressed protein to the plasma membrane in the heterologous system.
Our kinetic analysis in yeast indicated that the Km of OsSUT2 is about 1.9 mm at pH 4.0 (Fig. 3). Compared with other tonoplast SUTs, including LjSUT4, this represents a relatively high affinity for Suc (Weise et al., 2000; Weschke et al., 2000; Reinders et al., 2008). Analysis of [14C]Suc uptake into yeast is thought to underestimate Km because SUT activity depletes the proton motive force and membrane potential is not controlled (Chandran et al., 2003). This tends to decrease the Vmax measured and therefore decreases the apparent Km. Using voltage clamping to measure Km is more accurate; however, OsSUT2 did not show activity when expressed in Xenopus oocytes (Sun et al., 2010). Of the tonoplast SUTs, only LjSUT4 has been studied by expression in Xenopus oocytes and electrophysiology (Reinders et al., 2008).
OsSUT2 Functions Mainly in Photosynthetic Assimilatory Tissues
In dicot species, Arabidopsis AtSUT4 was found to be expressed predominantly in the minor veins in source leaves in promoter::GUS fusion experiments (Weise et al., 2000). Subsequently, AtSUT4 was confirmed to be expressed in the companion cells of minor veins (Schulze et al., 2003), thus suggesting a role in Suc loading into the phloem. In monocot species, barley HvSUT2 expression was reported to be highest in growing leaves and also to be at considerable levels in other tissues such as roots and caryopses, while that of HvSUT1, a member of the SUT3 clade, was highest in developing caryopses. Thus, HvSUT2 has been suggested to play a general housekeeping role (Weschke et al., 2000). In further studies, RT-PCR analysis revealed that both AtSUT4 and HvSUT2 transcripts are abundant in leaf mesophyll cells. In poplar, in situ hybridization was used to show that PtaSUT4 is expressed in leaf mesophyll cells and minor veins (Payyavula et al., 2011). The recent finding that all these transporters are localized to the tonoplast suggests that they may be involved in vacuolar transfer of photosynthetically derived Suc.
Our histochemical GUS assays revealed that OsSUT2 is highly expressed in leaf mesophyll cells but not in the vascular bundle itself (Fig. 2A). Interestingly, in developing caryopses, we found that OsSUT2 expression was detected preferentially in the cross cell layer of seed coats (Fig. 2J). It is known that this cross cell layer contains some chloroplasts and functions in supplying photoassimilates to the endosperm (Cochrane and Duffus, 1979; Ebenezer et al., 1990). The high expression of OsSUT2 in seed coats was confirmed by real-time PCR analysis (Supplemental Fig. S4). We were not able to observe any significantly altered expression of OsSUT2 in response to environmental stress stimuli such as drought, cold, or salt treatment (data not shown), suggesting that OsSUT2 does not likely function in stress responses. Previously reported results have shown that HvSUT2 is strongly expressed in pericarps (Weschke et al., 2000). In rice, barley, and wheat (Triticum aestivum), the pericarp supplies photoassimilates to the endosperm (Cochrane and Duffus, 1979). This suggests that cereals have a high demand for Suc transporters to facilitate the transport of reserve Suc across the tonoplast in photosynthetic assimilatory cells. It is noteworthy that the regions around emerging lateral roots (Fig. 2D) and the pedicel parts of spikelets after fertilization (Fig. 2F) also exhibited high GUS activity. This suggests that OsSUT2 function is required for efficient transport of reserve Suc in tissues with a high metabolic turnover.
The tonoplast monosaccharide transporters (TMTs) have been characterized in Arabidopsis (Wormit et al., 2006; Wingenter et al., 2010) and rice (Cho et al., 2010) and are thought to function in the transport of monosaccharides, Glc and Fru, from the cytosol to the vacuole lumen using a H+-sugar antiport mechanism. Based on their expression patterns, the Arabidopsis TMTs are thought to play a major role in osmotic stress responses, whereas rice TMTs mainly function in sugar retrieval during sugar translocation. Our results here indicate that OsSUT2 functions as a H+-symporter rather than an antiporter. Tonoplast H+-Suc antiporter activity has been characterized previously (Briskin et al., 1985; Greutert and Keller, 1993), but the corresponding genes remain to be identified. It is likely that Suc symporters and antiporters function together to control Suc flux across the tonoplast and that a regulatory system must be involved to avoid a futile cycle of H+ and Suc transport.
OsSUT2 Function Is Essential for the Normal Growth of Rice Plants
Suppression of PtaSUT4 in RNAi transgenic poplar had no obvious effect on plant growth but led to altered biomass partitioning, with a higher ratio of leaf area to stem volume compared with wild-type plants. Thus, PtaSUT4 appeared to modulate Suc efflux and utilization (Payyavula et al., 2011). However, no null mutant for a tonoplast SUT has been described to date. Here, we characterize a rice ossut2 mutant in detail and describe its function in planta. Rice plants lacking OsSUT2 display severe growth defects such as reduced tiller number, plant height, and grain and root weights compared with the wild type and OsSUT2-complemented lines (Figs. 4 and 5). Hence, OsSUT2 function is necessary for normal plant growth in rice. In the ossut2 mutant, the level of Suc was found to be increased by about 2-fold in the source leaves (Fig. 6), mostly because of the loss of Suc transport ability across the tonoplast to the cytosol. Thus, it is probable that the accumulation of sugar in ossut2, due to the decreased transport of Suc from the vacuole lumen to the cytosol, reduces the metabolic flux, thereby interfering with efficient energy supply for normal plant growth. In addition, we found that ossut2 has a significantly reduced sugar export ability (Fig. 7), which indicates that the reduced transport of Suc to the cytosol in ossut2 likely also interferes with Suc translocation to sink organs, thus affecting plant growth.
It is noteworthy that the net photosynthetic activity of the ossut2 mutant was not altered (Supplemental Fig. S5). In this regard, the higher levels of Glc and Fru could be the result of the hydrolysis of accumulated Suc by vacuolar invertase (Sturm and Tang, 1999; Ji et al., 2007). Given that soluble sugar levels in the cytosol control photosynthesis (Rolland et al., 2002), considerable amounts of Glc and Fru might be accumulated in the vacuole lumen in ossut2, which thereby do not affect photosynthetic capacity.
In summary, we have characterized the expression and function of the rice tonoplast Suc transporter OsSUT2 in detail. Our results demonstrate that OsSUT2 is most likely to be involved in Suc transport across the tonoplast from the vacuole to the cytosol, a process that is required to support normal plant growth in rice.
MATERIALS AND METHODS
Plant Materials
Wild-type rice (Oryza sativa ‘Hwayoung’), the OsSUT2 mutant ossut2, and complemented transgenic rice lines were used in these experiments. Rice plants were grown in a greenhouse at 30°C during the day and 20°C at night in a light/dark cycle of 14 h/10 h or in an experimental field plot of Kyung Hee University under natural environmental conditions during the summer. Leaves were sampled from 4-week-old plants grown in the greenhouse to analyze primary carbon metabolites and sugar export ability. Roots were collected from 4-week-old plants grown in the greenhouse to determine dry weights. Plant height, tiller number, and 1,000-grain weight were analyzed from mature plants grown in a rice paddy field.
Cloning of the OsSUT2 Full-Length cDNA Clone
The full-length cDNA of OsSUT2 was isolated by RT-PCR from the leaves of 4-week-old rice (cv Dongjin) plants using gene-specific primers encompassing the translation start codon and 3′ untranslated region: 5′-CTCTTCTGAACTAACCCAAAGAT-3′ and 5′-AACTCCTGCAACTTTTATTCACTA-3′. The cDNA was then subcloned into the pGEM-T Easy vector (Promega) and confirmed by sequencing. The OsSUT2 cDNA sequence has been deposited in the National Center for Biotechnology Information database with the accession number HQ540307.
Sequence Alignment and Phylogenetic Tree Construction
The deduced amino acid sequence of the OsSUT2 gene was aligned with previously reported SUT genes from other plant species using the ClustalW program (Thompson et al., 1994). A phylogenetic tree was constructed using MEGA software version 4.0 (Tamura et al., 2007) via the neighbor-joining method.
Construction of the OsSUT2-GFP Fusion Vector and Subcellular Localization Analysis
To examine the subcellular localization of OsSUT2 in rice, the entire open reading frame without the stop codon was amplified by PCR. The amplified product was digested with XbaI and XhoI and was cloned into the region between the CaMV35S promoter and sGFP of the JJ461 binary vector (Cho et al., 2009). The primers used were 5′-GCTCTAGACTCTTCTGAACTAACCCAAAGAT-3′ and 5′-CCCTCGAGATCGGTGACCTCTCCTCCTTGAT-3′ (the underlined sequences indicate XbaI and XhoI sites, respectively). The resulting CaMV35S::OsSUT2-GFP (pJJ1747) construct was used to transform rice. Leaves or isolated protoplasts from the CaMV35S::OsSUT2-GFP transgenic rice plants were examined using a confocal microscope (LSM 510 META; Zeiss). Chlorophyll autofluorescence was used as a chloroplast marker. The plasma membrane was stained with a lipophilic styryl dye, FM4-64 (Molecular Probes).
Construction of the pOsSUT2::GUS Fusion Vector
About 2 kb of the 5′ upstream region of the OsSUT2 gene was amplified by PCR using genomic DNA as a template. The primers used were 5′-TACAATCCAATACTCCAGTAGAC-3′ and 5′-CTTCTTCTCGTGTTTGCCTCCG-3′. The amplified product was subcloned into a pGEM-T Easy vector (Promega) and confirmed by sequencing. The cloned fragment was then inserted in front of the promoterless GUS reporter gene of pBI101 (Jefferson, 1987). As a result, the promoter region of the OsSUT2 gene was fused to a GUS gene linked to the terminator of the nopaline synthase gene (Nos). This construct was designated pOsSUT2::GUS.
Agrobacterium-Mediated Rice Transformation
To produce transgenic rice (cv Hwayoung) plants expressing CaMV35S::OsSUT2-GFP and pOsSUT2::GUS constructs, Agrobacterium tumefaciens LBA4404 strains harboring each of these vectors were grown on Agrobacterium broth medium supplemented with 25 mg L−1 kanamycin for 3 d at 28°C, and transgenic calli were obtained via the Agrobacterium-mediated cocultivation method as described previously (Jeon et al., 2000). Transgenic rice plants were regenerated from the transformed calli on selection medium containing 50 mg L−1 hygromycin and 250 mg L−1 cefotaxime.
Histochemical GUS Analysis
Various tissues from pOsSUT2::GUS transgenic plants were collected and stained in GUS reaction solution containing 0.2 m sodium phosphate (pH 7.0), 10 mm EDTA, 20% methanol, 2% dimethyl sulfoxide, and 0.1% 5-bromo-4-chloro-3-indole-β-glucuronide, as described previously by Cho et al. (2010). Briefly, samples were incubated at 37°C overnight and then transferred to 70% ethanol to remove chlorophyll. After staining, samples were fixed in a solution containing 50% ethanol, 5% acetic acid, and 3.7% formaldehyde. The stained samples were observed with a dissecting microscope (SZX12; Olympus). For longitudinal and transverse sectioning, the stained samples were embedded in Paraplast Plus (Sigma), and 10-μm sections were prepared using a microtome (Microm HM360). The sections mounted on slides were observed with a compound microscope.
Suc Transport Experiment in Yeast
An expression-optimized OsSUT2 clone (GenBank accession no. HQ830493) was synthesized by Blue Heron Biotechnology and inserted into the Gateway entry vector pENTR221. The resulting amino acid sequence of this clone is identical to the sequence determined from cDNA isolated from rice. For expression in yeast, the entry clone was recombined in vitro with the Gateway destination vector pDR196/GW (provided by Dr. Wolf Frommer, Carnegie Institution). This construct, named OsSUT2syn, as well as the native rice OsSUT2 coding region, the potato (Solanum tuberosum) Suc transporter StSUT1 (Schulze et al., 2000), and the empty vector were used to transform the yeast strain SEY6210 (MATα leu2-3,112 ura3-52 his3-Δ200 lys2-801 trpl-Δ901 suc2-Δ9; Brown and Bussey, 1993). The uptake of radiolabeled [14C]Suc into yeast cells was determined as described previously (Reinders and Ward, 2001).
Isolation of the OsSUT2 Mutant
A T-DNA-tagged OsSUT2 mutant, ossut2, was identified by searching the rice T-DNA Insertion Sequence Database (An et al., 2005a, 2005b; Jeong et al., 2006; http://www.postech.ac.kr/life/pfg/risd/index.html). A homozygous mutant (cv Hwayoung) was isolated by genomic DNA PCR analysis. The gene-specific primers used for the genotyping of ossut2 were 5′-TGCTGTATCCTAAAATAACCTGTTGATGG-3′ and 5′-CATTTCATACACACACCTTAACACAGCAC-3′. The T-DNA-specific primer used was 5′-GACCAGAAAGCCCATAAACCTTGGAGGC-3′.
Genetic Complementation Experiment
To complement the ossut2 mutant, the full-length OsSUT2 cDNA was amplified by PCR using the primers 5′-GGATCCCTCTTCTGAACTAACCCAAAGAT-3′ and 5′-GGTACCAACTCCTGCAACTTTTATTCACT-3′ (the underlined sequences indicate BamHI and KpnI sites, respectively). The PCR product was then subcloned into the pGEM-T Easy vector (Promega) and confirmed by sequencing. The insert digested with BamHI and KpnI was further subcloned between the maize (Zea mays) Ubi1 promoter and the Nos terminator of the Ubi/NC4300 binary vector carrying the Phosphomannose Isomerase gene as a selectable marker (provided by Dr. Pamela Ronald, University of California, Davis). The resulting construct, Ubi::OsSUT2 (pJJ2521), was used to transform the ossut2 mutant as described previously (Lucca et al., 2001).
RNA Isolation and PCR Analysis
For RT-PCR analysis of ossut2 and transgenic lines, total RNA was prepared from harvested samples using Trizol reagent and reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad). First-strand cDNAs were used in RT-PCR with gene-specific primers and control primers for the housekeeping genes Act1 (McElroy et al., 1990) and OsUBQ5 (Jain et al., 2006). The gene-specific primers used were as follows: for OsSUT2, 5′-CTATCATGATGGTGTGAGAATG-3′ and 5′-CTACCCAGTGACACAATAACCT-3′; for Act1, 5′-GGAACTGGTATGGTCAAGGC-3′ and 5′-AGTCTCATGGATACCCGCAG-3′; and for OsUBQ5, 5′-GACTACAACATCCAGAAGGAGTC-3′ and 5′-TCATCTAATAACCAGTTCGATTTC-3′. RT-PCR analyses were repeated at least three times, giving similar results using previously described methods (Cho et al., 2005).
For quantitative real-time PCR, endosperm and seed coat cDNA was prepared from seeds harvested 6 to 10 DAF according to the method described by Cho et al. (2005). The expression level of OsUBQ5 was used for the normalization of real-time PCR results (Jain et al., 2006). All reactions were performed in triplicate using the HotStart-IT SYBR Green qPCR Master Mix (USB) and 7500 Real-Time PCR System (Applied Biosystems), according to the manufacturers’ instructions. Changes in gene expression were analyzed by the comparative cycle threshold method and Sequence Detector Systems version 1.2 software (Applied Biosystems). The gene-specific primers used for quantitative real-time PCR were as follows: for OsSUT2, 5′-TGTGGCAAAGAATATGGATTAT-3′ and 5′-CTACCCAGTGACACAATAACCT-3′; and for OsUBQ5, 5′-GACTACAACATCCAGAAGGAGTC-3′ and 5′-TCATCTAATAACCAGTTCGATTTC-3′.
Determination of Soluble Sugars and Starch
Approximately 100 mg of leaf blades was harvested at the end of day and night from 4-week-old plants grown in the greenhouse. Suc, Glc, Fru, and starch were measured using NAD(P)H-coupled enzymatic methods in the soluble and residual fractions of ethanol-water extracts (Lee et al., 2005). The measured metabolite contents were normalized to the leaf fresh weights.
Sugar Export Assay
Sugar export assays were essentially carried out as described by Wingenter et al. (2010) with slight modifications suitable for rice. Briefly, mature leaf blades of 4-week-old plants grown in the greenhouse were collected by cutting the bottom of the leaf blades at the end of day, and the cut regions were soaked in a 15 mm EDTA solution (pH 7.25). To avoid blockage of the cut end regions, the end-most parts of each 1 mm were consecutively recut off four times at 1-min intervals under the buffer solution. The leaf blade ends were then immediately transferred into reaction tubes filled with 500 μL of the same buffer solution and incubated for 16 h under dark conditions. Subsequently, the solutions containing the exudates were collected to quantify total soluble sugars including Suc, and also Glc and Fru, since Suc is hydrolyzed into its monosaccharides.
Sugars were analyzed using a HPLC system (Agilent Technologies 1200 series) equipped with a temperature-controlled column chamber and autosampler and a refractive index detector (Agilent Technologies). The HPLC system and detectors were controlled, and results were calculated using the ChemStation software (Agilent Technologies). The carbohydrate column (5 μm, 4.6 × 250 mm; Agilent Technologies) was used for separation of the sugars in the exudates. Samples prepared were injected in a volume of 100 μL with a mobile phase in a 75% acetonitrile solution at a speed of 1.4 mL min−1 after mixing with acetonitrile (1:1, v/v) and filtration through a 0.2-μm SmartPor syringe filter (Woongi Science). Peaks were identified by comparing the retention times with those of standard sugars. This experiment was carried out in triplicate.
Measurement of Net Photosynthetic Activity
Net photosynthetic activity was measured on the most recent fully expanded leaves of 8-week-old rice plants using a portable gas-exchange system (Li-6400; Li-Cor) at sequentially changed photon flux density, leaf temperature of 25°C, and reference CO2 concentration of 350 μmol mol−1.
Statistical Analysis
Student’s t test was performed using Microsoft Excel to determine the significance of the differences in the morphological data and metabolite contents among wild-type, ossut2, and complemented rice lines.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HQ540307 (OsSUT2) and HQ830493 (OsSUT2syn).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of SUT genes constructed using the neighbor-joining method.
Supplemental Figure S2. Comparison of the deduced amino acid sequences for the rice tonoplast Suc transporter OsSUT2 and the previously identified tonoplast-localized SUTs, HvSUT2, LjSUT4, AtSUT4, and PtaSUT4.
Supplemental Figure S3. Subcellular localization of the OsSUT2-GFP fusion protein in leaf mesophyll protoplasts isolated from transgenic rice plants expressing OsSUT2-GFP.
Supplemental Figure S4. Expression levels of OsSUT2 in endosperms and seed coats determined by quantitative real-time PCR.
Supplemental Figure S5. Net photosynthetic activity of the ossut2 mutant.
Acknowledgments
We thank Dr. Pamela Ronald and Dr. Wolf Frommer for providing the binary vector Ubi/NC4300 carrying the Phosphomannose Isomerase gene as a selectable marker and the Gateway destination vector pDR196/GW, respectively.
References
- An G, Jeong DH, Jung KH, Lee S. (2005a) Reverse genetic approaches for functional genomics of rice. Plant Mol Biol 59: 111–123 [DOI] [PubMed] [Google Scholar]
- An G, Lee S, Kim SH, Kim SR. (2005b) Molecular genetics using T-DNA in rice. Plant Cell Physiol 46: 14–22 [DOI] [PubMed] [Google Scholar]
- Aoki N, Hirose T, Scofield GN, Whitfeld PR, Furbank RT. (2003) The sucrose transporter gene family in rice. Plant Cell Physiol 44: 223–232 [DOI] [PubMed] [Google Scholar]
- ap Rees T, Hill SA. (1994) Metabolic control analysis of plant metabolism. Plant Cell Environ 17: 587–599 [Google Scholar]
- Briskin DP, Thornley WR, Wyse RE. (1985) Membrane transport in isolated vesicles from sugarbeet taproot. II. Evidence for a sucrose/H-antiport. Plant Physiol 78: 871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Bussey H. (1993) The yeast KRE9 gene encodes an O glycoprotein involved in cell surface β-glucan assembly. Mol Cell Biol 13: 6346–6356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkle L, Hibberd JM, Quick WP, Kühn C, Hirner B, Frommer WB. (1998) The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiol 118: 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D, Reinders A, Ward JM. (2003) Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. J Biol Chem 278: 44320–44325 [DOI] [PubMed] [Google Scholar]
- Chincinska IA, Liesche J, Krügel U, Michalska J, Geigenberger P, Grimm B, Kühn C. (2008) Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiol 146: 515–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JI, Burla B, Lee DW, Ryoo N, Hong SK, Kim HB, Eom JS, Choi SB, Cho MH, Bhoo SH, et al. (2010) Expression analysis and functional characterization of the monosaccharide transporters, OsTMTs, involving vacuolar sugar transport in rice (Oryza sativa). New Phytol 186: 657–668 [DOI] [PubMed] [Google Scholar]
- Cho JI, Lee SK, Ko S, Kim HK, Jun SH, Lee YH, Bhoo SH, Lee KW, An G, Hahn TR, et al. (2005) Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.). Plant Cell Rep 24: 225–236 [DOI] [PubMed] [Google Scholar]
- Cho JI, Ryoo N, Eom JS, Lee DW, Kim HB, Jeong SW, Lee YH, Kwon YK, Cho MH, Bhoo SH, et al. (2009) Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol 149: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane MP, Duffus CM. (1979) Morphology and ultrastructure of immature cereal grains in relation to transport. Ann Bot (Lond) 44: 67–72 [Google Scholar]
- Ebenezer GAI, Amirthalingam M, Ponsamuel J, Davanandan P. (1990) Role of palea and lemma in the development of rice caryopsis. J Indian Bot Soc 69: 245–250 [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT, Scofield GN, Hirose T, Wang XD, Patrick JW, Offler CE. (2001) Cellular localisation and function of a sucrose transporter OsSUT1 in developing rice grains. Aust J Plant Physiol 28: 1187–1196 [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greutert H, Keller F. (1993) Further evidence for stachyose and sucrose/H+ antiporters on the tonoplast of Japanese artichoke (Stachys sieboldii) tubers. Plant Physiol 101: 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. (2004) Codon bias and heterologous protein expression. Trends Biotechnol 22: 346–353 [DOI] [PubMed] [Google Scholar]
- Hirose T, Takano M, Terao T. (2002) Cell wall invertase in developing rice caryopsis: molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol 43: 452–459 [DOI] [PubMed] [Google Scholar]
- Hirose T, Zhang Z, Miyao A, Hirochika H, Ohsugi R, Terao T. (2010) Disruption of a gene for rice sucrose transporter, OsSUT1, impairs pollen function but pollen maturation is unaffected. J Exp Bot 61: 3639–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345: 646–651 [DOI] [PubMed] [Google Scholar]
- Jefferson RA. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, et al. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22: 561–570 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al. (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45: 123–132 [DOI] [PubMed] [Google Scholar]
- Ji X, Van den Ende W, Schroeven L, Clerens S, Geuten K, Cheng S, Bennett J. (2007) The rice genome encodes two vacuolar invertases with fructan exohydrolase activity but lacks the related fructan biosynthesis genes of the Pooideae. New Phytol 173: 50–62 [DOI] [PubMed] [Google Scholar]
- Kühn C, Grof CP. (2010) Sucrose transporters of higher plants. Curr Opin Plant Biol 13: 288–298 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Wipf D, Frommer WB. (2004) Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu Rev Plant Biol 55: 341–372 [DOI] [PubMed] [Google Scholar]
- Lauterbach C, Niedermeier M, Besenbeck R, Stadler R, Sauer N. (2007) Immunolocalization of the PmSUC1 sucrose transporter in Plantago major flowers and reporter-gene analyses of the PmSUC1 promoter suggest a role in sucrose release from the inner integument. Plant Biol (Stuttg) 9: 357–365 [DOI] [PubMed] [Google Scholar]
- Lee JW, Lee DS, Bhoo SH, Jeon JS, Lee YH, Hahn TR. (2005) Transgenic Arabidopsis plants expressing Escherichia coli pyrophosphatase display both altered carbon partitioning in their source leaves and reduced photosynthetic activity. Plant Cell Rep 24: 374–382 [DOI] [PubMed] [Google Scholar]
- Lee SK, Jeon JS, Börnke F, Voll L, Cho JI, Goh CH, Jeong SW, Park YI, Kim SJ, Choi SB, et al. (2008) Loss of cytosolic fructose-1,6-bisphosphatase limits photosynthetic sucrose synthesis and causes severe growth retardations in rice (Oryza sativa). Plant Cell Environ 31: 1851–1863 [DOI] [PubMed] [Google Scholar]
- Lemoine R, Bürkle L, Barker L, Sakr S, Kühn C, Regnacq M, Gaillard C, Delrot S, Frommer WB. (1999) Identification of a pollen-specific sucrose transporter-like protein NtSUT3 from tobacco. FEBS Lett 454: 325–330 [DOI] [PubMed] [Google Scholar]
- Lim JD, Cho JI, Park YI, Hahn TR, Choi SB, Jeon JS. (2006) Sucrose transport from source to sink seeds in rice. Physiol Plant 126: 572–584 [Google Scholar]
- Linka N, Weber AP. (2010) Intracellular metabolite transporters in plants. Mol Plant 3: 21–53 [DOI] [PubMed] [Google Scholar]
- Lucca P, Ye X, Potrykus I. (2001) Effective selection and regeneration of transgenic rice plants with mannose as selective agent. Mol Breed 7: 43–49 [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE. (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R. (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie EH, Yang J, Hubbart S, Horton P, Peng S. (2002) Are there associations between grain-filling rate and photosynthesis in the flag leaves of field-grown rice? J Exp Bot 53: 2217–2224 [DOI] [PubMed] [Google Scholar]
- Nakano H, Makino A, Mae T. (1995) Effects of panicle removal on the photosynthetic characteristics of the flag leaf of rice plants during the ripening stage. Plant Cell Physiol 36: 653–659 [Google Scholar]
- Neuhaus HE. (2007) Transport of primary metabolites across the plant vacuolar membrane. FEBS Lett 581: 2223–2226 [DOI] [PubMed] [Google Scholar]
- Payyavula RS, Tay KHC, Tsai C-J, Harding SA. (2011) The sucrose transporter family in Populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J 65: 757–770 [DOI] [PubMed] [Google Scholar]
- Reinders A, Sivitz AB, Starker CG, Gantt JS, Ward JM. (2008) Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lotus japonicus. Plant Mol Biol 68: 289–299 [DOI] [PubMed] [Google Scholar]
- Reinders A, Ward JM. (2001) Functional characterization of the α-glucoside transporter Sut1p from Schizosaccharomyces pombe, the first fungal homologue of plant sucrose transporters. Mol Microbiol 39: 445–454 [DOI] [PubMed] [Google Scholar]
- Rennie EA, Turgeon R. (2009) A comprehensive picture of phloem loading strategies. Proc Natl Acad Sci USA 106: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riens B, Lohaus G, Heineke D, Heldt HW. (1991) Amino acid and sucrose content determined in the cytosolic, chloroplastic, and vacuolar compartments and in the phloem sap of spinach leaves. Plant Physiol 97: 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB. (1994) Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J 13: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N. (2007) Molecular physiology of higher plant sucrose transporters. FEBS Lett 581: 2309–2317 [DOI] [PubMed] [Google Scholar]
- Schulze W, Weise A, Frommer WB, Ward JM. (2000) Function of the cytosolic N-terminus of sucrose transporter AtSUT2 in substrate affinity. FEBS Lett 485: 189–194 [DOI] [PubMed] [Google Scholar]
- Schulze WX, Reinders A, Ward J, Lalonde S, Frommer WB. (2003) Interactions between co-expressed Arabidopsis sucrose transporters in the split-ubiquitin system. BMC Biochem 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield GN, Hirose T, Aoki N, Furbank RT. (2007) Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J Exp Bot 58: 3155–3169 [DOI] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Ward JM. (2008) Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol 147: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski TL, Meeley R, Braun DM. (2009) Sucrose transporter1 functions in phloem loading in maize leaves. J Exp Bot 60: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Truernit E, Gahrtz M, Sauer N. (1999) The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J 19: 269–278 [DOI] [PubMed] [Google Scholar]
- Sturm A, Tang GQ. (1999) The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci 4: 401–407 [DOI] [PubMed] [Google Scholar]
- Sun AJ, Xu HL, Gong WK, Zhai HL, Meng K, Wang YQ, Wei XL, Xiao GF, Zhu Z. (2008) Cloning and expression analysis of rice sucrose transporter genes OsSUT2M and OsSUT5Z. J Integr Plant Biol 50: 62–75 [DOI] [PubMed] [Google Scholar]
- Sun Y, Reinders A, LaFleur KR, Mori T, Ward JM. (2010) Transport activity of rice sucrose transporters OsSUT1 and OsSUT5. Plant Cell Physiol 51: 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevanion SJ. (2002) Regulation of sucrose and starch synthesis in wheat (Triticum aestivum L.) leaves: role of fructose 2,6-bisphosphate. Planta 215: 653–665 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Weise A, Barker L, Kühn C, Lalonde S, Buschmann H, Frommer WB, Ward JM. (2000) A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Sauer N, Wang Q, Neubohn B, Weber H, Wobus U. (2000) Sucrose transport into barley seeds: molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J 21: 455–467 [DOI] [PubMed] [Google Scholar]
- Williams LE, Lemoine R, Sauer N. (2000) Sugar transporters in higher plants: a diversity of roles and complex regulation. Trends Plant Sci 5: 283–290 [DOI] [PubMed] [Google Scholar]
- Winder TL, Sun J, Okita TW, Edwards GE. (1998) Evidence for the occurrence of feedback inhibition of photosynthesis in rice. Plant Cell Physiol 39: 813–820 [Google Scholar]
- Wingenter K, Schulz A, Wormit A, Wic S, Trentmann O, Hoermiller II, Heyer AG, Marten I, Hedrich R, Neuhaus HE. (2010) Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis. Plant Physiol 154: 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Robinson DG, Heldt HW. (1993) Subcellular volumes and metabolite concentrations in barley leaves. Planta 191: 180–190 [Google Scholar]
- Wormit A, Trentmann O, Feifer I, Lohr C, Tjaden J, Meyer S, Schmidt U, Martinoia E, Neuhaus HE. (2006) Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18: 3476–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]