Abstract
The BRASSINOSTEROID INSENSITIVE1 (BRI1) receptor kinase has recently been shown to possess tyrosine kinase activity, and preventing autophosphorylation of the tyrosine-831 regulatory site by site-directed mutagenesis enhances shoot growth. In this study, we characterized the increased leaf growth of Arabidopsis (Arabidopsis thaliana) plants expressing BRI1(Y831F)-Flag compared with BRI1-Flag (both driven by the native promoter and expressed in the bri1-5 weak allele background) and provide insights into the possible mechanisms involved. On average, relative leaf growth rate was increased 16% in the Y831F plants (in the bri1-5 background), and the gain of function of the Y831F-directed mutant was dominant in the wild-type background. Leaves were larger as a result of increased cell numbers and had substantially increased vascularization. Transcriptome analysis indicated that genes associated with brassinolide biosynthesis, secondary cell wall biosynthesis and vascular development, and regulation of growth were altered in expression and may contribute to the observed changes in leaf architecture and whole plant growth. Analysis of gas exchange and chlorophyll fluorescence indicated that Y831F mutant plants had higher rates of photosynthesis, and metabolite analysis documented enhanced accumulation of starch, sucrose, and several amino acids, most prominently glycine and proline. These results demonstrate that mutation of BRI1 can enhance photosynthesis and leaf growth/vascularization and may suggest new approaches to increase whole plant carbon assimilation and growth.
Brassinosteroids (BRs) are essential plant steroid hormones that regulate multiple aspects of growth and development, including cell elongation, cell division, vascular differentiation, seed germination, timing of senescence, male fertility, and organ formation (Clouse and Sasse, 1998; Altmann, 1999; Nakaya et al., 2002; Gonzalez et al., 2010). It is known that BRs bind to the BRASSINOSTEROID-INSENSITIVE1 (BRI1) receptor kinase, which functions in conjunction with the coreceptor BRASSINOSTEROID-ASSOCIATED KINASE1 (BAK1) in hormone perception and signal transduction (Li et al., 2002; Nam and Li, 2002). The BR signal transduction pathway ultimately controls the phosphorylation status of the transcription factors BZR1 and BZR2/BES1 in the nucleus (Kim et al., 2009) and thereby regulates the expression of more than 700 genes in Arabidopsis (Arabidopsis thaliana; Goda et al., 2002; Müssig et al., 2002; Vert et al., 2005). Many specific components of the signal transduction pathway have been identified and have recently been reviewed (Kim and Wang, 2010; Tang et al., 2010).
BRs are now considered essential chemical signals and plant hormones, and accordingly, manipulation of endogenous BR content or BR signaling has a profound effect on plant growth. For example, overexpression of DWARF4, which encodes an enzyme that catalyzes a rate-limiting step in BR biosynthesis, enhances plant growth and seed yield in Arabidopsis (Choe et al., 2001), while down-regulation of BR biosynthesis by RNA interference of biosynthetic genes results in a semidwarf phenotype (Chung et al., 2010). Similarly, transgenic rice plants overexpressing a sterol C-22 hydroxylase that catalyzes a key step in BR biosynthesis had increased levels of BR hormone intermediates downstream of 6-deoxo-cathasterone, and plants had higher rates of CO2 assimilation, more tillers, and increased biomass and seed yields by 14% to 44% (Wu et al., 2008). Likewise, exogenous application of BRs has been shown to increase photosynthetic rate and photochemical efficiency (Yu et al., 2004; Xia et al., 2009; Wang et al., 2010). Another potential approach to increase grain yield in rice is to control plant architecture by altering BR signaling. Transcriptional down-regulation of OsBRI1 (Morinaka et al., 2006) or OsBAK1 (Li et al., 2009) altered plant architecture, and both were shown to have the potential to increase grain yield at high planting densities; however, in neither case was the grain yield benefit demonstrated to occur.
A common mechanistic property associated with the activation of many animal and plant receptor kinases is ligand-dependent homodimerization or heterodimerization, followed by activation of the cytoplasmic kinase domains via autophosphorylation and subsequent transphosphorylation of downstream components involved in the specific signaling pathway (Becraft, 2002; Schlessinger, 2002). For example, Wang et al. (2005a) showed that BRI1 and BAK1 activation in vivo follows a sequential transphosphorylation model in which BRI1 controls signaling specificity by directly binding the hormone ligand followed by some autophosphorylation. The coreceptor BAK1 is then activated by BRI1-dependent transphosphorylation, and BAK1 subsequently enhances signaling output through reciprocal BRI1 transphosphorylation (Wang et al., 2008). Numerous sites of Ser and Thr phosphorylation have been identified on both BRI1 and BAK1 (Oh et al., 2000; Wang et al., 2005a, 2005b, 2008), consistent with their classification as Ser/Thr-protein kinases (Shiu and Bleecker, 2001). Interestingly, Tyr phosphorylation of BRI1 (Oh et al., 2009b) and BAK1 (Oh et al., 2010) was recently reported, indicating that both kinases have dual specificity, thereby introducing the possibility that Tyr phosphorylation plays a role in BR signaling. Indeed, transgenic plants expressing BRI1(Y831F)-Flag in the bri1-5 weak allele background are larger than those expressing wild-type BRI1-Flag (Oh et al., 2009b), suggesting that phosphorylation of Tyr-831 attenuates plant growth. To further understand the role of Tyr phosphorylation in BR signaling, we compared transgenic Arabidopsis plants expressing BRI1(Y831F)-Flag with plants expressing wild-type BRI1-Flag, both in the bri1-5 weak allele background. We characterized these plants in terms of leaf morphology and relative growth rate (RGR), photosynthetic parameters, and gene expression in an effort to understand the basis for the increased growth. The results obtained confirm the importance of Tyr phosphorylation in plant receptor kinase signaling and indicate the potential for improving plant performance by engineering receptor kinase function.
RESULTS
Analysis of Leaf Morphology and Plant Growth
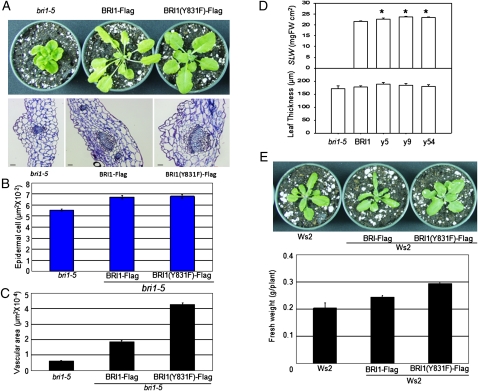
A prominent feature of transgenic plants expressing BRI1(Y831F)-Flag in the bri1-5 background is increased leaf size and shoot biomass relative to plants expressing wild-type BRI1-Flag (Oh et al., 2009a, 2009b), and an important question to address is the basis for the enhanced growth (Fig. 1A). Observations of leaf cross-sections of bri1-5, BRI1-Flag, and BRI1(Y831F)-Flag plants established small differences in epidermal cell size (Fig. 1B) and leaf thickness and specific leaf weight (Fig. 1D), suggesting that leaves of the Y831F mutant plants are larger as a result of more cells rather than larger cells. More cells were also apparent in the vascular tissue associated with the leaf midrib. As shown in Figure 1A (bottom panel) and quantified in Figure 1C, the Y831F mutant plants had an approximately 2-fold greater vascular cross-sectional area compared with BRI1-Flag plants. In vascular development, procambial cells can be formed by divisions of the procambial stem cells themselves or by recruitment of cells from adjacent regions (Esau, 1965). Because increased vascularization of the Y831F plants is associated with increased leaf size, it appears that cell divisions within the procambium have been increased; thus, the Y831F growth phenotype is distinct from the thickvein mutant, which displays increased vascularization but reduced leaf size as a result of increased outside cell recruitment (Clay and Nelson, 2005). Control of cell division, along with cell expansion, by BRs is well known (Nakaya et al., 2002; Gonzalez et al., 2010).
Figure 1.
Plants expressing BRI1(Y831F)-Flag are larger than plants expressing wild-type BRI1-Flag protein in the bri1-5 mutant (A–D) or the wild-type Ws2 (E) background. A, Thirty-three-day-old plants (bri1-5 background) grown in short days (8 h of light/16 h of dark). The bottom panel shows leaf cross-sections through the midrib region of the three genotypes stained with toluidine blue. B, Epidermal cell area. C, Vascular area measured from images similar to those in A. D, Specific leaf weight (SLW) and leaf thickness. Results with three independent transgenic lines (y5, y9, and y54) of Y831F mutants are shown. FW, Fresh weight. E, Transgenes expressed in the Ws2 background. The bottom panel shows shoot fresh weight.
The prominent Y831F phenotype of increased shoot growth was not dependent on the bri1-5 background, as indicated by several lines of evidence. First, expression of BRI1(Y831F)-Flag in wild-type ecotype Wassilewskija (Ws2) plants also produced a growth increase and altered leaf shape (Fig. 1E), indicating that the Y831F mutation appears to be at least semidominant. Second, the content of BRI1-Flag protein and endogenous BRI1-5 protein was similar for transgenic plants expressing native sequence BRI1-Flag or the Y831F mutant (Supplemental Fig. S1). This rules out the possibility, for example, that the Y831F mutant protein somehow specifically stabilizes the BRI1-5 protein, which is a functional BRI1 receptor that is normally retained in the endoplasmic reticulum, where it is degraded (Hong et al., 2008; Belkhadir et al., 2010). Thus, the Y831F phenotype is not background specific, but for simplicity the remainder of these studies are focused on comparisons between wild-type BRI1-Flag and the Y831F mutant in the bri1-5 background.
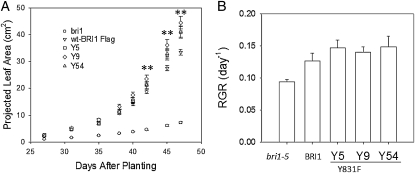
Leaf-area development of the genotypes was examined as an indicator of plant growth (Fig. 2). Three independent lines of the Y831F mutant (designated Y5, Y9, and Y54 in Fig. 2) grew faster and had larger cumulative leaf areas than BRI1-Flag plants. The last three times the plants were imaged, the differences in leaf area per plant were significantly different (P < 0.05; Fig. 2A). The average RGRs were about 15% higher for the Y831F mutants compared with BRI1-Flag plants (Fig. 2B). The bri1-5 mutant grew much more slowly than the Y831F mutants or BRI1-Flag plants. Interestingly, even though the bri1-5 mutant had dramatically less total leaf area compared with the other genotypes (Fig. 2A), RGR was surprisingly high (Fig. 2B), indicating that small differences in RGR can have a dramatic impact on final plant size. Thus, the 15% increase in RGR of the Y831F mutants reflects a substantial increase in growth capacity.
Figure 2.
A, Plant growth measured as leaf area increase over time of bri1-5, BRI-Flag, and three independent transgenic lines of BRI1(Y831F)-Flag (y5, y9, and y54). B, Calculated growth rates averaged over the period of growth from 27 to 47 d after planting. n = 30; ** P < 0.01.
It is noteworthy that the Y831F mutation had no significant effect on seed size (Supplemental Fig. S2). This is important because seed size can influence plant growth (El-Lithy et al., 2004). However, effects of seed size on growth are restricted to early stages of development (e.g. 10 d post sowing), while the growth advantage of the Y831F mutant was most apparent at later stages (e.g. more than 40 d post sowing). Thus, differences in growth rate of the Y831F mutant were not associated with altered seed size.
Many Genes Are Differentially Expressed in Y831F Plants
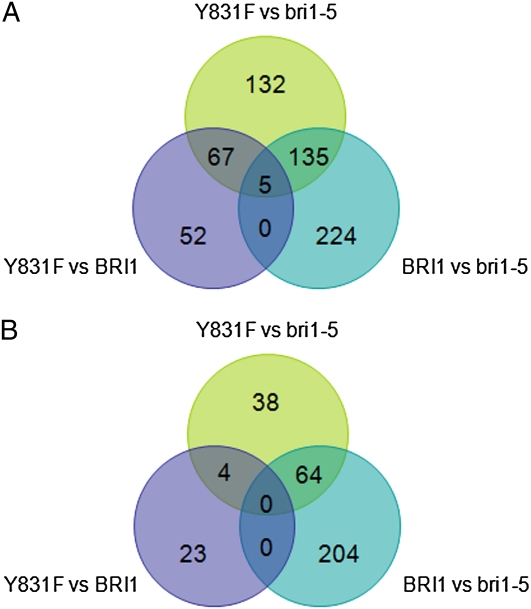
We conducted a genome-wide analysis of the BRI1(Y831F)-responsive transcriptome using Affymetrix ATH1 gene arrays. An overall view of differential gene expression among the three genotypes is presented as a Venn diagram in Figure 3. Comparisons were made between the two transgenics (the Y831F mutant and wild-type BRI1-Flag plants) and bri1-5. Genetic up-regulation of BR signaling by expression of BRI1-Flag in the bri1-5 background resulted in increased expression of 364 genes and decreased expression of 268 genes (Fig. 3, BRI1 versus bri1-5 comparison), similar in magnitude to the changes in gene expression that occur in wild-type plants in response to exogenous brassinolide (BL; Vert et al., 2005). Likewise, expression of BRI1(Y831F)-Flag in the bri1-5 background resulted in up-regulation of 339 genes, but only 106 genes were down-regulated (Fig. 3, Y831F versus bri1-5 comparison), indicating significant changes in gene expression when phosphorylation of Tyr-831 is prevented. Comparisons between Y831F and BRI1-Flag plants indicated that 124 genes were up-regulated and 27 genes were down-regulated in Y831F (Fig. 3, Y831F versus BRI1 comparison). These differentially regulated genes are presented in Supplemental Table S1 and may help explain the basis for the enhanced growth of the Y831F plants. For example, many of the genes up-regulated in Y831F plants are involved in secondary cell wall biosynthesis or are associated with vascular development (Table I), which may partially explain the increased vascularization observed (Fig. 1, A and C). It is recognized that three cellulose synthase (CESA) genes (CESA4, CESA7, and CESA8) are essential for cellulose synthesis for secondary cell wall development in vascular bundles (Taylor et al., 2003; Persson et al., 2005). These CESA genes are responsible for the irregular xylem (irx) mutants irx1, irx3, and irx5 (Taylor et al., 1999, 2000; Gardiner et al., 2003) and are thought to function together as subunits of a cellulose synthesis protein complex (Gardiner et al., 2003), and all were up-regulated in the Y831F mutants. Additional secondary cell wall biosynthetic genes were also up-regulated in Y831F plants, including the COBRA-like protein 4 (IRX6), the glycosyl transferases IRX8 and IRX9, and laccase (IRX12). The involvement of these genes in secondary cell wall development was originally discovered based on strong coexpression with CESA7 (IRX3) and a clear irx phenotype when mutated (Brown et al., 2005). In addition, a number of vascular-related genes, which are up-regulated in response to an auxin transport inhibitor that induces vascular overgrowth, were also up-regulated in the Y831F plants. These genes included the cell wall-associated PEROXIDASE64 (PRX64), two lipid-transfer proteins, and an auxin response protein (AT3G25290). Finally, several transcription factors that appear to be involved in secondary cell wall biogenesis were up-regulated, including IXR11 (Brown et al., 2005), SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 (SND1), the SND1-regulated MYB transcription factor MYB58 (Zhou et al., 2009), and the homeodomain-containing protein IRX11 (Table I). It is noteworthy that none of the genes described above are up-regulated by BL in wild-type plants (Table I). However, BL is well known to up-regulate numerous genes involved in cell expansion, including cellulose biosynthesis and the cytoskeleton machinery (Vert et al., 2005). Interestingly, several genes involved in primary cell wall biosynthesis, including CESA1, CESA3, and CESA6, which encode proteins that likely function together as subunits of a hexameric plasma membrane complex (Desprez et al., 2007), are up-regulated by BL (Vert et al., 2005) but were not differentially expressed in Y831F mutants relative to BRI1-Flag plants.
Figure 3.
Venn diagrams based on microarray analysis showing the number of genes up- and down-regulated (FDR P < 0.05, fold change > 2.0) in transgenic Arabidopsis expressing BRI1-Flag or BRI1(Y831F)-Flag in the bri1-5 background. A, Genes up-regulated among a total of 16,035 genes monitored. B, Genes down-regulated among a total of 16,327 genes monitored. The microarray chips used in this study were Affymetrix ATH1 chips. [See online article for color version of this figure.]
Table I. Genes up-regulated in BRI1(Y831F)-Flag relative to BRI1-Flag transgenic plants that are associated with vascular development or secondary cell wall biosynthesis (FDR P < 0.05, fold change > 2.0).
Many lignin biosynthetic genes were up-regulated approximately 1.2-fold.
| Symbol | Description | GenBank Accession No. | Y831F/BRI1 | BL/Mocka_ |
| fold change | ||||
| Secondary cell wall biosynthesis | ||||
| AtCESA4 | Cellulose synthase A4 (IRX5) | AT5G44030 | 3.72 | – |
| AtCESA7 | Cellulose synthase A7 (IRX3) | AT5G17420 | 3.78 | – |
| AtCESA8 | Cellulose synthase A8 (IRX1) | AT4G18780 | 5.55 | – |
| COBL4 | COBRA-like protein 4 (IRX6) | AT5G15630 | 8.00 | – |
| IRX8 | Glycosyl transferase family 8 | AT5G54690 | 5.26 | – |
| IRX9 | Xylosyltransferase (IRX9) | AT2G37090 | 3.13 | – |
| IRX12 | Laccase | AT2G38080 | 4.24 | – |
| Vascular-related genes | ||||
| PRX4 | Peroxidase 64 | AT5G42180 | 78.6 | – |
| LTP4 | Lipid transfer protein 4 | AT5G59310 | 26.7 | – |
| LTP12 | Lipid transfer protein 12 | AT3G53980 | 28.6 | – |
| Auxin response protein | AT3G25290 | 4.88 | – | |
| Transcription factors | ||||
| IXR11 | Homeodomain protein | AT1G62990 | 2.03 | – |
| SND1 | NAC domain transcription factor | AT1G32770 | 3.42 | – |
| MYB58 | MYB transcription factor | AT1G16490 | 2.04 | – |
Data from Vert et al. (2005).
A number of genes involved in BR biosynthesis were also differentially regulated in Y831F plants (Table II). In Arabidopsis, BRs are synthesized from campesterol via a series of oxidative reactions catalyzed by cytochrome P450 (P450) enzymes, with some P450s catalyzing multiple steps. Two parallel pathways exist for the production of C28-BRs, referred to as the early and late C-6 oxidation pathways (Kim et al., 2005). In addition, a novel shortcut pathway has been proposed that involves C-23 hydroxylation of early 22-hydroxylated intermediates to directly form 3-dehydro-6-deoxoteasterone and 6-deoxotyphasterol (Ohnishi et al., 2006). The shortcut pathway involves CYP90C1/ROTUNDIFOLIA3 (ROT3) and CYP90D1, which generates intermediates that can be utilized by CYP85A2 (Kim et al., 2005) to form castasterone and BL, both of which are considered to be biologically active BRs. The genes encoding all three of these P450s were up-regulated in the Y831F mutants, along with CYP90A1/CPD, which encodes another steroid hydroxylase (Szekeres et al., 1996) that is thought to catalyze reactions just upstream of the intermediates formed in the shortcut pathway. It is generally accepted that the synthesis of BRs is regulated to a large extent at the transcriptional level (Vert et al., 2005); thus, the changes in transcript abundance observed in Y831F plants would predict increased levels of BRs, which could contribute to the increased growth observed.
Table II.
BR biosynthetic genes altered in expression in BRI1(Y831F)-Flag relative to BRI1-Flag transgenic plants (FDR P < 0.05, fold change > 2.0)
| Symbol | Description | GenBank Accession No. | Y831F/BRI1 | BL/Mocka |
| fold | ||||
| CYP90A1 | P450, CPD | AT5G05690 | 2.11 | – |
| CYP90C1 | P450, ROT3 | AT4G36380 | 1.14 | −1.5 |
| CYP90D1 | 3-Epi-6-deoxocathasterone, 23-monooxygenase | AT3G13730 | 3.15 | −1.8 |
| CYP85A2 | P450, BR6OX2 | AT3G30180 | 2.98 | −7.2 |
Data from Vert et al. (2005).
We also observed differential expression in Y831F plants of three genes that have been correlated with growth. One gene is GA20OX1, which encodes a GA 20-oxidase that has been identified as an intrinsic yield gene. When GA20OX1 is overexpressed in transgenic Arabidopsis, plants have higher endogenous GA content (Huang et al., 1998) and leaf growth is enhanced (Gonzalez et al., 2010). The expression of GA20OX1 was increased 1.8-fold in Y831F plants (Table III). Two other growth-associated genes were identified by transcript profiling of 21 Arabidopsis accessions differing in growth (Sulpice et al., 2009). It was reported that transcript levels of IPS1, which encodes myoinositol 1-phosphate synthase, are negatively correlated with biomass, whereas a Kelch repeat F-box protein (AT1G23390) is positively correlated with biomass (Sulpice et al., 2009). Interestingly, IPS1 was down-regulated relative to BRI1-Flag plants, while the F-box protein was up-regulated 2.6-fold (Table III). Thus, all three of these critical growth-associated genes were altered in expression in a manner that would predict increased growth of the Y831F mutant plants. Only IPS1 is regulated by BR signaling in wild-type plants (Table III), but even with this gene the expression is modulated to a greater extent in the Y831F plants.
Table III.
Growth-related genes altered in expression in BRI1(Y831F)-Flag relative to BRI1-Flag transgenic plants (FDR P < 0.05, fold change > 2.0)
| Symbol | Description | Gene | Y831F/BRI1 | +BL/Mocka |
| fold | ||||
| Increased leaf size when overexpressed (Gonzalez et al., 2010) | ||||
| GA20OX1 | GA 20-oxidase | AT4G25420 | 1.84 | – |
| Negatively correlated with growth (Sulpice et al., 2009) | ||||
| IPS1 | Myoinositol 1-phosphate synthase | AT4G39800 | −1.39 | −2.1 |
| Positively correlated with growth (Sulpice et al., 2009) | ||||
| F-box Kelch domain protein | AT1G23390 | 2.60 | – | |
Data from Vert et al. (2005).
Because microarrays can be very reliable for identifying differentially expressed genes when fold changes are generally large (Shi et al., 2008), as in this study, we did not consider it essential to evaluate changes in gene expression by quantitative real-time PCR using the same samples. In future studies, it will certainly be of interest to examine the expression of key genes identified in this study under a variety of different conditions.
BRI1(Y831F)-Flag Plants Have Enhanced Photosynthetic Rates
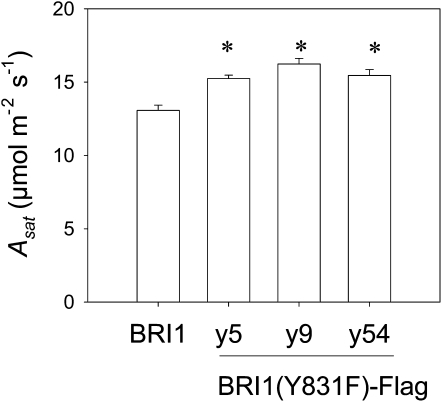
Gas exchange was measured on Y831F mutants and BRI1-Flag transgenic plants in order to determine whether the genotypes differed in CO2 assimilation capacity. Light-saturating CO2 assimilation rates (Asat) were measured under ambient [CO2] (Fig. 4). In these experiments, three independent transgenic lines expressing BRI1(Y831F)-Flag in the bri1-5 background were compared, and as shown, Asat values were about 20% higher in the Y831F mutants compared with the BRI1-Flag transgenics. The values of maximum Rubisco carboxylation rates (Vcmax), maximum electron transport rates (Jmax), and respiration rates in the light were calculated from A/intercellular CO2 concentration curves (Supplemental Fig. S3). The average values of Vcmax and Jmax were 18% and 19% higher, respectively, in Y831F mutants compared with BRI1-Flag transgenics. Because both Vcmax and Jmax were higher in the Y831F mutants, it can be derived and expected that net leaf photosynthetic rates will be greater than BRI1-Flag transgenics under growth conditions, regardless of whether photosynthesis is Rubisco or ribulose 1,5-bisphosphate (RuBP) regeneration limited (Farquhar et al., 1980; Sharkey, 1985; Long and Bernacchi, 2003; Sun et al., 2009). Because leaf thickness (Fig. 1D) and chlorophyll content (Supplemental Fig. S4) did not differ significantly among the genotypes tested, the differences in CO2 assimilation rate cannot be simply ascribed to changes in leaf architecture. Consistent with these results, chlorophyll fluorescence indicated that photochemical quenching and electron transport rate of individual leaves were on average 9% higher in the Y831F mutant compared with the BRI1-Flag transgenics (Supplemental Fig. S5). In parallel studies, we monitored carbon exchange rate under ambient conditions of whole plants rather than individual leaves and confirmed that CO2 assimilation under growth conditions was quite constant throughout majority of the day and night, except for the first approximately 20 min after switching off the lights, when a clear postillumination burst of CO2 was observed (Supplemental Fig. S6). Importantly, the results suggest that differences in CO2 assimilation rate were maintained throughout the photoperiod and thus could contribute to the increased growth observed in the Y831F mutant plants.
Figure 4.
Leaf Asat of Y831F mutants (lines y5, y9, and y54) and BRI1-Flag plants measured at 400 μL L−1 [CO2]. n = 24; * P < 0.05.
Diurnal Carbohydrate and Amino Acid Levels in Leaves
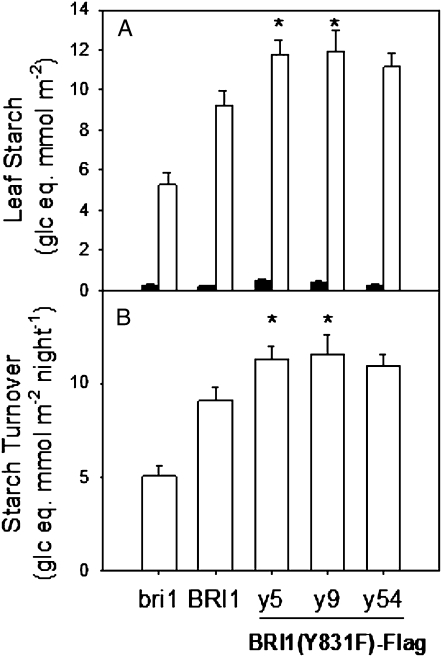
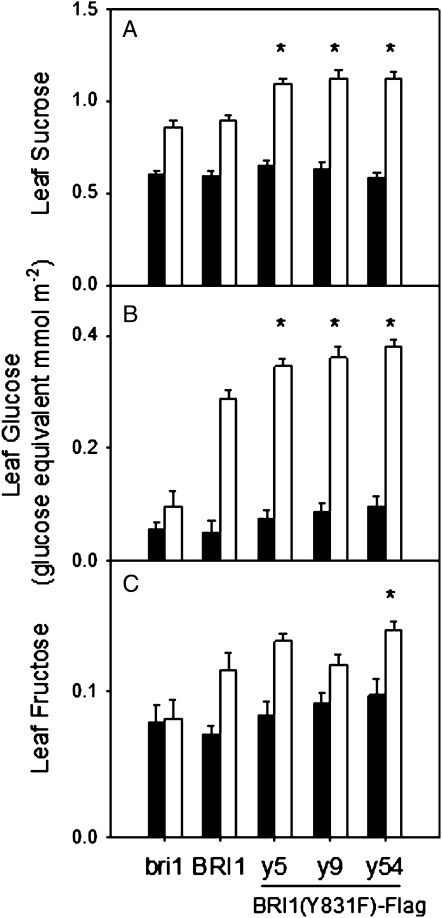
As expected, all genotypes tested accumulated starch and soluble sugars in leaves during the photoperiod. Leaves of the Y831F mutant accumulated higher levels of starch (Fig. 5), Suc (Fig. 6A), and Glc (Fig. 6B) at the end of the light period compared with BRI1-Flag and bri1-5 plants. In contrast, leaf Fru contents were much lower and did not differ between the Y831F mutants and BRI1-Flag plants (Fig. 6C). Starch was effectively mobilized in all genotypes, and calculated values of leaf starch turnover per night period were about 25% higher in the Y831F mutants compared with BRI1-Flag plants (Fig. 5B). Interestingly, at the end of the night period, the leaf contents of soluble sugars did not differ among the genotypes (Fig. 6), indicating that higher contents of starch and sugars in leaves of Y831F mutants at the end of the photoperiod cannot be attributed to differences in leaf thickness or architecture.
Figure 5.
BRI1(Y831F)-Flag mutants have higher leaf starch levels and higher starch turnover. A, Leaf starch levels at the end of the light period (white bars) and the dark period (black bars). B, Nocturnal leaf starch turnover, calculated from the values in A. n = 16; * P < 0.05.
Figure 6.
Soluble sugar content in leaves of Y831F mutants (lines y5, y9, and y54), BRI1-Flag, and bri1-5 plants at the end of the dark period (black bars) and the light period (white bars). A, Suc. B, Glc, C, Fru. n = 16; * P < 0.05.
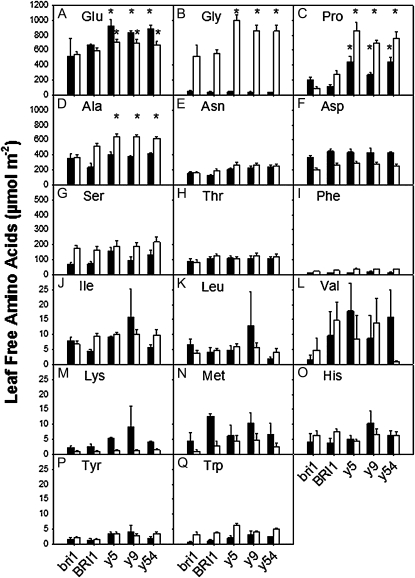
The leaf contents of many amino acids also fluctuated diurnally (Fig. 7). It is well known that the total free amino acid pool increases in leaves during the photoperiod and is remobilized at night (Hummel et al., 2010). In our study, Gly (Fig. 7B), Pro (Fig. 7C), Ala (Fig. 7D), Ser (Fig. 7G), Phe (Fig. 7I), and Trp (Fig. 7Q) were higher at the end of the light period compared with the end of the night period, whereas several other amino acids, such as Glu (Fig. 7A), Asp (Fig. 7F), Lys (Fig. 7M), and Met (Fig. 7N), showed the opposite behavior. Of particular note is the overall increased content of Pro in Y831F mutants compared with BRI1-Flag transgenic and bri1-5 plants. With the exception of the bri1-5 mutant, Pro accumulated in leaves during the day and decreased at night, and content in the Y831F mutant was substantially greater compared with the other genotypes (Fig. 7C). Pro accumulation is typically associated with osmotic stress, and a requirement for light has also been noted in previous studies (Hanson and Tully, 1979; Joyce et al., 1984; Hayashi et al., 2000). Accordingly, we monitored leaf relative water content (RWC) to determine whether the Pro accumulation observed in leaves of the Y831F mutants was the result of inadvertent water stress, but as shown in Supplemental Figure S7, the RWC of the Y831F mutants was on average slightly higher than the BRI1-Flag transgenic plants. Moreover, the transcriptome analysis gave no indication that the Y831F mutants were experiencing water stress, as there was no differential expression of biosynthetic or catabolic genes (data not shown).
Figure 7.
Leaf free amino acid content at the end of the dark period (black bars) and the light period (white bars). A, Glu. B, Gly. C, Pro. D, Ala. E, Asn. F, Asp. G, Ser. H, Thr. I, Phe. J, Ile. K, Leu. L, Val. M, Lys. N, Met. O, His. P, Tyr. Q, Trp. n = 8; * P < 0.05.
DISCUSSION
One important area for biotechnological improvement is the boosting of intrinsic yield and biomass production with a minimum input of water, fertilizers, and agrochemicals. In spite of the importance of yield and biomass, surprisingly little is known about the molecular networks controlling plant growth (Gonzalez et al., 2010). In this study, we show that engineering site-specific Tyr phosphorylation of BRI1 enhances the growth promotion associated with BR signaling. Transgenic plants expressing the Y831F mutant of BRI1-Flag have increased leaf growth (Oh et al., 2009b), whether expressed in the bri1-5 background or in the Ws2 background (Fig. 1). In this study, we provide further characterization of the growth response and identify some of the cellular and molecular mechanisms that may be responsible.
Factors Contributing to Increased Growth of the BRI1(Y831F)-Flag Mutants
The Y831F mutants are larger than BRI1-Flag transgenics primarily as a result of more cells (rather than larger cells) and have prominently increased vascular development (Fig. 1). RGRs were 15% higher in the Y831F mutants compared with BRI1-Flag transgenic plants (Fig. 2), and transcriptome analysis suggested that three factors may contribute to the observed changes in rosette growth rate and leaf architecture. First, 14 genes that are involved in secondary cell wall biosynthesis or are associated with vascular development were up-regulated in the Y831F mutants (Table I), which could certainly contribute to the increased vascular development that was observed (Fig. 1). Because gene expression was generally increased to a greater extent than the 2-fold increase in vascular area, we believe that the changes in gene expression may be contributing to the increase in vascularization (as opposed to simply reflecting the greater vascular development). Interestingly, while BL signaling up-regulates many growth-related genes, those involved in cell wall biosynthesis function predominantly in primary (rather than secondary) wall formation (Vert et al., 2005). Thus, altering the Tyr-831 phosphorylation site on BRI1 up-regulated a set of genes that is not normally affected by BR signaling. A second factor that may contribute to vascular development and the increased overall growth rate of the Y831F mutants is altered BR homeostasis. In wild-type plants, BR homeostasis is maintained through feedback regulation of multiple genes involved in sterol biosynthesis and BR-specific biosynthesis and degradation. In response to exogenous BL application, four BR-specific synthesis genes (DWF4, CPD, BR6ox1, and ROT3) and a sterol synthesis gene (DWF7) are strongly down-regulated (Tanaka et al., 2005). Under steady-state conditions, DWF4, CPD, BR6ox2 (a paralog of BR6ox1), and ROT3 were expressed at higher levels in the Y831F-directed mutant compared with BRI1-Flag transgenic plants, suggesting that BR homeostasis was shifted to favor increased BL biosynthesis. These changes in expression would be expected to increase the content of bioactive BRs (Tanaka et al., 2005; Ohnishi et al., 2006), which could help to explain the increased cell division, vascular development, and growth of the Y831F mutants. Third, three genes that are often correlated with growth were altered in their expression in the Y831F mutants in a direction that could explain the increased growth rate observed (Table III). Only one of the growth-related genes is regulated by exogenous BL in wild-type plants, indicating again that the transcriptional reprogramming that results from engineering of the Tyr-831 site involves genes that are not normally considered to be responsive to BR signaling.
BRI1(Y831F)-Flag Mutants Have Enhanced Rubisco Carboxylation and RuBP Regeneration
Gas-exchange measurements and chlorophyll fluorescence analysis suggested that Y831F mutant plants had higher rates of CO2 assimilation compared with BRI1-Flag transgenic plants under both ambient and saturated [CO2] (Fig. 4; Supplemental Figs. S3, S5, and S6). The analysis of photosynthetic parameters collectively suggests that Y831F mutants have enhanced photosynthetic rates regardless of whether photosynthesis is limited by Rubisco or RuBP regeneration. The higher rates of photosynthesis in the Y831F mutant plants were also evident in the increased accumulation of starch (Fig. 5), Suc (Fig. 6A), and Glc (Fig. 6B) in leaves during the day. If the molecular basis for the enhanced rates of photosynthesis in the Y831F mutants involves transcriptional reprogramming, the changes involved are extremely subtle. About 16 genes encoding components of the thylakoid membrane involved in light harvesting and electron transport were up-regulated from 10% to 100% in the Y831F mutants (data not shown), which is an extremely modest change. None of these genes are up-regulated by treatment of wild-type plants with exogenous BL (Vert et al., 2005). However, because chlorophyll content per unit of leaf area was unchanged, it would suggest that none of the chlorophyll-binding components of the electron transport chain were altered at the protein level. Thus, the modest changes in gene expression may not have been translated to the protein level. Nonetheless, whether the transcriptional changes observed in the Y831F plants actually play a role in the increased rates of photosynthesis observed remains an important question for the future. Another general question is whether the increase in photosynthesis is part of the driving force resulting in increased growth, or whether photosynthetic rates increase in response to elevated growth capacity (Supplemental Fig. S8). Whichever scenario is correct, our results would be consistent with previous observations that exogenous BL treatment increases photosynthetic rate and photochemical efficiency and leaf contents of Suc and starch (Yu et al., 2004; Wu et al., 2008; Xia et al., 2009) if, as discussed above, the endogenous content of bioactive BRs is increased in the Y831F mutants.
BRI1(Y831F)-Flag Mutants Accumulate Pro in the Absence of Apparent Water Stress
In addition to the increased accumulation of nonstructural carbohydrates during the photoperiod, the Y831F mutant plants also accumulated higher levels of several amino acids compared with the BRI1-Flag transgenic plants (Fig. 7). In particular, Pro, Gly, and Ala were significantly higher in the Y831F mutants, and all accumulated in leaves during the day. Levels of Gly were extremely low at the end of the night and accumulated during the day to levels equivalent to that of Suc. A similar light-dependent increase in the Gly content of barley (Hordeum vulgare) leaves was reported previously (Sicher, 2008) and was strongly suppressed by elevated [CO2], suggesting that Gly was formed primarily by photorespiratory metabolism. Increased accumulation of Gly in Y831F mutants would be consistent with higher rates of both photosynthesis and photorespiration. Also of interest is the substantial light-dependent accumulation of Pro in leaves of the Y831F mutant plants (Fig. 7C). While the accumulation of Pro is typically associated with environmental stress such as drought (Yoshiba et al., 1995), the Y831F mutant plants used in this study were apparently unstressed, as evidenced by the maintenance of leaf RWC (Supplemental Fig. S7) and photosynthetic activity at the end of the photoperiod (Supplemental Fig. S6). Moreover, the Y831F plants were also larger than the BRI1-Flag transgenic plants; thus, growth is not impaired. These observations make it unlikely that an unrecognized stress was triggering the accumulation of Pro; nonetheless, the stimulus that results in Pro accumulation is not readily apparent.
The molecular basis for Pro accumulation is also unclear. Typically, Pro accumulates when biosynthesis is activated and catabolism is repressed (Yoshiba et al., 1995; Kiyosue et al., 1996; Xue et al., 2009). However, none of the genes involved in Pro biosynthesis or catabolism (Szabados and Savouré, 2010) were altered in expression in Y831F mutants relative to BRI1-Flag plants that could explain the increased Pro accumulation (data not shown). Thus, we tentatively conclude that the regulation of Pro accumulation in the Y831F mutant plants may occur primarily at the posttranscriptional level. Indeed, phosphorylation of at least one enzyme involved in both Pro biosynthesis (P5CS1; Reiland et al., 2009) and Pro catabolism (PDH2; Durek et al., 2010) has been demonstrated in vivo and could conceivably play a role in the regulation of fluxes. Regardless of the regulatory mechanisms involved, it is interesting that among the genotypes tested, the increase in Pro content during the day was correlated positively with the decrease in Glu content (r2 = 0.855; Supplemental Fig. S9). This is consistent with the notion that Pro is synthesized mainly from Glu and that catabolism converts Pro back to Glu (Szabados and Savouré, 2010) and suggests that biosynthesis is promoted during the day while catabolism is promoted at night. However, how the pool sizes of Pro and Glu are determined and why these fluxes occur to a much greater extent in the Y831F mutant plants is not clear at present, and these are interesting questions for future studies.
It is also important to consider the possible significance of Pro accumulation in the Y831F mutant plants. While Pro accumulation is often correlated with abiotic stress tolerance, there are exceptions that indicate that Pro likely does not act in a simple manner. Rather, Pro may act as a signaling molecule that can modulate mitochondrial function and influence gene expression in the nucleus and thereby increase a plant’s ability to tolerate stress (Szabados and Savouré, 2010, and refs. therein). Conceivably, induction of Pro accumulation in response to BR signaling may play a role in plant adaptation to stress (Krishna, 2003; Kagale et al., 2007), and the results of this study suggest that regulation by phosphorylation of Tyr-831 may be involved. Thus, abiotic stress tolerance of the Y831F mutant plants emerges as another important topic for future study.
CONCLUSION
We demonstrate that Y831F mutant plants have increased growth rates compared with plants expressing wild-type BRI1-Flag, confirming that Tyr phosphorylation plays an important role in BR signaling. Enhanced growth is suggested to result from altered expression of key genes regulating (1) growth potential, (2) BL biosynthesis, and (3) vascular development. In addition, the Y831F mutants had increased rates of net photosynthesis, providing additional resources for growth. However, it remains to be determined whether increased photosynthetic activity contributes to the driving force behind increased growth or is a consequence of the increased growth potential (Supplemental Fig. S8). Understanding these altered growth properties may yield new insights for the regulation of the molecular basis of biomass accumulation in plants and may have applications for agriculture.
MATERIALS AND METHODS
Plant Material and Growth Conditions
BRI1-Flag transgenic Arabidopsis (Arabidopsis thaliana) lines (native sequence and Y831F-directed mutant; Oh et al., 2009b) with its native promoter were expressed in the bri1-5 mutant background or in the Ws2 background as described (Wang et al., 2005a). Plants were grown in controlled environmental growth chambers with a short photoperiod (8 h of light/16 h of dark) to obtain larger rosettes and delay flowering or with a long photoperiod (16 h of light/8 h of dark) with photosynthetic photon flux density of 200 μmol m−2 s−1 provided by fluorescent lamps. Day and night temperatures were 23°C ± 1°C and 18°C ± 1°C, respectively, and relative humidity was 70%. For gas-exchange analysis, seeds were planted in Sunshine LC1 soil mixture in 38-mm Cone-tainers (LI-COR). After 2 weeks, plants were thinned to one plant per tube. All plants were irrigated with modified Hoagland solution (Hoagland and Arnon, 1950), which consisted of 0.625 mm K2SO4, 0.5 mm MgSO4, 0.25 mm KH2PO4, 3 mm calcium, 20 μm Fe-EDTA, 35 μm 330 iron (Sequestrene 330; Ciba-Geigy), 46 μm H3BO3, 9 μm MnCl2, 0.76 μm ZnSO4, 0.32 μm CuSO4, 0.12 μm NaMoO4, and 12 mm nitrogen. Healthy 6- to 9-week-old plants before bolting were typically used for experiments.
Plant Growth Analysis
Arabidopsis plants were imaged several times per week with a digital camera. The projected areas of the plants were determined from the images of the plants with WinRhizo Pro 2007d software (Regent Instruments). RGR was calculated as RGR = (LnA2 − LnA1)/(t2 − t1), where A2 and A1 are the leaf areas at time t2 and t1, respectively, and results are presented in Figure 2B. Plant growth curves were also fitted with the logistic growth curve: Y = C/(1 + ae−rx), where C is the maximum rosette area and r is the specific growth rate. Both calculations gave similar results.
Leaf thickness was measured on young fully expanded leaves with a precision Starrett T230 μm (L.S. Starrett Co.). The midrib was avoided for measurements, which were made on 16 plants of each genotype.
Isolation of Total RNA, and Microarray Analysis
Plants were grown under long-day conditions (16 h of light/8 h of dark) for 25 d in soil. Total RNA was isolated from shoots harvested at the rosette stage (before bolting) at the middle of the photoperiod and cleaned using the RNeasy Plant Mini Kit (Qiagen). Total RNA was extracted from triplicate samples of bri1-5, wild-type BRI1-Flag, and BRI11(Y831F)-Flag in the bri1-5 background, with each array indicating the gene expression of a single plant. RNA microarray analysis was performed by the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign. RNA was prepared and hybridized to the Affymetrix GeneChip Arabidopsis ATH1 Genome Array in the Functional Genomics unit utilizing the GeneChip 3′ Express Kit (Ambion) according to the manufacturer’s instructions. Additional details are provided in Supplemental Text S1.
Histological Analysis
Preparation of samples for leaf tissue sectioning was performed according to standard procedures using a Leica ASP300 (Langham Creek). Fully expanded leaf tissues were fixed in 10% neutral buffered formalin, dehydrated in a graded ethanol and xylene series, and infiltrated with paraffin. Sections were cut on a Leica RM2255 microtome at 5 μm and applied to charged slides (Fisher Scientific). Slides were deparaffinized in xylene and hydrated in a graded ethanol series. Sections were further stained in 0.1% Toluidine Blue Solution (Electron Microscopy Sciences).
Gas Exchange
Gas exchange was measured using a portable gas-exchange system (LI-COR 6400LCF). The CO2 sensors and water vapor sensors of the gas-exchange system were calibrated using gas of a known [CO2] with 21% oxygen and nitrogen as balance and known water vapor concentrations generated with a controlled humidification system, respectively (LI-610 Portable Dew Point Generator; LI-COR). Leaf temperatures were set at 25°C, relative humidity was maintained at 60% to 70% in the leaf chamber, and photosynthetic photon flux density levels were adjusted up to 1,500 μmol m−2 s−1 using a chamber-integrated red-blue light source with 10% blue light for all of the measurements. Additional details of the gas-exchange analysis are provided in Supplemental Text S1.
Leaf Pigment Content
Leaf samples were extracted in 95% ethanol in the dark for 2 d at 4°C. Chlorophyll a and b and carotenoids were determined by measuring absorbance at 470, 648.6, and 664.2 nm with a microplate spectrophotometer (Biotek) using the extinction coefficients determined by Lichtenthaler (1987).
Carbohydrate Analysis and Amino Acid Profiling
Leaf discs (approximately 2 cm2) were extracted with 80% (v/v) ethanol several times until the leaf discs were colorless, and starch, Suc, and hexose sugars were measured by enzymatic analysis as described previously (Sun et al., 2002).
Free amino acids in the ethanol-soluble fractions were measured using the EZ:Faast amino acid analysis kit (Phenomenex). Amino acids extracted in ethanol together with a nor-Val internal standard were subjected to gas chromatography-mass spectrometry analysis for profiling. Amino acid derivatives were separated on a ZB-AAA gas chromatography capillary column (10 m × 0.18 mm i.d., with a 0.18-μm film thickness; Phenomenex) using an Agilent 6890N gas chromatograph system and detected with Agilent 5975B insert mass spectrometry detector (Agilent Technologies). For more details, see Supplemental Text S1.
Statistical Analysis
All data were analyzed by use of SAS ANOVA, and means and se values were calculated by use of SAS MEANS (SAS System 9.1; SAS Institute). Significant probability values were set at P < 0.05.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: BRI1 (At4g39400), BAK1 (At4g33430), BZR1 (At1g75080), BZR2/BES1 (At1g19350), DWARF4 (At3g50660), and DWARF7 (At3g02580).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The growth phenotype of Y831F plants cannot be explained by increased content of BRI1 protein.
Supplemental Figure S2. Seed size of the genotypes used in this study.
Supplemental Figure S3. Photosynthetic parameters of Y831F mutants and BRI1-Flag plants calculated from A/intercellular CO2 concentration curves.
Supplemental Figure S4. Leaf chlorophyll a, chlorophyll b, carotenoids, and ratio of chlorophyll a to chlorophyll b.
Supplemental Figure S5. Chlorophyll fluorescence analysis of photochemical quenching and maximum electron transport rates of Y831F mutants and BRI1-Flag plants.
Supplemental Figure S6. Diurnal CO2 exchange rate for whole plants over two diurnal cycles (14 h of light/10 h of dark).
Supplemental Figure S7. Leaf RWC in the mutants.
Supplemental Figure S8. Two contrasting models depicting the factors contributing to increased growth of the Y831F mutant plants and the relationship between growth and photosynthetic activity.
Supplemental Figure S9. Correlation between diurnal changes in Glu and Pro contents in leaves of bri1-5 and BRI1-Flag in the bri1-5 background and three independent transgenic lines expressing BRI1(Y831F)-Flag in the bri1-5 background.
Supplemental Table S1. Genes up-regulated in BRI1(Y831F)-Flag plants relative to wild-type BRI1-Flag transgenics, both expressed in the bri1-5 weak allele background.
Supplemental Text S1. Additional experimental methods.
Acknowledgments
We thank Dr. Joanne Chory for the kind gift of anti-BRI1 antibodies used in the experiment presented in Supplemental Figure S1.
References
- Altmann T. (1999) Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta 208: 1–11 [DOI] [PubMed] [Google Scholar]
- Becraft PW. (2002) Receptor kinase signaling in plant development. Annu Rev Cell Dev Biol 18: 163–192 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Durbak A, Wierzba M, Schmitz RJ, Aguirre A, Michel R, Rowe S, Fujioka S, Tax FE. (2010) Intragenic suppression of a trafficking-defective brassinosteroid receptor mutant in Arabidopsis. Genetics 185: 1283–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26: 573–582 [DOI] [PubMed] [Google Scholar]
- Chung HY, Fujioka S, Choe S, Lee S, Lee YH, Baek NI, Chung IS. (2010) Simultaneous suppression of three genes related to brassinosteroid (BR) biosynthesis altered campesterol and BR contents, and led to a dwarf phenotype in Arabidopsis thaliana. Plant Cell Rep 29: 397–402 [DOI] [PubMed] [Google Scholar]
- Clay NK, Nelson T. (2005) Arabidopsis thickvein mutation affects vein thickness and organ vascularization, and resides in a provascular cell-specific spermine synthase involved in vein definition and in polar auxin transport. Plant Physiol 138: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX. (2010) PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res 38: D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Lithy ME, Clerkx EJM, Ruys GJ, Koornneef M, Vreugdenhil D. (2004) Quantitative trait locus analysis of growth-related traits in a new Arabidopsis recombinant inbred population. Plant Physiol 135: 444–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. (1965) Vascular Differentiation in Plants. Holt, Rinehart and Winston, New York [Google Scholar]
- Farquhar GD, Caemmerer S, Berry JA. (1980) A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149: 78–90 [DOI] [PubMed] [Google Scholar]
- Gardiner JC, Taylor NG, Turner SR. (2003) Control of cellulose synthase complex localization in developing xylem. Plant Cell 15: 1740–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Tully RE. (1979) Light stimulation of proline synthesis in water-stressed barley leaves. Planta 145: 45–51 [DOI] [PubMed] [Google Scholar]
- Hayashi F, Ichino T, Osanai M, Wada K. (2000) Oscillation and regulation of proline content by P5CS and ProDH gene expressions in the light/dark cycles in Arabidopsis thaliana L. Plant Cell Physiol 41: 1096–1101 [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. (1950) The water-culture method for growing plants without soil. California Agricultural Experiment Station Circular 347: 1–32 [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J. (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM. (1998) Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol 118: 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel I, Pantin F, Sulpice R, Piques M, Rolland G, Dauzat M, Christophe A, Pervent M, Bouteillé M, Stitt M, et al. (2010) Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol 154: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce PS, Paleg LG, Aspinall D. (1984) The requirement for low-intensity light in the accumulation of proline as a response to water deficit. J Exp Bot 35: 209–218 [Google Scholar]
- Kagale S, Divi UK, Krochko JE, Keller WA, Krishna P. (2007) Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225: 353–364 [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY. (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Hwang JY, Kim YS, Joo SH, Chang SC, Lee JS, Takatsuto S, Kim SK. (2005) Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17: 2397–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K. (1996) A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell 8: 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P. (2003) Brassinosteroid-mediated stress responses. J Plant Growth Regul 22: 289–297 [DOI] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K. (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7: 791–806 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Long SP, Bernacchi CJ. (2003) Gas exchange measurements, what can they tell us about the underlying limitations to photosynthesis? Procedures and sources of error. J Exp Bot 54: 2393–2401 [DOI] [PubMed] [Google Scholar]
- Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M. (2006) Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol 141: 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T. (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M, Tsukaya H, Murakami N, Kato M. (2002) Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol 43: 239–244 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Oh MH, Clouse SD, Huber SC. (2009a) Tyrosine phosphorylation in brassinosteroid signaling. Plant Signal Behav 4: 1182–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Ray WK, Huber SC, Asara JM, Gage DA, Clouse SD. (2000) Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol 124: 751–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Wang X, Kota U, Goshe MB, Clouse SD, Huber SC. (2009b) Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci USA 106: 658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M-H, Wang X, Wu X, Zhao Y, Clouse SD, Huber SC. (2010) Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc Natl Acad Sci USA 107: 17827–17832 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ohnishi T, Szatmari AM, Watanabe B, Fujita S, Bancos S, Koncz C, Lafos M, Shibata K, Yokota T, Sakata K, et al. (2006) C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18: 3275–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J. (2002) Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110: 669–672 [DOI] [PubMed] [Google Scholar]
- Sharkey TD. (1985) O2-insensitive photosynthesis in C3 plants: its occurrence and a possible explanation. Plant Physiol 78: 71–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Jones WD, Jensen RV, Harris SC, Perkins RG, Goodsaid FM, Guo L, Croner LJ, Boysen C, Fang H, et al. (2008) The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics (Suppl 9) 9: S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicher RC. (2008) Effects of CO2 enrichment on soluble amino acids and organic acids in barley primary leaves as a function of age, photoperiod and chlorosis. Plant Sci 174: 576–582 [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Gibson KM, Kiirats O, Okita TW, Edwards GE. (2002) Interactions of nitrate and CO2 enrichment on growth, carbohydrates, and Rubisco in Arabidopsis starch mutants: significance of starch and hexose. Plant Physiol 130: 1573–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yang L, Wang Y, Ort DR. (2009) FACE-ing the global change: opportunities for improvement in photosynthetic radiation use efficiency and crop yield. Plant Sci 177: 511–522 [Google Scholar]
- Szabados L, Savouré A. (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15: 89–97 [DOI] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S. (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Deng Z, Wang ZY. (2010) Proteomics shed light on the brassinosteroid signaling mechanisms. Curr Opin Plant Biol 13: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible W-R, Cutler S, Somerville CR, Turner SR. (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. (2005) Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol 21: 177–201 [DOI] [PubMed] [Google Scholar]
- Wang M, Jiang W, Yu H. (2010) Effects of exogenous epibrassinolide on photosynthetic characteristics in tomato (Lycopersicon esculentum Mill) seedlings under weak light stress. J Agric Food Chem 58: 3642–3645 [DOI] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD. (2005a) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang X, Li X, Meisenhelder J, Hunter T, Yoshida S, Asami T, Chory J. (2005b) Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al. (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XJ, Huang LF, Zhou YH, Mao WH, Shi K, Wu JX, Asami T, Chen Z, Yu JQ. (2009) Brassinosteroids promote photosynthesis and growth by enhancing activation of Rubisco and expression of photosynthetic genes in Cucumis sativus. Planta 230: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Xue X, Liu A, Hua X. (2009) Proline accumulation and transcriptional regulation of proline biosynthesis and degradation in Brassica napus. BMB Rep 42: 28–34 [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K. (1995) Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J 7: 751–760 [DOI] [PubMed] [Google Scholar]
- Yu JQ, Huang LF, Hu WH, Zhou YH, Mao WH, Ye SF, Nogués S. (2004) A role for brassinosteroids in the regulation of photosynthesis in Cucumis sativus. J Exp Bot 55: 1135–1143 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]